Abstract
O-Glycosylation of collagen is a unique type of posttranslational modifications (PTMs), involving the attachment of galactose (Gal) or glucose-galactose (Glc-Gal) moieties to hydroxylysine (HyK). Also, hydroxyproline (HyP) result from the posttranslational hydroxylation of some proline residues in collagen. Here, LC-MS/MS was effectively employed to identify 23 O-glycosylation sites and a large number of HyP residues associated with bovine type II collagen α-1 chain (CO2A1). The modifications of the 23 O-glycosylation sites varied qualitatively and quantitatively. Both Gal and Glc-Gal moieties occupied 22 of the identified glycosylation sites while K773 was observed as unmodified. A large number of HyP residues at Yaa positions of Gly-Xaa-Yaa motif were detected. HyP residues at Xaa positions of Gly-HyP-HyP, Gly-HyP-Ala, and Gly-HyP-Val motifs were also observed. Notably, HyP residue of Gly-HyP-Gln motif was detected, which has not been previously reported. Moreover, the deamidation of 8 Asn residues was identified, of which 2 Asp residues were observed at different retention times because of isomerization (Asp vs. isoAsp). Partial macroheterogeneities of some CO2A1 glycosylation sites were revealed by LC-MS/MS analysis. ETD experiments revealed partial macroheterogeneities associated with K299-K308, K452-K464, K464-K470 and K857-K884 glycosylation sites. Semi-quantitative data suggest that the glycosylation of hydroxylysine residues is site-specific.
Keywords: type II collagen, CO2A1, hydroxylation, glycosylation, glycopeptides, LC-MS/MS, quantitation
Graphical Abstract
Introduction
Collagen is a large family of extracellular matrix proteins. It is the most abundant protein in vertebrates. Type II collagen (CO2A1) is the main component in cartilage and vitreous humor, forming a fibril assembly.1, 2 It consists of triple α-1 helix chains of repeating Gly-Xaa-Yaa sequences, where Gly is glycine while Xaa and Yaa are any amino acids. During biosynthesis, the extensive posttranslational modifications endured by this protein include hydroxylation of proline and lysine residues and O-glycosylation of hydroxylysine (HyK) in a Gly-Xaa-HyK motif.3-5 The pattern/extent of these modifications influences many biological functions, including fibrillogenesis,6, 7 cross-linking,8, 9 and matrix mineralization.10, 11
Hydroxylation of proline residues in collagens includes 4-hydroxyproline (4HyP) and 3-hydroxyproline (3HyP).4 4HyP occurs at Yaa position of Gly-Xaa-Yaa motif.4 It is well established that 4HyP stabilizes triple helix of collagen by forming intramolecular hydrogen bonding.12-14 3HyP is also found in Xaa position of Gly-Xaa-Yaa motif, but its role is unclear.4 Moreover, Gly-Xaa-Yaa motif containing both 3HyP and 4HyP in Xaa and Yaa, respectively, also stabilizes triple helix of collagen.15, 16 However, Gly-HyP-Pro motif destabilizes triple helix of collagen.17
O-Glycosylation of collagen is a unique type of posttranslational modification (PTM) that involves the attachment of galactose (Gal) or glucose-galactose (Glc-Gal) moieties to HyK residues.18-20 These modifications may regulate or control the formation of stable intermolecular cross-linking of collagen fibrils.5, 18-20 The extent of hydroxylation on lysine varies among tissues and collagen types. In the 1970s, the CO2A1 sequence was determined by automated Edman degradation.19, 21-23 In these studies, “blanks” were assigned as hydroxylysine.
Liquid chromatography interfaced to mass spectrometry (LC-MS) is now routinely utilized for the identification and quantification of protein PTMs, such as glycosylation. Glycoproteins are enzymatically digested with proteases such as trypsin prior to chromatographic separation and MS and MS/MS analyses.24, 25 Several studies have shown that typical N or O-glycosylation could be successfully identified and quantified through LC-MS and LC-tandem MS (LC-MS/MS).26-28 The applicability of these methods to investigate aberrations in glycosylation associated with disease samples has been demonstrated.29-31 Enrichment techniques, such as lectin affinity chromatography, or hydrazide chemistry, and peptide labeling have also been used in conjunction with LC-MS/MS to characterize and quantify the glycosylation sites of glycoproteins.32-34
Tandem MS permits a reliable characterization of O-glycopeptides. Collision induced dissociation (CID) MS/MS of glycopeptides produces glycan fragments with minimal peptide backbone fragmentation.35, 36 Since electron transfer dissociation (ETD) occurs along the peptide backbone in a sequence-independent manner, preserving PTMs.37, 38 ETD offers complementary information in sequencing peptide backbone retaining the glycan moiety (ties) intact on the glycosylation sites.39, 40 Several studies have demonstrated the effectiveness of ETD along with CID for a better characterization of glycopeptides.41, 42 Also, MS3 which enables effective sequencing of the peptide backbone has been utilized along with CID and/or ETD.43-45
Several studies have reported the characterization of hydroxylation or glycosylation of different types of collagen using LC-MS or LC-MS/MS. Different modifications associated with human type II collagen have recently been analyzed by Edman degradation and ion trap mass spectrometer in conjunction with gelatinase B.46 The identification of glycopeptides from type I and II collagen using hydrazide-based chemistry enrichment in conjunction with LC-MS/MS was recently described.47 Also, the levels of proline hydroxylation in recombinant collagen variants was recently investigated by LC-MS.48 Yang et al. 49 have comprehensively characterized the hydroxylation and glycosylation of type V collagen by LC-MS/MS, employing CID, ETD, higher-energy collision dissociation (HCD), and high order tandem MS. They identified a large number of HyP residues in Gly-Xaa-Yaa at both Xaa and Yaa positions. Unusual hydroxyproline of Gly-HyP-Val and Gly-HyP-Ala motifs were also reported.
Another modification of collagen that has been recently reported is the deamidation of asparagines residues.50 Deamidation occurs as non-enzymatic post-translational modification resulting in the creation of aspartic acid (Asp) and isoaspartic acid (isoAsp) isomers. Unique diagnostic c/z ions formed in high resolution ETD as a result of cleaving Asp or isoAsp N-Cα bonds allowed the distinction between such isomers.
Herein, we report the identification and quantification of O-glycosylation sites of CO2A1 using LC-MS/MS. We also report the identification of hydroxylation of proline residues and deamidation of asparagine residues. This was achieved using different fragmentation experiments, including CID, ETD and HCD. The extent of glycosylation of each hydroxylysine residue was semi-quantitatively assessed. Evaluating the macroheterogeneity of some glycosylation sites was achieved through ETD analyses.
Experimental Section
Materials
A pepsinized immunization grade bovine collagen α-1 (II) chain (CO2A1) was acquired from Chondrex (Redmond, WA). Ammonium bicarbonate, sodium phosphate, and MS-grade formic acid were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade solvents, including methanol and isopropanol, were procured from Fisher Scientific (Pittsburgh, PA). HPLC grade water was acquired from Mallinckrodt (Hazelwood, MO). HPLC grade acetonitrile was acquired from J.T.Baker (Phillipsburg, NJ). Mass spectrometry grade Trypsin Gold was obtained from Promega (Madison, WI) while endoproteinase GluC was purchased from New England Biolabs (Ipswich, MA).
