ABSTRACT
Sphingomyelin is an essential cellular lipid that traffics between plasma membrane and intracellular organelles until directed to lysosomes for SMPD1 (sphingomyelin phosphodiesterase 1)-mediated degradation. Inactivating mutations in the SMPD1 gene result in Niemann-Pick diseases type A and B characterized by sphingomyelin accumulation and severely disturbed tissue homeostasis. Here, we report that sphingomyelin overload disturbs the maturation and closure of autophagic membranes. Niemann-Pick type A patient fibroblasts and SMPD1-depleted cancer cells accumulate elongated and unclosed autophagic membranes as well as abnormally swollen autophagosomes in the absence of normal autophagosomes and autolysosomes. The immature autophagic membranes are rich in WIPI2, ATG16L1 and MAP1LC3B but display reduced association with ATG9A. Contrary to its normal trafficking between plasma membrane, intracellular organelles and autophagic membranes, ATG9A concentrates in transferrin receptor-positive juxtanuclear recycling endosomes in SMPD1-deficient cells. Supporting a causative role for ATG9A mistrafficking in the autophagy defect observed in SMPD1-deficient cells, ectopic ATG9A effectively reverts this phenotype. Exogenous C12-sphingomyelin induces a similar juxtanuclear accumulation of ATG9A and subsequent defect in the maturation of autophagic membranes in healthy cells while the main sphingomyelin metabolite, ceramide, fails to revert the autophagy defective phenotype in SMPD1-deficient cells. Juxtanuclear accumulation of ATG9A and defective autophagy are also evident in tissues of smpd1-deficient mice with a subsequent inability to cope with kidney ischemia-reperfusion stress. These data reveal sphingomyelin as an important regulator of ATG9A trafficking and maturation of early autophagic membranes.
KEYWORDS: acid sphingomyelinase, ATG9A, autophagy, ischemia-reperfusion injury, kidney, lysosome, Niemann Pick disease, sphingomyelin, trafficking
Introduction
Autophagy is a lysosomal catabolic pathway that ensures the turnover of old and damaged cellular organelles and macromolecules thereby promoting the survival of starved and stressed cells.1-3 During autophagy, cytosolic cargoes are sequestered by expanding phagophores that arise from endoplasmic reticulum (ER)-derived omegasomes, form autophagosomes upon closure and finally fuse with endosomes and lysosomes to become degradative autophagic vacuoles. This process is tightly controlled by a family of autophagy-related (ATG) proteins (reviewed in 4–6). Briefly, the ULK (unc-51 like autophagy activating kinase) and class III PtdIns3K (Vps34 in yeast) lipid kinase complexes translocate to the ER, where the latter produces phosphatidylinositol 3-phosphate that recruits various lipid-binding effector proteins including WIPI1 and WIPI2. WIPI2 recruits ATG12–ATG5-ATG16L1 complex to the forming omegasome,7 where the latter promotes the conversion of MAP1LC3-I (microtubule associated protein 1 light chain 3-I) to MAP1LC3-phosphatidylethanolamine (MAP1LC3-II), which associates with autophagic membranes and promotes their maturation. ATG9A, the only multipass transmembrane ATG protein, assists the growth of autophagic membranes by a poorly defined mechanism that may involve trafficking of ATG9A-containing vesicles between the trans-Golgi network (TGN), plasma membrane, recycling endosomes (REs) and autophagic membranes.8,9 Finally, ATG2A and ATG2B promote the closure of the autophagic membrane.10
Sphingomyelin (SM) is an essential cellular lipid enriched in plasma membrane, REs and TGN, and degraded to ceramide and phosphocholine by SMPD1 in the lumen of lysosomes or by neutral sphingomyelinases located in the plasma membrane, Golgi and ER.11 Together with cholesterol, SM forms lipid rafts that serve as platforms for transmembrane signaling proteins and membrane budding.12 Proper SM organization is required for trafficking of TFRC (transferrin receptor) from the plasma membrane to REs,13 retrograde transport from endosomes to TGN,14 and vesicle biogenesis and cargo sorting at the TGN.15 Cancer cells have generally low SM levels and accumulation of SM is selectively toxic to them, possibly due to the destabilizing effect of SM on their fragile lysosomal membranes.16-18 Even though normal cells tolerate higher SM levels than cancer cells, inactivating mutations in the SMPD1 gene lead to Niemann-Pick type A (NPA) disease characterized by disturbed tissue homeostasis and death at infancy or early childhood.19 NPA is associated with an autophagy defect that has been suggested to be due to the failure of autophagosomes to fuse with lysosomes,20 and reduced lysosomal membrane stability.21
In order to enlighten the mechanism by which SM regulates autophagy, we undertook a detailed study of the autophagic process in NPA patient fibroblasts, SM-treated fibroblasts, MCF7 breast cancer cells depleted for SMPD1 by RNAi or clustered regularly interspaced short palindromic repeats (CRISPr)-Cas9 technology and tissues of smpd1−/− mice. Using an array of state-of-the-art autophagy assays, transmission electron microscopy (TEM) and super resolution 3D structured illumination microscopy (3D-SIM), we demonstrate that excess SM induces a severe autophagy defect characterized by altered trafficking of ATG9A and accumulation of omegasomes and phagophores.
Results
NPA fibroblasts accumulate immature autophagic membranes
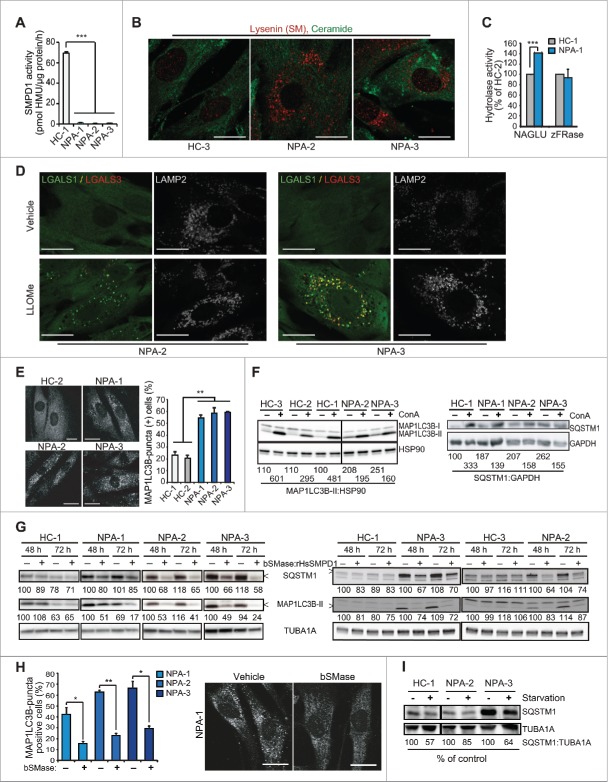
To enlighten the impact of SM metabolism on autophagy, we first investigated primary NPA patient fibroblasts with low residual SMPD1 enzyme activities and corresponding intracellular accumulation of SM (Fig. 1A and B). Notably, the activities of lysosomal cysteine cathepsins and NAGLU (N-acetyl-glucosaminidase, alpha) were not compromised in NPA cells (Fig. 1C and data not shown). In line with this, the lysosomal membranes of NPA fibroblasts were intact as shown by the absence of LGALS1/galectin-1- or LGALS3/galectin-3-positive lysosomes (Fig. 1D). Indicative of reduced autophagic flux, all NPA cells accumulated MAP1LC3B-positive puncta, and had increased levels of MAP1LC3B -II and an autophagic cargo protein SQSTM1/p62, which were only marginally increased upon autophagy inhibition by concanamycin A (ConA) (Fig. 1E and F). The autophagy defect could be effectively reverted by recombinant human (Homo sapiens) SMPD1 (rHsSMPD1) and bacterial neutral sphingomyelinase (bSMase) and partially overcome by amino acid starvation (Fig. 1G-I).
Figure 1.
