ABSTRACT
Human cytomegalovirus (HCMV) is a major cause of morbidity and mortality in transplant patients and is the leading viral cause of birth defects after congenital infection. HCMV infection relies on the recognition of cell-specific receptors by one of the viral envelope glycoprotein complexes. Either the gH/gL/gO or the gH/gL/UL128/UL130/UL131A (Pentamer) complex has been found to fulfill this role, accounting for HCMV entry into almost all cell types. We have studied the UL116 gene product, a putative open reading frame identified by in silico analysis and predicted to code for a secreted protein. Virus infection experiments in mammalian cells demonstrated that UL116 is expressed late in the HCMV replication cycle and is a heavily glycosylated protein that first localizes to the cellular site of virus assembly and then inserts into the virion envelope. Transient-transfection studies revealed that UL116 is efficiently transported to the plasma membrane when coexpressed with gH and that gL competes with UL116 for gH binding. Further evidence for gH/UL116 complex formation was obtained by coimmunoprecipitation experiments on both transfected and infected cells and biochemical characterization of the purified complex. In summary, our results show that the product of the UL116 gene is an HCMV envelope glycoprotein that forms a novel gH-based complex alternative to gH/gL. Remarkably, the gH/UL116 complex is the first herpesvirus gH-based gL-less complex.
IMPORTANCE HCMV infection can cause severe disease in immunocompromised adults and infants infected in utero. The dissection of the HCMV entry machinery is important to understand the mechanism of viral infection and to identify new vaccine antigens. The gH/gL/gO and gH/gL/UL128/UL130/UL131 (Pentamer) complexes play a key role in HCMV cell entry and tropism. Both complexes are formed by an invariant gH/gL scaffold on which the other subunits assemble. Here, we show that the UL116 gene product is expressed in infected cells and forms a heterodimer with gH. The gH/UL116 complex is carried on the infectious virions, although in smaller amounts than gH/gL complexes. No gH/UL116/gL ternary complex formed in transfected cells, suggesting that the gH/UL116 complex is independent from gL. This new gH-based gL-free complex represents a potential target for a protective HCMV vaccine and opens new perspectives on the comprehension of the HCMV cell entry mechanism and tropism.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus infecting 40 to 60% of the human population (1). HCMV infection usually is mild or asymptomatic in immunocompetent individuals, while it can cause severe disease in immunocompromised adults. Infants infected in utero can suffer from disseminated HCMV disease and impaired neurological development (2). Transplant patients do not adequately control HCMV infection or reactivation, often resulting in graft rejection and death (3). Preventing primary HCMV infection could significantly decrease the frequency of organ rejection in seronegative recipients of solid-organ transplants as well as prevent congenital infection (4). The Institute of Medicine has identified the development of an anti-HCMV vaccine protecting against congenital infection as a top priority (5).
The HCMV genome from clinical isolates encodes a minimum of 165 functional open reading frames (ORFs) (6–9). Out of the total number of proteins composing the HCMV virion, approximately 20 have been identified as envelope-associated proteins (10–12). Notably, all glycoproteins involved in HCMV cell entry were shown to be essential for virus replication in vitro in different cell types: glycoproteins B (gB), M (gM), N (gN), L (gL), H (gH), UL128, UL130, and UL131A (13). Although not originally included in this list, studies conducted with viruses deleted of the UL74 gene, whose product is known as gO, have shown that this glycoprotein is a crucial component of the fusion-promoting machinery (14–16).
HCMV infection begins with the low-affinity tethering of the virion to cell surface heparin sulfate proteoglycans (HSPGs), an event mediated by both gB and the gM/gN complex (13, 17). gB and gH/gL are part of the conserved herpesvirus fusion machinery and are required for cell fusion and viral entry (18). Current evidence suggests that the binding of a complex between gH, gL, and other viral envelope glycoproteins to a cell surface receptor triggers conformational changes in gB, the viral fusogen, resulting in membrane fusion (13). Compositions of the gH/gL-based complexes and cell-specific receptors determine viral tropism and mechanism of entry (19–22). The gH/gL/gO complex appears to be sufficient for HCMV entry and replication into fibroblasts (23, 24) but still is required for proper growth and for fusion events in all cell types (15, 16, 25). The more recently discovered gH/gL/UL128/UL130/UL131A complex (Pentamer) instead is required for endothelial/epithelial cell, leukocyte, and monocyte tropism (26–28). On the cellular side, several putative HCMV receptors have been described, but none of them completely fulfills the characteristics of an entry receptor (17, 29).
It has been estimated that in sera of HCMV-seropositive individuals, approximately 50% of antibodies neutralizing fibroblast infection are directed against gB (30, 31). gB has been tested, together with the adjuvant MF59, in human trials as a potential HCMV vaccine (30, 32, 33). The vaccine conferred partial protection in healthy seronegative subjects (34). The observation that MF59-adjuvanted gB was able to partially protect from primary infection suggests that a subunit HCMV vaccine is feasible, but it also has renewed interest for new targets among the exposed viral envelope glycoproteins. Recently, the analysis of human plasma from seropositive subjects has identified the pentamer complex as the main target of antibodies neutralizing infection in epithelial/endothelial cells (31). Moreover, immunization with Pentamer-containing vaccines elicited strong and broadly neutralizing responses in small animals and nonhuman primates (NHPs) (35–37). Therefore, the pentamer complex represents a promising candidate for a potent HCMV vaccine (35–41).
HCMV structural glycoproteins can be divided into those that are conserved among members of the Herpesviridae (including gB, gH, gL, gM, and gN) and subgenus-specific glycoproteins without homology to other herpesviruses. The latter include, among others, gpRL13 (11, 42), gpTRL10 (43), gpUL132 (44), UL74-encoded gO (45), UL4-encoded gp48 (46), US27 (47), and UL33 (48). In contrast to the conserved glycoproteins, knockout of many of the HCMV-specific structural proteins does not prevent in vitro replication in fibroblast, epithelial, or endothelial cells and presumably participates in cell and tissue tropism or pathogenicity (23, 24). The functional characterization of these proteins may be beneficial for the dissection of the mechanism of HCMV entry into the broad range of cell types susceptible to HCMV infection.
Here, we report the characterization of the betaherpesvirus-specific product of the UL116 gene from the HCMV TR strain as an envelope glycoprotein forming a stable complex with gH. In infected cells, UL116 showed late kinetics of expression, underwent extensive glycosylation, and localized at the site of virus assembly and secondary envelopment. Electron microscopy (EM) and phase separation experiments indicated that UL116 is part of the virion envelope. Coimmunoprecipitation experiments and multiangle light scattering (MALS) analysis of purified proteins confirmed the formation of a complex between UL116 and gH. Finally, cotransfection experiments and negative-staining EM coupled with single-particle analysis of purified complexes suggest that UL116 and gL form alternative, mutually exclusive complexes with gH. Overall, we identify a novel HCMV gH-based complex potentially playing a role in viral infection and dissemination.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses.
A bacterial artificial chromosome (BAC) containing the genome of the HCMV TR strain was obtained from Oregon Health Science University. TR, a clinical HCMV strain derived from an ocular vitreous fluid sample from a patient with HIV disease (49), was cloned into a BAC after limited passage in fibroblasts (50). HCMV strain TR and recombinant UL116-Flag-TR virus were propagated in human foreskin fibroblasts (HFF-1; ATCC SCRC-1041) grown in minimal essential medium (Invitrogen) supplemented with 10% fetal calf serum, glutamine (100 mg/liter), and gentamicin (350 mg/liter). Virions were isolated by glycerol-tartrate gradient centrifugation as previously described (51). HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, glutamine, and gentamicin. Lipofectamine 2000 (Invitrogen) was used to transfect HEK293T cells. Human codon-optimized UL116, gH (UL75), gL (UL115), and gB (UL55) HCMV genes based on the TR strain sequence were synthesized by GeneArt and cloned into the pcDNA3.1(−)/myc-His C vector (Invitrogen) in frame with C-terminal myc and the six-histidine tag sequences. Single-point mutations were introduced with the QuikChange mutagenesis kit (Stratagene), resulting in the mono-tag versions and the tag-less versions of these genes.
Generation of UL116 polyclonal antiserum.
Mouse antiserum recognizing UL116 was developed in-house. Briefly, His-tagged peptides encompassing the amino acid regions of residues 32 to 265 of UL116 from the TR strain were produced in Escherichia coli and purified through metal ion affinity chromatography. Purified peptides were used to immunize mice.
Construction and generation of UL116-Flag TR BAC.
Insertion of the Flag tag at the C terminus of the UL116 ORF in the TR BAC was achieved using the two-step red-mediated recombination method (52). The primer pair used to amplify the kanamycin insertion cassette was 5′-TTC GGC GCC AAC TGG CTC CTT ACC GTC ACA CTC TCA TCG TGC CGC AGA CTG ATT ACA AGG ATG ACG ACG ATA AG-3′ and 5′-TAT CAC CGG TCC AGG TGA GAA AGA GAA GCC GCA ATC CGG GCG GCG GCA CAT CA CTT ATC GTC GTC ATC CTT GTA ATC AGT CTG CGG CAC GAT GAG CAA CCA AAT TAA ACCA ATT CTG ATT TAG-3′, where the underlined base pairs encode the Flag peptide.