Enzymatic Digestion
CO2A1 samples were digested with GluC and trypsin. Two 20-μg aliquots of CO2A1 were prepared in 50mM ammonium bicarbonate buffer (ABC) while a 20-μg aliquot was prepared in 50mM phosphate-buffered saline (PBS). In both cases, denaturation was performed at 65° C for 2 hours. The samples prepared in PBS and one of the samples prepared in ABC were subjected to GluC digestion by adding a 1-μl aliquot of the enzyme (enzyme/substrate ratio of 1:20 w/w) to each sample and incubating at 37.5°C for 16 hours. The second sample prepared in ABC was subjected to tryptic digestion by adding a 0.4-μl aliquot of trypsin (enzyme/substrate ratio of 1:50 w/w) at 37.5° C for 16 hours. To ensure complete enzymatic digestion, microwave digestion was performed at 45°C and 50W for 30 min.51 Both GluC and tryptic digestion were quenched through the addition of 0.5-μl aliquots of neat formic acid to the samples.
LC-MS/MS
Trypsin and GluC digested peptides were subjected to LC-MS/MS analysis using Dionex 3000 Ultimate nano-LC system (Dionex, Sunnyvale, CA) interfaced to LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) equipped with a nano-ESI source. The samples were initially online-purified using a PepMap 100 C18 cartridge (3 μm, 100Å, Dionex). The purified peptides were then separated using a PepMap 100 C18 capillary column (75 μm id × 150 mm, 2 μm, 100Å, Dionex). The separation of peptides was achieved at 350 nl/min flow rate, using the following gradient: 0-10 min 3% solvent B (98% ACN with 0.1% formic acid), 10-35 min ramping solvent B 3-10%, 35-40 min ramping solvent B 10-15%, 40-43 min ramping solvent B 15-25%, 43-46 min ramping solvent B 25-80%, 46-50 min maintaining solvent B at 80%, 50-51 min decreasing solvent B 80-3%, and 51-60 min sustaining solvent B at 3%. Solvent A was a 2% ACN aqueous solution containing 0.1 % formic acid. The separation and scan time was set to 60 min.
The LTQ Orbitrap Velos mass spectrometer was operated with three scan events. The first scan event was a full MS scan of 380-2000 m/z at a mass resolution of 15,000. The second scan event was CID MS/MS of parent ions selected from the first scan event with an isolation width of 3.0 m/z, a normalized collision energy (CE) of 35%, and an activation Q value of 0.250. The third scan event was set to acquire HCD MS/MS of the parent ions selected from the first scan event. The isolation width of HCD experiment was set to 3.0 m/z while the normalized CE was set to 45% with an activation time of 0.1ms. The CID and HCD MS/MS were performed on the 8 most intense ions observed in the MS scan event. In a separate LC-MS/MS, ETD MS/MS was conducted in conjunction with CID and HCD. The first scan event was a full MS scan and 15 scan events were followed alternating between CID, HCD, and ETD. This enabled MS/MS of the 5 most intense ions observed in the first scan. For ETD, the isolation width was set to 4.0 m/z and the default charge state was set to 4. The reaction time was set to 150 ms with a supplemental activation. The LTQ Orbitrap Velos mass spectrometer was externally calibrated, permitting < 2ppm mass accuracy.
Data Processing and Quantitation
The identification of CO2A1 peptides/glycopeptides was achieved using MASCOT database and Scaffold 3 software. Proteome Discoverer version 1.2 software (Thermo Scientific, San Jose, CA) was used to generate a mascot generic format file (*.mgf) which was subsequently employed for database searching using MASCOT version 2.3.2 (Matrix Science Inc., Boston, MA) and Scaffold 3 (Proteome Software, Inc., Portland, OR). Parent ions were selected from a mass range of 350-5000Da with a minimum peak count of 1. The parameters from Mascot Daemon were set to search against the Swissprot database. Oxidation of methionine was set as a variable modification. Additionally, several PTMs of CO2A1 were set as variable modifications, including hydroxylation of proline and lysine, and glycosylation of hydroxylysine with 1 Hex and 2 Hex. An m/z tolerance of 5 ppm was set for the identification of peptide/glycopeptide with 2 missed cleavages. Also, tandem MS ions were searched within 0.8 Da mass tolerance. Additionally, manual searching of theoretical m/z was performed using mass accuracy < 5ppm. The m/z values were based on every theoretical peptide backbone sequence using the PeptideMass tool from the ExPASy website. Then, hydroxylation and glycosylation modifications were applied, and theoretical m/z values were generated. These m/z values were employed to create extracted ion chromatograms (EICs) using Xcalibur Qual Broswer 2.1 (Thermo Scientific, San Jose, CA). A mass accuracy of 5ppm or better was then employed to confirm ions prior to manual evaluation of tandem MS data. Manual inspection of tandem MS data was employed for these cases. We only considered and assigned a fragment ion if its S/N is equal or higher than 3. Moreover, ions with such S/N are only considered if they appear in more than one tandem mass spectra associated with a particular m/z value. From multiple MS/MS spectrum, the unique y/b ions or c/z ions of specific modification were subsequently confirmed. CID/HCD (/ETD) MS/MS was repeated using the mass list of the theoretical m/z values. Assignments of peptide modifications were attained using both MASCOT database searching and manual interpretations.
For quantitation, peak heights were acquired using Xcalibur Qual Browser. The software was employed to generate EICs. Mass range was set to full FTMS scan with smoothing of 7 points enabled and mass tolerance of 10 ppm allowed. Peak apex was determined as peak height with a complete peak shape. Invariably detected peptides were selected as internal standards against which all data were normalized to offset ESI spray variations. Three tryptic peptides with the following amino acid sequences GEAGAQGPMGPAGPAGAR, GFpGLpGPSGEPGK, and GFTGLQGLpGPpGPSGDQGASGPAGPSGPR were selected to normalize quantitative data. These peptides were detected at 18.66 min, 34.67min, and 46.44 min, respectively. Every modified peptide sequences possessing a glycosylation site was considered for semi-quantitation.
Results and Discussion
GluC and trypsin were used for the digestion of CO2A1. The enzymatic activity of GluC depends on the buffer used (phosphate vs. bicarbonate), while trypsin cleaves C-terminus of lysine and arginine. GluC only cleaves C-terminus of glutamic acid in a bicarbonate buffer (pH 8.5) while it cleaves C-terminal glutamic acid and aspartic acid in a phosphate buffer (pH 7). The rate of cleavage at aspartic acid is rather slow relative to glutamic acid. The use of multiple enzymes and different digestion conditions was utilized to attain higher sequence coverage.