Sphingomyelinases and starvation revert autophagy defect in NPA patient fibroblasts. (A) SMPD1 hydrolytic activity in lysates of fibroblasts from a healthy controls (HC) and NPA patients (NPA). Bars, SD for a quadruplicate representative (n = 3) experiment. (B) Representative confocal images of the indicated fibroblasts permeabilized and stained with lysenin plus anti-lysenin and anti-ceramide antibodies to visualize intracellular SM and ceramide pools. (C) NAGLU and cysteine cathepsin (zFRase) activities in lysates of the indicated fibroblasts. Bars, SD for 3 triplicate experiments. (D) Representative confocal images of NPA-2 and NPA-3 fibroblasts stained for LGALS1 (green), LGALS3 (red) and LAMP2 (white, converted from blue). When indicated, cells were treated with 2 mM LLOMe for 2 h to induce lysosomal membrane permeabilization and LGALS1/3 puncta formation. (E) Representative confocal images of the indicated fibroblasts stained with anti-MAP1LC3B antibody (left) and the quantification of MAP1LC3B-puncta positive cells (≥10 puncta/cell) in these samples (right). Bars, SEM for ≥2 experiments with >50 cells analyzed/sample. (F) Representative immunoblots of MAP1LC3B-I and -II (LC3B-I and LC3B-II) and SQSTM1 from the indicated fibroblasts left untreated (−) or treated (+) for 16 h with 1 nM ConA. HSP90 and GAPDH served as loading controls. The values below represent MAP1LC3B-II:HSP90 and SQSTM1:GAPDH ratios as a percentage of the ratio in untreated HC-1 cells (upper row) or as a percentage of values in untreated cells from the same individual (lower row). (G) Representative immunoblots of MAP1LC3B-II and SQSTM1 from the indicated fibroblasts treated with vehicle (–) or 5 µg/ml of bSMase (left) or rHsSMPD1 (right) (+) every 24 h for the indicated times. TUBA1A served as a loading control. The values below the blots represent SQSTM1:TUBA1A (upper row) and MAP1LC3B-II:TUBA1A (lower row) ratios as a percentage of values in untreated cells from the same individual. (H) Quantification of LC3-puncta positive NPA fibroblasts (≥10 puncta/cell) after treatment with vehicle (−) or 5 µg/ml/24 h bSMase (+) for 48 h and staining with anti-MAP1LC3B antibody (left). Representative images of NPA-1 fibroblasts are shown (right). Bars, as in (C). (I) Representative SQSTM1 immunoblots of proteins from the indicated fibroblasts cultured in the normal growth medium (−) or in HBSS plus 1 g/L glucose (+) for 8 h. TUBA1A served as a loading control. All lanes originate from the same blot. The values represent SQSTM1:TUBA1A ratios as a percentage of values in untreated cells from the same individual. Scale bars: 20 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
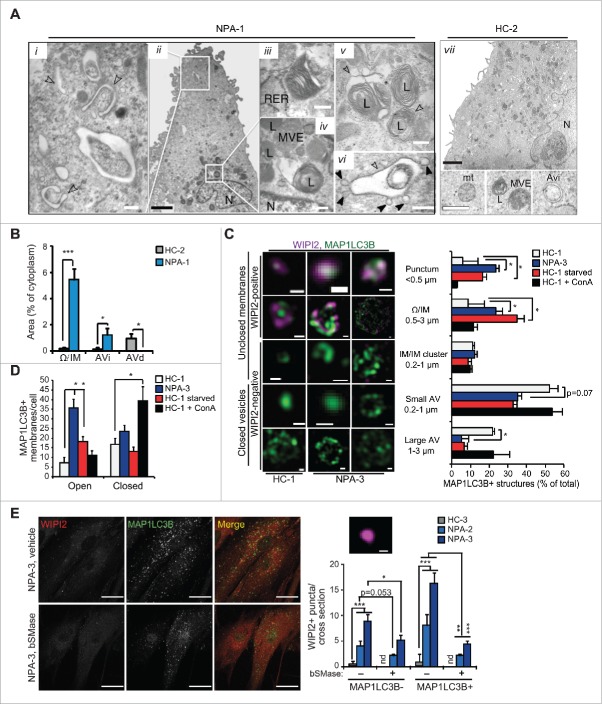
Analysis of the ultrastructure of NPA fibroblasts by TEM revealed a complete lack of normal initial (AVi) or degradative (AVd) autophagic vesicles (Fig. 2A and B). Instead, they had numerous unclosed and elongated membranes resembling omegasomes and phagophores that were only infrequently found in control fibroblasts (Fig. 2A i, ii, vii and B). Some of them formed vesicle-like structures with swollen intermembrane space and small vesicles frequently attached to their outer membranes (Fig. 2A vi). Lysosomes were enlarged and contained inclusions of electron-dense and lamellar structures indicative of membrane stacking and accumulation (Fig. 2A iv, v). Super resolution 3D-SIM confirmed the accumulation of WIPI2- and MAP1LC3B-positive immature membranes in NPA fibroblasts (Fig. 2C and D, and movies 1 to 4). Induction of autophagosome biogenesis by starvation resulted also in an increase in early autophagic membranes in control cells (Fig. 2C and D). They were, however, less abundant and generally smaller and more compact than in NPA fibroblasts. In contrast, ConA, which inhibits autophagic cargo degradation and triggers autophagosome biogenesis,22 increased the number of MAP1LC3B positive structures in control cells without altering the proportions of immature and mature vesicles (Fig. 2C and D). Confocal microscopy confirmed the accumulation of WIPI2 and MAP1LC3B-positive phagophores in NPA fibroblasts and revealed an additional accumulation of MAP1LC3B-negative WIPI2 puncta with a diameter of 200 to 400 nm corresponding to the size of early omegasomes (Fig. 2E). Akin to MAP1LC3B puncta (Fig. 1F), the accumulation of WIPI2-positive autophagic membranes was effectively reverted by bSMase (Fig. 2E).
Figure 2.
NPA fibroblasts accumulate unclosed autophagic membranes. (A) Representative TEM images of NPA-1 (i to vi) and HC-2 (vii) fibroblasts. Black arrowheads indicate small vesicles in contact with an outer membrane of AVi (vi). Insets in (vii) show representative images of a normal AVi, lysosome (L), multivesicular endososome (MVE) and mitochondrion (mt) in HC-2 fibroblasts. N, nucleus; RER, rough ER. White scale bars: 0.2 µm (i, iii to vi) or 0.5 µm (vii). Black scale bars: 2 µm. (B) Quantification of the cytoplasmic area covered by the indicated autophagic structures in TEM sections of HC-2 and NPA-1 fibroblasts. Bars, SD for 15 randomly chosen cross-sections from 2 independent blocks. Ω/IM, omegasomes and phagophores. (C and D) Representative super resolution SIM images of the indicated MAP1LC3B-positive autophagic membranes in HC-1 and NPA-3 fibroblasts stained for MAP1LC3B and WIPI2 (C, left) and their relative (C, right) and absolute (D) quantification. When indicated, cells were starved in HBSS plus 1 g/L glucose for 2 h or treated with 2 nM ConA for 4 h. Bars, SEM for 4 (HC-1), 10 (NPA-3) or 6 (HC-1, starved and ConA) randomly chosen stacks with 1 to 5 cells each. Scale bars: 0.2 µm. See also 3D reconstructions of a typical omegasome (Movie 1) and tubular phagophore (Movie 2) in starved HC-3 cells and compact (Movie 3) and diffuse (Movie 4) phagophores in NPA-3 cells. (E) Representative confocal images of NPA-3 fibroblasts treated with vehicle (−) or 5 µg/ml/24 h bSMase (+) for 48 h and stained for WIPI2 and MAP1LC3B (left), and quantification of puncta in similarly treated NPA-2 and NPA-3 fibroblasts (right). HC-3 control cells are shown as a negative control. Super resolution 3D-SIM image shows a typical WIPI2-positive puncta in NPA fibroblasts. Bars, SEM for a representative experiment with >20 cells/sample (n = 2). Scale bars: 20 µm (left) and 0.2 µm (right). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Student t test.
Collectively, these data reveal NPA as a severe autophagy disorder manifested by impaired maturation of early autophagic membranes.
