Abstract
Graft versus host disease (GVHD) is the most common complication of hematopoietic stem cell transplant (HCT). However, our understanding of the molecular pathways that cause this disease remains incomplete, leading to inadequate treatment strategies. To address this, we measured the gene expression profile of non-human primate (NHP) T cells during acute GVHD. Utilizing microarray technology, we measured the expression profiles of CD3+ T cells from five cohorts: allogeneic transplant recipients receiving 1) no immunoprophylaxis (No Rx); 2) sirolimus monotherapy (Siro); 3) tacrolimus-methotrexate (Tac-Mtx); as well as 4) autologous transplant recipients (Auto) and 5) healthy controls (HC). This comparison allowed us to identify transcriptomic signatures specific for allo-reactive T cells and determine the impact of both mTOR and calcineurin inhibition on GVHD-mediated pathway dysregulation. We found that the transcriptional profile of unprophylaxed GVHD was characterized by significant perturbation of pathways regulating T cell proliferation, effector function and cytokine synthesis. Within these pathways we discovered potentially druggable targets not previously implicated in GVHD, prominently including aurora kinase A (AURKA). Utilizing a murine GVHD model, we demonstrated that pharmacologic inhibition of AURKA could improve survival. Moreover, we found enrichment of AURKA transcripts both in allo-proliferating T cells and in sorted T cells from patients with clinical GVHD. These data provide a comprehensive elucidation of the T cell transcriptome in primate acute GVHD. This transcriptome enables a systems-based approach to the identification of targets for disease control, many of which are immediately amenable for clinical evaluation. The first such target identified, AURKA should now be considered a lead target for prevention of GVHD, which, given the many available AURKA inhibitors in clinical development, could be quickly deployed for the prevention of GVHD.
One Sentence Summary
We have performed transcriptomic profiling of primate T cells during acute graft versus host disease which has lead to the discovery of signaling pathways that when inhibited, ameliorate disease.
Introduction
Although allogeneic hematopoietic stem cell transplant (HCT) is capable of curing many otherwise fatal malignant and non-malignant diseases, a major challenge in this field remains that many HCT recipients develop, and often die from, its most serious complication, graft-versus-host-disease (GVHD). Despite the prevalence and severity of GVHD, few new prevention or treatment strategies have been adopted in over 20 years, and the field remains largely reliant on approaches developed decades ago. Current strategies include calcineurin inhibition (with either tacrolimus or cyclosporine) + methotrexate, a combination that can potently down-regulate immune activation through combined inhibition of the Nuclear Factor of Activated T cells (NFAT) transcription factor (through calcineurin inhibition)(1) and proliferation (by methotrexate). Although this approach can block TCR signaling and resultant T cell activation, it may be antagonistic to immune tolerance induction(2), and breakthrough acute and chronic GVHD occurs in a large proportion of HCT patients prophylaxed with this regimen. A more recent addition to GVHD prevention is mechanistic Target of Rapamycin (mTOR) inhibition with sirolimus, which has been shown to be more pro-tolerogenic than calcineurin inhibition(3). The best agents to pair with sirolimus, however, remain to be determined. Sirolimus continues to be explored for upfront GVHD prophylaxis, but remains a second-line GVHD therapy without clear superiority over calcineurin inhibition(4). Given the considerable inadequacies of current GVHD prevention strategies, the identification of novel targetable pathways and molecules represents one of the major challenges in the field. To propel the field past the candidate-molecule approach for the identification of new therapeutics, we have developed a non-human primate (NHP) model of GVHD(5, 6) and have used this model to perform whole transcriptome analysis of donor T cells, to identify the molecular pathways activated in these T cells during acute GVHD (GVHD). The NHP model offers several advantages compared with small animal models of GVHD, including the high degree of cross-reactivity of clinical reagents with NHP targets(7, 8), as well as the close similarity of pharmacokinetic/pharmacodynamic parameters in NHP compared to patients. In addition, the close functional similarity of the immune systems in NHP and humans facilitates rapid clinical translation of insights made in this model, with many clinical trials in both solid organ and HCT based directly on preceding NHP studies(9–15).
Using this model, we now describe the primate GVHD T cell transcriptome, controlled to identify pathways specifically enriched during allo-transplantation (using autologous HCT controls), and the partial normalization of this transcriptome with calcineurin + methotrexate- or monotherapy with mTOR- inhibition. This analysis has allowed us to identify molecular pathways active in GVHD, many of which represent druggable targets for which candidate interventions are immediately available. One of the most prominent pathways identified within the NHP GVHD transcriptome, the aurora kinase A pathway, encodes proteins controlling cell cycle progression, cell growth, differentiation and survival(16, 17), and inhibitors of this pathway are in clinical trials as targeted cancer therapies(18, 19). Our results identify Aurora Kinase A as a lead pathway for allo-specific T cell activation, representing a novel targetable strategy for controlling this disease. Importantly, we describe a paradigm for identifying new druggable targets for allo-specific T cell activation in the context of the intense systemic inflammatory GVHD environment, which utilizes NHP and mouse models as well as clinical GVHD patient samples.
Results
Sirolimus and Tac-Mtx provide graded disease protection in a NHP model of GVHD
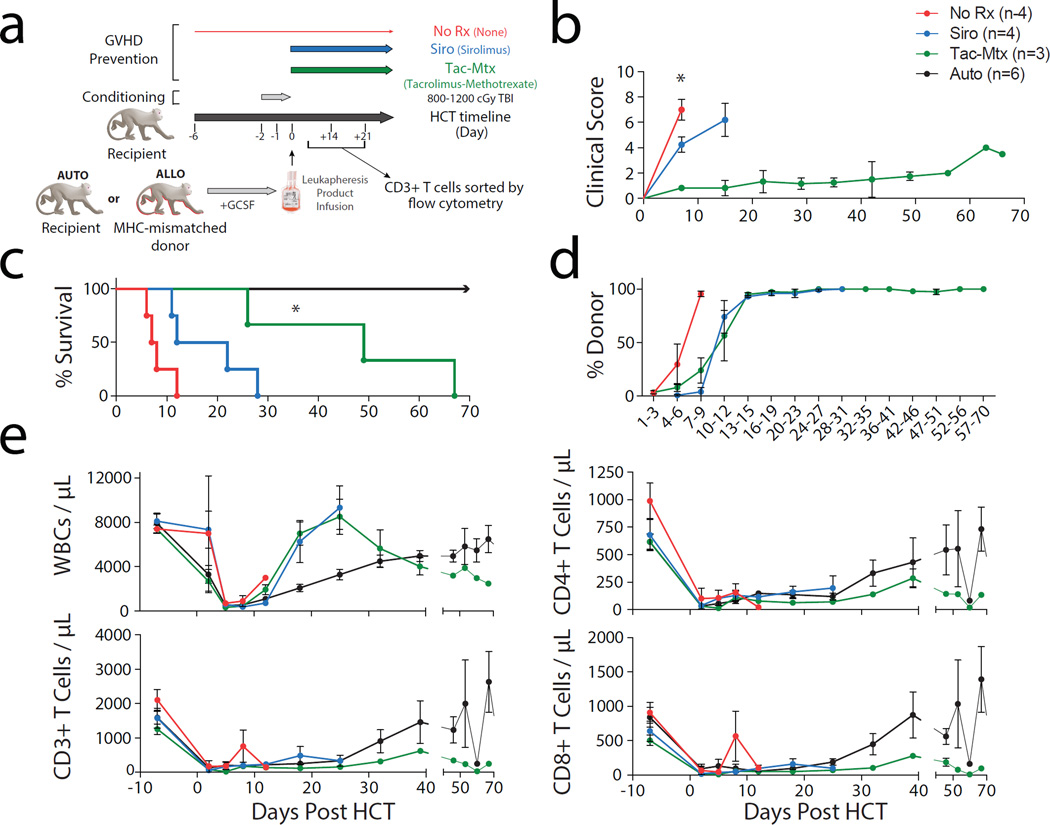
Figure 1a shows our previously described strategy(5) for MHC-mismatched allogeneic-HCT in NHP, in addition to the GVHD prophylaxis strategies employed in the current study. As shown in the figure, this study compared untransplanted healthy control animals (‘HC’, n = 34) as well as autologously transplanted controls (‘Auto’, n = 6 for flow cytometric data and n = 3 for gene expression profiling) to MHC-mismatched allogeneic transplant recipients receiving no immunoprophylaxis (‘No Rx’, n = 4), those receiving sirolimus(20, 21) (‘Siro’, n = 4) those receiving tacrolimus-methotrexate combination immunoprophylaxis(4) (‘Tac-Mtx’, n = 3). All transplants utilized myeloablative TBI as a pre-transplant preparative regimen, transplantation of T cell-replete peripheral blood stem cell products obtained after G-CSF-mobilized leukapheresis, and supportive care with anti-bacterial, anti-fungal and anti-viral prophylaxis. Figure 1b demonstrates longitudinal analysis of clinical GVHD scores for the allo-transplant recipients, using our previously described scoring system(5). Briefly, the GVHD clinical score increases with cumulative GI-specific abnormalities (e.g. diarrhea), liver-specific abnormalities (e.g. hyperbilirubinemia) and skin-specific abnormalities (extent and character of rash). These scores demonstrate the rapid and severe clinical syndrome that accompanied unprophylaxed GVHD, the partial protection and delay in onset of severe GVHD in the recipients treated with sirolimus monotherapy and the more substantial protection from clinical disease observed in the dual therapy tac-mtx cohort. As expected, significantly shorter overall survival paralleled an increase in clinical GVHD scores with median survival times (MST) of 7.5, 17 and 49 days observed in the No Rx, Siro and Tac-Mtx cohorts, respectively, with all autologous transplant recipients surviving long-term (Figure 1c). We found a significant difference between the MST of the Tac-Mtx cohort relative to the No Rx cohort (P = 0.018, log rank test) and a difference approaching significance between the Siro and No Rx cohort (P = 0.055, log rank test). Donor engraftment (Figure 1d) occurred in all recipients, and hematopoietic reconstitution was notable for a rapid expansion of CD4+ and CD8+ T cells in the unprophylaxed recipients, that occurred faster than in either autologous controls or prophylaxed cohorts (Figure 1e).
Figure 1. Sirolimus and Tac-Mtx provide graded disease protection in a NHP model of GVHD.
a) Experimental schema detailing the transplant protocol and immunoprophylaxis regimens used throughout this study.
b) Clinical score from a cohort of animals after allogeneic or autologous (‘Auto’, black) HCT. Allogeneic transplants received the following immunoprophylactic regimens: 1) none (‘No Rx’, red), sirolimus monotherapy (‘Siro’, blue) and tacrolimus-mtx (‘Tac-Mtx’, green). Scoring was based on our previously described NHP GvHD clinical scoring system(5). Significance of clinical scores for animals with GHVD was determined by unpaired t-test. Tac-Mtx score was found to be significantly lower on Day 7 than both No Rx (P = 0.0013) and Siro scores (P = 0.033).
c) Comparison of survival curves between all groups undergoing HCT. The Kaplan-Meier product-limit method was used to calculate survival. Significance between the immunoprophylaxis and No Rx groups was determined using log-rank statistics with * representing P < 0.05. A near significant difference in survival was observed between sirolimus and No Rx cohort (P = .0.055).
d) Donor chimerism as determined by donor-specific microsatellite-based analysis in the allogeneic HCT cohorts(5).
e) Engraftment of WBC (by Complete Blood Count), T cells (by flow cytometric analysis of CD3+/CD20- cells), CD4+ T cells (by flow cytometric analysis of CD3+/CD20-/CD4+/CD8- cells), and CD8+ T cells (by flow cytometric analysis of CD3+/CD20-/CD4-/CD8+ cells) in all HCT cohorts.