Reconstitution of infectious virus.
To reconstitute the virus, 2 μg of the BAC-HCMV DNA and 1 μg of a pp71 expression plasmid were transfected into MRC-5 cells by electroporation. Culture medium was changed 24 h later, and the cells were split and cultured until the appearance of cytopathic effect. The virus stock was prepared by harvesting cell-free culture supernatant when extensive cytopathic effect was visible.
Purification of HCMV TR virions.
HCMV particles in cell supernatants were separated into virion, dense-body, and noninfectious enveloped particle (NIEP) fractions by positive density/negative viscosity gradient centrifugation as described previously (53). Particle concentrations in the preparations were estimated by counting negatively stained samples by EM in relation to a standard concentration of latex beads. To separate virion envelope proteins from capsid and tegument proteins, 108 particles were mixed 1:1 with envelope stripping buffer (2% Nonidet-P40 in phosphate-buffered saline [PBS]) and incubated for 15 min at 4°C. Particles were pelleted (12,000 × g for 5 min at 4°C), and the soluble envelope fraction was harvested. The insoluble capsid/tegument material was washed twice with envelope-stripping buffer and once in PBS before being solubilized in SDS-PAGE sample buffer.
Flow cytometry.
For the detection of membrane-exposed UL116, HEK293T cells transiently transfected with vectors coding for UL116, gH, gB, and empty vector were trypsin detached 48 h posttransfection, incubated for 30 min at room temperature (RT) with Live/Dead Aqua (Invitrogen), diluted 1:400 in PBS, and incubated with different dilutions of anti-UL116 polyclonal mouse sera for 60 min on ice. After three washes in PBS, the Alexa Fluor 647-conjugated goat anti-mouse secondary antibody was added at a 1:200 dilution and incubated for 30 min on ice. A total of 104 cells were analyzed for each histogram using FACSCanto II (Becton Dickinson, Heidelberg, Germany). Experiments were performed in triplicate for statistical consistency; means and standard deviations were analyzed and plotted using GraphPad Prism software.
Immunogold EM.
Purified virions were air dried to the surface of Formvar-coated EM grids. The grids were treated with mouse anti-Flag antibody (F3165; Sigma) for 4 h at RT, washed 3 times with PBS, and incubated with goat anti-mouse antibody conjugated to 5-nm gold particles for 1 h at RT. After further washing in PBS, the grids were negatively stained with phosphotungstic acid and subjected to EM analysis.
Purification of gH/UL116 and gH/gL-C144A Fab complexes.
The gH/UL116 complex was purified from supernatants of HEK293 cells doubly transfected with plasmids encoding UL116 and gH containing a double Strep-tag and 6His tag at their C termini, respectively (UL116-strep and gH-His). The medium then was applied to StrepTrap HP 1-ml columns (GE Life Sciences), and the complex was eluted with the same buffer containing 2.5 mM desthiobiotin. The gH/gL complex harboring the gL-C144A mutation was purified from supernatants of HEK293 cells doubly transfected with plasmids encoding gH-His and untagged gL-C144A as previously described (54). Purified gH/gL-C144A or gH/UL116 was mixed at 1:1.2 with either 3G16 or MSL-109 Fabs. The ternary complexes were isolated by size exclusion chromatography (SEC).
MALS analysis.
Purified gH/UL116 complex, at two different dilutions, was analyzed by SEC on a Superose-6 10/30 column (GE Healthcare) using 25 mM Tris, pH 8.0, 150 mM NaCl as a running buffer. The SEC system was coupled in-line with the following calibrated detection systems: (i) HP1 1050 Hewlett-Packard UV detector (Norwalk, CT); (ii) MiniDawn Treos multiangle light scattering (MALS) detector (Wyatt Corporation, CA); (iii) quasielastic light scattering detector (Wyatt Corporation, CA); and (iv) Optilab T-reX refractive index (RI) detector (Wyatt Corporation). Briefly, these combined measurements allow both the absolute molar mass of an eluting glycoprotein and the individual contributions made by protein and carbohydrate components to be determined (55–57). The carbohydrate contribution to the molar mass can be assessed because glycans contribute directly to the RI signal but not the UV signal.
Competition between gL and UL116 for gH binding and formation of higher-order complexes.
HEK293T cells were transfected with DNA encoding gH-His and either gL, UL116-strep, or both. The samples involving UL116 were purified by Strep-tag affinity, while the gH/gL complex was purified by Ni affinity as described above. The elution fractions were run on SDS-PAGE with previously purified gH/gL and gH/UL116 as controls. To detect higher-order complex formation, HEK293T cells were transfected with DNA encoding soluble gH extracellular domain in four different combinations: (i) gH/gL/UL128/strep-UL130/UL131, (ii) gH/strep-UL116/UL128/UL130/UL131, (iii) gH/gL/His-gO, and (iv) gH/strep-UL116/gO. The formation of pentamer in supernatant was assessed by Western blotting with anti-UL128 antibody, whereas anti-His was used to reveal secreted gO. The samples involving UL116 were purified by Strep-tag affinity, and associated proteins were revealed by Coomassie staining of the gel.
Negative-staining electron microscopy and single-particle analysis.
Five microliters of purified gH/UL116/3G16, gH/UL116/MSL-109, or gH/gL-C144A/3G16 samples (approximately 10 ng) were placed onto a freshly glow-discharged holey carbon grid covered with a thin layer of continuous carbon. The grid was stained with sequential 75-μl drops of a freshly prepared 2% (wt/vol) uranyl formate solution. Samples were imaged using a Tecnai Spirit T12 transmission electron microscope operating at 120 keV at a nominal magnification of ×49,000 (1.57 Å/pixel at the detector level) using a defocus range of −0.8 to −1.2 μm. Images were recorded under low-dose conditions on a Gatan 4,096- by 4,096-pixel charge-coupled-device (CCD) camera. Particles were manually picked using e2boxer (EMAN2) (58) and extracted using a 224-pixel box size. The two data sets were band-pass filtered with a 200-Å high-pass cutoff and a 20-Å low-pass cutoff. For two-dimensional (2D) classification, we used iterative multivariate statistical analysis (MSA) and multireference alignment (MRA) in Imagic (59). 2D classes from gH/gL and gH/UL116 Fab complexes were aligned and compared by cross-correlation using the SPIDER AP SH command.
UL116 expression kinetics and glycosidase digestion.
Infected cells were harvested at the indicated time points and lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma) according to the manufacturer's protocol. Late-phase protein expression was inhibited by the use of phosphonoacetic acid (PAA; Sigma-Aldrich). A total of 250 μg/ml of PAA was added to the medium at the time of infection and maintained throughout infection. For deglycosylation treatments, 20 μg of protein extract was incubated with 2.5 μl of endoglycosidase H (Endo H; NEB), 2.5 μl peptide-N-glycosidase F (PNGase F; NEB), or buffer only for 3 h at 37°C according to the manufacturer's protocol. Samples were analyzed by Western immunoblotting.
SDS-PAGE and immunoblotting.
Proteins were separated by SDS-PAGE on 10 to 15% polyacrylamide gels under standard conditions. Proteins were transferred to nitrocellulose membranes, and membranes were blocked with PBS containing 0.1% Tween 20 and 5% powdered milk. Incubations with primary antibodies and sera, diluted in PBS–0.1% Tween 20 (PBST), were performed at RT for 1 h, followed by 3 washes in PBST before incubation with horseradish peroxidase-conjugated secondary antibodies (PerkinElmer) for 1 h. Membranes were washed 5 times in PBST prior to detection with an enhanced chemiluminescence detection system (Pierce).
Immunofluorescence.
Cells were plated on glass coverslips and infected with HCMV. At day 7 postinfection, cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, preblocked with HCMV seronegative human IgG, and incubated with primary antibody for 1 h at RT. Following washing in PBS, secondary antibodies were incubated for 1 h at 37°C, washed again, and mounted with 4′,6-diamidino-2-phenylindole (DAPI) ProLong safe stain mounting media (Invitrogen). Primary antibodies were rabbit anti-Flag (Sigma), mouse anti-Flag (Sigma), sheep anti-human TGN 46 (Serotec), mouse monoclonal antibody (MAb) anti-pp28 (Abcam), mouse anti-gH (AbD Serotech), and rabbit anti-gL (OHSU). Secondary antibodies were Alexa Fluor 488-, 568-, and 647-conjugated goat anti-mouse and anti-rabbit (Invitrogen). The intracellular locations of antibody-tagged proteins were examined under laser illumination in a Zeiss LMS 700 confocal microscope, and images were captured using ZEN 2009 software. Image analysis was performed with Fiji (NIH) and Imaris 8.1 (Bitplane). Deconvolution was performed using Autoquant X3 (MediaCybernetics) with fixed point spread function (PSF) modeling.
Immunoprecipitations.