SMPD1 is essential for autophagosome formation in MCF7 breast cancer cells
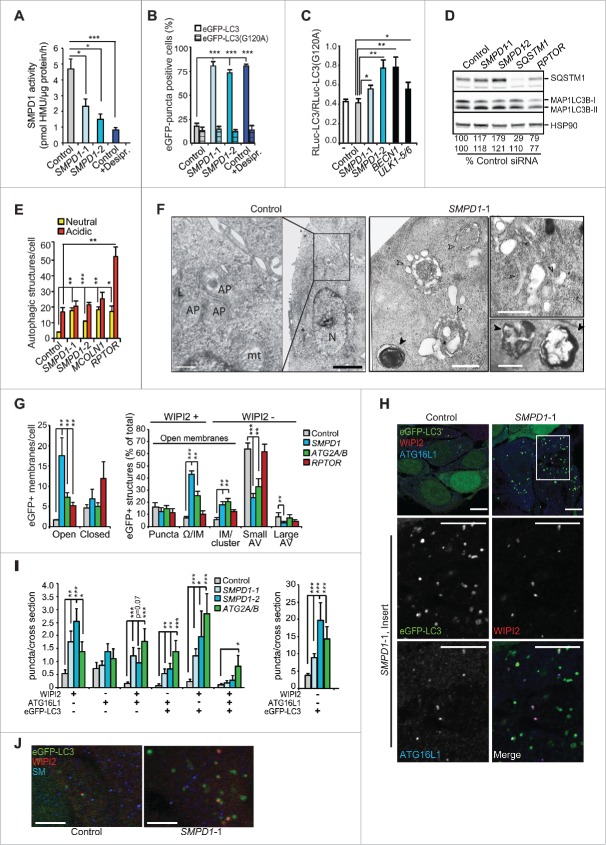
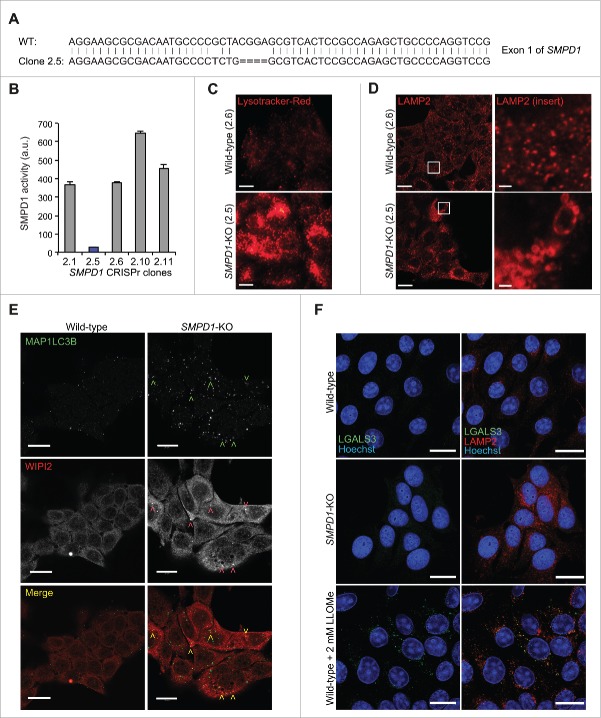
Chronic SM accumulation in NPA patient cells has numerous secondary effects on cellular functions.23 Thus, we investigated the more direct effects of SM on autophagy in SMPD1 siRNA-treated MCF7 breast cancer cells expressing various MAP1LC3B reporter constructs. As expected, SMPD1 siRNAs effectively reduced the SMPD1 activity and increased the volume of the lysosomal compartment with only small effect on other lysosomal hydrolases (Fig. 3A and S1A to 1C). SMPD1 siRNAs increased the number of enhanced green fluorescence-MAP1LC3B (eGFP-LC3B)-positive autophagic puncta but reduced autophagic flux as analyzed by MAP1LC3B-II and SQSTM1 immunoblots and a Renilla luciferase reporter-based MAP1LC3B turnover assay (Figs. 3A to D and S1D). The autophagy inhibitory effect of SMPD1 siRNAs was further supported by the accumulation of predominantly yellow (neutral pH) puncta in MCF7 cells expressing a tandem fluorescent MAP1LC3B fusion protein, tfLC3B (Figs. 3E and S1F), whose acid-sensitive green fluorescence is lost upon fusion of autophagosomes and lysosomes while the red fluorescence remains.24 Similar to NPA cells, the ultrastructural analysis revealed abnormal, elongated and enlarged phagophores, and super resolution 3D-SIM verified the abundance of unclosed WIPI2- and eGFP-LC3B-positive omegasomes and phagophores in SMPD1 siRNA-treated MCF7-eGFP-LC3B cells (Fig. 3F and G). Staining of SMPD1-depleted MCF7-eGFP-LC3B cells for WIPI2 and ATG16L1 verified an increase in early autophagic membranes, especially those positive for WIPI2 alone or together with ATG16L1 or eGFP-LC3B (Figs. 3H, 3I and S1G). Notably, the accumulating phagophores did not contain SM at levels detectable by a SM-binding protein, lysenin (Fig. 3J). CRISPr-Cas9-mediated homozygous deletion of SMPD1 induced a similar phenotype regarding the enlargement of the lysosomal compartment and accumulation of WIPI2- and MAP1LC3B-positive puncta (Fig. 4A to E). Notably, even the total SMPD1 deficiency did not destabilize lysosomal membranes as demonstrated by the lack of LGALS3 puncta formation (Fig. 4F).
Figure 3.
SMPD1 depletion impairs autophagosome closure in MCF7 cells. (A) SMPD1 activity in lysates of MCF7 cells transfected with the indicated siRNAs. Desipramine (25 µM for 1 h) served as a control for SMPD1 inhibition. (B) eGFP-puncta formation in MCF7-eGFP-LC3 cells transfected with the indicated siRNAs. Lipidation-defective mutant eGFP-LC3G120A served as a negative control. Quantification of cells with a minimum of 5 (eGFP-LC3) or one (LC3G120A) puncta are shown. Desipramine (25 µM for 6 h) served as a control. (C) Autophagic flux assessed by comparing the ratios of luciferase activities in lysates of MCF7-RLuc-LC3 and MCF7-RLuc-LC3G120A cells transfected with the indicated siRNAs. BECN1/Beclin 1 and ULK1 siRNAs served as positive controls for autophagy inhibition. (D) Representative immunoblots of the indicated proteins extracted from MCF7 cells transfected with the indicated siRNAs. RPTOR and SQSTM1 siRNAs served as positive controls for enhanced autophagic flux and SQSTM1 immunoblot, respectively, and HSP90 staining was used as a loading control. SQSTM1:HSP90 (upper row) and MAP1LCB3-II:HSP90 (lower row) ratios as a percentage of the ratio in control siRNA-treated cells are shown below. (E) Quantification of fluorescent puncta corresponding to neutral (yellow) and acidic (red) autophagic structures in MCF7-tfLC3 cells transfected with the indicated siRNAs. MCOLN1 (mucolipin 1) and RPTOR/raptor siRNAs served as controls for inhibited and enhanced autophagic flux, respectively.54,63 (F) Representative TEM images of MCF7 cells transfected with the indicated siRNAs. Arrowheads indicate elongated phagophores and swollen autophagosomes (open) and lamellar lysosomes (closed). AP, autophagosome; mt, mitochondria; N, nucleus. (G) Quantification of the indicated eGFP-positive membranes in MCF7-eGFP-LC3 cells transfected as indicated, stained for WIPI2 and analyzed by super resolution 3D-SIM. ATG2A/B and RPTOR siRNAs served as controls for disturbed and normal autophagy, respectively. (H and I) Representative confocal images (H) and quantification (I) of autophagic puncta in MCF7-eGFP-LC3 cells transfected with the indicated siRNAs and stained for WIPI2 and ATG16L1. Insert in the SMPD1-1 panel is enlarged and individual channels are shown in black and white, and the merged image in color. ATG2A/B siRNA served as a positive control for disturbed maturation of autophagic membranes. (J) Representative confocal images of MCF7 cells transfected with the indicated siRNAs and stained with LC3, WIPI2 and SM. Scale bars: 0.5 µm (F, white), 2 µm (F, black), or 10 µm (H and J). Bars, SD for 3 independent experiments (b,c,e), or a representative (n = 3) quadruplicate experiment (A), or SEM for 5–7 randomly chosen stacks with 2 to 10 cells/stack (G) or for ≥ 18 randomly chosen cells (H). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Student t test. See also Fig. S1.
Figure 4.
SMPD1-deficient MCF7 cells accumulate early autophagic membranes. (A) Sequence of SMPD1 exon1 in wild-type (WT) MCF7 cells and CRISPr-Cas9 clone 2.5. (B) Total SMPD1 activity in lysates of single cell clones of MCF7 cells transfected with SMPD1 guide-RNA. Bars, SD for a representative triplicate experiment. (C) Representative images of clones 2.6 (wild type) and 2.5 (SMPD1-KO) stained with 50 nM LysoTracker Red for 5 min were acquired by an epifluorescence microscope using Cellˆp software (Olympus). (D to F) Representative images of wild-type and SMPD1-KO MCF7 cells stained for LAMP2 (D), MAP1LC3B (green) and WIPI2 (red) (E), or LGALS3 (green) and LAMP2 (red) (F) were acquired by confocal laser scanning microscope 510 using Zen2009 software (Zeiss). When indicated, cells were treated with 2 mM LLOMe as a positive control for lysosomal membrane permeabilization and LGALS3 puncta formation (F). Scale bars: 20 µm ((C)to F) and 2 µm for inserts in (D).