Identification of O-glycosylation and Hydroxylation of Amino Acid Residues
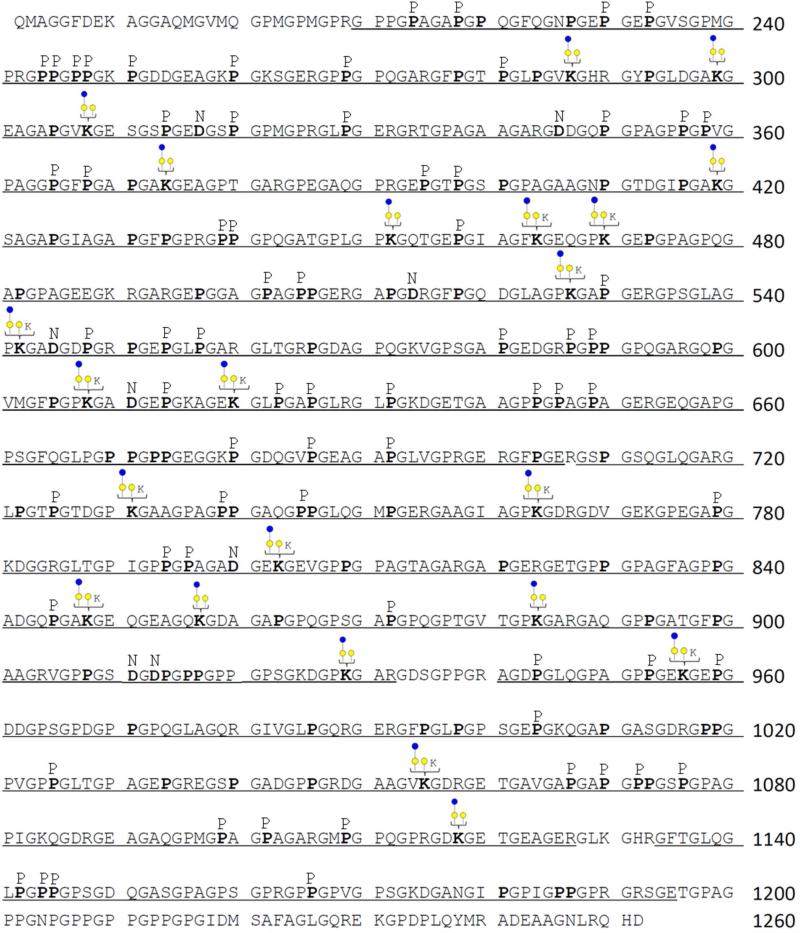
CO2A1 possesses 24 potential glycosylation sites with a Gly-Xaa-HyK motif.1, 2, 19 Here, LC-MS/MS analyses of trypsin and GluC digested bovine CO2A1 allowed the identification of 23 identified glycosylation sites with sequence coverage of ca. 90% as shown in Figure 1. Of the 24 potential glycosylation sites associated with CO2A1, only K1130 was not detected. We believe that peptide sequences generated by the protease containing K1130 were either too short or too large to be effectively observed by LC-MS/MS. The abovementioned sequence coverage does not include signal and propeptides from 1 to 201 and the C-telopeptide region from 1253 to 1487, since the sample used in this study is pepsinized. In Figure 1, detected sequences are underlined, and hydroxylated P/K are designated with bold letters. Letters P or K listed above the sequence indicate the detection of the residue as both modified and unmodified. 104 HyP residues were Yaa residue in Gly-Xaa-Yaa motif while 14 HyP residues were Xaa residue. Previously, two of Xaa positions of Gly-HyP-HyP from CO2A1 have been reported which are found at P1144 and P1186.52 We also determined 7 unusual HyP residues in Gly-HyP-Ala and a HyP residue in Gly-HyP-Val motifs, which were also reported by Yang et al,49. However, we here observed a HyP in Gly-HyP-Gln at P220 which has not been previously reported. As suggested by Yang et al49 and observed here, it appears that unusual HyPs at Xaa position would occur in different motifs associated with different types of collagens. The ETD or CID mass spectra of 9 peptides with unusual hydroxyproline residues are included as Supplementary Spectra. The glycosylation sites of CO2A1 identified here were compared to what has been reported by Butler and Francis et al,21-23 Seyer et al,53 and Taga et al47 (see Supplementary Table 1).
Figure 1.
Mapping different modifications of CO2A1 protein with sequence coverage of ca. 90%. Signal and propeptides (1~201), and C-telopeptide (1253~1487) are italicized since the sample was already pepsinized by the vendor. Identified amino acids from MASCOT database search are underlined. Hydroxylated proline or lysine residues are bold while unmodified forms are not. If modified and unmodified were detected, additional letters P or K are added in the upper line of the sequence. Glc-Gal or Gal modified lysines are represented.
There is a 98% amino acid sequence overlap (homology) between bovine and human CO2A1. Both bovine and human CO2A1 have 24 Gly-Xaa-HyK glycosylation motifs. In Supplementary Figure 1, the different PTMs associated with bovine CO2A1 as determined in this study were compared to those of human CO2A1, which were recently reported by Van den Steen et al.46 They have detected 19 Gly-Xaa-HyK glycosylation motifs in human CO2A1 while peptides containing K419, K452, K731, K929, and K1130 as part of Gly-Xaa-HyK glycosylation motifs were not observed (Supplementary Figure 1). Lysine residues at K308, K374, K527, K764, and K1055 in human CO2A1 were observed to be unmodified; however, these same residues were observed to be modified in bovine CO2A1. K287, K299, K470, and K542 in both human and bovine CO2A1 were observed as hydroxylated and modified with either Glc-Gal or Gal moieties. The PTMs of the remaining lysine residues with Gly-Xaa-HyK glycosylation motifs in human and bovine CO2A1 were not comparable. For example, K773 in bovine CO2A1, as determined here, was detected as unmodified, but K773 in human CO2A1 was observed as hydroxylated and modified with either Glc-Gal or Gal moieties. Although bovine and human CO2A1 proteins have the same Gly-Xaa-HyK motifs, their PTMs were not exactly the same.
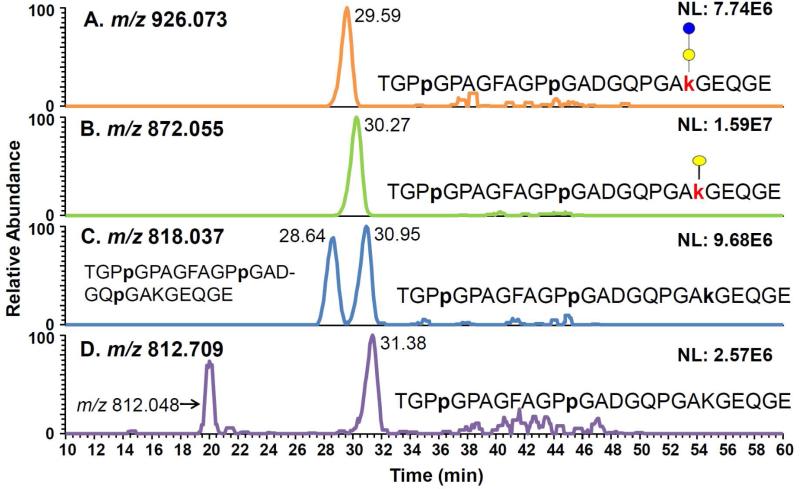
In some cases, the identification of PTMs associated with a specific lysine residue was facilitated by differences in the retention times. For example, the EICs of TGPPGPAGFAGPPGADGQPGAKGEQGE peptide with different modifications of proline and lysine residues are shown in Figure 2. The lysine residue at 848 (K848) of this peptide sequence is detected as unmodified, hydroxylated or glycosylated with either Glc-Gal or Gal moieties. Two of the proline residues (P839 and P830) associated with this peptide sequence were only observed as hydroxylated. The abovementioned peptide sequence modified with Glc-Gal moiety has an m/z value of 926.073 (+3) and retention time of 29.59 min (Figure 2A). The same peptide with a modification of Gal moiety has an m/z value of 872.055 (+3) and retention time of 30.27 min (Figure 2B). At m/z 818.037 (+3), two isomers were observed (Figure 2C). The peptide eluted at 28.64 min was the peptide sequence TGPPGPAGFAGPPGADGQPGAKGEQGE with the hydroxylation of 2 consensus proline residues (P839 and P830) and an additional proline residue (P845). The other isomer observed at 30.95 min was the same peptide sequence with the hydroxylation of 2 consensus proline residues (P839 and P830) and the lysine residue (K848). The retention time of TGPPGPAGFAGPPGADGQPGAKGEQGE peptide with HyK was higher than that with HyP. This is in agreement with what has been previously reported.54 The peptide sequence containing the unmodified K848 was the most retained structure (31.38 min) as shown in Figure 2D. Accordingly, it appears that the hydroxylation and glycosylation of this peptide sequence reduces hydrophobicity of the structures and subsequently decreases retention times.
Figure 2.
EICs of TGPpGPAGFAGPpGADGQPGAkGEQGE peptide with modifications of Glc-Gal(A), Gal(B), and HyK(B) and unmodified K(D). At m/z 818.037(+3), two isomers were observed. The peptide eluted at 28.64 min was determined to be the peptide sequence TGPPGPAGFAGPPGADGQPGAKGEQGE with hydroxylations of 3 prolines while the one observed at 30.95 min was hydroxylated on 2 prolines and K848.