HFF-1 cells were infected with HCMV TR and UL116-Flag-TR. Protein expression was allowed to proceed for 72 h, and then cells were washed in 1× PBS and lysed with a lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, pH 7.4) in the presence of protease inhibitors (Complete EDTA-free; Roche). Five hundred micrograms of total protein was incubated overnight at 4°C with 5 μg of the anti-gH MSL-109 monoclonal antibody. Complexes were immunoprecipitated using Protein G Dynabeads (Invitrogen) according to the manufacturer's protocol. The beads were washed four times in lysis buffer and then boiled for 5 min in SDS-PAGE loading buffer. Proteins were separated on SDS-PAGE, and immunoblotting was performed as described above. A similar procedure was adopted for immunoprecipitation from supernatants of transiently transfected HEK293T cells. Complexes were captured in parallel experiments with covalently linked anti-His antibody magnetic beads (GenScript) and anti-c-myc magnetic beads (Pierce) to avoid background signals in the eluted materials.
RESULTS
Primary structure of UL116 gene product.
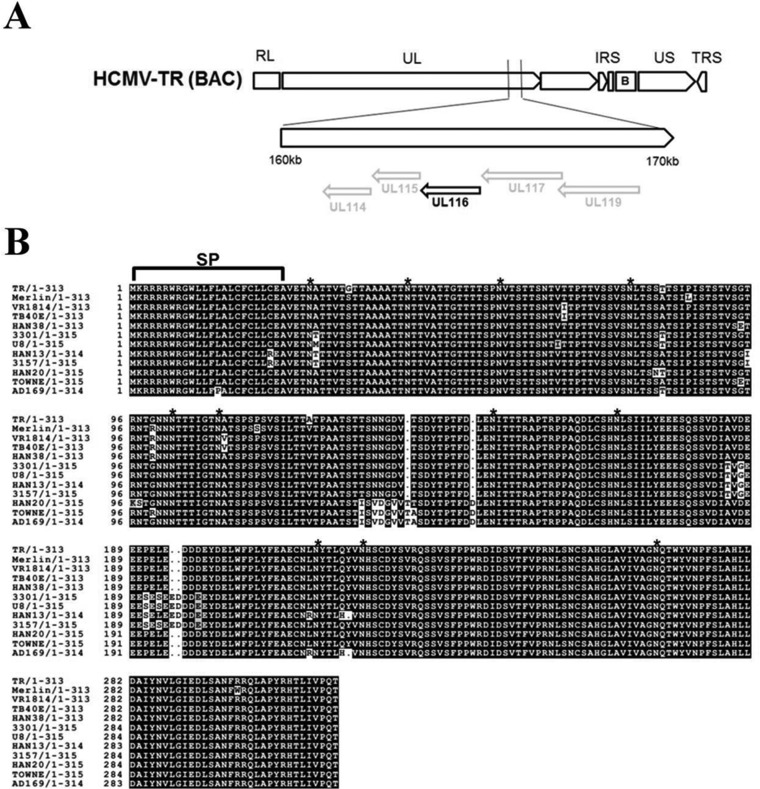
In all sequenced HCMV genomes, the UL116 gene is located in the unique-long (UL) region between the UL115 (gL) and UL117 genes on the antisense coding strand (Fig. 1A). The UL116 mRNA was previously shown to arise in the true-late stage of AD169 infection as part of the UL119-UL115 transcription unit, but the gene product was never analyzed (60). The UL116 gene is not conserved among alpha and gamma herpesviruses; thus, it does not belong to the core herpesvirus genes (61). Orthologous genes are present in cytomegaloviruses infecting nonhuman primates (62), mouse, rat, and guinea pig (63, 64). The UL116 ORF from the HCMV TR strain is predicted to encode a 313-amino-acid glycoprotein comprising a signal peptide (amino acid positions 1 to 24) (Fig. 1B, SP) and a threonine-rich domain (amino acids 27 to 157), with the resulting polypeptide backbone predicted to have a molecular mass of 34.2 kDa. Fourteen consensus sites for N-linked glycosylation are predicted, whereas the O-glycosylation acceptor potential sites are very numerous (>70). Moreover, UL116 lacks membrane anchor sequences and therefore is expected to be a secreted protein. A multisequence alignment of UL116 from clinical and laboratory-adapted HCMV strains reveals 98% sequence identity (Fig. 1B), suggesting a conserved functional role.
FIG 1.
UL116 position in the HCMV TR genome and sequence conservation among laboratory-adapted and clinical HCMV strains. (A) ORF map of the TR BAC clone used in this work. Arrows indicate the relative orientations of the repeated and unique ORF blocks. The UL116 gene, in boldface, is located between UL115 (gL) and UL117 genes on the antisense strand. (B) T-Coffee primary amino acid sequence multialignment showing 98% UL116 gene conservation among members of a consistent pool of HCMV strains. Asterisks indicate the predicted 14 N-glycosylation sites. SP indicates the N-terminal predicted signal peptide. RL, repeat long; UL, unique long; IRS, internal repeat short; US, unique short; TRS, terminal repeat short; B, BAC inserts.
Kinetics of UL116 expression during HCMV replication.
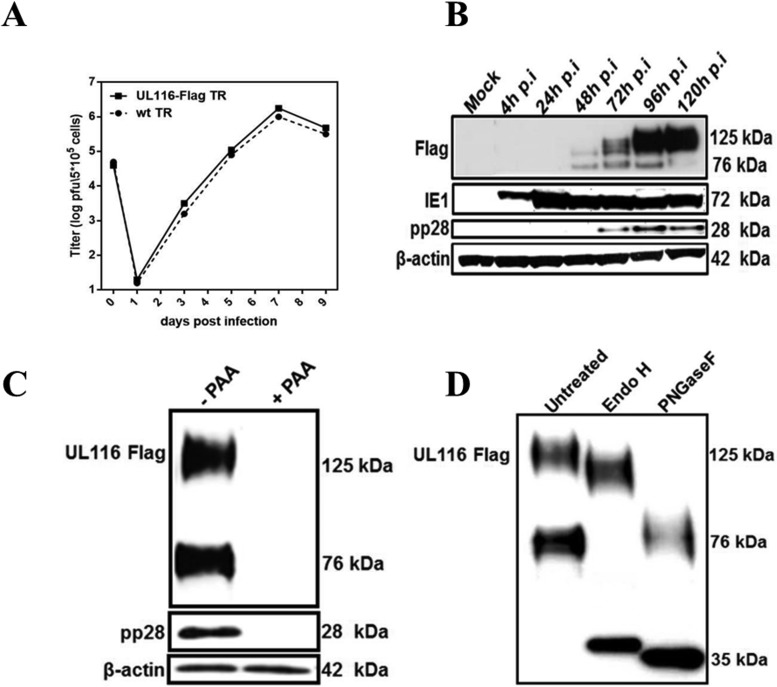
To investigate the expression kinetics of UL116 during productive HCMV infection, we generated a recombinant HCMV, derived from the BAC clone of the TR strain, with a Flag tag fused to the UL116 C-terminal end. The reconstituted TR UL116-Flag virus, which showed growth kinetics identical to those of the parental virus (Fig. 2A), was used to infect HFF-1 cells. Extracts were prepared from infected cells at different time points, ranging from 4 to 120 h postinfection (hpi), and the expression kinetics of UL116 were monitored by immunoblotting using an anti-Flag antibody. UL116 migrated as two species, a faster-migrating protein of 76 kDa and a slower-migrating protein of about 125 kDa (Fig. 2B). The latter appeared to be the mature product of the 76-kDa species, as indicated by the increased accumulation of the 125-kDa species from 72 through 120 hpi compared to that of the 76-kDa band (Fig. 2B). As a control, expression of the immediate-early protein IE1 pp72 (UL123) and the late phosphoprotein pp28 (UL99) were monitored in the same samples (Fig. 2B). The detection of UL116 expression paralleled that of pp28, a late-phase marker (65). Consistent with the observed kinetic pattern, the inhibition of viral DNA synthesis, a prerequisite for late gene expression, with PAA resulted in the disappearance of the UL116 bands (Fig. 2C).
FIG 2.
UL116 expression kinetics and carbohydrate addition in HCMV-infected fibroblasts. (A) Multiple-step growth curve analysis of the reconstituted virus UL116-Flag TR and of the parental HCMV strain TR. Time point 0 titers represent input inocula, and each data point represents averages from three independent wells. (B) Uninfected (mock) and TR-UL116-Flag-infected HFF-1 cells at an multiplicity of infection of 5 were harvested at the indicated times postinfection. Equivalent amounts of cell lysates were subjected to SDS-PAGE under reducing conditions and analyzed by immunoblotting with anti-Flag, HCMV IE1, and pp28 protein antibodies as indicated on the left. Actin detection was used as a protein loading control. (C) TR-UL116-Flag infections of HFF cells were performed in the presence (right) or absence (left) of phosphonoacetic acid (PAA), an inhibitor of HCMV late-phase protein expression. Five days postinfection, lysates were prepared from infected cells, and UL116 and pp28 expression was detected by Western blotting using anti-Flag and anti-pp28 antibodies, respectively. (D) Equal amounts of cell lysates of TR UL116-Flag-infected HFF-1 cells (72 h p.i.) were subjected to glycosidase treatments: left lane, untreated control; middle and right lanes, endoglycosidase H (Endo H) and PNGase F treatments, respectively. Proteins were separated on SDS-PAGE under reducing conditions, and UL116 was detected by immunoblot analysis using an anti-Flag antibody.