Unsupervised analysis identifies treatment cohort as the major contributor of transcriptional variation during GVHD
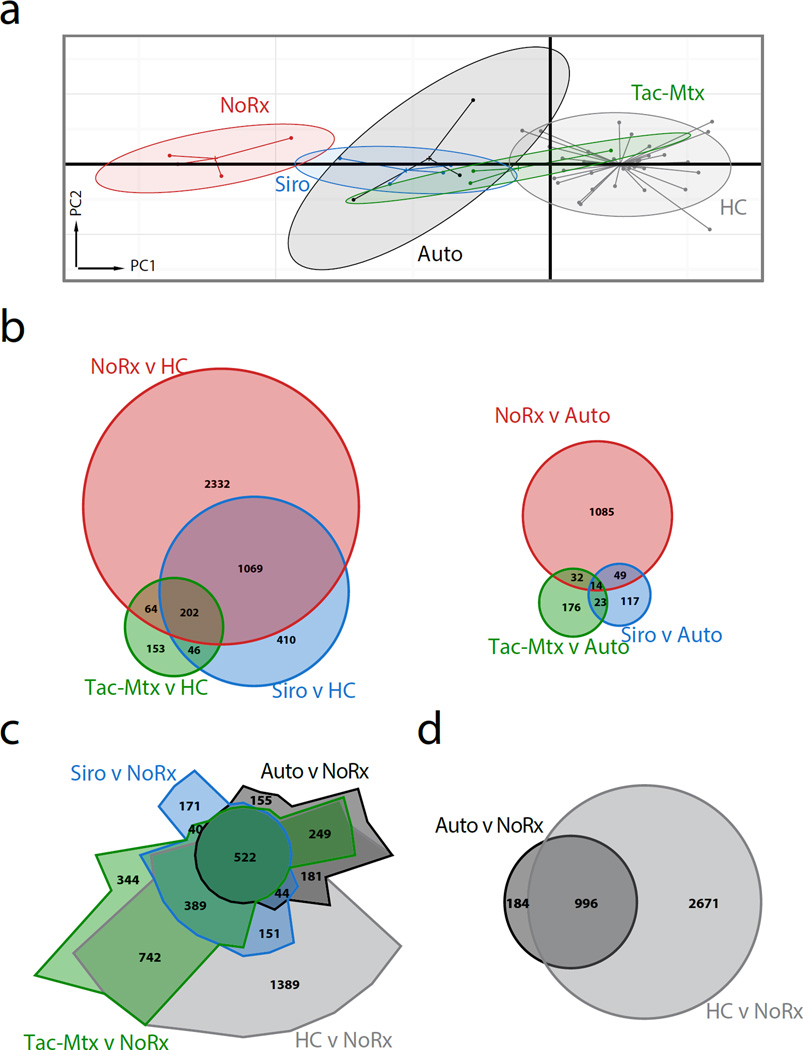
To facilitate a comprehensive, unsupervised analysis of the molecules and pathways that are aberrantly regulated during GVHD and how the expression of these genes changes with immunoprophylaxis, we profiled messenger RNA (mRNA) expression from sorted CD3+ T cells isolated from healthy controls (with no history of HCT), and on day +14 or at the time of sacrifice (if sacrifice occurred prior to day +14) from transplant recipients. Principal Component Analysis (PCA), a dimensionality reducing method that is used to identify the determinants of variation in complex data(22), was then performed. This allowed us to determine the degree to which the T cell transcriptome of the three allo-HCT cohorts differed from both the autologous transplant recipients and from healthy controls, and from each other, and the degree to which transcriptional variation was shared between cohorts. As shown in Figure 2a, projections of the first and second principal components showed discrete clustering when grouped by treatment cohort. Confidence ellipses (90%) of PCA projections showed no overlap between the No Rx cohort and either healthy controls or autologous recipients and independent tests of statistical significance confirmed that the first principal component projections were different between these groups (P < 0.001 – No Rx vs HC, P < 0.001 – No Rx vs Auto; unpaired t-test corrected for multiple hypothesis testing using BH procedure). In contrast, projections from the Tac-Mtx cohort exhibited substantially more overlap with those of autologous transplant recipients and healthy controls and the first principal component of the Tac-Mtx group was not significantly different than that of the autologous controls (P > 0.05). This analysis provided strong evidence that the treatment groups could be distinguished transcriptionally from each other, and that the No Rx group could be distinguished from both Autos and HCs, and as such, provided the foundation for further analysis of the gene expression pathways and networks that define each group.
Figure 2. Principal component analysis and differential expression show that the T cell transcriptome recapitulates clinical observations.
a) First and second principal component projections reveal clustering of transplanted animals by immunoprophylactic strategy and donor source. Each dot represents an array sample, the center of inertia ellipses correspond to the mean projections of the group, and the shaded area representing the 90% confidence interval.
b) Weighted Venn diagrams showing the number of transcripts differentially expressed in all allogeneic transplants compared to healthy controls (left) and compared to autologous controls (right). Overlapping areas in the Venn diagrams indicate over- or under-represented transcripts that are shared between the cohorts. The number of transcripts is indicated for each comparative group. Differential expression (DE) analysis used a significance threshold of 0.05 derived from an empirical Bayes moderated t-statistic, corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. An absolute fold-change threshold of 1.4 was employed.
c) Chow-Ruskey Venn diagram (weighted) showing all expression data compared to the No Rx cohort.
d) Weighted Venn diagram showing the Auto cohort and untransplanted Healthy Controls versus the No Rx cohort.
To develop a more detailed understanding of the transcriptional landscape among the treatment cohorts on a single gene level, we performed differential expression (DE) analysis. Confirming our PCA results, we found an inverse correlation between the number of DE transcripts (up- and down-regulated) compared to either HC or autologous transplant controls and the length of GVHD-free survival after allogeneic HCT (Figure 2b). Showing the complimentary comparison using a spatially-weighted, Venn diagram (Chow-Ruskey plot), Figure 2c depicts healthy controls, autologous controls and treated allogeneic HCT cohorts relative to the No Rx cohort. This analysis underscores the high degree of transcriptomic variation between unprophylaxed GVHD and all other groups, as well as the significant sharing of differentially expressed transcripts in the autologous and healthy controls (Figure 2d) when compared to the No Rx allo-HCT transcriptome. The comparative analysis of DE transcripts in Figures 2b–c demonstrated that while each treatment cohort displayed a unique transcriptional profile, there was also important sharing of DE transcripts between the cohorts.
Transcripts associated with immune effector function and cell turnover are over-represented in T cells during GVHD: Top gene analysis and Gene Set Enrichment Analysis (GSEA)
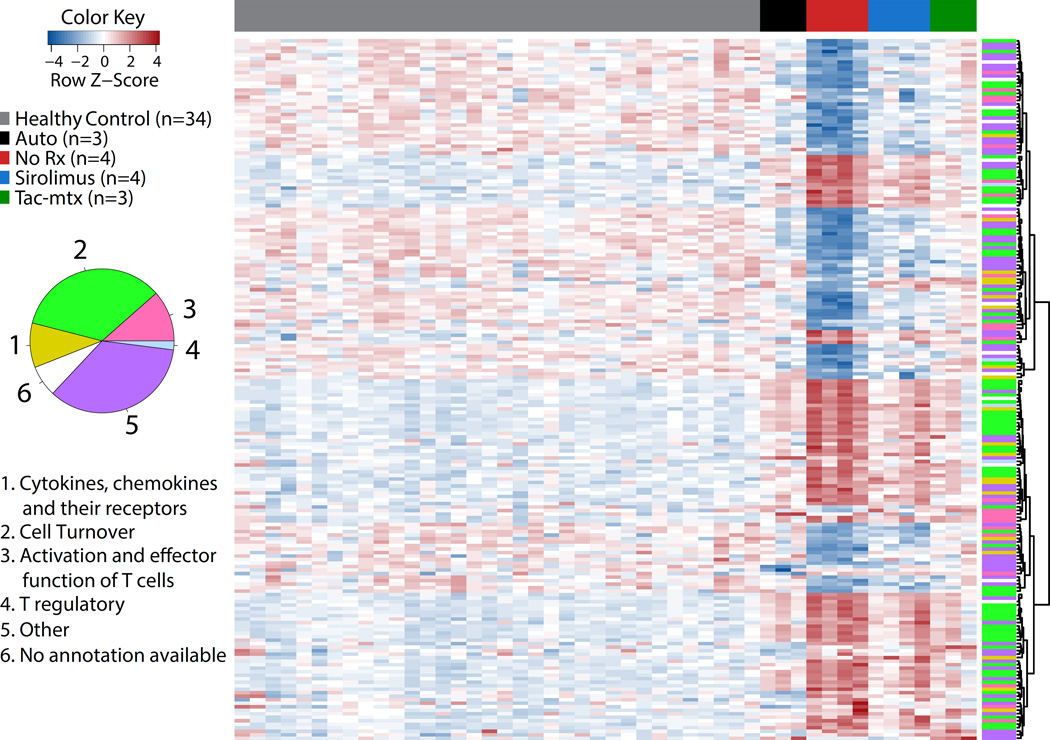
To specifically characterize the genes and pathways impacted by allo-activation, GVHD and its prophylaxis, we identified the top 100 transcripts over-represented in the No Rx group compared to Auto controls, as well as the top 100 transcripts that were comparatively under-represented in the No Rx group, (and thus enriched in the Auto controls). This analysis was performed using absolute fold change and statistical significance as ranking parameters (Figure 3 and Supplementary Table 2). The 200 identified transcripts could be largely grouped into 6 functional categories: 1) Cytokines, chemokines and their receptors; 2) Cell turnover; 3) Activation or effector function of T cells; 4) T regulatory; 5) Other; and 6) No annotation available. Figure 3 shows a heatmap of the Z-score of the expression values of these transcripts and their functional classification, with an accompanying graphic depicting the relative frequencies of the 6 categories. Supplementary Table 2 catalogues each of the 200 top transcripts with more detailed information of each transcript including fold change and statistical significance.
Figure 3. Top 200 differentially expressed transcripts from CD3+ T cells during GVHD reveals that cell turnover is a dominant expression theme during GVHD.
Heatmap of top 100 over-represented and top 100 under-represented transcripts (200 total) sorted on absolute fold change observed in the comparison between the No Rx and autologous transplant cohorts. Transcripts were categorized into six functional categories: 1) Cytokines, chemokines and their receptors (yellow), 2) Cell Turnover (green), 3) Activation or effector function of T cells (magenta), 4) T regulatory (light blue), 5) Other (dark green), 6) No annotation available (white).
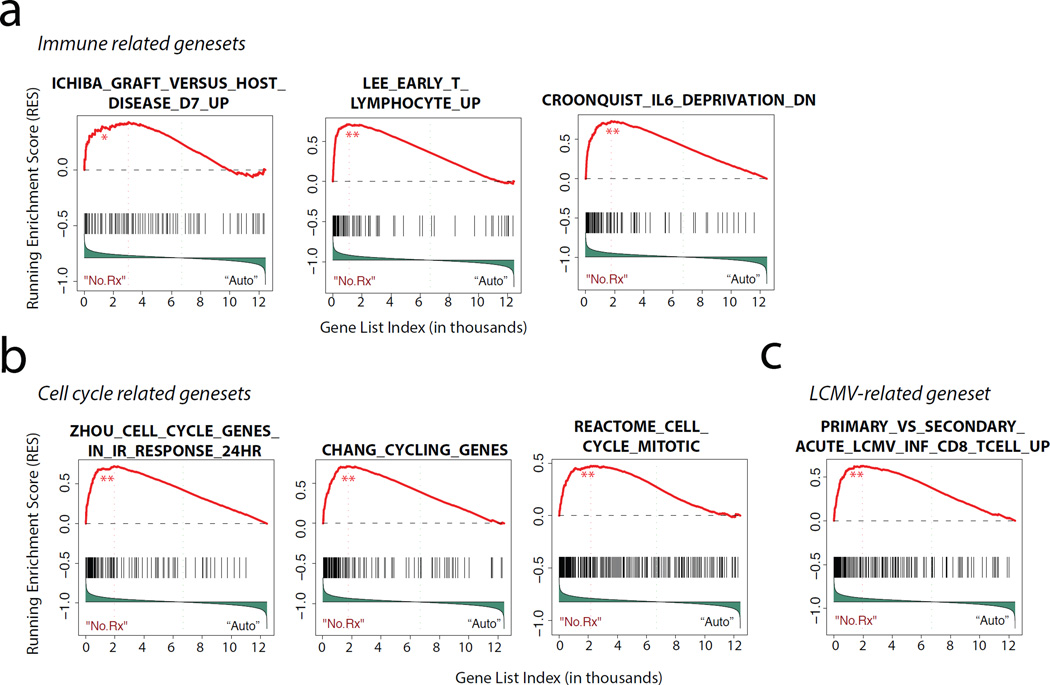
Working to parallel our Top Gene analysis using an approach that can overcome the bias associated with cut-off-based analyses, we performed GSEA(23–25). As depicted in the selected examples in Figure 4a–b and Supplementary Table 3, GSEA analysis identified multiple gene sets that were enriched in the leading edge (left side of figure) of the ranked gene list for No Rx vs Auto controls (transcripts enriched in T cells during GVHD-associated allo-activation). This analysis reinforced the concept that two broad categories of pathways were activated during GVHD on a system level: those involved in immune signaling (Figure 4a) and those involved in cell cycle progression and T cell proliferation (Figure 4b).
Figure 4. Gene set enrichment analysis (GSEA) identifies T cell immune dysregulation and proliferation as pathophysiologic drivers of primate GVHD.
a) GSEA was performed as previously described24, 25). Shown are representative enrichment plots of immune-related gene-sets over-represented in the No Rx allogeneic versus autologous transplant cohorts. The False Discovery Rate (FDR) q value for the observed enrichment score was derived using a null distribution of enrichment scores generated from 1000 permutations of gene set labels. **FDR q value < 0.001; *FDR q value < 0.05 but > 0.001. NS FDR q value > 0.05
b) Representative gene-sets related to proliferation and cell cycle control, with statistics computed as in (a).
c) Representative LCMV-associated gene-sets, with statistics computed as in (a).