These data support the hypothesis that the NPA-associated autophagy defect is a direct consequence of reduced SMPD1 activity rather than a secondary adaptive response, and that it is primarily caused by a failure to form proper autophagic vesicles rather than destabilization of lysosomal membranes as suggested previously.21
Altered trafficking of ATG9A in SMPD1-deficient cells
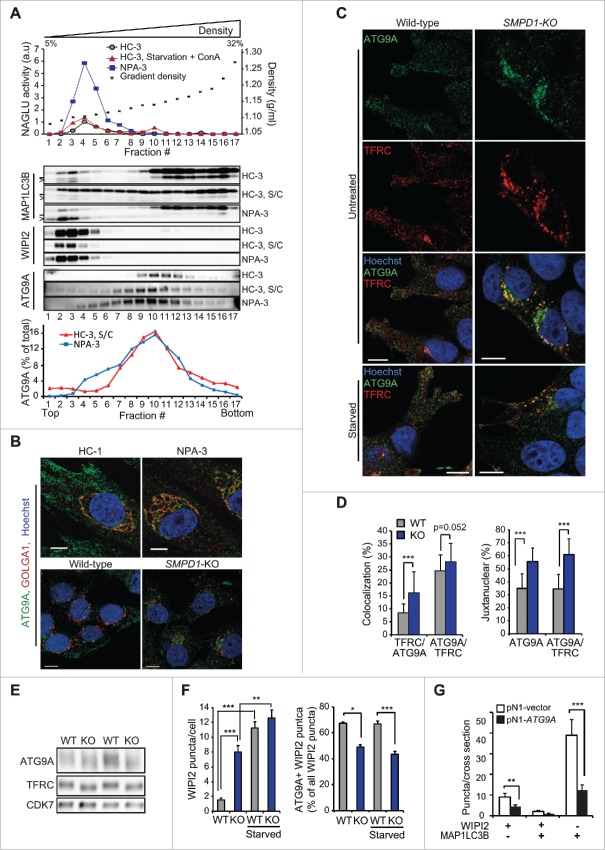
ATG9A and ATG2A being essential for the maturation and closure of phagophores, we investigated their codistribution with autophagic membranes in organelle fractions from fibroblasts from 2 controls and 2 NPA patients. Early autophagic membranes were found in light density fractions 1 and 2 as judged by the presence of WIPI2 and MAP1LC3B-II and the absence of lysosomal markers (Figs. 5A, S3A and S3B). ATG9A was not detected in these fractions in untreated control cells, but approximately 4% of total ATG9A appeared there after autophagy induction (Figs. 5A and S3B). In spite of the abundance of phagophores, fractions 1 and 2 from NPA cells were devoid of ATG9A, whereas no consistent difference in the distribution of ATG2A or ATG16L1 was observed between control and NPA cells (Figs. 5A, S2A and S2B).
Figure 5.
Altered trafficking of ATG9A in SMPD1-deficient cells. (A) NAGLU activities (top), indicated immunoblots (middle) and ATG9A distribution profiles of density gradient fractions of postnuclear supernatant fractions from HC-3 and NPA-3 fibroblasts (bottom). When indicated, HC-3 fibroblasts were starved in HBSS plus 1 g/L glucose and treated with 2 nM ConA for 2 h (S/C). The migration of MAP1LC3B-I and MAP1LC3B-II are marked with a line and greater-than symbol, respectively. ATG9A distribution in fractions is expressed as percentage of the total ATG9A in all fractions as quantified by densitometry and ImageJ software. See figures S2A and B for additional data. (B and C) Representative confocal images of HC-1 and NPA-3 fibroblasts and CRISPr-Cas-9 wild-type (WT) and SMPD1-KO (KO) MCF7 cells stained as indicated. When indicated, cells were starved in HBSS plus 1 g/L glucose for 2 h. Step size, 300 nm. See figures S2C and D for additional staining. (D) Manders coefficients for colocalization of TFRC with ATG9A and vice versa (left), and percentages of ATG9A and colocalized ATG9A-TFRC in the juxtanuclear area (4 µm from nucleus) in WT and KO cells. Bars, SEM for 20 randomly chosen cells. (E) Representative immunoblots of the indicated proteins in lysates of WT and KO cells. (F) Quantification of the number of WIPI2 puncta/cell (top) and the percentage of ATG9A-positive WIPI2 puncta of all WIPI2 puncta (bottom) in WT and KO cells. Bars, SEM for 20 randomly chosen cells (top) or 4–10 randomly chosen stacks with 2 to 9 cells/stack (bottom). Representative confocal images are shown in Fig. S2E. (G) Quantification of the number of WIPI2 and MAP1LCB3 puncta in MCF7 SMPD1-KO (KO) cells successfully transfected (green) with pN1 vector or pN1-ATG9A plasmids and stained 48 h later for WIPI2 and LC3. Bars, SEM 4 to 8 randomly chosen stacks with 2 to 9 cells/stack. Representative confocal images are shown in Fig. S2F. Scale bars: 10 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Student t test.
Contrary to the rather even punctate distribution throughout the cytosol and plasma membrane of control cells, ATG9A accumulated in the juxtanuclear Golgi region in NPA cells (Fig. 5B). The distribution of Golgi-resident protein GOLGA1 (golgin A1) was, however, largely unaltered and LAMP2 (lysosomal-associated membrane protein 2) that traffics from the Golgi to lysosomes did not concentrate in the Golgi region (Figs. 5B and S2C). Abundance and localization of EEA1 (early endosome antigen 1) were also unaltered in NPA cells (Fig. S2C). On the contrary, immunocytochemistry suggested NPA-associated alterations in the localization of REs but due to very heterogeneous staining patterns in primary fibroblasts, the data remained inconclusive. Thus, we continued the analysis in more homogenous MCF7-SMPD1-KO cells, in which ATG9A showed an NPA-like accumulation in the Golgi region with only a modest direct colocalization with Golgi markers (GOLGA1, TGOLN2/TGN46 and M6PR) (Figs. 5B and S2D). Instead, TFRC, which normally traffics in a SM-dependent manner between plasma membrane and juxtanuclear REs,13 displayed a significantly increased colocalization with ATG9A in the juxtanuclear area of SMPD1-KO cells (Fig. 5C and D). In line with the ability of starvation to partially overcome the autophagy defect in SMPD1-deficient cells (Fig. 1H), it reverted the juxtanuclear clustering of ATG9A and TFRC-positive puncta and induced their more peripheral distribution (Fig. 5C, bottom). The lack of SMPD1 did not affect the expression levels of ATG9A or TFRC but increased their mobility in SDS-PAGE (Fig. 5E), possibly reflecting their reduced phosphorylation.25,26 Additionally, the juxtanuclear clustering of ATG9A was associated with its significantly reduced colocalization with WIPI2-positive early autophagic membranes in unstressed and starved SMPD1-KO cells (Figs. 5F and S2E). Contrary to ATG9A, ATG16L1, which has been reported to interact with ATG9A in TFRC-positive REs,9 did not accumulate in the juxtanuclear area and its association with WIPI2 was increased in SMPD1-depleted cells (Figs. 3H, 3I and S1G). Importantly, ectopic expression of eGFP-ATG9A rescued the autophagy defect in SMPD1-KO cells as demonstrated by the significant reduction of WIPI2 and MAP1LC3B puncta (Figs. 5G and S2F). Contrary to the endogenous ATG9A, the ectopic eGFP-ATG9A did not cluster in the juxtanuclear area but was more evenly distributed in the cytoplasm and plasma membrane (Fig. S2F). These data strongly suggest that the trapping of ATG9A in the TFRC-positive RE compartment contributes to the maturation defect of autophagic membranes in SMPD1-deficient cells.
Excess SM disturbs autophagosome biogenesis
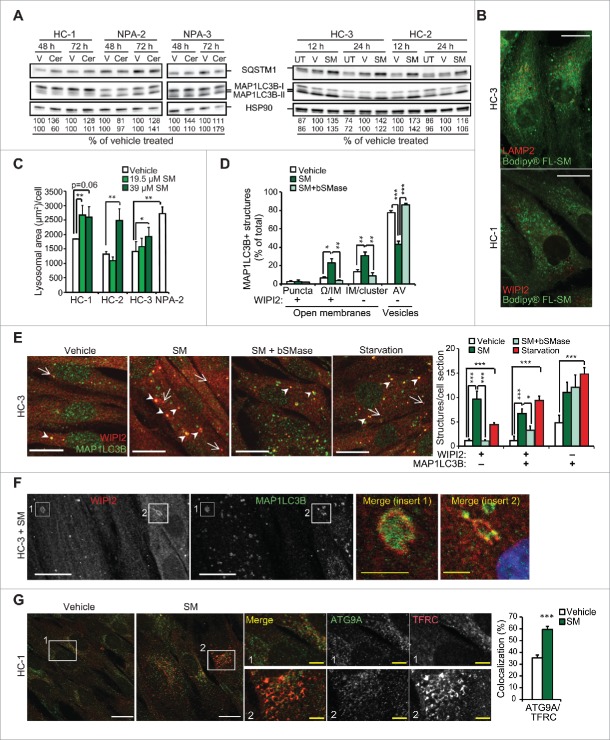
Next, we investigated whether the autophagosome maturation defect caused by reduced SMPD1 activity was due to the accumulation of SM or lack of its breakdown product ceramide. Supporting the former, C12-SM increased the levels of both SQSTM1/p62 and LC3-II in control fibroblasts, whereas C16-ceramide failed to revert the autophagy defect in patient fibroblasts (Fig. 6A). Similar results were obtained in control fibroblasts treated with BODIPY®-FL-labeled C12-SM (data not shown), which was effectively internalized and distributed mainly to nonlysosomal, WIPI2-negative membranes and accumulated in the juxtanuclear area (Fig. 6B). C12-SM treatment of control fibroblasts resulted in the enlargement of the acidic compartment and accumulation of WIPI2-, as well as WIPI2- and MAP1LC3B-positive unclosed autophagic membranes that resembled omegasomes and phagophores (Fig. 6C to F). This phenotype was effectively reverted by the addition of bSMase (Fig. 6D and E). Treatment with C12-SM and C16-SM also phenocopied the juxtanuclear clustering of ATG9A and increased ATG9A colocalization with TFRC (Fig. 6G and S3). Thus, the excess SM rather than lack of its metabolites, is the likely cause of the autophagy defect in SMPD1-deficient cells.