The high number of putative N- and O-linked glycosylation sites predicted on UL116 and the difference between the apparent molecular mass observed (125 kDa) by immunoblotting and the calculated molecular mass (34.2 kDa) of the polypeptide backbone suggested that UL116 undergoes an extensive posttranslational glycosylation process. The trafficking of glycoproteins to the Golgi apparatus is associated with the processing of N-linked oligosaccharides to complex oligosaccharides that are resistant to the action of Endo H (66). Endo H completely cleaves only endoplasmic reticulum (ER)-type N-linked carbohydrates, while it does not trim hybrid and complex high-mannose N-linked carbohydrates that form when the glycoproteins reach the Golgi apparatus. PNGase F is able to digest both ER and Golgi-type high-mannose carbohydrate moieties of glycoproteins (67). To analyze the glycosylation of UL116 during the virus life cycle, HFF-1 cells were infected with TR UL116-Flag for 3 days, followed by glycosidase digestion of the cell extract. As shown in Fig. 2D, digestion with Endo H had several effects: (i) the 125-kDa band was only slightly affected, and its apparent molecular mass was reduced by about 10 kDa; (ii) the 75-kDa band collapsed to ∼38 kDa, a value very close to the predicted mass based on sequence (Fig. 2C, compare lanes 1 and 2). PNGase F digestion caused the disappearance of the faster-migrating 76-kDa band that migrated to approximately 35 kDa, while the apparent mass of the 125-kDa band was reduced at ∼78 kDa (Fig. 2D, compare lanes 1 and 3). These results are consistent with the presence of an immature species entirely modified with ER-type glycan and of a mature form carrying mostly Golgi-type carbohydrates.
Confocal microscopy analysis of HCMV-infected human fibroblasts reveals colocalization of UL116 with the viral assembly complex.
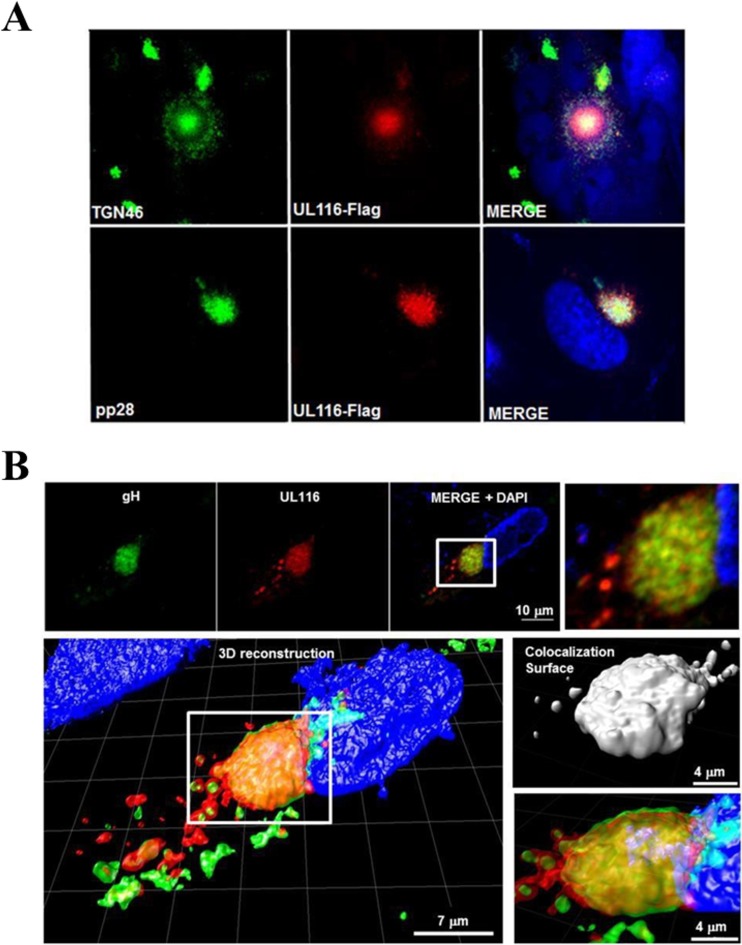
Confocal microscopy was used to investigate the localization of UL116 in HCMV-infected cells. An anti-Flag antibody was employed together with antibodies specific for cellular compartments to trace the subcellular distribution of UL116. Human fibroblasts were infected with TR UL116-Flag and at 72 hpi were fixed and stained for confocal analysis. Among the cellular markers used, UL116 showed colocalization with the trans-Golgi marker TGN 46 (Fig. 3A, upper). Based on the cytoplasmic compartmentalization of UL116, we asked whether the structural HCMV proteins colocalized with UL116. We found a distinct colocalization of UL116 with tegument phosphoprotein pp28 and gH in infected HFF-1 cells 72 h after infection with TR UL116-Flag (Fig. 3A and B, respectively, bottom). Image deconvolution and 3D reconstruction allowed us to better define the colocalization surface between UL116 and the glycoprotein gH. As shown in Fig. 3B, both proteins strongly accumulate in a cytoplasmic perinuclear inclusion and in smaller vesicles scattered in its close proximity. pp28 and gH previously were reported to localize with other tegument or envelope proteins at the virus assembly complex (AC) site during the late phase of the infectious cycle (68, 69) and to be acquired by the nascent virion.
FIG 3.
Intracellular localization of UL116 in infected HFF-1 cells. (A) HFF-1 cells were infected with the recombinant virus TR-UL116-Flag at an MOI of 5 for 72 h. Cells then were fixed, permeabilized, and stained. UL116 detection was achieved by anti-Flag (in red) in combination with either the cellular marker TGN 46 (upper) or the HCMV tegument protein pp28 (lower). Cell nuclei are stained blue. The merge panels show colocalization of the red and green signals. (B) Same experiment as that described for panel A, except that mouse anti-gH was used. A 3D reconstruction and the colocalization surface of the UL116 and gH signals are shown in the lower images.
These data are consistent with the trafficking of UL116 to the site of viral AC and suggest that UL116 can associate with other envelope glycoproteins.
UL116 is a component of the HCMV virion envelope.
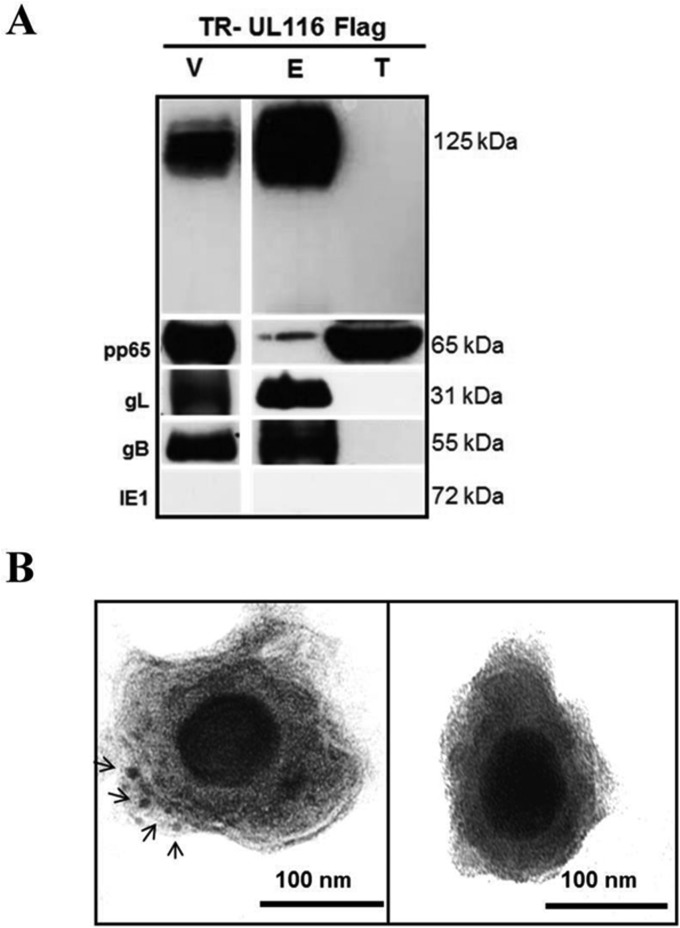
To verify if UL116 was incorporated into the virion and to establish its localization, we purified the recombinant TR UL116-Flag viral particles from the supernatant of infected HFF-1 cells and performed both Western blotting and immuno-EM analysis. TR UL116-Flag particles purified by positive density/negative viscosity glycerol/tartrate gradient were extracted with detergent to separate envelope and tegument fractions (51), which were probed by immunoblotting with different antibodies (Fig. 4A). pp65 was used as a marker of the tegument fraction, and the nonstructural HCMV protein IE1 was used to exclude samples that were contaminated by cellular extracts. As expected, the envelope proteins gB and gL were detected by specific antibodies in the envelope fraction (Fig. 4A). Notably, probing with the anti-Flag antibody showed that UL116 was present exclusively in the envelope fraction and was not detectable in the pp65-containing tegument fraction.