GSEA also allowed us to interrogate the degree to which T cell activation during GVHD resembled other T cell functional pathways. For this analysis, we focused on gene sets previously identified to be differentially regulated during the protective immune response to viral antigens, given the well-documented phenomenon of heterologous allo-immunity (where T cells activated by viruses have been observed to be serendipitously allo-reactive) and because of the potentially clinically significant overlap between effective downregulation of the alloimmune response, and unintended inhibition of protective immunity(26–29). The dataset for transcriptional control of the T cell response to the lymphocytic choriomeningitis virus (LCMV) is particularly robust, (30) and therefore, we performed a focused analysis of the degree to which GVHD-associated T cell activation overlapped with the immune activation triggered by LCMV. The Molecular Signatures Database (MSigDB) is a collection of annotated gene sets for use with GSEA; therefore, to perform this analysis, we probed the MSigDB database for genesets curated during both primary and secondary T cell responses in the setting of both acute and chronic LCMV infection (Gene Express Omnibus – GSE30962)(31). As shown in the representative example in Figure 4c and in Supplementary Figure 3, we found that 7 of 8 LCMV genesets curated in MSigDB were enriched during the comparison of the No Rx and auto-HCT cohorts. Pathway analysis performed on the combined leading edge genes in these enrichment plots provided key insights into the important nodes of overlap between allo- and virus-mediated T cell activation. As shown in Supplementary Table 4, this analysis demonstrated that of the nine statistically significant pathways identified (p <0.05 using Benjamini-Hochberg corrected P values), all were involved in cell proliferation. The gene lists associated with these pathways provide a new compendium of genes that are involved in both viral and allo—mediated antigen-driven T cell activation.
The gene dysregulation seen in CD3+ T cells during GVHD in NHP parallels current therapeutic and diagnostic efforts in patients
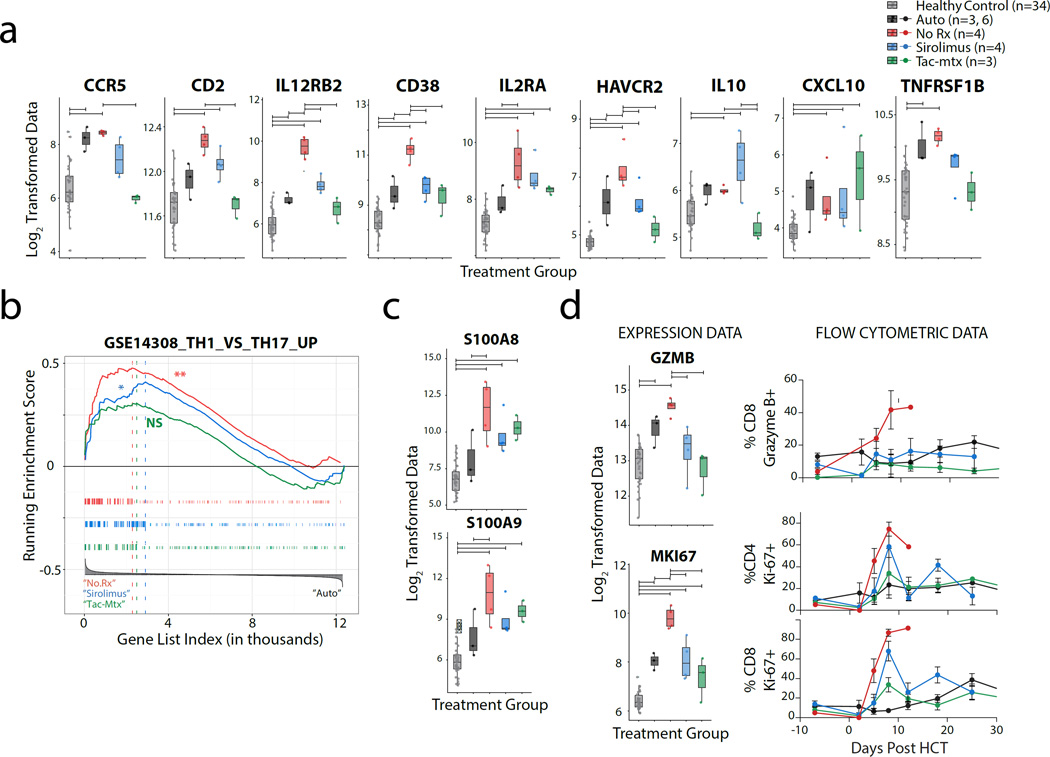
Having enumerated and classified the transcripts that were most highly over- or under-represented during GVHD compared to both HC (which identifies genes differentially expressed during both homeostatic reconstitution and allo-activation) and Auto controls (which specifically identifies those genes differentially expressed during allo-activation), we next queried our dataset individually for selected gene products that are currently, or have been recently, investigated as pharmacologic targets for GVHD prevention or therapy. Among the gene products queried were CCR5, CD2, IL12RB2 and CD38 (Figure 5a). In clinical studies, CCR5 blockade has been shown to be a potential strategy for visceral GVHD prevention(32). Consistent with this, CCR5 expression was highly enriched in the NHP GVHD transcriptome compared to HC; however, as shown in Figure 5a, CCR5 enrichment was also observed during autologous reconstitution, suggesting that its upregulation may be associated with both homeostatic and allo-mediated T cell expansion. This observation is consistent with previous studies in rhesus macaques, which have shown that CCR5 upregulation occurs during homeostatic reconstitution of CD4+T cells in animals treated with a depleting anti-CD4 antibody(33). Likewise, CD2 functions as a costimulatory molecule during antigen-specific lymphocyte activation and has been studied in a phase II trial as a potential target for the treatment of GVHD(34). Similar to what we observed for CCR5, while we found significant enrichment of CD2 in T cells during GVHD when compared to HC, this enrichment did not reach statistical significance when compared to Auto controls (Figure 5a), suggesting that while blocking CD2 may decrease T cell activation, it may not represent a specific target for allo-activated cells. In contrast, CD38, which has recently been shown to be highly correlated with GVHD in patient samples(35), was specifically enriched in NHP GVHD compared to both autologous transplants and healthy controls. Similarly, IL-12/23, which is also being investigated as a potential therapy for GVHD(36), was substantially and specifically enriched T cells during NHP GVHD. The enrichment of these receptor subunits is reflective of a high degree of Th1 skewing in the CD3 cells isolated from the No Rx cohort(37). GSEA also supported a Th1-skewed immune response, with significant enrichment for Th1 versus Th17 transcripts(38) (Figure 5b). Thus, while we did observe dysregulation of selected Th17-related transcripts in the No Rx cohort (including IL17F (but not IL17A), POU2AF1, and TSC22D3, Supplementary Figure 4), the overarching GVHD response in animals receiving no prophylaxis was Th1-polarized.
Figure 5. Gene expression data from NHP with aGvHD are consistent with recent therapeutic strategies for GvHD prophylaxis and with T cell biomarkers for GVHD.
a) Box-plots of expression data for individual transcripts recently being investigated as pharmacologic targets for GvHD prophylaxis (CCR5, CD2, IL12RB2, CD38) or as potential biomarkers (IL2RA, HAVCR2 (TIM3), IL-10, CXCL10, TNFRSF1B) in NHP HCT cohorts and healthy controls. Horizontal significance bars denote comparisons with a moderated t-statistic < 0.05 corrected for multiple hypothesis testing using Benjamini-Hochberg procedure.
b) GSEA was performed comparing No Rx (left, red), Tac-Mtx (left, green), Siro (left, blue) to Auto (right) cohorts. No Rx and Siro cohorts have a significant enrichment of transcripts previously shown to be upregulated in Th1 relative to Th17 cells(38). Hypothesis testing was performed as described in Figure 4a.
c) Box-plots of expression data for S100A8 and S100A9 in NHP HCT cohorts and healthy controls. Significance bars and color groups as described in Figure 5a, above.
d) Expression data (left) and longitudinal flow cytometric analysis (right) of granzyme B and Ki-67 expression on CD4+ and CD8+ in NHP HCT allo-transplant cohorts and controls. Horizontal bars denote significance as described in Figure 5a.
The addition of serum biomarkers to clinical GVHD staging and grading is being increasingly recognized to improve accuracy in diagnosis and prognostication. We set out to validate known, serum biomarkers of GVHD in our dataset and to provide additional information about the natural history of these markers during untreated GVHD as well as during treatment with current standards-of-care. Several of the key molecules identified from serum protein interrogation include elafin(39), ST-2(40, 41), Reg3A(42), HGF(43), IL2RA(43), TNFRSF1(43), CXCL10(44), IL-10(45), and TIM3(41), Of those proteins expressed by T cells (which excludes elafin, Reg3A and HGF) and are annotated for NHP (which excludes ST-2), we observed robust and specific upregulation of IL2RA and HAVCR2 (the gene encoding TIM3) during GVHD, with significant activation compared to both HC and Auto controls (Figure 5a). While we also observed modest upregulation of IL-10, CXCL10 and TNFRSF1B compared to healthy controls during GVHD (Figure 5a), the expression profiles of these molecules were not significantly different between the No Rx cohort and the Auto controls, suggesting that they may mark both homeostatic- and allo-proliferating T cells.
In addition to investigating genes that have been previously identified as important biomarkers or treatment targets in GVHD, this study has allowed us to begin to query for cross-over of molecules that have been previously recognized as important pathogenic biomarkers in other diseases caused by T cell activation. Consistent with the hypothesis that significant sharing of pathogenic mechanisms exists between diseases of auto- and allo-activation, as shown in Figure 5c and Supplementary Table 2, the two most highly enriched transcripts in our dataset are the ‘calprotectin’ genes, S100A8 and S100A9 (10.4- and 8.6-fold increase compared to Auto controls, respectively), which have been previously implicated as having an important pathogenic role in inflammatory bowel disease(46–48). These data provide support for the existence of common pathogenic mechanisms between these two immune-mediated disorders and point to the calprotectin gene transcripts in T cells as novel putative biomarkers of acute GHVD.
We have shown previously(5) that during NHP GVHD, T cells express high levels of Ki-67 and granzyme B. Consistent with these findings, we observed significant transcriptional upregulation of the gene encoding Ki-67 (MKI67) in the No Rx cohort compared to both HC and Auto controls, with normalization in prophylaxed animals (Figure 5d). Granzyme B gene expression was also found to be significantly upregulated in T cells during GVHD in the No Rx cohort, and was normalized by therapy. Its expression was also higher in the No Rx cohort compared to Auto controls, although this difference did not reach statistical significance. As shown in Figure 5d, flow cytometric measurements of Ki-67 and Granzyme B at the time of gene array analysis paralleled the expression data, and provided independent validation of our transcriptomic approach to defining T cell dysregulation during GVHD.
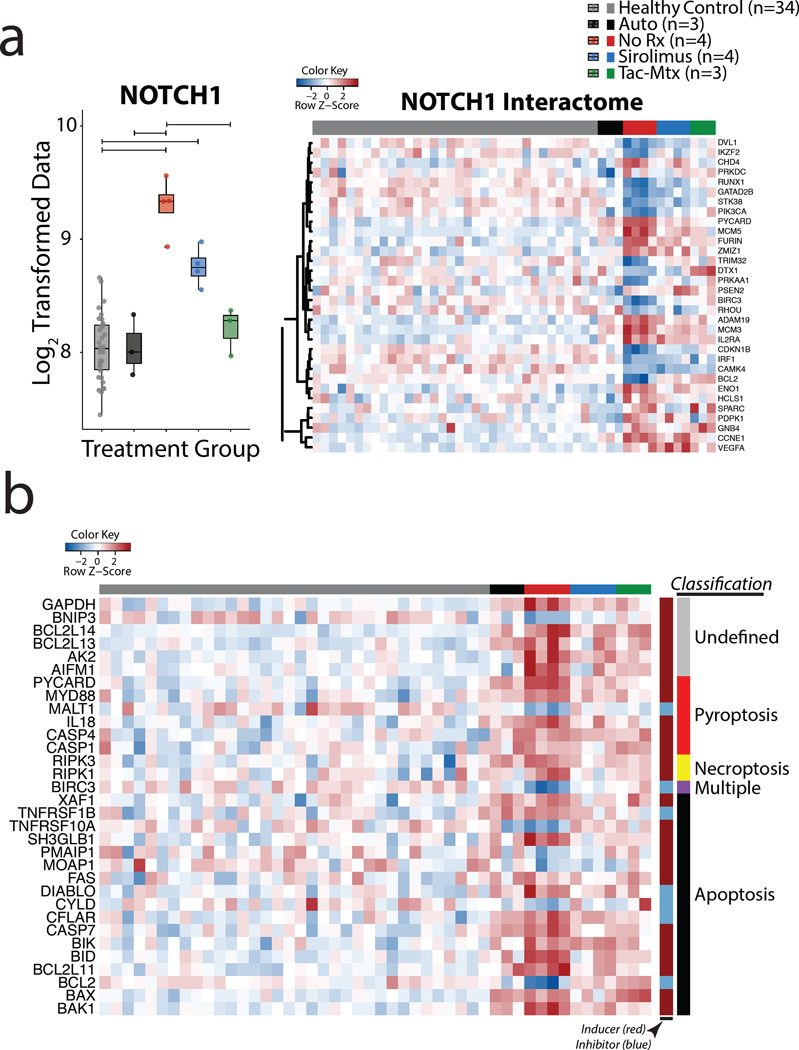
Pathophysiology of NHP aGVHD includes dysregulation of Notch, Notch targets, as well as evidence for T cell pyroptosis
Seeking to better understand the pathophysiologic drivers of primate GVHD and to provide further external validation to our systems approach, we also queried our dataset for known pathways that have been implicated in GVHD pathogenesis. These included Notch signaling and the inflammasome/pyroptosis pathways (Figure 6). Consistent with its role in T cell expansion during GVHD, the Notch pathway has been shown to be a potential target for GVHD therapy(49). In agreement with these previous studies, we observed significant and specific upregulation of NOTCH1 in CD3 T cells during acute GVHD compared to both HC and autologous controls (Figure 6a, top). Expanding this line of investigation further, we constructed an interactome, using Ingenuity Pathway Analysis (IPA), to identify molecules with which NOTCH1 has been mechanistically linked. We were able to identify a number of transcripts from the Notch interactome, (Figure 6a, bottom) (CCNE1, IL2RA, ADAM19, IFNG, VEGFA, GNB4, MCM5, TUBB4B, MCM3, PYCARD, GZMB, ENO1), that were over- or under-represented in the No Rx cohort, consistent with this pathway being a major driver of T cell allo-reactivity(50).