Isomeric separation deamidated Asn residues at N612 and N800 with different retention times was also observed (Supplementary Table 2). Such isomers are observed as either Asp or isoAsp. Although ETD spectrum did not provide enough information to assign the elution orders of these isomers, the isoAsp containing peptide are believed to elute before Asp counterpart similar to what have been previously reported.55-57 In other cases CID, HCD, and ETD tandem MS has facilitated the identification of many PTMs.
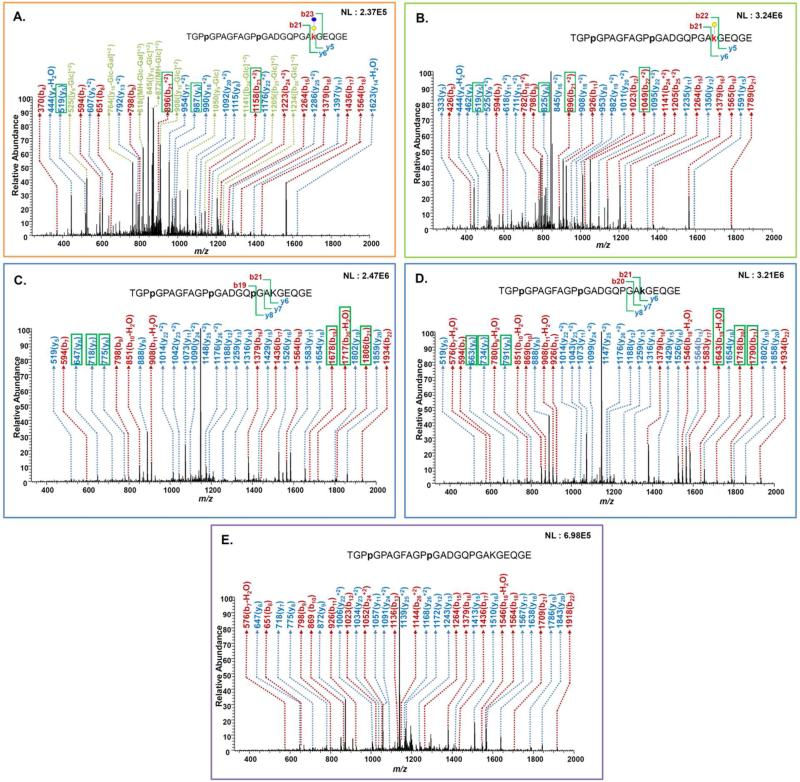
The identifications of the majority of the modified peptide sequences summarized in Supplementary Table 2 were achieved employing different fragmentation mechanisms, including CID, HCD and ETD. For example, Figure 3 depicts the CID tandem MS of the peptide sequence TGPPGPAGFAGPPGADGQPGAKGEQGE with different modifications. Figure 3A illustrates CID tandem MS of Glc-Gal-HyK modified peptide with m/z 926.073 (+3). This ion exhibited a very complex fragmentation pattern since Glc and Glc-Gal are easily lost during the CID process. For this ion, fragment ions originating from the loss of Glc and Glc-Gal were observed in the CID tandem MS. The fragment ion representative of a neutral loss of Glc was observed at m/z 872 (+3) while a fragment ion representative of neutral loss of Glc-Gal was also observed at m/z 818 (+3). Both fragment ions were observed at high intensities. As depicted in Figure 3B, the fragment ions originating from the Gal modified peptide sequence (m/z 872.055, +3) were mainly peptide fragments with intact Gal moiety. A very limited number of y or b ions with a loss of Gal were observed in the spectrum. Figure 3C exhibits CID tandem MS of the isomer observed at 28.64 min with a hydroxylation on a different proline and not on K848. On the other hand, Figure 3D is a CID tandem MS of the other isomer detected at 30.95 min, in which K848 is hydroxylated. The CID of unmodified K848 peptide is shown in Figure 3E.
Figure 3.
CID tandem MS of TGPpGPAGFAGPpGADGQPGAkGEQGE peptide modified with Glc-Gal, Gal, and HyK, and without modification on K848. CID tandem MS of the peptide modified with Glc-Gal moiety (A) and modified with Gal moiety (B). CID tandem MS of m/z 818.037(+3) representing peptide modified with HyP but not HyK(C) and modified with HyP and HyK(D). CID tandem MS of the peptide with no modification of K848(E).
There are many isomeric peptides/glycopeptides associated with CO2A1 because there are many possible combinations for the same masses with multiple hydroxyproline and hydroxylysine residues. As shown in Figure 2C, the retention times of two isomeric peptides were notably different, although they only differ in the hydroxylation of a single amino acid residue. They were unequivocally identified by tandem MS as depicted in Figures 3C and D. Each of these isomers has a unique series of y ions (y6, y7, and y8) and b ions (b19, b20, and b21) associated with the PGAK sequence. For example, the y6 fragment ion at m/z 647 (+1) corresponds to the N-terminus HyP fragment (Figure 3C) while the y6 fragment ion at m/z 663 (+1) corresponds to the N-terminus HyK fragment (Figure 3D). The ion with m/z 647 (+1) is a fragment ion unique for the HyP modified peptide, while the ion with m/z 663 (+1) is a fragment ion unique to the HyK modified peptide. The ETD or CID mass spectra of 48 peptides containing unmodified, glycosylated, or/and hydroxylated lysine residues are available as
Supplementary Spectra
Microheterogeneity of Glycosylation Sites
The assignment of glycosylation sites by CID tandem MS when multiple potential sites are present in a peptide sequence may not be feasible because of the limited number of b and y fragment ions detected. Also, fragments involving a loss of Glc and Gal prompt highly complex patterns, thus making the identification of the glycosylation sites quite challenging. Therefore, ETD experiments were employed to confirm the glycosylation sites of CO2A1 and their micro- and macroheterogeneities.
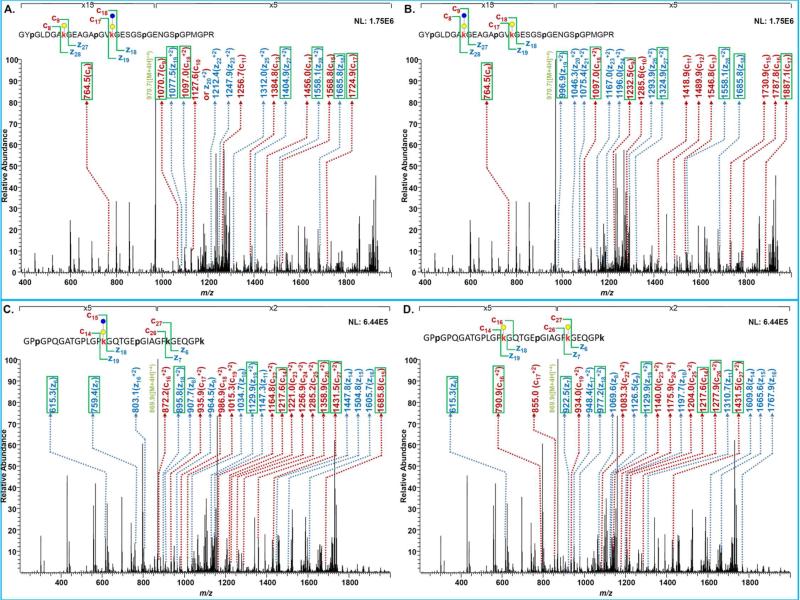
Figure 4A and 4B depict an ETD tandem mass spectrum of two glycoforms associated with a peptide sequence possessing two possible glycosylation sites (K299 and K308). The fragment ions originating from the first glycoform with a Gal moiety on K299 and Glc-Gal moiety on K308 are labeled in Figure 4A while the fragment ions originating from the second glycoform with Glc-Gal moiety on K299 and Gal moiety on K308 are labeled in Figure 4B. These two glycoforms co-eluted as suggested by the co-existence of both characteristic ions in the same spectrum. In Figure 4A, the m/z difference between c8 and c9 fragment ions and z27 and z28 fragment ions is 306, which corresponds to the presence of Gal moiety at K299 site. The m/z difference between c17 and c18 fragment ions and z18 and z19 fragment ions is 468, which corresponds to the presence of Glc-Gal moiety at K308 site. On the other hand, the characteristic fragment ions suggesting the other glycoform, including c8 and c9 fragment ions and z27 and z28 fragment ions (a difference of 468) as well as c17 and c18 fragment ions and z18 and z19 fragment ions (a difference of 306), are shown in Figure 4B. In addition to those diagnostic ions of glycosylated lysine residues, annotations of a series of different fragment ions between c9 and c17 and z19 and z27 confirmed the co-existence of two glycoforms on K299 and K308.