FIG 4.
Localization of UL116 on the virion envelope. (A) Western blotting was performed on purified virions from virus expressing Flag-tagged UL116. The virion total lysate (V), envelope fraction (E), and tegument/capsid fraction (T) were probed for the indicated antigens. (B) Immunogold EM staining for anti-Flag MAb of purified TR-UL116Flag and TR wild type (left and right, respectively). Black arrows indicate gold particles. Original magnification, ×40,000.
To confirm the presence of UL116 on the viral envelope, we performed immunoelectron microscopy on the purified TR UL116-Flag particles using a wild-type TR viral preparation as a control. The 10-nm gold-labeled anti-Flag secondary antibody displayed distinct labeling of the envelope (Fig. 4B). Taken together, these results are consistent with the localization of UL116 on the surface of the HCMV envelope.
Cotransfection of recombinant UL116 and gH.
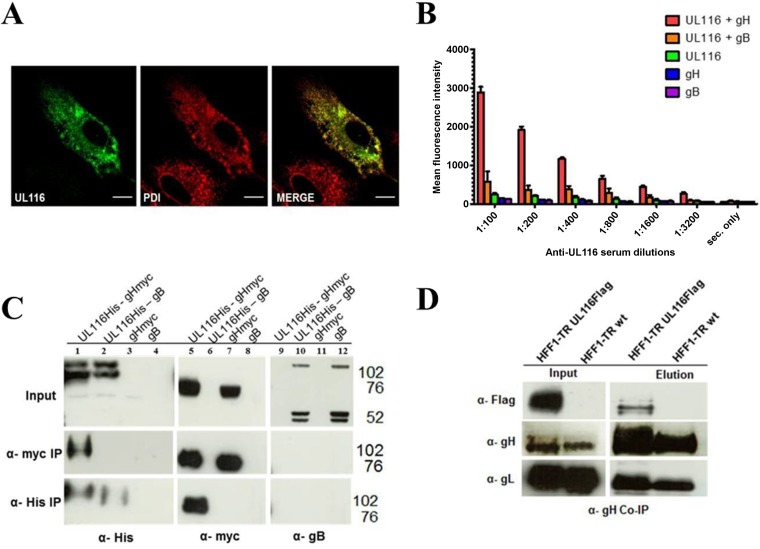
To better characterize the UL116 gene product, we generated a recombinant myc-His tagged version of the gene that was codon optimized for efficient expression and cloned in a eukaryotic expression vector. Surprisingly, transfection in MRC-5 or HEK293T cells did not result in secretion of the recombinant product or its transport to the cell surface (Fig. 5A). Confocal analysis of MRC-5-transfected cells showed that UL116 colocalized almost completely with protein disulfide-isomerase (PDI), a marker of the endoplasmic reticulum (Fig. 5A). We hypothesized that correct localization of UL116 can be achieved following association with one or more of the major HCMV envelope glycoproteins (70). To investigate this possibility, we performed binary cotransfection experiments in HEK293T cells of TR-UL116 with well-characterized HCMV envelope glycoproteins and analyzed the membrane localization of UL116 by cytofluorimetric assay on nonpermeabilized cells with a polyclonal mouse UL116 antisera and a fluorescein isothiocyanate (FITC)-conjugated secondary anti-mouse antibody. To exclude possible nonspecific detection, we included single-gene-transfected cells as negative controls. The results from a representative experiment using gH/UL116 and gB/UL116 are shown in Fig. 5B. A strong UL116 plasma membrane signal was observed only in gH/UL116-coexpressing cells. The cotransfection of UL116 with other HCMV proteins, such as gO and UL10, did not allow the transport of UL116 on the plasma membrane (unpublished results). These data are consistent with UL116 depending on the formation of a heterodimeric complex with gH for translocation to the cell surface.
FIG 5.
UL116 interaction with gH in HEK293T transfected cells. (A) ARPE-19 cells transiently expressing UL116-myc were fixed, permeabilized, and stained with anti-myc (green) and anti-PDI (red) specific antibodies prior to confocal immunofluorescence microscopy. Scale bars represent 10 μm. (B) Detection of UL116 by fluorescence-activated cell sorter (FACS) analysis on nonpermeabilized HEK293T cells. Cells were transfected with expression vectors for UL116 as well as gH and gB, both alone and in combination, as indicated. Forty-eight hours posttransfection, cells were stained at 4°C with anti-UL116 polyclonal mouse serum at different dilutions. Excess probe was removed by washing in PBS, and then cells were fixed and stained with Alexa Fluor 488-conjugated anti-mouse antibody. For each point, 10,000 cells were analyzed and the mean fluorescent intensity of Alexa Fluor 488-positive cells was reported. Secondary antibody only was used as a negative control. Experiments were performed in triplicate. (C) UL116-gH coimmunoprecipitation. Lysates from HEK293T cells transiently expressing UL116-his/gH-myc, UL116-his, gB, and UL116-his/gB were subjected to parallel immunoprecipitation (IP) experiments (antibodies used are specified on the left) with both covalently linked magnetic anti-His and anti-myc beads. Total lysates (input) and eluted samples were separated by SDS-PAGE and analyzed by immunoblotting for both the His and myc tag (indicated at the bottom). (D) Coimmunoprecipitation (Co-IP) of the UL116-gH complex in infected cells. Cell lysates were prepared from HFF-1 infected with HCMV-TR UL116 Flag and wt TR separately (5 dpi). Complexes were captured using the human monoclonal antibody MSL-109. Elutions and crude extracts were subjected to immunoblotting using a rabbit anti-gH polyclonal serum, an anti-Flag M2 clone monoclonal MAb, and a rabbit polyclonal serum specific for gL.
To test the formation of a gH/UL116 complex, we generated eukaryotic expression vectors carrying singly tagged gH and UL116 versions of the codon-optimized genes. We first performed coimmunoprecipitation experiments on extracts of HEK293T cells coexpressing gH-myc/UL116-His or gB/UL116-His. Extracts from single-gene transfection with gH-myc and gB were used as controls. Coimmunoprecipitations were performed with an anti-His antibody to detect proteins associated with UL116 and by anti-myc to reveal species associated with gH. Each immunoprecipitated sample was divided into three aliquots and treated for Western blotting using anti-His (for UL116), anti-myc (for gH), and anti-gB as probes. Pulldown of UL116 resulted in association with gH only and not with gB (Fig. 5C, bottom, lanes 5 and 10, respectively). Conversely, immunoprecipitation of gH resulted in the coimmunoprecipitation of UL116 (Fig. 5C, middle, lane 1). To confirm the formation of the gH-UL116 complex and to exclude potential artifacts induced by the overexpression system, we decided to test the interaction between the two proteins in HCMV-infected cells. To this aim, we performed coimmunoprecipitation experiments using the anti-gH human monoclonal antibody MSL-109. Extracts from TR UL116-Flag-infected HFF-1 cells at 96 hpi were prepared and submitted to anti-gH immunoprecipitation. Cell lysate from wild-type-TR-infected HFF-1 cells was used as a negative control. The gH-associated proteins were separated by SDS-PAGE, and as a positive control, gL was probed by Western blotting. Anti-Flag antibody was used as the probe to reveal the presence of UL116. As expected, gL was detected in extracts from cells infected with both viruses (Fig. 5D). Notably, a clear signal for UL116-Flag was detected in extracts from cells infected with TR UL116-Flag but not when wild-type TR virus was used for infection (Fig. 5D).
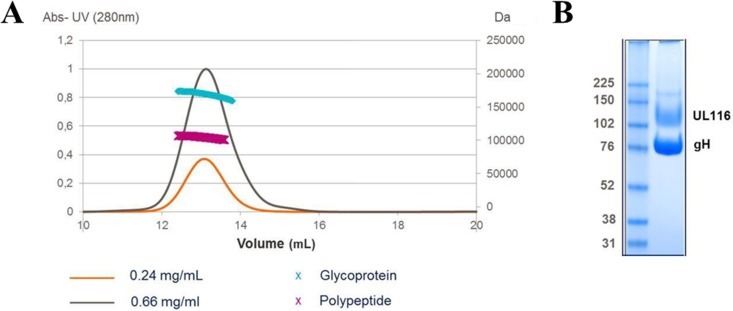
Finally, HEK293T cells were transfected with plasmids encoding a soluble form of gH and UL116 containing a Strep-tag at the C terminus. HEK293T cell supernatants were loaded on a Strep-Tactin resin, and the eluted material was analyzed using SEC, multiangle light scattering, and SDS-PAGE. SEC and SDS-PAGE revealed that gH and UL116 form a stable complex that migrates as a single peak. MALS analysis suggested that the mass of the gH/UL116 complex is 161 kDa (i.e., glycans account for ∼37% of the total mass of the complex), consistent with a 1-to-1 stoichiometry (Fig. 6A). The heterodimer is not disulfide linked and dissociates in single subunits in SDS-PAGE under nonreducing conditions (Fig. 6B).