Figure 6. Transcriptional analysis of primate GVHD pathophysiology.
a) Boxplot of expression data from NOTCH1 (top) and a heatmap showing Z-score genetic expression of the NOTCH interactome (bottom) in NHP HCT cohorts and healthy controls. The NOTCH interactome was constructed using Ingenuity software and was interrogated for differentially expressed transcripts that were significantly over- or under-represented in the No Rx cohort relative to the Autologous cohort..
b) Heatmap showing Z score transformed genetic expression data for cell death pathways in NHP HCT cohorts and healthy controls. Innermost row side colors indicate whether the gene has been shown to positively regulate (dark red) or inhibit (light blue) cell death. Outermost row side colors indicate the classification of the mechanism of cell death most associated with each gene product. The gene list was generated by interrogating DeathBase(51) for transcripts that were differentially expressed (adjusted P value < 0.05) between the No Rx cohort and healthy controls. Classification data was also taken from DeathBase.
In addition to identifying gene expression signatures of T cell activation and proliferation during GVHD, we also identified enrichment of a striking number transcripts associated with cell death. To better understand the mechanisms of cell death that may be active during GVHD, we classified those cell death-related transcripts that were significantly over- or under-represented (adjusted P value < 0.05) in the No Rx cohort relative to both Auto controls and to HC (Figure 6b) using DeathBase(51). As shown in the figure, we observed the enrichment of many transcripts known to be pro-apoptotic as well as a relative reduction in the number of inhibitors of apoptosis in our dataset. This is consistent with other published studies of accelerated cell death during aGVHD(52). In addition, as shown in Figure 6b, significant transcriptional activation of several of the canonical pyroptosis transcripts was also observed in the No Rx cohort, including PYCARD (ASC), CASP 1, 4, and their downstream target IL-18. While the inflammasome is most commonly attributed to innate immune cells(53), recent work has documented cleaved caspase-1 in LPS-stimulated leukocytes in patients with GVHD(54), caspase-1-dependent pyroptosis in CD4+ T cells of HIV+ patients,(55, 56) and that CD4+ expression of PYCARD impacts T cell proliferation(56, 57). Together, these observations suggest that the inflammasome is also active in adaptive immune cells. Our data echo these observations and further suggest that the inflammasome may play an important role in the initiation and/or progression of GVHD.
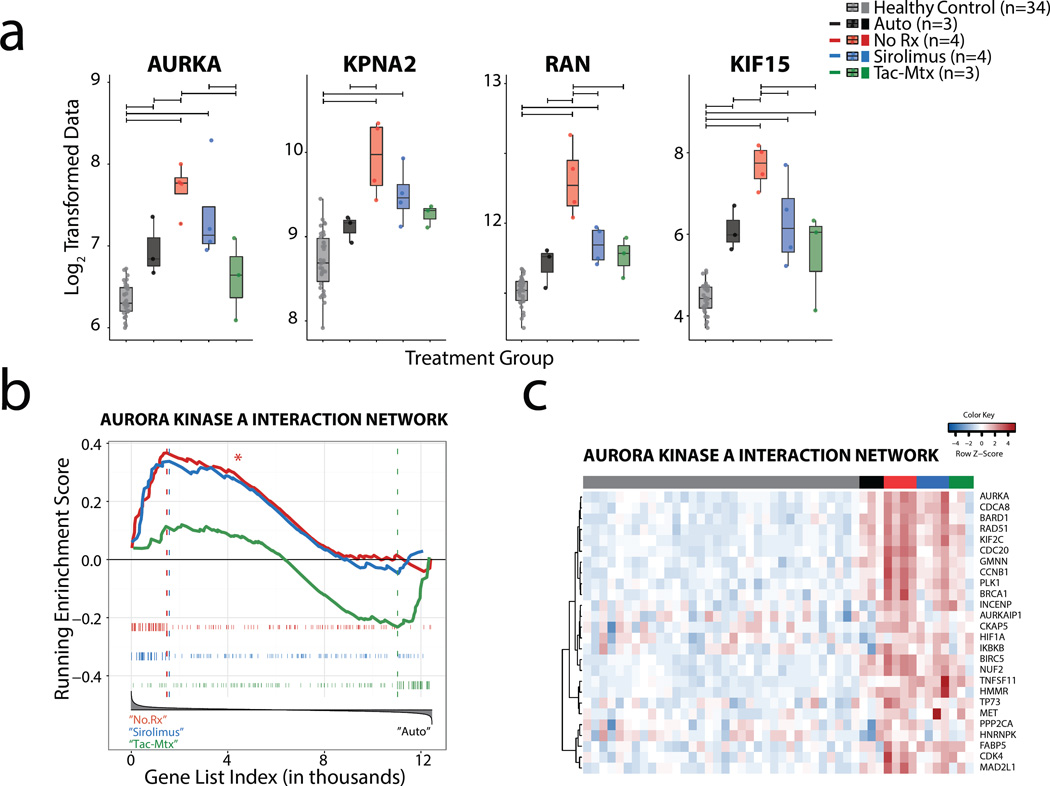
An analysis gene expression in NHP GVHD reveals aurora kinase A as a novel potential molecular target for GVHD
To discover new putative targets for GVHD that could complement current standard of care therapy, we performed a stepwise analysis of over- or under-represented pathways using KEGG(58), Panther(59), Biocarta(60), and Reactome(61) Pathway analysis. The focus of this analysis was to determine which pathways were specific to allo-activated T cells and thus GVHD, that were not present during autologous reconstitution. We found that the pathways involved with control of cell turnover predominated this comparison (Supplementary Table 5), and further found that 4 transcripts (AURKA, RAN, KPNA2 and KIF15) associated with the formation of the mitotic cell spindle(60, 62) were significantly and specifically enriched in T cells during GVHD (Figure 7a). Both to interrogate this relationship on a systems level, and to focus our efforts on genes for which clinically relevant inhibitors are available, we constructed an AURKA interactome using IPA, which included those genes with which AURKA has been shown to interact. We then used GSEA to evaluate whether this interactome showed enrichment in the leading edge of the No Rx cohort versus autologous controls. As predicted from the DAVID Pathway analysis, the aurora kinase A interactome showed significant enrichment (Figure 7b, False Discovery Rate (FDR) = 0.006). Moreover, a closer inspection of the leading edge genes in this analysis revealed that the most enriched transcripts in the aurora kinase A interactome were also highly enriched during immunoprophylaxis with sirolimus (Figure 7c), suggesting that this may be a pathway that is left un-corrected with mTOR inhibition and thus, represents a potentially important partner with sirolimus. As shown in Supplementary Figure 5, we further found that the aurora kinase A interactome displayed little overlap with those proliferation transcripts (discussed above, and shown in Figure 4c, Supplementary Figure 3a and Supplementary Table 4) that were shared during LCMV and allo— mediated T cell activation. Taken together, these results suggest that the expression of the aurora kinase A interaction network is dysregulated during NHP GVHD to a level not seen during autologous reconstitution, and that targeting this pathway may 1) complement residual gene dysregulation that occurs during sirolimus-based GVHD prophylaxis and 2) target cell turnover pathways that may be distinct from those shared with virus-mediated T cell activation.
Figure 7. Analysis of over- or under-represented transcripts during GVHD reveals enrichment of aurora kinase A interactomes.
a) Box-plots of expression data for four mitotic spindle-associated transcripts (AURKA, KPNA2, RAN, KIF15) that show enrichment in No Rx cohort relative to autologous controls. Horizontal significance bars denote comparisons with a moderated t-statistic < 0.05 corrected for multiple hypothesis testing using Benjamini-Hochberg procedure.
b) Enrichment of the aurora kinase A interactome in the leading edge of three gene lists ranked by expression differences between 1) NoRx and Auto (red traces), 2) Sirolimus and Auto (blue traces), 3) Tac-mtx and Auto (green traces). The aurora kinase A interactome was created using Ingenuity Pathway Analysis. Hypothesis testing was performed as in Figure 4a.
c) Heatmap showing Z score transformed genetic expression data for transcripts in the leading edge of the aurora kinase A GSEA enrichment plot (b above) from all animals.
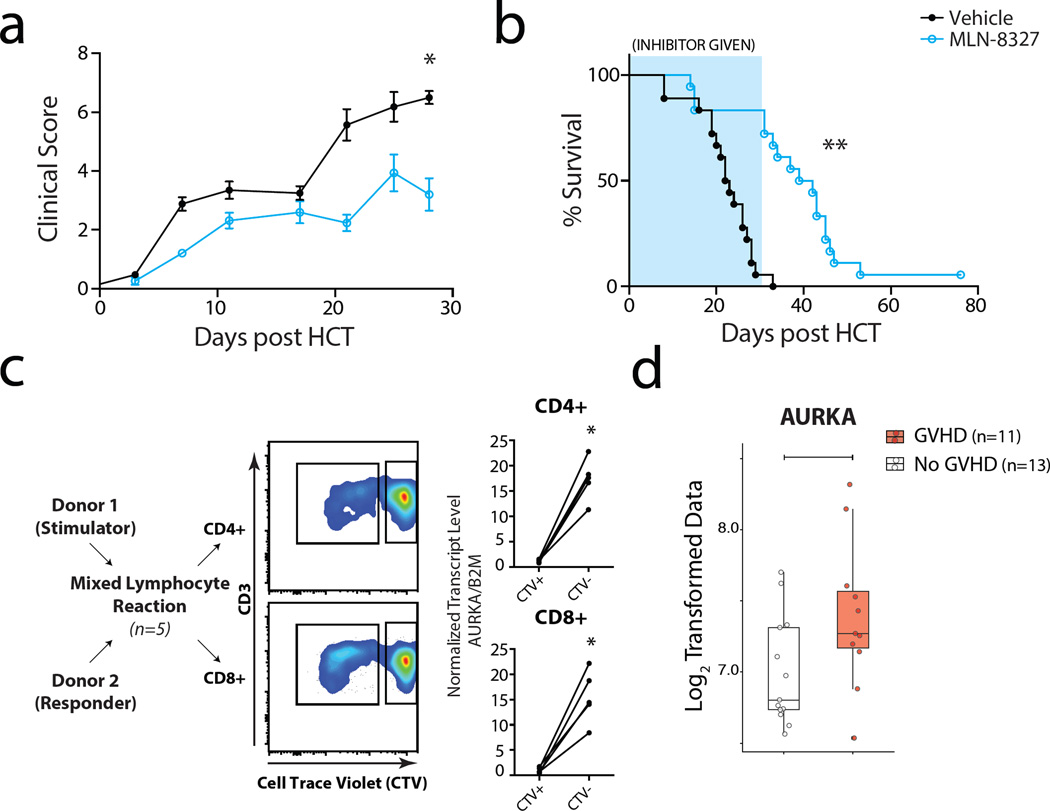
Inhibition of aurora kinase A mitigates GVHD and prolongs survival in a murine model of acute GVHD
To rigorously validate the Aurora Kinase A pathway, it was further explored using a multi-species analysis pipeline (Supplementary Figure 6). We first performed a proof-of-concept study in our established MHC-mismatched murine model of GVHD(63) using MLN8327, a commercially available selective, reversible small molecule inhibitor of aurora kinase A. As shown in Figure 8, inhibition of the aurora kinase A pathway with in vivo MLN-8327 administration was, indeed, able to mitigate clinical severity and lethality in the murine GVHD model, significantly extending MST from 22.5 (vehicle) to 40.5 days (MLN-8327), p <0.0001). These results predict that targeting aurora kinase A will mitigate GVHD, but that, as expected, for full protection it will likely need to be paired with adjunctive immunosuppression.