Figure 4.
ETD spectra of glycopeptides possessing multiple glycosylation sites of K299 and K308 showing assignments of Gal moiety on K299 with Glc-Gal moiety on K308 (A) and assignments of Glc-Gal moiety on K299 with Gal moiety on K308 (B). Other ETD spectra of glycopeptides possessing multiple glycosylation sites of K452 and K464 with assignment of Glc-Gal moiety on K452 while hydroxylated K464(C) and assignment of Gal moieties on both K452 and K464(D).
Figure 4C and 4D depict the ETD tandem mass spectrum of the glycoforms observed for another peptide sequence possessing two glycosylation sites (K452 and K464). The fragment ions labeled in Figure 4C originate from the occupation of K452 with Glc-Gal moiety and the hydroxylation of K464. These modifications were determined based on assignments of c14 and c15 fragment ions and z18 and z19 fragment ions (a difference of 468) while those of c26 and c27 fragment ions and z6 and z7 fragment ions (a difference of 144). In Figure 4D, the different m/z values of those fragment ions from the same ETD spectra support the occupancy of each K452 and K464 with a Gal moiety. Additionally, fragment ions suggesting the hydroxylation ofK452 and occupancy K464 with Glc-Gal were not observed.
Macroheterogeneity of Glycosylation Sites
The macroheterogeneity of a glycoprotein represents the variability of glycan occupancies at multiple glycosylation sites of the same protein, resulting in different copies of the protein. Enzymatic or chemical proteolysis, in bottom-up approach, facilitates MS or tandem MS analysis of modified amino acid sequences. However, it is quite challenging to study the macroheterogeneity of glycosylation sites since there is a lack of information on different copies of protein modification. Here in the case of CO2A1, the tryptic digestion generated several peptides containing two glycosylation sites, such as K299 and K308, K452 and K464, K464 and K470 as well as K857 and K884, thus enabling the assessment of the macroheterogeneity of these sites.
As shown in Figure 4A and 4B, K299 and K308 co-exist in a tryptic peptide with an m/z value of 969.9250 (+4) with different glycoforms. The tandem MS data suggest that there are two glycoforms associated with this m/z value. The first has K299 modified with Gal moiety and K308 modified with Glc-Gal moiety while the second has K299 modified with Glc-Gal moiety and K308 modified with Gal moiety. The chromatographic conditions employed here were not sufficient to resolve such isomers. Based on the intensities of fragment ions observed in the spectrum, the isoform with K299 modified with Gal moiety and K308 modified with Glc-Gal moiety appears to be more abundant than the other isoform. A tryptic peptide containing K299 modified with Glc-Gal moiety and K308 as hydroxylated was also detected at m/z value of 929.4114 (+4). ETD spectrum of this m/z value suggested the presence of only one form (data not shown). Another tryptic peptide with m/z value of 1010.4374 (+4) corresponds to both K299 and K308 modified with Glc-Gal moieties. Other peptides with the different number of HyP residues and with deamidation were also observed with the same macroheterogeneities (data not shown). Accordingly, the macroheterogeneity associated with those two sites are 2 Glc-Gal moieties on K299 and K308, Gal moiety on K299 with Glc-Gal moiety on K308, Glc-Gal moiety on K299 with Gal moiety on K308, and Glc-Gal moiety on K299 with hydroxylation on K308.
Isoforms associated with the glycosylation of K452 and K464 were observed at m/z 869.1708 (+4) as illustrated in Figure 4C and 4D. The tandem MS depicted in Figure 4C suggests K452 modification with Glc-Gal moiety and hydroxylation of K464. On the other hand, the tandem MS shown in Figure 4D suggests that both K452 and K464 are modified with a Gal moiety. The other possible isoform consisting of hydroxylated K452 and K464 modified with Glc-Gal moiety was not observed. By comparing intensities of fragment ions observed in ETD spectrum, both isoforms appear to be present at comparable levels. Another isoforms associated with K452 and K464 was detected at m/z value of 909.6833 (+4). These isoforms were determined to possess K452 modified with Gal moiety and K464 modified with Glc-Gal moiety as well as K452 modified with Glc-Gal moiety and K464 modified with Gal moiety. The levels of these isoforms appear to be comparable. Multiple glycosylation with 2 Glc-Gal moieties on K452 and K464 was also detected at m/z value of 946.1980 (+4) and 950.1942 (+4). These tryptic peptides contain unmodified and hydroxylated K470, respectively. Consequently, macroheterogeneities associated with K452-K464 appear to be 2 Glc-Gal moieties on K452 and K464, Gal moiety on K452 with Glc-Gal moiety on K464, Glc-Gal moiety on K452 with Gal moiety on K464, Glc-Gal moiety on K452 with hydroxylation on K464, and 2 Gal moieties on K 452 and K464.
A tryptic peptide possessing both K464 and K470 was observed. ETD tandem MS data suggested the presence of different forms, including 2 Glc-Gal moieties on both K464 and K470 (m/z 1057.4783, +4), Glc-Gal moiety on K464 and a Gal moiety on K470 (m/z 1016.9632, +4), and Glc-Gal moiety on K464 with hydroxylated K470 (m/z 976.4493, +4). No other combinations were observed. Another tryptic peptide with K857 and K884 was observed as having 2 Glc-Gal moieties on each K857 and K884 (m/z 1048.9772, +4), Gal moiety on K857 and Glc-Gal moiety on K884 (m/z 1008.4650, +4), and hydroxylation on K857 with Glc-Gal moiety on K884 (m/z 967.9489, +4) with 2HyPs and m/z 963.9534 (+4) with 1HyP). Accordingly, ETD tandem MS data have permitted the assignment of partial macroheterogeneity of several glycosylation sites.
Semi-quantitation of O-linked Glycopeptides from Collagen α-1 (II) Chain
Several glycoforms and isoforms were observed for each glycosylation site. Therefore, the peak heights of all observed peptides representative of a glycosylation site were added to depict the abundance of such site. We are aware of the fact that there are two major challenges associated with label-free quantitation: spray variability and the discrepancy in the ionization efficiencies of peptides/glycopeptides. Therefore, we have applied normalization and relative comparison of abundances to improve the reliability in quantitation.58-60 Raw intensities were normalized using invariably present peptides detected at different retention times to offset the spray variability. The quantitative data were compared as percentages of different modifications at the same glycosylation site, thus compensating for the different ionization efficiencies of peptides. Hence, this semi-quantitative approach can be employed to compare different modifications of each glycosylation site associated with different samples.