FIG 6.
MALS analysis of purified gH/ULL116 complex. (A) SEC profile of the gH/UL116 complex at two different concentrations (gray and orange traces) and molecular mass estimates for the polypeptide only (110 kDa) and for the glycosylated complex (160 kDa) determined by MALS (pink and cyan lines, respectively). The left y axis pertains to UV absorbance (Abs), while the right y axis represents the molecular weight. (B) SDS-PAGE analysis under nonreducing conditions of the gH/UL116 complex used in panel A.
These results demonstrate the formation of a gH/UL116 complex in transfected cells that does not require the presence of gL to form. Moreover, we also were able to confirm the formation of the complex during a productive HCMV infection.
gL competes with UL116 for gH binding in transfected cells.
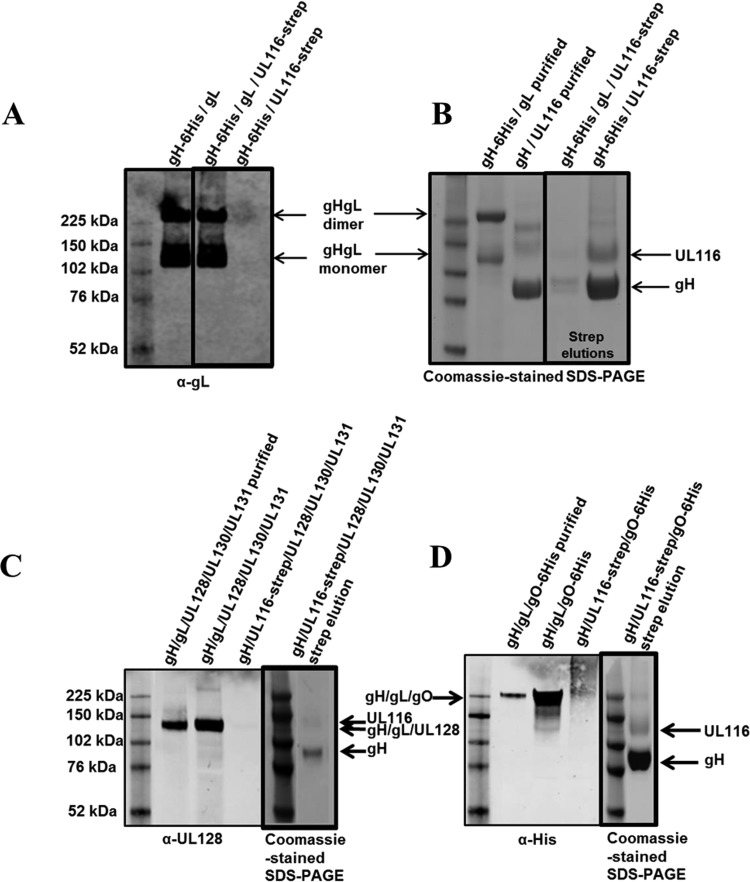
To assess if gH, gL, and UL116 can form a ternary complex, we transfected HEK293T cells with a construct corresponding to the soluble gH extracellular domain together with either gL or UL116-strep or both gL and UL116-strep. Western blot analysis of the cell supernatants with an anti-gL antibody showed the formation of monomers and homodimers of the gH/gL heterodimers in cells transfected with either gH and gL or gH, gL, and UL116 (Fig. 7A). The formation of disulfide-linked monomers and homodimers of soluble gH/gL is consistent with what has been described previously (35, 54, 71). The Strep-tag at the C terminus of UL116 allowed us to isolate gH/UL116-containing complexes from cell supernatants. SDS-PAGE analysis of the eluted fraction revealed that when gH, gL, and UL116 are transfected together, only a very minor amount of gH/UL116 complex forms and no ternary complex could be detected (Fig. 7B). Together, these data suggest that gL competes with UL116 for gH binding. It has to be noted that gH and gL form disulfide-linked heterodimers, and perhaps once this complex forms UL116 is not able to displace gL.
FIG 7.
Western blotting and SDS-PAGE for gH/gL, gH/gL/UL116, and gH/UL116 transfections. (A) Anti-gL Western blotting (nonreducing) on conditioned media from Expi293 cells transiently transfected with DNA encoding the indicated proteins. (B) gH-6His/UL116-strep and gH-6His/gL/UL116-strep samples were Strep-tag affinity purified from the medium, and the elution fraction was run in SDS-PAGE. Coomassie-stained nonreducing SDS-PAGE gels show purified gH/gL and gH/UL116 (controls) followed by strep elution samples. (C and D) Anti-UL128 (C) and anti-His (D) Western blotting (nonreducing) on purified protein alongside conditioned medium from Expi293 cells transiently transfected with the DNA encoding the indicated proteins. Samples containing UL116-strep then were Strep-tag affinity purified from the medium, and the elution fractions were run in SDS-PAGE. Coomassie-stained nonreducing SDS-PAGE gel showing elution fractions are in thickly lined boxes in panels C and D.
The gL subunit is not only disulfide linked to gH but also is engaged in disulfide bond formation with gO, in the gH/gL/gO complex, or with UL128 in the pentamer (54). Although the absence of gL from the gH/UL116 dimer suggested that neither gO nor UL128 can associate with this heterodimer, we sought to verify this hypothesis. To assess if UL116 could replace gL as a scaffold for the formation of these complexes, we transfected HEK293 cells with constructs encoding the soluble extracellular gH domain with the following combinations: (i) gH/gL/UL128/strep-UL130/UL131; (ii) gH/strep-UL116/UL128/UL130/UL131; (iii) gH/gL/His-gO; and (iv) gH/strep-UL116/His-gO. Secreted proteins then were detected by Western blotting, and Strep-tag pulldown was used to reveal subunits associated with UL116. As can be seen in Fig. 7B and C, UL128 is secreted only when gL is present and not when UL116 is expressed. Similarly, gO does not form a complex when UL116 replaces gL (Fig. 7D). Thus, the association of UL116 with gH competes with the formation of both pentamer and gO subunits.
Negative-stain EM analysis of the purified UL116/gH complex.
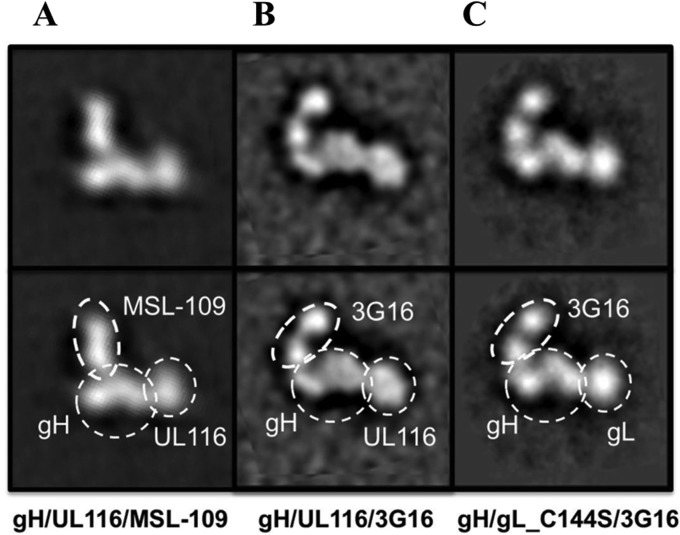
We previously used single-particle negative-stain EM to characterize gH/gL, gH/gL/gO, and Pentamer complexes bound to neutralizing antibodies (54, 72). Our data revealed that HCMV gH/gL has a boot-shaped structure similar to that of HSV-2 gH/gL (73) and that gO and the UL128/UL130/UL131A subunits bind to the N-terminal end of gH/gL in gH/gL/gO and Pentamer, respectively. We also engineered a gL mutant, C144A, which prevents gH/gL homodimerization.
Here, we extended the EM analysis to the purified gH/UL116 complex bound to either MSL-109 or 3G16 antibody (41, 72). The trimeric gH/UL116/3G16 and gH/UL116/MSL-109 complexes were analyzed by negative-stain EM and single-particle analysis. 2D class averages were generated and compared to the corresponding 2D class averages for the gH/gL-C144A/3G16 complex (Fig. 8). This analysis revealed that the gH/UL116 complex has a boot-shaped structure similar to that of gH/gL. 3G16 and MSL-109 Fabs bind to the C-terminal domain and to the “heel” of the gH molecule, respectively, as previously described for gH/gL (54, 72). Furthermore, the EM analysis and binding data (unpublished results) suggest that UL116 occupies a position similar to that of gL at the N-terminal end of gH (Fig. 8) and that the presence of UL116 does not affect the 3G16 and MSL-109 binding sites.
FIG 8.