Figure 8. Aurora kinase A inhibition represents a rational target for GVHD inhibition.
a) GvHD clinical score from transplant recipients: Mice were weighed and monitored for clinical signs of GvHD, with scores based on weight loss, posture, activity, fur texture and skin integrity twice weekly as previously described82). MLN8237 (aurora kinase inhibitor, blue lines) was given by gavage daily at a dose of 30mg/kg. Treatment with MLN8237 was begun on day 0 and was continued through day 30.
b) Percent survival of mice treated with MLN-8327. The Kaplan-Meier product-limit method was used to calculate survival. Differences between groups were determined using log-rank statistics.. ** P=0.0007 for MLN8237 vs vehicle.
c) Allo-proliferating CD4 and CD8 T cells in a Mixed Lymphocyte Reaction (MLR) exhibit significant enrichment of AURKA relative to non-proliferating cells. Bulk T cells were isolated from healthy donors and allowed to proliferate on T cell depleted, irradiated PBMC from an allogeneic donor. On day 4 of the MLR, T cells were harvested and sorted for CD4 and CD8 expression and proliferative status. Absolute AURKA expression was measured using drop-digital PCR and normalized to beta-2 microglobulin expression. This assay showed that allo-proliferative human CD4 and CD8 T cells were 15.3+/− 1.8-fold and 15.5+/− 2.3-fold enriched, respectively, for AURKA transcripts compared to non-proliferating CD4 and CD8 cells. *P value < 0.05 using paired t-test.
d) Transcriptome profiling was performed on sorted T cells from 11 patients with acute GVHD (GVHD - red) and 13 post-HCT patients without evidence of GVHD (No-GVHD -white). Patients with GVHD showed significant enrichment of AURKA transcripts. Horizontal bar indicates significance to a level < 0.05 using unpaired t-test.
Given the results of the NHP and murine studies, we further explored whether, in human cells and patient samples, the aurora kinase A pathway represented a robust target for GVHD prevention. Two assays were performed, both of which suggest that aurora kinase A represents a highly promising pathway for further clinical investigation. In the first assay, an ex-vivo mixed lymphocyte reaction (MLR) was performed with human T cells, and the relative expression of AURKA in the allo-proliferating versus non-proliferating cells was determined. As shown in Figure 8c, allo-proliferative human CD4 and CD8 T cells were 15.3+/− 1.8-fold and 15.5+/− 2.3-fold enriched, respectively, for AURKA transcripts compared to non-proliferating CD4 and CD8 cells. Finally, we compared Aurora Kinase A gene expression in T cells purified from 11 transplant patients who developed acute GVHD and 13 control transplant patients who did not. (Figure 8d). Closely mirroring the NHP transcriptome results, this analysis confirmed enrichment of AURKA gene transcripts in patients with GVHD.
Discussion
Despite the critical need to develop more successful prevention strategies for GVHD, there have been few additions to the field in the past two decades. Moreover, there are numerous limitations to the standard methodologies for understanding the mechanisms controlling T cell activation during GVHD, determining the molecular and mechanistic overlaps between autologous and allogeneic T cell reconstitution and activation, and for identifying novel druggable targets for GVHD. Thus, while standard methodologies such as flow cytometric immunophenotyping are increasingly able to track multiple T cell markers simultaneously(41, 64), they remain supervised analyses, only able to query previously identified proteins and pathways and confirm existing hypotheses. Similarly, serum biomarkers, while gaining increasing relevance for accurate GVHD diagnosis, staging, and prognostication(65), do not readily permit identification of the cells producing the secreted molecules and do not identify transcription pathways that link to GVHD. Furthermore, as most commonly reported to date, these biomarkers often represent later stage pathway perturbations, rather than identifying upstream events that may be especially amenable as potential targets for prevention of disease.
Recently, in vitro assays have been used to identify the transcriptional networks controlling the allogeneic response as opposed to non-specific receptor-mediated proliferation, and have been able to identify several potentially important targets(66). However, though they may be informative for some pathways, by their nature, these experiments are not able to fully recapitulate the complex post-transplant in vivo milieu, where tissue-specific antigens (that may not be represented via the typical in vitro assays) and conditioning-mediated inflammation may lead to more complex, multi-modal T cell activation not limited to purely allogeneic stimuli, and where both homeostatic and allogeneic mechanisms control T cell proliferation. It is very likely that all of these inputs lead to the final end-product of T cell proliferation and linked activation that leads to GVHD. Therefore, to most comprehensively identify novel molecules and pathways that could be targeted to control this disease, we developed an in vivo NHP GVHD system capable of modeling, to a greater extent, the complexity of the post-transplant immune landscape, but where a molecular definition of the mechanisms controlling GVHD (and a comparative determination of the impact that GVHD therapies make on these mechanisms), was feasible to attain.
Using this model, we found that NHP with unprophylaxed GVHD (No Rx) exhibited rampant cytokine secretion, as well as T cell activation, proliferation and apoptosis, with preferential expansion of memory/effector-phenotype CD4+ and CD8+ T cells exhibiting high cytotoxic potential. In order to move beyond the supervised analysis inherent in both flow cytometric and serum biomarkers, we have now developed a systems biology approach to study GVHD pathogenesis, similar to the systems approaches currently gaining prominence in the fields of infectious disease and vaccinology(67, 68). In the current study, we have used this approach to identify, for the first time, the transcriptional networks that are activated during NHP GVHD and the impact made on these networks by the current standard immunosuppression platforms.
Our results show that the pathways activated during unprophylaxed GVHD are concentrated on two axes of T cell function: those controlling immune effector function (including evidence for Th1 skewing), and those controlling entry into and progression through the cell cycle, including the activation of multiple kinase-driven pathways. From an immune standpoint, our transcriptomic approach identified genetic dysregulation that was consistent with a number of current GVHD therapeutic efforts. Importantly, the NHP transcriptome has also yielded several novel insights into the pathophysiology of GVHD, including the striking observation that several of the most prominently over-represented pathways identified in our network analysis involved molecules capable of driving entry into and progression through the cell cycle, using pathways that were often distinct from those in autologous reconstitution.
The inclusion of the autologous transplant cohort in our flow cytometric and transcriptome analysis has thus substantially enhanced our ability to probe the specificity of the T cell activation signature during GVHD, and to determine the extent to which this signature can be distinguished from that which occurs during homeostatic reconstitution. This is of particular importance to the field, given the clinical importance of preserving hematologic reconstitution and protective immunity while controlling GVHD in patients undergoing transplantation. While it is still not clear the extent to which interventions should be chosen for their allo-specificity, given that homeostatic expansion mechanisms could also contribute to T cell proliferation and the acquisition of effector and memory-like phenotypes(69, 70), a detailed molecular understanding of which pathways and molecules are specifically dysregulated during allo-activation may identify strategies for more restricted targeting of the T cell allo-response. Targeting these pathways may more effectively preserve reconstitution of T cells that can contribute to anti-viral and anti-leukemia effects and thus lead to an optimized risk: benefit ratio. The dataset that we present in the current study will permit an evidence-based approach for choosing interventions in terms of their degree of specificity for allo-activation.
Of the multiple components of the cell cycle machinery that were identified in our transcriptome analysis, one of the most salient observations that we made was the degree to which aurora kinase A gene expression, as well as its interaction network, were enriched during GVHD. While aurora kinase pathways have increasingly been shown to be active in a variety of malignancies, including those involving transformed B cells and T cells(18, 71), they have not previously been linked either mechanistically or therapeutically with GVHD. In the present study, we developed a rigorous, multi-species and multi-assay pipeline by which to validate candidate pathways that were identified through NHP transcriptome analysis (Supplementary Figure 6), and with this pipeline, confirmed aurora kinase A as tightly linked with GVHD in NHP, murine and human systems. It should be noted that a second potential pathway, the sonic hedgehog (SHH) pathway was also provisionally identified in the NHP transcriptome analysis, in that its interactome was significantly enriched in the No Rx vs Auto comparison (although the SHH gene itself was not statistically significantly enriched in the same comparison, see Supplementary Figure 7a). Given the interactome enrichment, SHH was carried through the full analysis pipeline but, unlike AURKA, did not uniformly show a positive signal in all assays (Supplementary Figure 7 c – f). The comparative analysis thus allowed us to “pick a winner” and supports prioritized clinical translation of aurora kinase A inhibition for GVHD prevention.
This study has provided a comprehensive description of the T cell transcriptome during NHP GVHD. NHP offer the critical advantage of being able to identify the genes and pathways dysregulated during unprophylaxed GVHD, such that the natural history of this disease in vivo in primates can be understood on a molecular level, with specificity of the analysis assured through the comparison to Auto controls. This comprehensive dataset also establishes a novel resource for comparative evaluation of the impact of both standard-of-care and novel immunomodulatory strategies on T cell gene expression, with specificity for both homeostatic reconstitution during autologous reconstitution, and for T cell activation driven specifically during allo-stimulation. This resource has already yielded insights about the potential impact of the aurora kinase A pathway on GVHD, and will serve as a platform for the dissection of the impact of additional agents on this disease.
Materials and Methods
Study Design
This was a prospective cohort study in NHP designed to compare the clinical and immunologic outcomes of transplantation, and to discern the transcriptome of autologous and allogeneic transplant recipients. Experiments were performed in 4 cohorts: (1) Autologous transplants; (2) Allogeneic transplants with no GVHD prophylaxis (No Rx); (3) Allogeneic transplants using sirolimus for GVHD prophylaxis (Siro); (4) Allogeneic transplants using tacrolimus + methotrexate for GVHD prophylaxis (Tac-Mtx). Group sizes of 3–6 were chosen based on power analysis, which assumed that survival in each of the treated cohorts would be at least two-times that in the No Rx cohort. In order to comply with the IACUC ‘reduction’ mandate, based on the “three Rs” of Replacement, Refinement, and Reduction(72), statistical significance of survival data for the prophylaxis cohorts was compared to the No Rx cohort after 3 animals were enrolled, and group sizes were initially limited to 3 animals if statistical significance was obtained at that point. If other immunologic analysis required additional animals to be added to any cohort, this was performed post-hoc. The treatment of animals with immunosuppression was performed in an open-label format (no blinding). However, blinding was performed on all pathologic analysis and on the initial analysis of flow cytometry data and transcriptome sample handling and data processing. Flow cytometric immune phenotyping as well as transcriptional analysis was performed on a prospectively-designed timed sample acquisition schedule.
NHP Ethics Statement
This study used specific pathogen-free, juvenile rhesus macaques that were housed at the Yerkes National Primate Research Center and the Washington National Primate Research Center. The study was conducted in strict accordance with USDA regulations and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. They were approved by the Emory University, University of Washington, and Seattle Children’s Research Institute’s Institutional Animal Care and Use Committees.
Rhesus HCT
We utilized our previously-described strategy for MHC-mismatched HCT in rhesus macaques(5). Briefly, apheresis was performed after G-CSF mobilization (Amgen, 50mcg/kg for 5 days), and an unmanipulated apheresis product was transplanted into MHC-mismatched recipients. For these studies, we utilized our rhesus macaque MHC typing system(23) to choose donor: recipient pairs, which were all partially MHC-matched, either as half-sibling haplo-identical pairs or as unrelated partially-matched pairs. In the case of the autologous controls, donors were harvested and apheresis products were cryopreserved for a minimum of 2 months prior to the onset of conditioning and subsequent reinfusion of the unmanipulated apheresis product. The transplanted total nucleated cell dose (TNC) and CD3+ cell doses are shown in Supplementary Table 1. The pre-HCT preparative regimen consisted of total body irradiation (TBI) of 9.6 Gy given in two fractions over two days. Irradiation was delivered with a Varian Clinac 23EX (Varian), at a dose rate of 7 cGy/min. Supportive care included antibacterial prophylaxis with polymyxin b and neomycin sulfate, (begun on Day −6 and continued until the absolute neutrophil count was greater than 500 cells/microliter) as well as enorofloxacin (Baytril®, Bayer Healthcare, 7mg/kg IM daily, begun on day −1 and continued until ANC > 500 cells/microliter). Antiviral prophylaxis (cidofovir, Gilead, 5 mg/kg IV weekly) and antifungal prophylaxis (fluconazole 5mg/kg orally, daily) were also employed. Leukoreduced (using an LRF10 leukoreduction filter, Pall Medical) and irradiated (2200 rad) platelet-rich plasma or whole blood was given for a peripheral blood platelet count of ≤ 50 × 103 per µL or a hemoglobin < 9 g/dL, respectively, or if clinically significant hemorrhage was noted. Blood product support adhered to ABO antigen matching principles.
Four transplant cohorts were compared in this analysis: (1) allo-transplant recipients who did not receive post-transplant immunoprophylaxis, abbreviated as ‘No-Rx’ (n = 4); (2) allo-transplant recipients treated with sirolimus (LC laboratories), given daily for the length of analysis as an intramuscular formulation with doses adjusted to achieve a serum trough of 5–15 ng/dL, abbreviated as ‘Siro’ (n = 4); (3) allo-transplant recipients treated with tacrolimus (given daily for the length of analysis, with doses adjusted to achieve a serum trough of 5–15 ng/dL) and methotrexate (10mg/m2 given on days +1, +3 +8, +11 post-transplant) abbreviated as ‘Tac-Mtx’ (n = 3); (4) autologous transplant recipients who did not receive any immunoprophylaxis abbreviated as ‘Auto’ (n = 6 for flow cytometric data and n = 3 for expression profiling). GVHD clinical score was assessed weekly for allo-transplant recipients as previously described.(5) Briefly, the GVHD clinical score increases with cumulative GI-specific abnormalities (e.g. diarrhea), liver-specific abnormalities (e.g. hyperbilirubinemia) and skin-specific abnormalities (extent and character of rash). Significance of clinical scores was determined by pairwise t-test corrected for multiple hypothesis testing using Benjamini-Hochberg procedure. The Kaplan-Meier product-limit method was used to calculate survival. Differences between groups were determined using log-rank statistics.