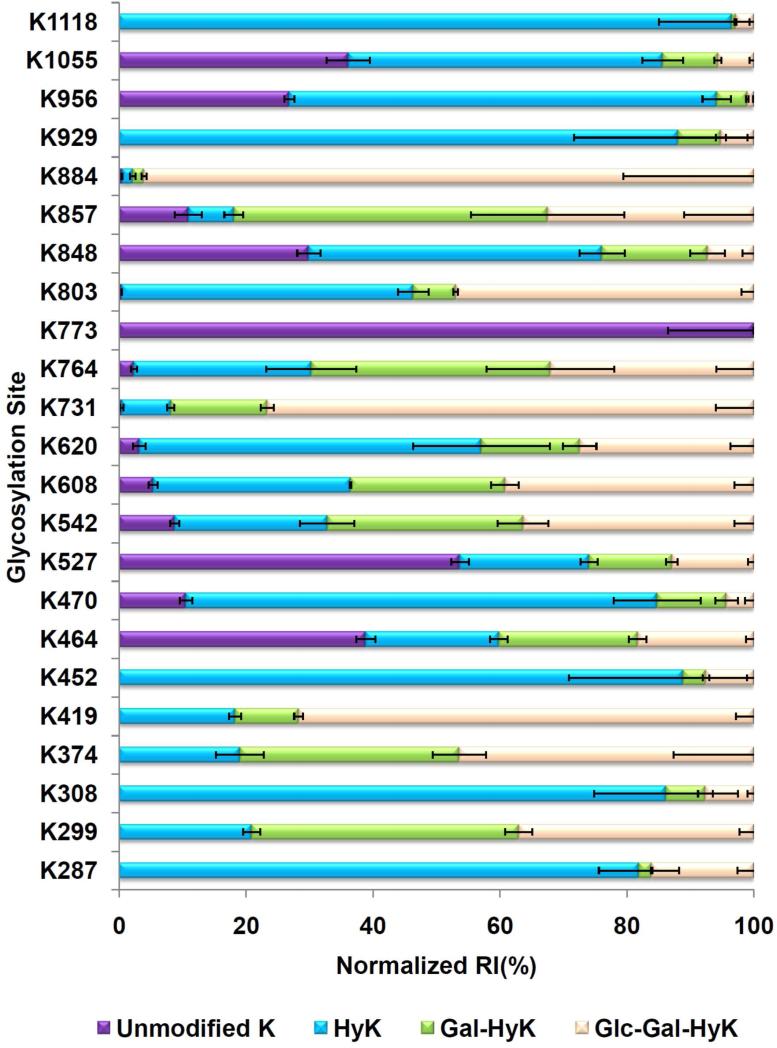
Table 1 and Figure 5 summarize the abundances of all the CO2A1 peptide sequences possessing Gly-Xaa-HyK motif with different posttranslational modifications. The sequences, m/z values, retention times, and mass accuracies of all the modified peptides used for quantitation are summarized in Supplementary Table 2. All modified peptide sequences were supported by MS/MS and/or at least 5ppm mass accuracy. Using the abovementioned semi-quantitative approach, the data indicate that the glycosylation of hydroxylysine residues is not only dependent on the extent of hydroxylation of the lysine residues but also dependent on other factors such as steric hindrance. As summarized in Table 1 and shown in Figure 5, K1118, K929, K452, K419, K374, K308, K299 and K287 were only observed as hydroxylated or glycosylated. Unmodified forms of K731 (<1%) and K803 (2%) were only observed at trace levels. K884 (97.7%), K731 (92%), K419 (82%), K374 (81%), K857 (81%), K299 (79%), K764 (70%), K542 (67%), and K608 (63%) were mainly detected as glycosylated with either Glc-Gal or Gal moieties. On the other hand, K1118 (3%), K1055 (14%), K956 (6%), K452 (11%) K308 (14%) and K287 (18%) exhibited low total glycosylation. K1118 (97%), K452 (89%), K929 (88%), K308 (86%), K287 (82%), K470 (74%), K848 (62%) and K620 (54%) are mainly observed as hydroxylated (Table 1 and Figure 5). K773 (100%) was only observed as unmodified while K527 (54%) and K464 (39%) were mainly observed as unmodified.
Table 1.
Summary of semi-quantitation results by normalized relative intensities (RI).
| Glycosylation Site | Glc-Gal-HyK Normalized RI | Gal-HyK Normalized RI | HyK Normalized RI | Unmodified K Normalized RI |
|---|---|---|---|---|
| K287 | 16±3 | 2.0±0.1 | 82±6 | N/D |
| K299 | 37±2 | 42±2 | 21±1 | N/D |
| K308 | 8±1 | 6±1 | 86±11 | N/D |
| K374 | 46±13 | 35±4 | 19±4 | N/D |
| K419 | 72±3 | 10.0±0.7 | 18±1 | N/D |
| K452 | 7.4±1.0 | 3.6±0.5 | 89±18 | N/D |
| K464 | 18±1 | 22±1 | 21±1 | 39±2 |
| K470 | 4±1 | 11±2 | 74±7 | 11±1 |
| K527 | 12.9±0.9 | 13.1±0.9 | 20±1 | 54±1 |
| K542 | 36±3 | 31±4 | 24±4 | 9.0±0.7 |
| K608 | 39±3 | 24±2 | 31.4±0.1 | 5.6±0.7 |
| K620 | 27±4 | 15±3 | 54±11 | 4±1 |
| K731 | 77±6 | 15±1 | 7.6±0.6 | 0.4±0.3 |
| K764 | 32±6 | 38±10 | 28±7 | 2.0±0.5 |
| K773 | N/D | N/D | N/D | 100±14 |
| K803 | 46±2 | 6.6±0.4 | 45±2 | 2.4±0.1 |
| K848 | 8±2 | 17±3 | 62±4 | 13±1 |
| K857 | 32±11 | 49±12 | 8±1 | 11±2 |
| K884 | 96±21 | 1.8±0.4 | 1.7±0.4 | 0.50±0.09 |
| K929 | 5.2±0.9 | 6.8±0.8 | 88±16 | N/D |
| K956 | 1.00±0.08 | 4.9±0.2 | 67±2 | 27.1±0.8 |
| K1055 | 5.4±0.6 | 8.6±0.6 | 50±3 | 36±3 |
| K1118 | 2.5±0.7 | 0.5±0.1 | 97±11 | N/D |
Figure 5.
Quantitative results of 23 glycosylation sites which have included all the modified peptide sequences of a glycosylation site and been normalized.
Semi-quantitation of micro- and macroheterogeneity of bovine CO2A1 glycosylation correlated to proline or lysine hydroxylations could help in better understanding the biological roles of these modifications in fibrillogenesis, cross-linking, or matrix mineralization. This comprehensive characterization could enable better understanding the biological roles of CO2A1 posttranslational modifications in several diseases, including rheumatoid arthritis or collagen-induced arthritis.
Conclusion
Here, LC-CID/HCD/ETD-MS/MS allowed comprehensive characterization of O-glycosylation and hydroxylation of amino acids residues associated with CO2A1 protein. 23 CO2A1 lysine residues were observed as unmodified, hydroxylated or glycosylated with Glc-Gal or Gal moieties. Employing different types of tandem MS permits the characterization of CO2A1 glycosylation sites. ETD experiments effectively helped in confirming the presence or absence of multiple K glycosylation sites on a peptide. Moreover, partial macroheterogeneities were described for K299-K308, K452-K464, K464-K470 and K857-K884. The 23 glycosylation sites were modified to different levels and with different modifications. Also, a high level of HyP residues was observed. 104 HyPs were found in Yaa position of Gly-Xaa-Yaa motif and 14 HyPs were in Xaa position of Gly-HyP-HyP motif. Unusual motifs with HyPs at Xaa positions were detected, such as Gly-HyP-Ala, Gly-HyP-Val, and Gly-HyP-Gln. It appears that HyPs at Xaa positions would be observed in different motifs associated with different types of collagen.