Figure 6.
Exogenous SM phenocopies NPA-associated lysosomal and autophagic pathologies. (A) Representative SQSTM1, MAP1LCB3-I and MAP1LCB3-II immunoblots of protein lysates from the indicated fibroblasts left untreated (UT) or treated with DMSO vehicle (V), 3 µM C16-ceramide (Cer) or 39 µM C12-SM (SM) for the indicated times. HSP90 served as a loading control. SQSTM1:HSP90 (upper row) and MAP1LCB3-II:HSP90 (lower row) ratios as a percentage of the ratio in vehicle-treated cells from the same individual are shown below. (B) Representative confocal images of the HC-3 (top) and HC-1 (bottom) fibroblasts treated with 39 µM of BODIPY® C12-SM (green) for 12 h and stained for LAMP2 and WIPI2 (red), respectively. Note juxtanuclear mainly accumulation of BODIPY-FL® C12-SM and lack of colocalization with either LAMP2 or WIPI2. Step size, 1 µm. (C) Lysosomal areas in the indicated fibroblasts treated for 12 h with vehicle (V) or 19.5 or 39 µM C12-SM (SM) were measured with Celigo® adherent cell cytometer after staining with LysoTracker®Red and Hoechst. Bars, SEM for a representative duplicate experiment (n = 3). (D) Quantification of the indicated autophagic membranes in HC-1 cells treated with vehicle or 39 µM C12-SM for 24 h with or without 10 µg/ml bSMase for the last 12 h, stained for WIPI2 and LC3, and analyzed by super resolution 3D-SIM. Bars, SEM for a representative experiment with 4 fields analyzed (n = 3). (E) Representative confocal images of HC-3 control fibroblasts treated and stained as in (D) (left), and quantification of WIPI2 and MAP1LC3B puncta (right; n ≥ 20). HC-3 fibroblasts starved for amino acids by 2 h treatment with HBSS plus 1 g/L glucose served as a positive control. White arrows and arrowheads indicate WIPI2-positive/MAP1LC3B-negative (red) and double-positive (yellow) structures, respectively. Similar data were obtained with fibroblasts from another healthy individual. (F) Representative confocal images of HC-3 fibroblasts treated with 39 µM C12-SM for 24 h and stained for WIPI2 and MAP1LC3B. Close-ups show a typical omegasome- (1) and phagophore membrane (2). (G) Representative confocal images of HC-1 fibroblasts treated with vehicle or 39 µM C12-SM for 12 h and stained for ATG9A and TFRC. Close-ups demonstrate the perinuclear colocalization of ATG9A and TFRC and the histogram shows Manders coefficients for the colocalization. Bars, SEM for minimum 20 randomly chosen cells. C16-SM induced a similar increase in the colocalization of ATG9A and TFRC (Fig. S3). Scale bars: 20 µm (white) or 2 µm (yellow). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Student t test.
Autophagy defect in smpd1-deficient mice
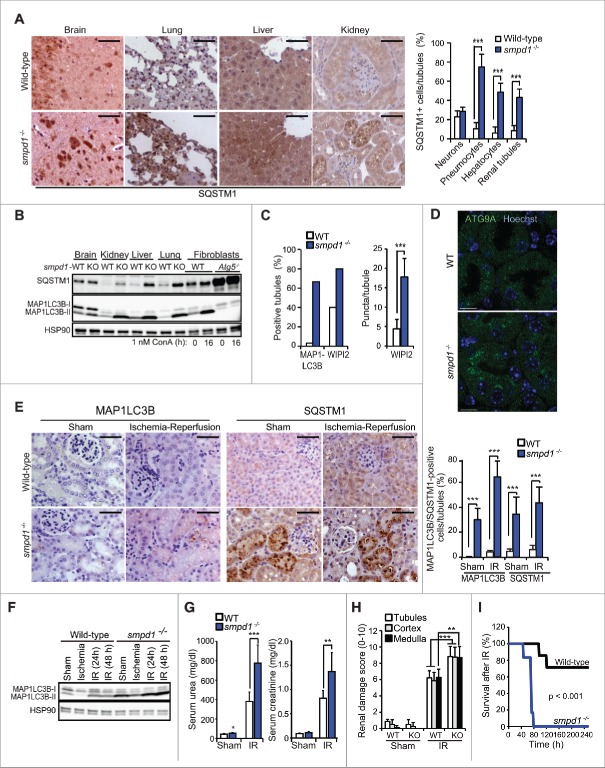
Corroborating a physiological role for SM in autophagy in vivo, SQSTM1 and MAP1LC3B-II accumulated in pneumocytes, hepatocytes and renal tubules of smpd1-deficient (smpd1−/−) mice (Fig. 7A and B). MAP1LC3B-II accumulated also in smpd1−/− neurons, where SQSTM1 was only marginally increased (Fig. 7A and B). More detailed analysis of smpd1−/− kidneys revealed a significant increase also in MAP1LC3B and WIPI2 puncta, and prominent juxtanuclear clustering of ATG9A (Figs. 7C, 7D and S4).
Figure 7.
smpd1−/− mice have defective autophagic flux and increased sensitivity to renal IR injury. (A) Representative images (left) and quantification (right) of SQSTM1 staining in tissues from wild-type (WT) and smpd1−/− mice. (B) Representative SQSTM1 and MAP1LC3B immunoblots of extracts from the tissues in (A). Extracts of WT and atg5−/− murine embryonic fibroblasts left untreated or treated with ConA served as controls. HSP90 served as a loading control. (C) Quantification of MAP1LC3B- and WIPI2-puncta positive tubules (>5 puncta/tubule) and WIPI2 puncta/tubule in kidneys from the indicated mice stained for MAP1LC3B and WIPI2 and analyzed by confocal microscopy. Confocal images of MAP1LC3B- and WIPI2-stained kidney sections are shown in Fig. S4. (D) Representative confocal projections of 5-µm thick areas of kidney sections from the indicated mice stained for ATG9A and DNA (Hoechst). (E) Representative images (left) and quantifications (right) of cells with punctuate MAP1LC3B staining and high intensity SQSTM1 immunostainings of paraffin sections of kidneys of the indicated mice exposed to a sham operation or 30 min ischemia followed by a 48 h reperfusion period. (F) Representative MAP1LC3B immunoblot of protein extracts from kidneys of wild-type (WT) and smpd1−/− mice exposed to a sham operation or 30 min ischemia followed by a 24 or 48 h reperfusion period (IR). HSP90 served as a loading control. (G) Serum urea (left) and creatinine (right) levels in the indicated mice treated as in (F). (H) Quantification of renal damage scores (right) of paraffin sections of kidneys from mice treated as in (E) and stained with periodic acid-Schiff (PAS) reagent. (I) Kaplan-Meyer survival plots for WT (n=7) and KO (n=6) mice exposed to a sham operation or 35 min renal ischemia followed by a 240-h reperfusion period. Scale bars: 10 µm (D) or 100 µm (A, E). Bars, SD for 4 independent sections (A), SEM for 52 wild-type and 45 smpd1−/− kidney tubules (C), or SD for 4 sham-operated mice and 8 (E) or 7 (G, H) IR-treated mice/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Student t test.