Flow cytometry
Longitudinal flow cytometric analysis
Multicolor flow cytometric analysis was performed using an LSRII flow cytometer (BD Biosciences) on all transplant recipients, using the following leukocyte phenotypic characteristics: T cells: CD3+/CD20-; B cells: CD20+/CD3-; CD4+ T cells: CD4+/CD3+/CD8-/CD20-; CD8+ T cells: CD8+/CD3+/CD4-/CD20-; In addition, the level of Ki-67 and granzyme B were determined on both CD4+ and CD8+ T cells. Relative percentages of each of these subpopulations were determined using FloJo software (TreeStar) and absolute numbers of each of the subpopulations were determined by calculations from the complete blood count and absolute lymphocyte count analysis. The sources and clones used for each of these antibodies are as follows: from BD Biosciences: CD3, Clone SP34-2; CD8, Clone: RPA-T8; from eBioscience: CD4, Clone OKT4; CD20, Clone: 2H7; from Dako: Ki67, Clone Ki-67. Granzyme B, Clone GB11.
Flow Cytometric CD3+ T cell sorting
Using a FACSAria Cell Sorter (BD), T cells were sorted on day +14 post-transplant, or at the time of terminal analysis, (if this occurred prior to day +14). T cells were identified as CD3+/CD20- lymphocytes and were >90% pure based on post-sorting flow cytometric analysis (Supplementary Figure 1a). One animal from the sirolimus monotherapy group had collection and purification of T cells at day +22, and one animal from this group had collection and purification on day +28, each for technical reasons. The impact of these two technical delays on the gene array profiles appeared to be minimal: As shown in the PCA analysis (Figure 2a), tight clustering of the gene expression profiles of the entire sirolimus cohort was observed.
NHP Microarray and data analysis
Following T cell purification, RNA was stabilized in T cell lysates with RLT buffer (Qiagen) and RNA was purified using RNEasy Column Kit (Qiagen). RNA was quantified using a Nanodrop Spectrophotometer (Thermo Scientific) and purity was confirmed with an RNA 6000 Nano Kit (Agilent). The purified RNA was sent to the Vanderbilt Technologies for Advanced Genomics Core and to the Oregon Health Sciences University Gene Profiling Shared Resource where RNA quantity and quality were verified, followed by cDNA/cRNA synthesis, and target hybridization to GeneChip Rhesus Macaque Genome Array (Affymetrix). The resultant fluorescent signals were processed and normalized using the Robust Multichip Averaging (RMA) Method(73). The microarrays were performed in four batches, with all batches containing samples from both healthy controls and transplanted animals. The “ComBat” algorithm was implemented to adjust for batch effects(74) (shown in Supplementary Figure 2) and probe-sets containing low signal-to-noise measurement were filtered out in order to enhance statistical testing power(75). Principal Component Analysis (PCA) was applied to summarize modes of gene array variance using the Bioconductor MADE4 package(76). Probe-sets were annotated using 1) annotation file from Dr. Robert B. Norgren Jr.(77) 2) the annotation file provided by the chip manufacturer (release 33, 10–30-12) and 3) data provided by Ingenuity Systems (Ingenuity Systems, www.ingenuity.com) for the small number of probe-sets that were not annotated by the chip manufacturer.
Analyses of gene differential expression (DE) was performed using an empirical Bayes moderated t-statistic, with a cutoff of 0.05, corrected for multiple hypothesis testing using Benjamini-Hochberg procedure and an absolute fold change cutoff > 1.4 with the Bioconductor limma package(78).
Using pathway analysis systems IPA (Ingenuity Systems, www.ingenuity.com) and DAVID(79) (encapsulating Biocarta, KEGG, and Reactome pathway analysis), we interrogated our DE gene lists for significantly enriched pathways. Significance was estimated using a right-tailed Fisher’s exact test using the Benjamini-Hochberg procedure to account for multiple testing.
Further analysis of differentiating characteristics between untreated allo-HCT and control arrays involved Gene Set Enrichment Analysis (GSEA) as described(23–25). GSEA can identify whether the members of a gene set (a collection of which are housed in the Molecular Signatures Database (MSig-DB) - Broad Institute, Boston, MA) are enriched in an independent rank-ordered profile of genes that are differentially expressed between two experimental groups. In this manner, GSEA is able to provide definitions of overrepresented biological functions without implicit bias associated with cut-off based analyses. In the current analysis, gene sets were ranked using a signal to noise ratio difference metric with 1000 permutations of gene set labels.
The gene list for cell death was obtained from DeathBase(51). The notch, aurora kinase A and sonic hedgehog interactomes were created using Ingenuity Systems. Unless otherwise noted, gene analysis was carried out using R: Bioconductor.
Murine HCT
Murine models and ethics statement: B10.BR/SgSnJ (H2k) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). B6 (H2b: CD45.2) mice were purchased from the National Institutes of Health (Bethesda, MD). Mice were housed at the University of Minnesota Hospital in a specific pathogen-free facility in micro-isolator cages. Donor and recipient mice were between 8 to 10 weeks of age at the time of HCT. Murine experiments were conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and were approved by the University of Minnesota Institutional Animal Care and Use Committee.
Transplant Regimen: Lethally irradiated (1000 cGy) B10.BR (Jackson Labs) recipients were transplanted with five different treatment regimens: (1) Bone marrow control: These animals received 1×107 bone marrow (BM) cells alone from fully MHC-mismatched B6 mice (NCI, n = 15 in two independent experiments). (2) BM + 5 × 106 splenocytes from B6 mice + MLN8237, a selective aurora kinase A inhibitor (Chemie Tek) (n = 18 in two independent experiments). MLN8327 was given by gavage daily at a dose of 30mg/kg from day 0→28 (80). (3) BM + splenocytes + LDE225, an inhibitor of the sonic hedgehog coreceptor Smoothened (Smo, Chemie Tek (n = 27 in three independent experiments). LDE225 was given by gavage daily at a dose of 40 mg/kg from day 0 →28 (81). (4) BM + splenocytes + the vehicle for MLN (10% 2-hydroxypropyl-b-cyclodextrin, Sigma, 0926) (n = 27 in three independent experiments). (5) BM + splenocytes + the vehicle for LDE225 (PEG300, Sigma, 91462) (n = 27 in three independent experiments). Mice were monitored for clinical signs of GVHD (with scores based on weight loss, posture, activity, fur texture and skin integrity) twice weekly as previously described(82). The Kaplan-Meier product-limit method was used to calculate survival. Differences between groups were determined using log-rank statistics.
MLR studies
PBMC were isolated from 5 healthy donors by Ficoll (GE Healthcare) fractionation. Bulk T cells were isolated using a negative immunomagnetic selection kit (Miltenyi Biotech). The negative fraction (“Responder” T cells) were labeled with Cell Trace Violet (Invitrogen) and were co-cultured with the irradiated (3500 cGy) positive immunomagnetic fraction from an allogeneic donor. MLRs were cultured for 4 days at 37°C in XVivo15 (Lonza) supplemented with 10% FCS, 2 mM glutamine, penicillin– streptomycin and gentamycin. On day 4, cells were stained for CD3, CD4, CD8 and sorted using the following gates 1) CD3+, CD4+, CTV+, 2) CD3+, CD4+, CTV-, 3) CD3+, CD8+, CTV+, 2) CD3+, CD8+, CTV−. Total RNA was obtained from cell lysates using RNAEasy Mini kit (Qiagen), RNA yields were measured using a Nanodrop Spectrophotometer (Thermo Scientific, Waltham, MA) and cDNA was synthesized using iScript kit (BioRad) normalizing the amount of input RNA across samples. AURKA, SHH, and beta-2 microglobulin (B2M) transcript levels were measured by drop digital PCR (QX200 Bio-Rad) as previously described(83) using exon-spanning primers dHsaEG5024289 for AURKA, dHsaEG5006220 for SHH, and dHsaEG5020739 for B2M (Bio-Rad, Hercules, CA). AURKA and SHH transcript levels were then normalized to B2M, and the ratio of the transcript levels in the allo-proliferating (CTV-) and non-proliferating (CTV+) cell fractions determined.
Transcriptional studies of patient samples
Human Studies Ethics Statement
The patients and healthy participants described in this manuscript were enrolled in clinical trials that were conducted according to the principles set forth in the Declaration of Helsinki, and which were approved by the institutional review boards (IRB). Written informed consent was received from all participants.
Transcriptional analysis
For gene array analysis, available cryopreserved patient peripheral blood mononuclear cell (PBMC) samples were obtained from patients enrolled in hematopoietic stem cell transplant clinical trials performed at Emory University and the University of Minnesota, with and without GVHD. Patients from Emory University were enrolled on two contemporaneous IRB-approved clinical trials: (1) The Bone Marrow Immune Monitoring Protocol and (2) The Abatacept Feasibility Study as previously described(9, 84). For the Emory samples, available cryopreserved PBMC that were collected prior to GVHD onset or in control patients without GVHD were analyzed. The patients from the University of Minnesota were enrolled on an immune monitoring protocol approved by the University of Minnesota IRB. Samples from patients at the University of Minnesota with acute GVHD were identified and then control samples were selected that were matched for sample collection day, preparative regimen intensity, disease, stem cell source, and GVHD prophylaxis. Clinical details pertaining to each of the samples included in the gene array analysis are shown in Supplementary Table 6.
To prepare for gene array, PBMC were thawed and labeled with CD4, CD8, CD11c, CD14, CD16, CD20, CD56. Cells positive for either CD4 or CD8 but negative for the remainder of markers were sorted (Supplementary Figure 1b) and subsequently preserved in RLT buffer (Qiagen). T cell lysates were then sent to Oregon Health Sciences University Gene Profiling Shared Resource where total RNA was obtained using the RNAeasy kit (Qiagen) and quantified using a Nanodrop Spectrophotometer (Thermo Scientific). Purity was confirmed with an RNA 6000 Nano Kit (Agilent, Santa Clara, CA) and samples were then hybridized to Human Transcriptome 2.0 Array (Affymetrix). CEL files were downloaded and fluorescent signals were normalized using the Robust Multi-array Averaging (RMA) method using Expression Console Software (Affymetrix). The resulting fluorescent intensities were then adjusted for batch effects (74) using the “sva” package (Bioconductor). Probe-sets containing low signal-to-noise measurement were filtered out in order to enhance statistical testing power. Arrays (n = 24) generated from patient samples (shown in Supplementary Table 6) were then queried for relative expression levels of AURKA and SHH, in a planned comparison between patients with GVHD to those without.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical assistance of Dr. Natalia Kozyr, the veterinary care provided by Dr. Elizabeth Strobert, Dr. Joe Jenkins, Dr. Annie Torrence, Dr. Keith Vogel, and Jessica Lewellen, in addition to the services provided by both the Vanderbilt Technologies for Advanced Genomics and the OHSU Gene Profiling Shared Resource.
Funding: This work was supported by Yerkes National Primate Research Center Base Grant, #RR00165. Washington National Primate Research Center at the University of Washington support was funded through the Office of Research Infrastructure Programs at the National Institutes of Health, Grant #P51 OD 010425. Funding also provided through Emory University ACTSI Pilot Grant (EKW), NIH 2 R01 HL56067, R01 AI 34495 and P01 AI 056299 (BRB), and NHLBI 5 R01 HL095791, NIAID 5U19-AI051731 (LSK).
Footnotes
Supplementary Figure 1: Sorting strategy and purity of T cells destined for microarray analysis.
Supplementary Figure 2: Successful batch effect correction for PCA.
Supplementary Figure 3: Gene set enrichment analysis (GSEA) performed using previously published gene sets related to LCMV infection reveals commonality between T cell activation in response to allo-activation and acute viral infection.
Supplementary Figure 4: Supplementary Figure 3: Selected dysregulation of Th17-associated transcripts.
Supplementary Figure 5: Aurora kinase A interactome shows little overlap with the leading edge genes in the LCMV GSEA analyses.
Supplementary Figure 6: Translational pipeline for GVHD pathway discovery.
Supplementary Figure 7: Investigation of SHH pathway enrichment in GVHD
Supplementary Table 1: Transplant cell doses for autologous and allogeneic transplant recipients.
Supplementary Table 2: Top Gene Analysis - Top 200 over- or under-represented transcripts during GVHD in NHP: Comparison between the No Rx and autologous controls.
Supplementary Table 3: GSEA Results - Gene sets enriched in No Rx vs autologous controls.
Supplementary Table 4: DAVID pathway analysis of the combined leading edge genes found in the No Rx vs autologous controls from the LCMV gene sets depicted in Figure 4C and Supplementary Figure 3.