Single-particle EM of the gH/UL116 complex. Purified gH/UL116/MSL-109 (A), gH/UL116/3G16 (B), and gH/gL_C144S/3G16 (C) ternary complexes were analyzed by single-particle electron microscopy. Individual particles, representing a specific 2D view of the complex, were aligned and used to generate 2D class averages. Comparison among the class averages of these 3 complexes and also with previous ones obtained for gH/gL and gH/gL/gO bound to MSL-109 or 3G16 (72) suggest the position of individual subunits and Fabs.
DISCUSSION
HCMV envelope glycoproteins are potential components of a prophylactic HCMV vaccine. Among them, gB is the first recombinant antigen used for clinical immunization studies (74). In more recent years the focus for preclinical vaccination studies has shifted to the components of the gH-based complexes gH, gL, gO, UL128, UL130, and UL131A. Here, we describe the initial characterization of the product of the HCMV UL116 gene from the TR strain that we have identified as a virion envelope glycoprotein. Our results suggest that UL116 forms a noncovalent heterodimer with gH, independently from gL, which is present on the virion surface.
Our analysis showed that the mature form of UL116 is heavily glycosylated, principally by Golgi-type carbohydrate chains. An ER-type glycosylated UL116 of lower mass is constantly present over the infection cycle, likely as an immature precursor, but it was not found to be present on the virion. Following translocation from the ER, UL116 is targeted to the intracellular virion AC and is incorporated into virions. Transient expression experiments with UL116 expressed as single HCMV species revealed that the UL116 protein is retained in the ER and not secreted or targeted to the plasma membrane. Among the HCMV envelope proteins we tried in cotransfection, gH was the only one that was able to release UL116 ER retention, suggesting the formation of a heterodimeric gH/UL116 complex. This hypothesis was confirmed by experiments in HCMV-infected cells.
So far the only dimeric complex known for HCMV gH is the disulfide-linked gH/gL. Although it was reported to be sufficient (with gB) for entry into fibroblasts (75), a more recent analysis has shown that on the surface of different HCMV strains there are two distinct complexes assembling on the gH/gL scaffold: the trimeric gH/gL/gO and the pentameric gH/gL/UL128/UL130/UL131A complex (76). The efficiency of infection of specific cell types, perhaps by distinct entry pathways, could depend on relative ratios between these complexes on the envelope (76). Replication in distinctive cell types generates viral populations carrying different ratios of these complexes on their envelope (77). The association of gH/gL with these alternative complexes relies on the formation of a disulfide bond with either gO or UL128. Both proteins target the same Cys residue on gL, thus forming mutually exclusive gH/gL complexes (54). Both gO and UL128/UL130/UL131A occupy the N-terminal region of gH and were proposed to govern the specificity of receptor interaction (54). EM analysis of purified gH/UL116 indicated that the complex has the same overall organization of gH/gL and that UL116 might share the gH binding site of gL, although gH binding to UL116 is not mediated by disulfide bond formation. Peculiarly, the UL116 gene is adjacent to that encoding gL (UL115). Although not a rule, it has been noticed that in the HCMV genome, genes with similar functions are in close proximity to each other (8). Complex formation analysis by the coexpression of gH, gL, and UL116 and subsequent SEC and immunoprecipitation experiments confirmed that gL did not associate with the gH/UL116 heterodimer. Furthermore, UL116 not only displaces gL from gH but also prevents binding of gO and UL128/130/131.
A very recent report described how the production of alternative gH-based complexes is regulated. The product of the UL148 gene is an ER glycoprotein that, in this compartment, interacts with the gH/gL heterodimer in the absence of either gO or UL128 and favors the formation of the gH/gL/gO complex (78). The association of UL116 with gH appears to take place in the ER, so we cannot exclude that a similar complex switch is driven by UL116 and that the heterodimer gH/UL116 represents the core scaffold for an unidentified complex that excludes gL but could recruit additional factors. However, we showed that unlike UL148, UL116 is present in the virion.
gH/UL116 is a novel gH-based complex, and it is tempting to speculate that it could act as a ligand for a different receptor, thus amplifying the potential for the specific recognition of human cell types. Intriguingly, the restoration of Pentamer expression in the AD169 laboratory strains induces the increased expression of UL116 (38), although the biological significance has not been further investigated. One can speculate that the increased expression of UL116 somehow is linked to the acquisition of a functional Pentamer and the concomitant expansion of cellular tropism. Our coimmunoprecipitation experiments performed in HFF-1 cells infected with the TR strain suggest that the amount of gH/UL116 complex is considerably lower than that of gH/gL complexes. Although we did not explore this observation further, it would be interesting to assess the ratio of gH/UL116 to gH/gL complexes in viruses with different genetic backgrounds or grown in different cell lines.
In conclusion, our results add a new layer of complexity by identifying for the first time a stable herpesvirus protein complex that contains gH but not gL. Given that gH-containing complexes have great potential as vaccine antigens, a better understanding of these antibody targets may guide the design of improved vaccine antigens or antibody-based therapies.
ACKNOWLEDGMENTS
The SEC-LS/UV/RI instrumentation was supported by NIH award 1S10RR023748-01. The MALS experiments and analysis were performed at the Biophysics Resource of the Keck Facility at Yale University.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Mocarski JE, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Cannon MJ. 2009. Congenital cytomegalovirus (CMV) epidemiology and awareness. J Clin Virol 46(Suppl 4):S6–S10. doi: 10.1016/j.jcv.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Topics Microbiol Immunol 325:417–470. [DOI] [PubMed] [Google Scholar]
- 4.Krause PR, Bialek SR, Boppana SB, Griffiths PD, Laughlin CA, Ljungman P, Mocarski ES, Pass RF, Read JS, Schleiss MR, Plotkin SA. 2013. Priorities for CMV vaccine development. Vaccine 32:4–10. doi: 10.1016/j.vaccine.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stratton KR, Durch JS, Lawrence RS. 2001. Vaccines for the 21st century: a tool for a decisionmaking. National Academy Press, Washington, DC. [PubMed] [Google Scholar]
- 6.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84:17–28. [DOI] [PubMed] [Google Scholar]
- 8.Murphy E, Shenk T. 2008. Human cytomegalovirus genome. Curr Topics Microbiol Immunol 325:1–19. [DOI] [PubMed] [Google Scholar]
- 9.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312. [DOI] [PubMed] [Google Scholar]
- 10.Shikhagaie M, Merce-Maldonado E, Isern E, Muntasell A, Alba MM, Lopez-Botet M, Hengel H, Angulo A. 2012. The human cytomegalovirus-specific UL1 gene encodes a late-phase glycoprotein incorporated in the virion envelope. J Virol 86:4091–4101. doi: 10.1128/JVI.06291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanton RJ, Baluchova K, Dargan DJ, Cunningham C, Sheehy O, Seirafian S, McSharry BP, Neale ML, Davies JA, Tomasec P, Davison AJ, Wilkinson GW. 2010. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Investig 120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG II, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaacson MK, Juckem LK, Compton T. 2008. Virus entry and innate immune activation. Curr Topics Microbiol Immunol 325:85–100. [DOI] [PubMed] [Google Scholar]
- 14.Jiang XJ, Adler B, Sampaio KL, Digel M, Jahn G, Ettischer N, Stierhof YD, Scrivano L, Koszinowski U, Mach M, Sinzger C. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J Virol 82:2802–2812. doi: 10.1128/JVI.01550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. 2010. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J Virol 84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Lanchy JM, Ryckman BJ. 2015. Human cytomegalovirus gH/gL/gO promotes the fusion step of entry into all cell types, whereas gH/gL/UL128-131 broadens virus tropism through a distinct mechanism. J Virol 89:8999–9009. doi: 10.1128/JVI.01325-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Compton T. 2004. Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends Cell Biol 14:5–8. doi: 10.1016/j.tcb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Pellett PE, Roizman B. 2013. Herpesviridae, p 1802–1822. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 19.Compton T, Nepomuceno RR, Nowlin DM. 1992. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 20.Haspot F, Lavault A, Sinzger C, Laib Sampaio K, Stierhof YD, Pilet P, Bressolette-Bodin C, Halary F. 2012. Human cytomegalovirus entry into dendritic cells occurs via a macropinocytosis-like pathway in a pH-independent and cholesterol-dependent manner. PLoS One 7:e34795. doi: 10.1371/journal.pone.0034795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Yu QC, Schroer J, Murphy E, Shenk T. 2007. Human cytomegalovirus uses two distinct pathways to enter retinal pigmented epithelial cells. Proc Natl Acad Sci U S A 104:20037–20042. doi: 10.1073/pnas.0709704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol 74:7720–7729. doi: 10.1128/JVI.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straschewski S, Patrone M, Walther P, Gallina A, Mertens T, Frascaroli G. 2011. Protein pUL128 of human cytomegalovirus is necessary for monocyte infection and blocking of migration. J Virol 85:5150–5158. doi: 10.1128/JVI.02100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Wilkie AR, Weller M, Liu X, Cohen JI. 2015. THY-1 cell surface antigen (CD90) has an important role in the initial stage of human cytomegalovirus infection. PLoS Pathog 11:e1004999. doi: 10.1371/journal.ppat.1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Britt WJ, Vugler L, Butfiloski EJ, Stephens EB. 1990. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol 64:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. 2012. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J Virol 86:7444–7447. doi: 10.1128/JVI.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cranage MP, Kouzarides T, Bankier AT, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell SE, Minson AC. 1986. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J 5:3057–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilja AE, Mason PW. 2012. The next generation recombinant human cytomegalovirus vaccine candidates-beyond gB. Vaccine 30:6980–6990. doi: 10.1016/j.vaccine.2012.09.056. [DOI] [PubMed] [Google Scholar]