Next, we tested the functional consequences of the smpd1−/−-associated autophagy defect in a kidney ischemia-reperfusion (IR) model, in which autophagy plays a protective role.27,28 The sham-operated smpd1−/− kidneys displayed a significantly higher number of strongly SQSTM1- and MAP1LC3B puncta-positive cells than the wild-type kidneys consistent with a basal autophagy defect (Fig. 7E). Ischemia decreased the levels of MAP1LC3B-II in wild-type and smpd1−/− kidneys, indicative of autophagosome turnover in both genotypes (Fig. 7F). Reperfusion resulted, however, in a greater increase in MAP1LC3B puncta- and SQSTM1-positive cells and MAP1LC3B-II levels in smpd1−/− kidneys than wild-type ones (Fig. 7E and F). These data suggest that the autophagy response of kidneys exposed to IR injury is defective and autophagosome maturation and turnover are delayed although not completely absent in smpd1−/− kidneys. Accordingly, smpd1 deficiency worsened renal IR injury considerably as indicated by significantly higher elevations in serum urea and creatinine, more severe renal tissue damage, and dramatically decreased survival when compared to similarly treated wild-type mice (Fig. 7G to I).
Discussion
The lipid composition varies largely between different cellular membranes, and there is compelling evidence that the properties of bulk lipids play a profound role in defining organelle identity and function.29 In line with this, the data presented above demonstrate that SM, one of the major bulk lipids of eukaryotic cells, controls the biogenesis of autophagosomes. This hypothesis originates from the TEM-based ultrastructural analysis that revealed an accumulation of unclosed vesicular structures and a complete lack of normal autophagic vesicles in SM-rich NPA patient fibroblasts. The autophagic nature and the open structure of these membranes were verified by super resolution 3D-SIM analysis of NPA cells stained for WIPI2 and MAP1LC3B. Treatment of healthy control cells with exogenous C12-SM as well as transient depletion of SMPD1 in MCF7 breast cancer cells induced a similar accumulation of immature autophagic membranes suggesting that the defective autophagosome formation in NPA cells is caused by the accumulation of SM rather than lysosomal dysfunction or other secondary effects of a long-term SMPD1 deficiency. The direct role of SM was further supported by the ability of bSMase, which functions in the neutral pH outside the lysosomal compartment, to revert the autophagy phenotype, and the corresponding inability of exogenously added C16-ceramide, the main SM metabolite, to do so. The accumulation of MAP1LC3B-II and SQSTM1 as well as MAP1LC3B- and WIPI2-positive puncta in smpd1-deficient murine tissues ensured that the autophagy defect observed was not a culture artifact. And finally, the increased sensitivity of smpd1-deficient murine kidneys to metabolic stress provided functional support for SM as a relevant regulator of stress-induced autophagy.
The autophagosomal membrane originates from multiple sources including ER, mitochondria, Golgi apparatus, REs and plasma membrane.4 The observed SM overload-induced accumulation of immature autophagic membranes suggests that the excess SM either disturbs the curvature and closure of the phagophores or inhibits the delivery of essential autophagosome components from their original source. Opposing the first alternative, neither the endogenous SM in SMPD1-deficient cells nor BODIPY® FL C12-SM loaded to control cells accumulated in immature autophagic membranes in detectable levels. Instead, the colocalization of ATG9A with the TFRC-positive juxtanuclear RE compartment and its significantly reduced colocalization with WIPI2-positive phagophores in response to SM overload point to the latter possibility, i.e. reduced membrane delivery from the RE compartment. Supporting this hypothesis the ectopic expression of murine Atg9a in SMPD1-deficient cells significantly decreased WIPI2- and MAP1LC3B-positive puncta. Notably, both ATG9A and REs are reported as essential for proper autophagosome biogenesis,30,31 and TFRC-positive REs serve as a primary membrane source for Streptococcus-targeting autophagic membranes.32 Furthermore, both plasma membrane and REs contain high levels of SM, which recycles between these compartments and accelerates the clathrin-mediated endocytosis and the subsequent trafficking of TFRC to REs.13,33 Similarly, trifluoperazine, an SMPD1-inhibiting cationic amphiphilic drug,34 induces TFRC internalization and delays its intracellular recycling.35 Because ATG9A and TFRC utilize a similar clathrin-dependent endocytosis pathway,9 it is tempting to speculate that SM regulates their trafficking in a similar manner, excess SM causing their enhanced endocytosis and accumulation in REs. Interestingly, the RE compartment has recently emerged as the site where ATG9A- and ATG16L1-positive vesicles fuse upon autophagy induction.9 We did not, however, observe juxtanuclear clustering of ATG16L1 in SMPD1-deficient cells. Instead, ATG16L1 was effectively recruited to early autophagic membranes. Thus, SM may specifically alter the recycling pathway utilized only by a subset of recycling proteins, including ATG9A and TFRC. In line with this, the ability of SM to direct TFRC to REs does not apply to all recycling receptors,11 and ATG16L1 and ATG9A follow different clathrin-dependent endocytic pathways on their way from plasma membrane to REs.9
Contrary to a previous report by Gabande-Rodiguez and coworkers suggesting that the NPA-associated autophagy defect is caused by destabilization of lysosomal membranes,21 we did not detect any signs of lysosomal leakage in NPA fibroblasts or SMPD1-KO MCF7 cells. We have, however, earlier shown that increased SM levels sensitize lysosomes to oxidative stress and other treatments that induce lysosomal membrane permeabilization.17,18 Thus, the controversy could be due to different culture conditions affecting the levels of lysosomal reactive oxygen species. Alternatively, it could be caused by the difference in methods used to detect lysosomal leakage. Gabande-Rodiguez and coworkers base their conclusion on monitoring CTSB (cathepsin B) in digitonin-extracted cytosols. While relatively sensitive, this method carries a risk of artifacts because digitonin permeabilizes membranes in a lipid-dependent manner. This is of special concern when extracting cytosol from cells with reduced SMPD1 activity because concentrations of digitonin optimized for the extraction of cathepsin-free cytosol from untreated cells cause unsolicited lysosomal membrane permeabilization in cells treated with SMPD1 inhibitors (unpublished data). To avoid this pitfall, we used highly sensitive LGALS1 and LGALS3 puncta assays to analyze the lysosomal integrity.36 The lack of LGALS1 and LGALS3 puncta in NPA fibroblasts and SMPD1-KO MCF7 cells strongly argues against spontaneous lysosomal leakage in these cells under normal culture conditions. It should be underlined that our data is limited to cultured NPA fibroblasts and SMPD1-KO MCF7 cells and do not exclude lysosomal leakage in other cell types or in tissues with reduced SMPD1 activity.
The early lethality of NPA is usually associated with degenerative disorders of the central nervous system, whereas Niemann-Pick disease type B (NPB) patients with higher residual SMPD1 activities often live to adulthood but suffer from disturbed homeostasis of various tissues including liver, lung, heart and kidneys.19 Considering the important role of autophagy in the maintenance of tissue homeostasis, the impaired autophagic flux is likely to contribute to tissue degeneration associated with Niemann-Pick diseases. Accordingly, cellular phenotypes in smpd1-deficient and autophagy-defective phenotypes carry many resemblances such as cytoplasmic inclusions, cellular hypertrophy and degeneration.28,37,38 Clearly, more work is needed to dissect the exact contribution of the autophagy defect on the symptoms of Niemann-Pick disease patients. However, our data demonstrating that strong autophagic stimuli can partially overcome the autophagy defect in NPA fibroblasts and smpd1-deficient tissues encourage the development of autophagy-inducing therapies for the treatment of Niemann-Pick diseases.
We show that smpd1-deficient mice are significantly sensitized to renal IR injury as evidenced by increased serum concentrations of urea and creatinine, tubular damage and mortality. Loss of kidney function following renal IR injury has been linked to necroptotic cell death,39 and autophagy has been shown to maintain proximal tubule cell homeostasis following IR injury.27,28 Thus, the severe autophagy defect in smpd1-deficient mice is likely to contribute to their inability to cope with renal IR stress. Contrary to kidneys, inhibition of SMPD1 or autophagy protects mice against hepatic IR injury.40,41 The different responses may reflect major differences in cell death pathways between these organs. Whereas necroptosis is the major cell death mechanism following IR injury in relatively apoptosis resistant kidneys,39 injured hepatocytes die mainly by apoptotic and lysosomal cell-death pathways.42 Thus, the ability of sphingomyelin metabolites, ceramide and sphingosine, to promote hepatocyte apoptosis and lysosomal cell death, respectively,43,44 may explain the protective effect of SMPD1 inhibition in hepatic IR injury, whereas the reported ability of autophagy to limit necroptosis may contribute to the opposite outcome in kidney.45
In summary, the data presented above identify NPA as a severe autophagosome maturation disorder and SMPD1 as an essential enzyme for ATG9A trafficking and the formation of autophagosomes. Together with the emerging knowledge of the enzymes and commonly used drugs that control sphingomyelin metabolism these data may open clinically relevant possibilities to regulate autophagy.