Supplementary Table 5: DAVID pathway analysis of transcripts differentially expressed between the No Rx and autologous controls.
Supplementary Table 6: Clinical characteristics of transplant patients
Supplementary Data 1: Spreadsheet with raw NHP clinical score data, NHP survival data, NHP chimerism data, murine clinical score data, murine survival data and human MLR gene expression data (Figures 1b, 1c, 1d, 8a, 8b and 8c).
Supplementary Data 2: Spreadsheet with raw flow cytometric data from Figures 1e and 5d.
Author Contributions: L.S.K. and B.R.B conceived the study and designed experiments. S.N.F., C.G.K.Z., D.E.L.P G.K.T., and S.E.B. developed computational methods. D.E.L.P performed statistical analysis. S.N.F. analyzed the data. B.W., S.R., K.S., C.G., K.H., L.S, J.C., K.B, K.B., S.C., J.S.M. V.T. and A.G. conducted the NHP and human experiments, or provided clinical trial samples for analysis. R.F and B.R.B conducted murine experiments. S.N.F, B.R.B, E.K.W. and L.S.K wrote the paper with input from all the authors.
Competing Interests: The authors report no competing interests with any of the data presented in this work. There are no current or pending patents or material transfer agreements related to this work.
References
- 1.Macian F. NFAT proteins: key regulators of T-cell development and function. Nature Reviews Immunology. 2005;5:472–484. doi: 10.1038/nri1632. PMID: 15928679. [DOI] [PubMed] [Google Scholar]
- 2.Izawa A, Sayegh MH, Chandraker A. The antagonism of calcineurin inhibitors and costimulatory blockers: fact or fiction? Transplantation Proceedings. 2004;36:S570–S573. doi: 10.1016/j.transproceed.2004.01.020. PMID: 15041407. [DOI] [PubMed] [Google Scholar]
- 3.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. PMID: 15746082. [DOI] [PubMed] [Google Scholar]
- 4.Pulsipher MA, Langholz B, Wall DA, Schultz KR, Bunin N, Carroll WL, Raetz E, Gardner S, Gastier-Foster JM, Howrie D, Goyal RK, Douglas JG, Borowitz M, Barnes Y, Teachey DT, Taylor C, Grupp SA. The addition of sirolimus to tacrolimus/methotrexate GVHD prophylaxis in children with ALL: a phase III COG/PBMTC trial. Blood. 2014 doi: 10.1182/blood-2013-10-534297. PMID: 24497539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller WP, Srinivasan S, Panoskaltsis-Mortari A, Singh K, Sen S, Hamby K, Deane T, Stempora L, Beus J, Turner A, Wheeler C, Anderson DC, Sharma P, Garcia A, Strobert E, Elder E, Crocker I, Crenshaw T, Penedo MCT, Ward T, Song M, Horan J, Larsen CP, Blazar BR, Kean LS. GVHD after haploidentical transplantation: a novel MHC-defined rhesus macaque model identifies CD28- CD8+ T cells as a reservoir of breakthrough T-cell proliferation during costimulation blockade and sirolimus-based immunosuppression. Blood. 2010;116:5403–5418. doi: 10.1182/blood-2010-06-289272. PMID: 20833977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaliyaperumal S, Watkins B, Sharma P, Furlan S, Ramakrishnan S, Giver C, Garcia A, Courtney C, Knight H, Strobert E, Elder E, Crenshaw T, Blazar BR, Waller EK, Westmoreland S, Kean LS. CD8-predominant T-cell CNS infiltration accompanies GVHD in primates and is improved with immunoprophylaxis. Blood. 2014;123:1967–1969. doi: 10.1182/blood-2014-01-547612. PMID: 24652969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kean LS, Singh K, Blazar BR, Larsen CP. Nonhuman primate transplant models finally evolve: detailed immunogenetic analysis creates new models and strengthens the old. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12:812–819. doi: 10.1111/j.1600-6143.2011.03873.x. PMID: 22177005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirk AD. Crossing the bridge: large animal models in translational transplantation research. Immunological reviews. 2003;196:176–196. doi: 10.1046/j.1600-065x.2003.00081.x. PMID: 14617205. [DOI] [PubMed] [Google Scholar]
- 9.Koura DT, Horan JT, Langston AA, Qayed M, Mehta A, Khoury HJ, Harvey RD, Suessmuth Y, Couture C, Carr J, Grizzle A, Johnson HR, Cheeseman JA, Conger JA, Robertson J, Stempora L, Johnson BE, Garrett A, Kirk AD, Larsen CP, Waller EK, Kean LS. In vivo T cell costimulation blockade with abatacept for acute graft-versus-host disease prevention: a first-in-disease trial. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2013;19:1638–1649. doi: 10.1016/j.bbmt.2013.09.003. PMID: 24047754. [DOI] [PubMed] [Google Scholar]
- 10.Vincenti F, Charpentier B, Vanrenterghem Y, Rostaing L, Bresnahan B, Darji P, Massari P, Mondragon-Ramirez GA, Agarwal M, Di Russo G, Lin C-S, Garg P, Larsen CP. A phase III study of belatacept-based immunosuppression regimens versus cyclosporine in renal transplant recipients (BENEFIT study) American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2010;10:535–546. doi: 10.1111/j.1600-6143.2009.03005.x. PMID: 20415897. [DOI] [PubMed] [Google Scholar]
- 11.Kirk AD, Mannon RB, Kleiner DE, Swanson JS, Kampen RL, Cendales LK, Elster EA, Wakefield T, Chamberlain C, Hoffmann SC, Hale DA. Results from a human renal allograft tolerance trial evaluating T-cell depletion with alemtuzumab combined with deoxyspergualin. Transplantation. 2005;80:1051–1059. doi: 10.1097/01.tp.0000174341.49741.8f. PMID: 16278585. [DOI] [PubMed] [Google Scholar]
- 12.Vincenti F, Larsen C, Durrbach A, Wekerle T, Nashan B, Blancho G, Lang P, Grinyo J, Halloran PF, Solez K, Hagerty D, Levy E, Zhou W, Natarajan K, Charpentier B, Group BS. Costimulation blockade with belatacept in renal transplantation. The New England journal of medicine. 2005;353:770–781. doi: 10.1056/NEJMoa050085. PMID: 16120857. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, Shaffer J, Preffer FI, Ding R, Sharma V, Fishman JA, Dey B, Ko DSC, Hertl M, Goes NB, Wong W, Williams WW, Colvin RB, Sykes M, Sachs DH. HLA-mismatched renal transplantation without maintenance immunosuppression. The New England journal of medicine. 2008;358:353–361. doi: 10.1056/NEJMoa071074. PMID: 18216355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Perez SD, Cheeseman J, Mehta AK, Kirk AD. The allo-viral-specific immunosuppressive effect of belatacept, but not tacrolimus attenuates with progressive T cell maturation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14:319–332. doi: 10.1111/ajt.12574. PMID: 24472192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turgeon NA, Avila JG, Cano JA, Hutchinson JJ, Badell IR, Page AJ, Adams AB, Sears MH, Bowen PH, Kirk AD, Pearson TC, Larsen CP. Experience with a novel efalizumab-based immunosuppressive regimen to facilitate single donor islet cell transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2010;10:2082–2091. doi: 10.1111/j.1600-6143.2010.03212.x. PMID: 20883542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nature reviews Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. PMID: 15630414. [DOI] [PubMed] [Google Scholar]
- 17.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. PMID: 23719536. [DOI] [PubMed] [Google Scholar]
- 18.Friedberg JW, Mahadevan D, Cebula E, Persky D, Lossos I, Agarwal AB, Jung J, Burack R, Zhou X, Leonard EJ, Fingert H, Danaee H, Bernstein SH. Phase II study of alisertib a selective Aurora A kinase inhibitor in relapsed and refractory aggressive B- and T-cell non-Hodgkin lymphomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:44–50. doi: 10.1200/JCO.2012.46.8793. PMID: 24043741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheikh A, Alvi AA, Aslam HM, Haseeb A. Hedgehog pathway inhibitors -current status and future prospects. Infectious agents and cancer. 2012;7:29. doi: 10.1186/1750-9378-7-29. PMID: 23116301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cutler C, Antin JH. Sirolimus for GVHD prophylaxis in allogeneic stem cell transplantation. Bone marrow transplantation. 2004;34:471–476. doi: 10.1038/sj.bmt.1704604. PMID: 15273708. [DOI] [PubMed] [Google Scholar]
- 21.Alyea EP, Li S, Kim HT, Cutler C, Ho V, Soiffer RJ, Antin JH. Sirolimus tacrolimus and low-dose methotrexate as graft-versus-host disease prophylaxis in related and unrelated donor reduced-intensity conditioning allogeneic peripheral blood stem cell transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2008;14:920–926. doi: 10.1016/j.bbmt.2008.05.024. PMID: 18640576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringnér M. What is principal component analysis? Nature Biotechnology. 2008;26:303–304. doi: 10.1038/nbt0308-303. PMID: 18327243. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ZP, Eady RD, Ahmad SF, Agravat S, Morris T, Else J, Lank SM, Wiseman RW, O’Connor DH, Penedo MCT, Larsen CP, Kean LS. Immunogenetic Management Software: a new tool for visualization and analysis of complex immunogenetic datasets. Immunogenetics. 2012;64:329–336. doi: 10.1007/s00251-011-0587-8. PMID: 22080300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. PMID: 12808457. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. PMID: 16199517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams AB, Pearson TC, Larsen CP. Heterologous immunity: an overlooked barrier to tolerance. Immunological Reviews. 2003;196:1–14. doi: 10.1046/j.1600-065x.2003.00082.x. PMID: 14617203. [DOI] [PubMed] [Google Scholar]
- 27.Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, Kaech SM, Wherry EJ, Onami T, Lanier JG, Kokko KE, Pearson TC, Ahmed R, Larsen CP. Heterologous immunity provides a potent barrier to transplantation tolerance. Journal of Clinical Investigation. 2003;111:1887–1895. doi: 10.1172/JCI17477. PMID: 12813024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urbani S. Heterologous T cell immunity in severe hepatitis C virus infection. Journal of Experimental Medicine. 2005;201:675–680. doi: 10.1084/jem.20041058. PMID: 15753202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsh RM, Selin LK, Szomolanyi-Tsuda E. Immunological Memory toViral nfections. Annual Review of Immunology. 2004;22:711–743. doi: 10.1146/annurev.immunol.22.012703.104527. PMID: 15032594. [DOI] [PubMed] [Google Scholar]
- 30.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. PMID: 17950003. [DOI] [PubMed] [Google Scholar]
- 31.West EE, Youngblood B, Tan WG, Jin H-T, Araki K, Alexe G, Konieczny BT, Calpe S, Freeman GJ, Terhorst C, Haining WN, Ahmed R. Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35:285–298. doi: 10.1016/j.immuni.2011.05.017. PMID: 21856186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reshef R, Luger SM, Hexner EO, Loren AW, Frey NV, Nasta SD, Goldstein SC, Stadtmauer EA, Smith J, Bailey S, Mick R, Heitjan DF, Emerson SG, Hoxie JA, Vonderheide RH, Porter DL. Blockade of Lymphocyte Chemotaxis in Visceral Graft-versus-Host Disease. The New England journal of medicine. 2012;367:135–145. doi: 10.1056/NEJMoa1201248. PMID: 22784116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nature Medicine. 2011;17:830–836. doi: 10.1038/nm.2395. PMID: 21706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Przepiorka D, Phillips GL, Ratanatharathorn V, Cottler-Fox M, Sehn LH, Antin JH, LeBherz D, Awwad M, Hope J, McClain JB. A phase II study of BTI-322 a monoclonal anti-CD2 antibody for treatment of steroid-resistant acute graft-versus-host disease. Blood. 1998;92:4066–4071. PMID: 9834211. [PubMed] [Google Scholar]
- 35.Khandelwal P, Lane A, Chaturvedi V, Owsley E, Davies SM, Marmer D, Filipovich AH, Jordan MB, Marsh RA. Peripheral Blood CD38 Bright CD8+ Effector Memory T Cells Predict Acute Graft-versus-Host Disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2015;21:1215–1222. doi: 10.1016/j.bbmt.2015.04.010. PMID: 25881755. [DOI] [PubMed] [Google Scholar]