The present results were compared with previous studies in terms of the number of identified glycosylation sites and quantitation of the levels of modification of such sites. Additional studies are needed to assess the correlation between glycosylation of K and hydroxylation of adjacent P/K associated with the conformations and 3-D structure of this glycoprotein.
Supplementary Material
Acknowledgments
This work was supported by the Office of the Vice President for Research at Texas Tech University and partially by NIH (1R01GM093322-01). We would like to thank Dr. Jeremy Sokolove from Stanford University School of Medicine for providing the sample.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Miller EJ, Lunde LG. Isolation and characterization of the cyanogen bromide peptides from the alpha 1(II) chain of bovine and human cartilage collagen. Biochemistry. 1973;12(17):3153–9. doi: 10.1021/bi00741a003. [DOI] [PubMed] [Google Scholar]
- 2.Chung E, Miller EJ. Collagen polymorphism: characterization of molecules with the chain composition (alpha 1 (3)03 in human tissues. Science. 1974;183(130):1200–1. doi: 10.1126/science.183.4130.1200. [DOI] [PubMed] [Google Scholar]
- 3.Gelse K, Poschl E, Aigner T. Collagens--structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55(12):1531–46. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–58. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi M, Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem. 2012;52:113–33. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notbohm H, Nokelainen M, Myllyharju J, Fietzek PP, Muller PK, Kivirikko KI. Recombinant Human Type II Collagens with Low and High Levels of Hydroxylysine and Its Glycosylated Forms Show Marked Differences in Fibrillogenesis in Vitro. J. Biol. Chem. 1999;274(13):8988–8992. doi: 10.1074/jbc.274.13.8988. [DOI] [PubMed] [Google Scholar]
- 7.Yang CL, Rui H, Mosler S, Notbohm H, Sawaryn A, Muller PK. Collagen II from articular cartilage and annulus fibrosus. Structural and functional implication of tissue specific posttranslational modifications of collagen molecules. Eur J Biochem. 1993;213(3):1297–302. doi: 10.1111/j.1432-1033.1993.tb17881.x. [DOI] [PubMed] [Google Scholar]
- 8.Eyre DR, Glimcher MJ. Analysis of a crosslinked peptide from calf bone collagen: evidence that hydroxylysyl glycoside participates in the crosslink. Biochem Biophys Res Commun. 1973;52(2):663–71. doi: 10.1016/0006-291x(73)90764-x. [DOI] [PubMed] [Google Scholar]
- 9.Robins SP. Cross-linking of collagen. Isolation, structural characterization and glycosylation of pyridinoline. Biochem J. 1983;215(1):167–73. doi: 10.1042/bj2150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamauchi M, Katz EP. The post-translational chemistry and molecular packing of mineralizing tendon collagens. Connect Tissue Res. 1993;29(2):81–98. doi: 10.3109/03008209309014236. [DOI] [PubMed] [Google Scholar]
- 11.Parisuthiman D, Mochida Y, Duarte WR, Yamauchi M. Biglycan modulates osteoblast differentiation and matrix mineralization. J Bone Miner Res. 2005;20(10):1878–86. doi: 10.1359/JBMR.050612. [DOI] [PubMed] [Google Scholar]
- 12.Berg RA, Prockop DJ. The thermal transition of a non hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun. 1973;52(1):115–20. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 13.Sakakibara S, Inouye K, Shudo K, Kishida Y, Kobayashi Y, Prockop DJ. Synthesis of (Pro-Hyp-Gly) n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim Biophys Acta. 1973;303(1):198–202. doi: 10.1016/0005-2795(73)90164-5. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki E, Fraser RDB, MacRae TP. Role of hydroxyproline in the stabilization of the collagen molecule via water molecules. International Journal of Biological Macromolecules. 1980;2(1):54–56. [Google Scholar]
- 15.Kawahara K, Nishi Y, Nakamura S, Uchiyama S, Nishiuchi Y, Nakazawa T, Ohkubo T, Kobayashi Y. Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10. Biochemistry. 2005;44(48):15812–22. doi: 10.1021/bi051619m. [DOI] [PubMed] [Google Scholar]
- 16.Improta R, Berisio R, Vitagliano L. Contribution of dipole-dipole interactions to the stability of the collagen triple helix. Protein Sci. 2008;17(5):955–61. doi: 10.1110/ps.073301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye K, Kobayashi Y, Kyogoku Y, Kishida Y, Sakakibara S, Prockop DJ. Synthesis and physical properties of (hydroxyproline-proline-glycine)10: hydroxyproline in the X-position decreases the melting temperature of the collagen triple helix. Arch Biochem Biophys. 1982;219(1):198–203. doi: 10.1016/0003-9861(82)90149-7. [DOI] [PubMed] [Google Scholar]
- 18.Spiro RG. The structure of the disaccharide unit of the renal glomerular basement membrane. J Biol Chem. 1967;242(20):4813–23. [PubMed] [Google Scholar]
- 19.Butler WT, Miller EJ, Finch JE, Jr., Inagami T. Homologous regions of collagen alpha1(I) and alpha1(II) chains: apparent clustering of variable and invariant amino acid residues. Biochem Biophys Res Commun. 1974;57(1):190–5. doi: 10.1016/s0006-291x(74)80375-x. [DOI] [PubMed] [Google Scholar]
- 20.Ryhanen L, Kivirikko KI. Hydroxylation of lysyl residues in native and denatured protocollagen by protocollagen lysyl hydroxylase in vitro. Biochim Biophys Acta. 1974;343(1):129–37. doi: 10.1016/0304-4165(74)90244-x. [DOI] [PubMed] [Google Scholar]
- 21.Butler WT, Miller EJ, Finch JE., Jr. The covalent structure of cartilage collagen. Amino acid sequence of the NH2-terminal helical portion of the alpha 1 (II) chain. Biochemistry. 1976;15(14):3000–6. doi: 10.1021/bi00659a010. [DOI] [PubMed] [Google Scholar]
- 22.Butler WT, Finch JE, Jr., Miller EJ. The covalent structure of cartilage collagen. Evidence for sequence heterogeneity of bovine alpha1(II) chains. J Biol Chem. 1977;252(2):639–43. [PubMed] [Google Scholar]
- 23.Francis G, Butler WT, Finch JE., Jr. The covalent structure of cartilage collagen. Amino acid sequence of residues 552-661 of bovine alpha1(II) chains. Biochem J. 1978;1753:921–30. doi: 10.1042/bj1750921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mechref Y, Novotny MV. Structural investigations of glycoconjugates at high sensitivity. Chem Rev. 2002;102(2):321–69. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 25.Bond MR, Kohler JJ. Chemical methods for glycoprotein discovery. Curr Opin Chem Biol. 2007;11(1):52–8. doi: 10.1016/j.cbpa.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 26.Harazono A, Kawasaki N, Itoh S, Hashii N, Ishii-Watabe A, Kawanishi T, Hayakawa T. Site-specific N-glycosylation analysis of human plasma ceruloplasmin using liquid chromatography with electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;348(2):259–68. doi: 10.1016/j.ab.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 27.Borges CR, Jarvis JW, Oran PE, Nelson RW. Population studies of Vitamin D Binding Protein microheterogeneity by mass spectrometry lead to characterization of its genotype-dependent O-glycosylation patterns. J Proteome Res. 2008;7(9):4143–53. doi: 10.1021/pr8002936. [DOI] [PubMed] [Google Scholar]