Materials and methods
Cell culture and treatments
Fibroblasts were cultured in DMEM supplemented with glutamax, 12% fetal calf serum (FCS), nonessential amino acids and antibiotics (Life Technologies, 31966–021, 10270–106, 11140–050, 15140–122), and MCF7 breast cancer cells and their derivatives in RPMI-1640 supplemented with 10% FCS and antibiotics (Life Technologies, 61870–010, 10270–106, 15140–122). NPA-1 fibroblasts from a 5-mo-old NPA patient with hepatosplenomegaly were kindly provided by Professor Konrad Sandhoff (University of Bonn, Bonn, Germany). NPA-2 (GM13205) and NPA-3 (GM00112) fibroblasts from NPA patients as well as HC-1 (GM05659), HC-2 (GM06808) and HC-3 (GM00200) fibroblasts from healthy individuals were obtained from Coriell Cell Repositories. The origin of the S1 clone of MCF7 human breast carcinoma cells,46 and its transfected derivatives expressing eGFP-LC3B,47 mRFP-eGFP-LC3B,48 and RLuc-LC3B49 has been described previously.
Amino acid starvation was performed in Hank's balanced salt solution (HBSS) containing 1 g/L glucose (Gibco, 14025–050). C16-Ceramide (d18:1/6:0) C12-SM (d18:1/12:0) and C16-SM (d18:1/16:0) were from Avanti Polar Lipids (860516P, 860583P, 860584P) and BODIPY® FL C12-SM from Life Technologies (D-7711). Neutral bSMase was purified from Bacillus cereus (Sigma-Aldrich, S9396). Recombinant human SMPD1 (stock 4g/L) was provided by Dr Petra Ross-Macdonald (Genzyme). L-leucyl-L-leucine-O-methylester (LLOMe) and concanamycin A were from Santa Cruz Biotechnology (sc-285992) and Sigma-Aldrich (27689), respectively.
CRISPr-Cas9
The guide RNA sequence to target exon 1 of the human SMPD1 was chosen from the “Bioinformatically computed genome-wide resource of candidate unique gRNA targets in human exons” (http://arep.med.harvard.edu/human_crispr).50 SMPD1-gRNA-f (ACCGGCGGAGTGACGCTCCGTAG) and SMPD1-gRNA-r (AAACCTACGGAGCGTCACTCCGC) oligos were annealed and inserted into the guideRNA plasmid digested with Bsa1. MCF7 cells were transfected with guide-RNA vector, cas9 expression vector and pBabe-puro and selected for 72 h in 1 µg/ml puromycin (Sigma-Aldrich, P-7255). Single cell clones were established and tested for SMPD1 activity. Clone 2.5 with very low activity was chosen for further study and the target site was amplified using following primers: CRISPr-SMPD1-f: GAGAGATCAGCTGTCAGAGA and CRISPr-SMPD1-r: CCATCAGGGATGCATTCTGA. Sequencing revealed a homozygous deletion of 4 base pairs deletion and one substitution at the target site (Fig. S2A).
Subcellular fractionation
Pellets of approximately 10 × 106 cells washed in ice cold Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, 14190–094) were suspended in 1 vol of homogenization buffer (10 mM Tris-HCl [Sigma-Aldrich, T1378], pH 7.4, 1 mM EDTA [Riedel de Haen, 27270], and 0.25 M sucrose [Sigma-Aldrich, S0389]) containing 5x cOmplete protease inhibitor cocktail (Roche, 11697498001). Postnuclear supernatant fractions were collected by centrifugation at 1500 g for 10 min and top-layered on an 8 ml preformed continuous iodixanol 0 to 32% gradient (Sigma-Aldrich, D1556).51 Organelle and membrane separation by sedimentation was performed at 100000 g for 17 h after which 500-µl fractions were manually collected from the top. The whole procedure was performed at 4°C. The linearity of the gradient and the integrity of the purified organelles were confirmed by measuring in each fraction OD244 and lysosomal activity (NAGLU activity after digitonin extraction), respectively.
Transfections
siRNAs were transfected with Oligofectamine (Invitrogen, 12252–011) at 20 nM according to the manufacturer's protocol and cells were analyzed 72 h later if not otherwise indicated. ATG2A/B,10 ATG5,52 BECN1,22, MCOLN1,53 RPTOR,48 SMPD1-1 and SMPD1-2,18 ULK1-5 and ULK1-6,45 and nontargeting control54 siRNAs were purchased from Qiagen or Invitrogen and ATG4B siRNA (A-005786-13) was from Dharmacon. The pN1 vector encoding eGFP and the pN1-ATG9A plasmid encoding eGFP fused to ATG9A were kindly provided by Professor Tamotsu Yoshimori (Osaka University, Japan).55 Plasmids were transfected to subconfluent cells with a carrier DNA (1:5) using FuGENE® HD reagent (Promega, E2311) according to the manufacturer's instructions. The transfected cells were analyzed 48 h after the transfection.
Hydrolase activity assays
Total SMPD1 activity in cultured cells and tissue samples was measured by the cleavage of HMU-PC using the SMPD1 activity kit (Echelon Bioscience, K-3200) as recommended by the manufacturer. Total cysteine cathepsin and NAGLU/β-N-acetyl-glucosaminidase activities were determined as previously described.56
Volume of acidic compartment (VAC)
Cells plated in 96-well plates (10,000 cells per well) and treated as indicated were incubated with 0.1 µg/ml Hoechst 33342 (Life Technologies, B2261) and 25 nM LysoTracker Red DND-99 (Molecular Probes, L-7528) for 15 min at 37°C. Thereafter, both fluorescence intensities were measured by Celigo® Cell Imaging Cytometer (Brooks Automation, Chelmsford, MA, USA) and the resulting images were analyzed by ImageJ.
Antibodies
Primary antibodies used included murine monoclonal antibodies against ceramide (Sigma-Aldrich, C8104), HSP90 (HSP90AA1 and HSP90AB1; BD Transduction Laboratories, 610418), MAP1LC3B (NanoTools, 0231–100), LAMP2 (Developmental Studies Hybridoma Bank, clone H4B4), M6PR (Thermo Fisher Scientific Pierce, MA1-066), TFRC (Life Technologies, 13–6800), TGOLN2/TGN46 (Abcam, ab2809) and WIPI2 (Abcam, ab105459), rabbit antibodies against SQSTM1/p62 (Enzo Scientific, BML-PW960), TUBA1A/alpha-tubulin (Abcam, ab15246), ATG2A (MBL Life Science, PD041), ATG2B (Sigma-Aldrich, HPA019665), ATG9A for immunoblotting (Cell Signaling Technology, D4O9D), ATG9A for staining (Abcam, ab71795), ATG16L1 (Cell Signaling Technology, D6D5), CAT (catalase; Abcam, ab1877), EEA1 (Abcam, ab2900), GOLGA1 (golgin A1; Life Technologies, A21270), LAMP1 (Abcam, ab24170), MAP1LC3B (Cell Signaling Technology, 3868), lysenin (Peptides International, NLY-14802-v), RAB11 (Cell Signaling Technology, 5589), WIPI2 (Abcam, ab131271), and rabbit anti-LGALS1/galectin-1 (Abcam, ab25138), rat anti-LGALS3/galectin-3 supernatant (anti-Mac-2, kindly provided by Dr. Hakon Leffler, Lund University, Sweden) and guinea pig polyclonal antibodies against SQSTM1/p62 (Progen Biotech, GP62-C). Appropriate peroxidase-conjugated secondary antibodies were from Vector Laboratories, DAKO A/S and Abcam (anti-rabbit IgG, Vector Laboratories, PI-1000; anti-mouse IgG, DAKO, P0260; anti-guinea pig IgG, Abcam, ab6771), and Alexa Fluor 488-, Alexa Fluor 555-, Alexa Fluor 568-, Alexa Fluor 594- and Alexa Fluor 647-conjugated secondary antibodies for were from Invitrogen (A-21202, A-21206, A-21422, A-11077, A-21043, A-21207, A-21235, A-21245).
Immunoblotting
Cells were lysed in Laemmli sample buffer (125 mM Tris, pH 6.7, 20% glycerol, 140 mM SDS [Sigma-Aldrich, G5516 and L3771]) supplemented with 3x complete protease inhibitor cocktail (Roche, 11697498001) and 40 µg/ml pepstatin A (Roche, 10359053001) at 10000 cells/µl. After addition of 0.1 M dithiothreitol and bromophenol blue (Sigma-Aldrich, D9779 and B0126), boiling and separation by 4–20% gradient SDS-PAGE, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes using Bio-Rad Trans-Blot Turbo system. Membranes were blocked with DPBS containing 5% milk and 0.1% Tween-20 (Sigma-Aldrich, 274348) and labeled with the antibodies listed above. The signal was detected with Clarity Western ECL Substrate (Bio-Rad, 170–5061), and Luminescent Image Reader (LAS-1000Plus, Fujifilm, Tokyo, Japan), and quantified by densitometry with ImageJ software.