- 34.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, Corey L, Hill J, Davis E, Flanigan C, Cloud G. 2009. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 360:1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen Y, Monroe J, Linton C, Archer J, Beard CW, Barnett SW, Palladino G, Mason PW, Carfi A, Lilja AE. 2014. Human cytomegalovirus gH/gL/UL128/UL130/UL131A complex elicits potently neutralizing antibodies in mice. Vaccine 32:3796–3804. doi: 10.1016/j.vaccine.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Wussow F, Chiuppesi F, Martinez J, Campo J, Johnson E, Flechsig C, Newell M, Tran E, Ortiz J, La Rosa C, Herrmann A, Longmate J, Chakraborty R, Barry PA, Diamond DJ. 2014. Human cytomegalovirus vaccine based on the envelope gH/gL pentamer complex. PLoS Pathog 10:e1004524. doi: 10.1371/journal.ppat.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabanova A, Perez L, Lilleri D, Marcandalli J, Agatic G, Becattini S, Preite S, Fuschillo D, Percivalle E, Sallusto F, Gerna G, Corti D, Lanzavecchia A. 2014. Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc Natl Acad Sci U S A 111:17965–17970. doi: 10.1073/pnas.1415310111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freed DC, Tang Q, Tang A, Li F, He X, Huang Z, Meng W, Xia L, Finnefrock AC, Durr E, Espeseth AS, Casimiro DR, Zhang N, Shiver JW, Wang D, An Z, Fu TM. 2013. Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proc Natl Acad Sci U S A 110:E4997–E5005. doi: 10.1073/pnas.1316517110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lilleri D, Kabanova A, Revello MG, Percivalle E, Sarasini A, Genini E, Sallusto F, Lanzavecchia A, Corti D, Gerna G. 2013. Fetal human cytomegalovirus transmission correlates with delayed maternal antibodies to gH/gL/pUL128-130-131 complex during primary infection. PLoS One 8:e59863. doi: 10.1371/journal.pone.0059863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilleri D, Kabanova A, Lanzavecchia A, Gerna G. 2012. Antibodies against neutralization epitopes of human cytomegalovirus gH/gL/pUL128-130-131 complex and virus spreading may correlate with virus control in vivo. J Clin Immunol 32:1324–1331. doi: 10.1007/s10875-012-9739-3. [DOI] [PubMed] [Google Scholar]
- 41.Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, Lanzavecchia A. 2010. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol 84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortese M, Calo S, D'Aurizio R, Lilja A, Pacchiani N, Merola M. 2012. Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One 7:e50166. doi: 10.1371/journal.pone.0050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spaderna S, Blessing H, Bogner E, Britt W, Mach M. 2002. Identification of glycoprotein gpTRL10 as a structural component of human cytomegalovirus. J Virol 76:1450–1460. doi: 10.1128/JVI.76.3.1450-1460.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spaderna S, Kropff B, Kodel Y, Shen S, Coley S, Lu S, Britt W, Mach M. 2005. Deletion of gpUL132, a structural component of human cytomegalovirus, results in impaired virus replication in fibroblasts. J Virol 79:11837–11847. doi: 10.1128/JVI.79.18.11837-11847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber MT, Compton T. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol 72:8191–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CP, Vesole DH, Nelson J, Oldstone MB, Stinski MF. 1989. Identification and expression of a human cytomegalovirus early glycoprotein. J Virol 63:3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218–232. doi: 10.1034/j.1600-0854.2002.030307.x. [DOI] [PubMed] [Google Scholar]
- 48.Margulies BJ, Browne H, Gibson W. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith IL, Taskintuna I, Rahhal FM, Powell HC, Ai E, Mueller AJ, Spector SA, Freeman WR. 1998. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch Ophthalmol 116:178–185. [DOI] [PubMed] [Google Scholar]
- 50.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talbot P, Almeida JD. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J Gen Virol 36:345–349. [DOI] [PubMed] [Google Scholar]
- 52.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 53.Irmiere A, Gibson W. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 54.Ciferri C, Chandramouli S, Donnarumma D, Nikitin PA, Cianfrocco MA, Gerrein R, Feire AL, Barnett SW, Lilja AE, Rappuoli R, Norais N, Settembre EC, Carfi A. 2015. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc Natl Acad Sci U S A 112:1767–1772. doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arakawa T, Langley KE, Kameyama K, Takagi T. 1992. Molecular weights of glycosylated and nonglycosylated forms of recombinant human stem cell factor determined by low-angle laser light scattering. Anal Biochem 203:53–57. doi: 10.1016/0003-2697(92)90042-6. [DOI] [PubMed] [Google Scholar]
- 56.Hayashi Y, Matsui H, Takagi T. 1989. Membrane protein molecular weight determined by low-angle laser light-scattering photometry coupled with high-performance gel chromatography. Methods Enzymol 172:514–528. doi: 10.1016/S0076-6879(89)72031-0. [DOI] [PubMed] [Google Scholar]
- 57.Folta-Stogniew E. 2006. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. Methods Mol Biol 328:97–112. [DOI] [PubMed] [Google Scholar]
- 58.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 59.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J Struct Biol 116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 60.Leatham MP, Witte PR, Stinski MF. 1991. Alternate promoter selection within a human cytomegalovirus immediate-early and early transcription unit (UL119-115) defines true late transcripts containing open reading frames for putative viral glycoproteins. J Virol 65:6144–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mockarski JE, Shenk T, Griffiths P, Pass R. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 62.Malouli D, Nakayasu ES, Viswanathan K, Camp DG II, Chang WL, Barry PA, Smith RD, Fruh K. 2012. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J Virol 86:8959–8973. doi: 10.1128/JVI.01132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanai K, Yamada S, Yamamoto Y, Fukui Y, Kurane I, Inoue N. 2011. Re-evaluation of the genome sequence of guinea pig cytomegalovirus. J Gen Virol 92:1005–1020. [DOI] [PubMed] [Google Scholar]
- 64.Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kohler CP, Kerry JA, Carter M, Muzithras VP, Jones TR, Stenberg RM. 1994. Use of recombinant virus to assess human cytomegalovirus early and late promoters in the context of the viral genome. J Virol 68:6589–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson DC, Spear PG. 1983. O-linked oligosaccharides are acquired by herpes simplex virus glycoproteins in the Golgi apparatus. Cell 32:987–997. doi: 10.1016/0092-8674(83)90083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maley F, Trimble RB, Tarentino AL, Plummer TH Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem 180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez V, Sztul E, Britt WJ. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J Virol 74:3842–3851. doi: 10.1128/JVI.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, Johnson DC. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol 82:60–70. doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loomis RJ, Lilja AE, Monroe J, Balabanis KA, Brito LA, Palladino G, Franti M, Mandl CW, Barnett SW, Mason PW. 2013. Vectored co-delivery of human cytomegalovirus gH and gL proteins elicits potent complement-independent neutralizing antibodies. Vaccine 31:919–926. doi: 10.1016/j.vaccine.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 72.Ciferri C, Chandramouli S, Leitner A, Donnarumma D, Cianfocco MA, Gerrein R, Friedrich K, Aggarwal Y, Palladino G, Aebersold R, Norais N, Settembre EC, Carfi A. 2015. Antigenic characterization of the HCMV gH/gL/gO and Pentamer cell entry complexes reveals binding sites for potently neutralizing human antibodies. PLoS Pathog 11:e1005230. doi: 10.1371/journal.ppat.1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonczol E, Berensci K, Pincus S, Endresz V, Meric C, Paoletti E, Plotkin SA. 1995. Preclinical evaluation of an ALVAC (canarypox)–human cytomegalovirus glycoprotein B vaccine candidate. Vaccine 13:1080–1085. doi: 10.1016/0264-410X(95)00048-6. [DOI] [PubMed] [Google Scholar]
- 75.Vanarsdall AL, Chase MC, Johnson DC. 2011. Human cytomegalovirus glycoprotein gO complexes with gH/gL, promoting interference with viral entry into human fibroblasts but not entry into epithelial cells. J Virol 85:11638–11645. doi: 10.1128/JVI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou M, Yu Q, Wechsler A, Ryckman BJ. 2013. Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J Virol 87:9680–9690. doi: 10.1128/JVI.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7:e1001256. doi: 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li G, Nguyen CC, Ryckman BJ, Britt WJ, Kamil JP. 2015. A viral regulator of glycoprotein complexes contributes to human cytomegalovirus cell tropism. Proc Natl Acad Sci U S A 112:4471–4476. doi: 10.1073/pnas.1419875112. [DOI] [PMC free article] [PubMed] [Google Scholar]