- 36.Das R, Komorowski R, Hessner MJ, Subramanian H, Huettner CS, Cua D, Drobyski WR. Blockade of interleukin-23 signaling results in targeted protection of the colon and allows for separation of graft-versus-host and graft-versus-leukemia responses. Blood. 2010;115:5249–5258. doi: 10.1182/blood-2009-11-255422. PMID: 20382845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manetti R, Parronchi P, Giudizi MG, Piccinni MP, Maggi E, Trinchieri G, Romagnani S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. The Journal of experimental medicine. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. PMID: 8096238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh T-Y, Watford WT, Schones DE, Peng W, Sun H-w, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. PMID: 19144320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, Coffing B, Olsen S, Choi SW, Wang H, Faca V, Pitteri S, Zhang Q, Chin A, Kitko C, Mineishi S, Yanik G, Peres E, Hanauer D, Wang Y, Reddy P, Hanash S, Ferrara JLM. Elafin is a biomarker of graft-versus-host disease of the skin. Science translational medicine. 2010;2:13ra12. doi: 10.1126/scitranslmed.3000406. PMID: 20371463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vander Lugt MT, Braun TM, Hanash S, Ritz J, Ho VT, Antin JH, Zhang Q, Wong C-H, Wang H, Chin A, Gomez A, Harris AC, Levine JE, Choi SW, Couriel D, Reddy P, Ferrara JLM, Paczesny S. ST2 as a Marker for Risk of Therapy-Resistant Graft-versus-Host Disease and Death. The New England journal of medicine. 2013;369:529–539. doi: 10.1056/NEJMoa1213299. PMID: 23924003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald GB, Tabellini L, Storer BE, Lawler RL, Martin PJ, Hansen JA. Plasma biomarkers of acute GVHD and non-relapse mortality: predictive value of measurements before GVHD onset and treatment. Blood. 2015 doi: 10.1182/blood-2015-03-636753. PMID: 68D50D7B-1CAB-4035-BDA8-093BAE0AACA7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrara JLM, Harris AC, Greenson JK, Braun TM, Holler E, Teshima T, Levine JE, Choi SWJ, Huber E, Landfried K, Akashi K, Vander Lugt M, Reddy P, Chin A, Zhang Q, Hanash S, Paczesny S. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118:6702–6708. doi: 10.1182/blood-2011-08-375006. PMID: 21979939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, Misek DE, Cooke KR, Kitko CL, Weyand A, Bickley D, Jones D, Whitfield J, Reddy P, Levine JE, Hanash SM, Ferrara JLM. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113:273–278. doi: 10.1182/blood-2008-07-167098. PMID: 18832652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piper KP, Horlock C, Curnow SJ, Arrazi J, Nicholls S, Mahendra P, Craddock C, Moss PAH. CXCL10-CXCR3 interactions play an important role in the pathogenesis of acute graft-versus-host disease in the skin following allogeneic stem-cell transplantation. Blood. 2007;110:3827–3832. doi: 10.1182/blood-2006-12-061408. PMID: 17766680. [DOI] [PubMed] [Google Scholar]
- 45.Visentainer JEL, Lieber SR, Persoli LBL, Vigorito AC, Aranha FJP, de Brito Eid KA, Oliveira GB, Miranda ECM, de Souza CA. Serum cytokine levels and acute graft-versus-host disease after HLA-identical hematopoietic stem cell transplantation. Experimental Hematology. 2003;31:1044–1050. doi: 10.1016/j.exphem.2003.08.005. PMID: 14585368. [DOI] [PubMed] [Google Scholar]
- 46.Meuwis MA, Vernier-Massouille G, Grimaud JC, Bouhnik Y, Laharie D, Piver E, Seidel L, Colombel JF, Louis E. Serum calprotectin as a biomarker for Crohn's disease. Journal of Crohn's & colitis. 2013;7:e678–e683. doi: 10.1016/j.crohns.2013.06.008. PMID: 23845231. [DOI] [PubMed] [Google Scholar]
- 47.Guardiola J, Lobaton T, Rodriguez-Alonso L, Ruiz-Cerulla A, Arajol C, Loayza C, Sanjuan X, Sanchez E, Rodriguez-Moranta F. Fecal Level of Calprotectin Identifies Histologic Inflammation in Patients With Ulcerative Colitis in Clinical and Endoscopic Remission. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2014;12:1865–1870. doi: 10.1016/j.cgh.2014.06.020. PMID: 24993368. [DOI] [PubMed] [Google Scholar]
- 48.Molander P, Farkkila M, Ristimaki A, Salminen K, Kemppainen H, Blomster T, Koskela R, Jussila A, Rautiainen H, Nissinen M, Haapamaki J, Arkkila P, Nieminen U, Kuisma J, Punkkinen J, Kolho KL, Mustonen H, Sipponen T. Does fecal calprotectin predict short-term relapse after stopping tnfalpha-blocking agents in inflammatory bowel disease patients in deep remission? Journal of Crohn's & colitis. 2015;9:33–40. doi: 10.1016/j.crohns.2014.06.012. PMID: 25052347. [DOI] [PubMed] [Google Scholar]
- 49.Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, Shan GT, Radojcic V, Friedman A, Gridley T, Shelton A, Reddy P, Samuelson LC, Yan M, Siebel CW, Maillard I. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. Journal of Clinical Investigation. 2013;123:1590–1604. doi: 10.1172/JCI65477. PMID: 23454750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Sandy AR, Wang J, Radojcic V, Shan GT, Tran IT, Friedman A, Kato K, He S, Cui S, Hexner E, Frank DM, Emerson SG, Pear WS, Maillard I. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood. 2011;117:299–308. doi: 10.1182/blood-2010-03-271940. PMID: 20870902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Díez J, Walter D, Muñoz-Pinedo C, Gabaldón T. DeathBase: a database on structure evolution and function of proteins involved in apoptosis and other forms of cell death. Cell death and differentiation. 2010;17:735–736. doi: 10.1038/cdd.2009.215. PMID: 20383157. [DOI] [PubMed] [Google Scholar]
- 52.Lin MT, Tseng LH, Frangoul H, Gooley T, Pei J, Barsoukov A, Akatsuka Y, Hansen JA. Increased apoptosis of peripheral blood T cells following allogeneic hematopoietic cell transplantation. Blood. 2000;95:3832–3839. PMID: 10845917. [PubMed] [Google Scholar]
- 53.Martinon F, Gaide O, Pétrilli V, Mayor A, Tschopp J. NALP inflammasomes: a central role in innate immunity. Seminars in Immunopathology. 2007;29:213–229. doi: 10.1007/s00281-007-0079-y. PMID: 17703304. [DOI] [PubMed] [Google Scholar]
- 54.Jankovic D, Ganesan J, Bscheider M, Stickel N, Weber FC, Guarda G, Follo M, Pfeifer D, Tardivel A, Ludigs K, Bouazzaoui A, Kerl K, Fischer JC, Haas T, Schmitt-Gräff A, Manoharan A, Müller L, Finke J, Martin SF, Gorka O, Peschel C, Ruland J, Idzko M, Duyster J, Holler E, French LE, Poeck H, Contassot E, Zeiser R. The Nlrp3 inflammasome regulates acute graft-versus-host disease. Journal of Experimental Medicine. 2013;210:1899–1910. doi: 10.1084/jem.20130084. PMID: 23980097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steele AK, Lee EJ, Manuzak JA, Dillon SM, Beckham JD, McCarter MD, Santiago ML, Wilson CC. Microbial exposure alters HIV-1-induced mucosal CD4+ T cell death pathways Ex vivo. Retrovirology. 2014;11:14. doi: 10.1186/1742-4690-11-14. PMID: 24495380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayan S, Kolly L, So A, Busso N. Increased interleukin-10 production by ASC-deficient CD4+ T cells impairs bystander T-cell proliferation. Immunology. 2011;134:33–40. doi: 10.1111/j.1365-2567.2011.03462.x. PMID: 21718313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jankovic D, Ganesan J, Bscheider M, Stickel N, Weber FC, Guarda G, Follo M, Pfeifer D, Tardivel A, Ludigs K, Bouazzaoui A, Kerl K, Fischer JC, Haas T, Schmitt-Gräff A, Manoharan A, Müller L, Finke J, Martin SF, Gorka O, Peschel C, Ruland J, Idzko M, Duyster J, Holler E, French LE, Poeck H, Contassot E, Zeiser R. The Nlrp3 inflammasome regulates acute graft-versus-host disease. The Journal of experimental medicine. 2013;210:1899–1910. doi: 10.1084/jem.20130084. PMID: 23980097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. PMID: 10592173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function other gene attributes in the context of phylogenetic trees. Nucleic Acids Research. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. PMID: 23193289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2:117–120. PMID. [Google Scholar]
- 61.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Research. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. PMID: 18981052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nature Reviews Molecular Cell Biology. 2003;4:842–854. doi: 10.1038/nrm1245. PMID. [DOI] [PubMed] [Google Scholar]
- 63.Aoyama K, Saha A, Tolar J, Riddle MJ, Veenstra RG, Taylor PA, Blomhoff R, Panoskaltsis-Mortari A, Klebanoff CA, Socie G, Munn DH, Murphy WJ, Serody JS, Fulton LM, Teshima T, Chandraratna RA, Dmitrovsky E, Guo Y, Noelle RJ, Blazar BR. Inhibiting retinoic acid signaling ameliorates graft-versus-host disease by modifying T-cell differentiation and intestinal migration. Blood. 2013;122:2125–2134. doi: 10.1182/blood-2012-11-470252. PMID: 23814022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brinkman RR, Gasparetto M, Lee S-JJ, Ribickas AJ, Perkins J, Janssen W, Smiley R, Smith C. High-content flow cytometry and temporal data analysis for defining a cellular signature of graft-versus-host disease. Biology of Blood and Marrow Transplantation. 2007;13:691–700. doi: 10.1016/j.bbmt.2007.02.002. PMID: 17531779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paczesny S. Discovery and validation of graft-versus-host disease biomarkers. Blood. 2013;121:585–594. doi: 10.1182/blood-2012-08-355990. PMID: 23165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y, Tawara I, Zhao M, Qin ZS, Toubai T, Mathewson N, Tamaki H, Nieves E, Chinnaiyan AM, Reddy P. Allogeneic T cell responses are regulated by a specific miRNA-mRNA network. Journal of Clinical Investigation. 2013 doi: 10.1172/JCI70013. PMID: 24216511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G-M, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. PMID: 21743478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology. 2009;10:116–125. doi: 10.1038/ni.1688. PMID: 19029902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyman O, Létourneau S, Krieg C, Sprent J. Homeostatic proliferation and survival of naïve and memory T cells. European Journal of Immunology. 2009;39:2088–2094. doi: 10.1002/eji.200939444. PMID: 19637200. [DOI] [PubMed] [Google Scholar]
- 70.Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nature Immunology. 2011;12:478–484. doi: 10.1038/ni.2018. PMID: 21739670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanagal-Shamanna R, Lehman NL, O'Donnell JP, Lim MS, Schultz DS, Chitale DA, Bueso-Ramos CE, Medeiros LJ, Inamdar KV. Differential expression of aurora-A kinase in T-cell lymphomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:640–647. doi: 10.1038/modpathol.2012.211. PMID: 23411487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guide for the Care and Use of Laboratory Animals. 8th. Washington (DC): National Academies Press (US; 2011. [PubMed] [Google Scholar]
- 73.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration normalization and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. PMID: 12925520. [DOI] [PubMed] [Google Scholar]
- 74.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. PMID: 16632515. [DOI] [PubMed] [Google Scholar]
- 75.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. PNAS. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. PMID: 20460310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21:2789–2790. doi: 10.1093/bioinformatics/bti394. PMID: 15797915. [DOI] [PubMed] [Google Scholar]
- 77.Duan F, Pauley MA, Spindel ER, Zhang L, Norgren RB. Large scale analysis of positional effects of single-base mismatches on microarray gene expression data. BioData Mining. 2010;3:2. doi: 10.1186/1756-0381-3-2. PMID: 20429935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gentleman R. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science+Business Media; 2005. [Google Scholar]
- 79.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. PMID: 19131956. [DOI] [PubMed] [Google Scholar]
- 80.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shinde V, Huck JJ, Wysong DR, Janowick DA, Hyer ML, Leroy PJ, Gershman RE, Silva MD, Germanos MS, Bolen JB, Claiborne CF, Sells TB. Characterization of Alisertib (MLN8237)an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. PMID: 22016509. [DOI] [PubMed] [Google Scholar]
- 81.Zerr P, Palumbo-Zerr K, Distler A, Tomcik M, Vollath S, Munoz LE, Beyer C, Dees C, Egberts F, Tinazzi I, Del Galdo F, Distler O, Schett G, Spriewald BM, Distler JHW. Inhibition of hedgehog signaling for the treatment of murine sclerodermatous chronic graft-versus-host disease. Blood. 2012;120:2909–2917. doi: 10.1182/blood-2012-01-403428. PMID: 22915638. [DOI] [PubMed] [Google Scholar]
- 82.Cooke KR, Kobzik L, Martin TR, Brewer J, Delmonte J, Crawford JM, Ferrara JL. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88:3230–3239. PMID: 8963063. [PubMed] [Google Scholar]
- 83.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Analytical chemistry. 2012;84:1003–1011. doi: 10.1021/ac202578x. PMID: 22122760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suessmuth Y, Mukherjee R, Watkins B, Koura DT, Finstermeier K, Desmarais C, Stempora L, Horan JT, Langston A, Qayed M, Khoury HJ, Grizzle A, Cheeseman JA, Conger JA, Robertson J, Garrett A, Kirk AD, Waller EK, Blazar BR, Mehta AK, Robins HS, Kean LS. CMV reactivation drives posttransplant T-cell reconstitution and results in defects in the underlying TCRβ repertoire. Blood. 2015;125:3835–3850. doi: 10.1182/blood-2015-03-631853. PMID: 25852054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. PMID: 11752295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.