- 28.Tajiri M, Ohyama C, Wada Y. Oligosaccharide profiles of the prostate specific antigen in free and complexed forms from the prostate cancer patient serum and in seminal plasma: a glycopeptide approach. Glycobiology. 2008;18(1):2–8. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- 29.Mechref Y, Hu Y, Garcia A, Hussein A. Identifying cancer biomarkers by mass spectrometry based glycomics. Electrophoresis. 2012;33(12):1755–67. doi: 10.1002/elps.201100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whelan SA, Lu M, He J, Yan W, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Mass spectrometry (LC MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J Proteome Res. 2009;8(8):4151–60. doi: 10.1021/pr900322g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada Y, Kadoya M, Okamoto N. Mass spectrometry of apolipoprotein C-III, a simple analytical method for mucin-type O-glycosylation and its application to an autosomal recessive cutis laxa type-2 (ARCL2) patient. Glycobiology. 2012;22(8):1140–4. doi: 10.1093/glycob/cws086. [DOI] [PubMed] [Google Scholar]
- 32.Madera M, Mechref Y, Klouckova I, Novotny MV. High-sensitivity profiling of glycoproteins from human blood serum through multiple-lectin affinity chromatography and liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;845(1):121–37. doi: 10.1016/j.jchromb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 34.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–29. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirgorodskaya E, Hassan H, Clausen H, Roepstorff P. Mass spectrometric determination of O-glycosylation sites using beta elimination and partial acid hydrolysis. Anal Chem. 2001;73(6):1263–9. doi: 10.1021/ac001288d. [DOI] [PubMed] [Google Scholar]
- 36.Balog CI, Mayboroda OA, Wuhrer M, Hokke CH, Deelder AM, Hensbergen PJ. Mass spectrometric identification of aberrantly glycosylated human apolipoprotein C III peptides in urine from Schistosoma mansoni-infected individuals. Mol Cell Proteomics. 2010;9(4):667–81. doi: 10.1074/mcp.M900537-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelleher NL, Zubarev RA, Bush K, Furie B, Furie BC, McLafferty FW, Walsh CT. Localization of labile posttranslational modifications by electron capture dissociation: the case of gamma-carboxyglutamic acid. Anal Chem. 1999;71(19):4250–3. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 38.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mormann M, Paulsen H, Peter-Katalinic J. Electron capture dissociation of O-glycosylated peptides: radical site induced fragmentation of glycosidic bonds. Eur J Mass Spectrom (Chichester, Eng) 2005;11(5):497–511. doi: 10.1255/ejms.738. [DOI] [PubMed] [Google Scholar]
- 40.Sihlbom C, van Dijk Hard I, Lidell ME, Noll T, Hansson GC, Backstrom M. Localization of O-glycans in MUC1 glycoproteins using electron-capture dissociation fragmentation mass spectrometry. Glycobiology. 2009;19(4):375–81. doi: 10.1093/glycob/cwn144. [DOI] [PubMed] [Google Scholar]
- 41.Thomsson KA, Karlsson H, Hansson GC. Sequencing of sulfated oligosaccharides from mucins by liquid chromatography and electrospray ionization tandem mass spectrometry. Anal Chem. 2000;72(19):4543–9. doi: 10.1021/ac000631b. [DOI] [PubMed] [Google Scholar]
- 42.Christiansen MN, Kolarich D, Nevalainen H, Packer NH, Jensen PH. Challenges of determining O-glycopeptide heterogeneity: a fungal glucanase model system. Anal Chem. 2010;82(9):3500–9. doi: 10.1021/ac901717n. [DOI] [PubMed] [Google Scholar]
- 43.Deguchi K, Ito H, Baba T, Hirabayashi A, Nakagawa H, Fumoto M, Hinou H, Nishimura S. Structural analysis of O-glycopeptides employing negative- and positive ion multi-stage mass spectra obtained by collision-induced and electron-capture dissociations in linear ion trap time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(5):691–8. doi: 10.1002/rcm.2885. [DOI] [PubMed] [Google Scholar]
- 44.Zauner G, Hoffmann M, Rapp E, Koeleman CA, Dragan I, Deelder AM, Wuhrer M, Hensbergen PJ. Glycoproteomic analysis of human fibrinogen reveals novel regions of O-glycosylation. J Proteome Res. 2012;11(12):5804–14. doi: 10.1021/pr3005937. [DOI] [PubMed] [Google Scholar]
- 45.Halim A, Nilsson J, Ruetschi U, Hesse C, Larson G. Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol Cell Proteomics. 2012;11(4):M111 013649. doi: 10.1074/mcp.M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van den Steen PE, Proost P, Brand DD, Kang AH, Van Damme J, Opdenakker G. Generation of glycosylated remnant epitopes from human collagen type II by gelatinase B. Biochemistry. 2004;43(33):10809–16. doi: 10.1021/bi0493665. [DOI] [PubMed] [Google Scholar]
- 47.Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Development of a Novel Method for Analyzing Collagen O-glycosylations by Hydrazide Chemistry. Mol Cell Proteomics. 2012;11(6):M111 010397. doi: 10.1074/mcp.M111.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan SW, Greaves J, Da Silva NA, Wang SW. Assaying proline hydroxylation in recombinant collagen variants by liquid chromatography-mass spectrometry. BMC Biotechnol. 2012;1251 doi: 10.1186/1472-6750-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, Park AC, Davis NA, Russell JD, Kim B, Brand DD, Lawrence MJ, Ge Y, Westphall MS, Coon JJ, Greenspan DS. Comprehensive mass spectrometric mapping of the hydroxylated amino acid residues of the alpha1(V) collagen chain. J Biol Chem. 2012;287(48):40598–610. doi: 10.1074/jbc.M112.406850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurtado PP, O'Connor PB. Deamidation of collagen. Anal Chem. 2012;84(6):3017–25. doi: 10.1021/ac202980z. [DOI] [PubMed] [Google Scholar]
- 51.Segu ZM, Hammad LA, Mechref Y. Rapid and efficient glycoprotein identification through microwave-assisted enzymatic digestion. Rapid Commun Mass Spectrom. 2010;24(23):3461–8. doi: 10.1002/rcm.4774. [DOI] [PubMed] [Google Scholar]
- 52.Weis MA, Hudson DM, Kim L, Scott M, Wu JJ, Eyre DR. Location of 3 hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J Biol Chem. 2010;285(4):2580–90. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seyer JM, Hasty KA, Kang AH. Covalent structure of collagen. Amino acid sequence of an arthritogenic cyanogen bromide peptide from type II collagen of bovine cartilage. Eur J Biochem. 1989;181(1):159–73. doi: 10.1111/j.1432-1033.1989.tb14707.x. [DOI] [PubMed] [Google Scholar]
- 54.Langrock T, Garcia-Villar N, Hoffmann R. Analysis of hydroxyproline isomers and hydroxylysine by reversed-phase HPLC and mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847(2):282–8. doi: 10.1016/j.jchromb.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 55.Hao P, Qian J, Dutta B, Cheow ES, Sim KH, Meng W, Adav SS, Alpert A, Sze SK. Enhanced separation and characterization of deamidated peptides with RP-ERLIC-based multidimensional chromatography coupled with tandem mass spectrometry. J Proteome Res. 2012;11(3):1804–11. doi: 10.1021/pr201048c. [DOI] [PubMed] [Google Scholar]
- 56.Chelius D, Rehder DS, Bondarenko PV. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal Chem. 2005;77(18):6004–11. doi: 10.1021/ac050672d. [DOI] [PubMed] [Google Scholar]
- 57.Johnson BA, Shirokawa JM, Hancock WS, Spellman MW, Basa LJ, Aswad DW. Formation of isoaspartate at two distinct sites during in vitro aging of human growth hormone. J Biol Chem. 1989;264(24):14262–71. [PubMed] [Google Scholar]
- 58.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75(18):4818–26. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 59.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4(4):1442–50. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 60.Andreev VP, Li L, Cao L, Gu Y, Rejtar T, Wu S-L, Karger BL. A New Algorithm Using Cross-Assignment for Label-Free Quantitation with LC-LTQ-FT MS. J Proteome Res. 2007;6(6):2186–2194. doi: 10.1021/pr0606880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.