Immunocytochemistry
Cells were grown on glass coverslips, fixed with 4% paraformaldehyde (Ampliqon, 43226.1000) or ice-cold methanol (Fluka 34966), quenched with 50 mM ammonium chloride (Sigma-Aldrich, A0171) in DPBS, permeabilized and blocked in 5% goat serum (DAKO, X0907), 1% BSA (Amresco, E531), 0.3% Triton-X-100 (Sigma-Aldrich, T9284) in DPBS, and stained with the antibodies listed above. Nuclei were labeled with 5 µg/ml Hoechst 33342 and coverslips were mounted with Prolong Gold Antifade mounting medium (Life Technologies, P36930). Images were acquired using a Zeiss LSM510 microscope with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective or a Zeiss LSM700 microscope with ECPlan-Neofluar 40x/1.30 Oil DIC M27 or Plan-Apochromat 63x/1.40 Oil DIC M27 objective and Zen 2010 software (all equipment and software from Carl Zeiss, Jena, Germany). Pinholes were set so that the section thickness was equal for all channels and ≤ 1AU. For colocalization analyses, z-stacks (300-nm slices) were acquired. Cell contours (n > 20) were defined manually and green and red thresholds were set up in single channel mode and retained for all samples in an experiment. Manders coefficients were obtained with the colocalization module of the Zen 2012 (Black Edition) software using one slice per stack.
SM staining
To label intracellular SM, cells growing on cover slips were fixed for 10 min in ultrapure 3.7% paraformaldehyde (TH.Geyer Polysciences, 18814–20), quenched for 10 min in 50 mM NH4Cl, permeabilized with 50 µg/ml digitonin (Sigma-Aldrich, D141) for 15 min, blocked for 30 min in DPBS containing 5% goat serum and 2% BSA at 20°C and incubated with 1 µg/ml lysenin (PeptaNova GmbH, 4802-v) in the blocking buffer for 18 h at 4°C. After washing, lysenin was visualized with rabbit anti-lysenin antiserum (PeptaNova GmbH, 14802-v) and anti-rabbit Alexa Fluor 594 secondary antibody (Invitrogen, A-21207).
Autophagy assays
The puncta formation in MCF7 cells expressing fluorescent MAP1LC3B constructs was assessed after fixation in 3.7% formaldehyde (Sigma-Aldrich, 252549) applying a Zeiss LSM510 microscope. In fibroblasts, MAP1LC3B-puncta formation was analyzed after methanol permeabilization and antibody staining. A minimum of 200 randomly chosen cells was counted for each condition. Autophagic flux was analyzed by measuring the ratio of luciferase activities in lysates of MCF7 cells expressing RLuc fused to either wild-type LC3B or its autophagy deficient mutant, LC3BG120A, essentially as described previously.49
Confocal analysis of autophagic membranes
Confocal images were acquired by confocal laser scanning microscope 700 using Zen2009 software (Carl Zeiss, Jena, Germany). After subtraction of the image background (rolling ball algorithm) and thresholding, colocalization of 2 molecules was determined using the ImageJ Colocalization plugin, and the colocalization of a third molecule by determining the extent of colocalized spots using the RG2B plugin.
Super resolution microscopy—3d-structured illumination microscopy (3D-SIM)
SIM was performed using a 63x, 1.4NA, oil immersion objective lens (Carl Zeiss, Jena, Germany) and an sCMOS camera (PCO.edge, Germany) mounted on an Elyra PS.1 microscope (Carl Zeiss, Jena, Germany). Samples were illuminated with 488 and 561 nm lasers passed through diffraction gratings of 34 and 42 µm, respectively. The illumination pattern was shifted 5 times laterally and rotated 3 (MCF7) or 5 (fibroblasts) times laterally. Processing was performed with Zen software black edition 2012 and the channels were aligned in x, y, z according to a matrix calculated with an image of 100 nm beads recorded in the same conditions as the sample. Autophagic structures were classified according to their morphology after 3-D modelization using Zen software (black edition 2012) and the indicated markers.57 WIPI2 and LC3-positive puncta with diameter less than 100 nm (resolution limit) were excluded from quantification.
TEM
TEM was performed as described previously.58 Fifteen sections (primary magnification ×10,000) were chosen randomly from 2 independent blocks of each sample. The volume of autophagic vesicles was estimated by morphometry using the Visilog program (Noesis Vision, Inc., France). Autophagic structures were classified as described previously.59
LGALS1/3 puncta assay
Lysosomal membrane permeabilization was detected by staining leaky lysosomes with anti-LGALS1 and/or anti- LGALS3 antibody as described previously.37
Mice and treatments
Mice used included 6- to 8-wk-old male C57Bl/6 mice (Charles River, Sulzfeld, Germany) or smpd1−/− mice on a C57Bl/6 × 129/SVEV background and backcrossed over 10 generations to C57Bl/6 background.60 The broadly used technique of IR acute kidney injury has been described in detail previously.61 The mice were either sacrificed 48 h after reperfusion for blood and kidney collection or observed for survival. The time of ischemia was 35 min (survival experiments) or 30 min (all other experiments). Sham-operated mice underwent the identical procedure without clamp settings. All in vivo experiments were performed according to protocols approved by the Protection of Animals Act and approved by the institutional review board.
Immunohistochemistry
Murine organs were fixed in 4% neutral-buffered formaldehyde (Surgipath, Leica 00600E) for 24 h and dehydrated in a graded ethanol series and xylene (Sigma-Aldrich, 32221 and 534056) and finally embedded in paraffin. For chromogenic stainings, paraffin sections (3 to 5 µm) were stained with periodic acid-Schiff reagent (Sigma-Aldrich, 395B) and the indicated antibodies according to a standard routine protocol. Stained sections were analyzed using an Axio Imager microscope (Zeiss, Oberkochen, Germany) at 400x magnification. Micrographs were digitalized using an AxioCam MRm Rev 3 FireWire camera and AxioVision Rel. 4.5 software (Carl Zeiss, Jena, Germany). Experienced pathologists quantified the percentage of SQSTM1-positive or MAP1LC3B-positive cells and the organ damage score (scale ranging from 0 (unaffected tissue) to 10 (severe organ damage) in a double-blinded manner as previously described.62
For staining with fluorescent antibodies, 4-µm sections were deparaffinized in xylene and antigen retrieval was performed by boiling in Tris EDTA buffer, pH 9.0, for 10 min in a microwave oven. After blocking with 5% FCS in DPBS, sections were stained with the indicated antibodies, nuclei labeled with 2.5 µg/ml Hoechst 33342 and coverslips mounted with Prolong Gold Antifade mounting medium. Z-stacks (1-µm slices) were acquired with an LSM 700 confocal microscope using the Plan-Apochromat 63x/1.40 Oil DIC M27 objective and Zen 2010 software (Carl Zeiss, Jena, Germany). Maximum projections were created from the z-stacks using ImageJ analysis software (Fiji).
Statistical analysis
Statistical significance was evaluated using the 2-tailed, unpaired Student t test. The degree of colocalization between fluorophores was quantified by Manders overlap coefficient.
Abbreviations
- ATG
autophagy related
- AV
autophagic vesicle
- AVd
degradative autophagic vesicle
- AVi
initial autophagic vesicle
- bSMase
bacterial sphingomyelinase
- ConA
concanamycin A
- CRISPr
clustered regularly interspaced short palindromic repeats
- DPBS
Dulbecco's phosphate-buffered saline
- eGFP
enhanced green fluorescent protein
- eGFP-LC3B
eGFP fused with MAP1LC3B/LC3B
- ER
endoplasmic reticulum
- HBSS
Hank's balanced salt solution
- HC
healthy control
- HSP90
heat shock protein 90 proteins HSP90AA1 (heat shock protein 90kDa alpha family class A member 1) and HSP90AB1 (heat shock protein 90kDa alpha family class B member 1)
- IR
ischemia-reperfusion
- LAMP
lysosomal-associated membrane protein
- LGALS
galectin, lectin, galactoside-binding, soluble
- MAP1LC3B
microtubule-associated protein 1 light chain 3 beta
- NAGLU
N-acetyl-glucosaminidase, alpha
- NPA
Niemann-Pick disease type A
- NPB
Niemann-Pick disease type B
- RE
recycling endosome
- rHsSMPD1
recombinant human (Homo sapiens) SMPD1
- RLuc
Renilla luciferase
- RLuc-LC3B
RLuc fused with MAP1LC3B
- SM
sphingomyelin
- SMPD1
acid sphingomyelinase, sphingomyelin phosphodiesterase 1
- SQSTM1/p62
sequestosome 1
- SR-SIM
super resolution structured illumination microscopy
- TFRC
transferrin receptor
- TGN
trans-Golgi network
- TEM
transmission electron microscopy
- ULK1
unc-51 like autophagy activating kinase
- WIPI
WD repeat domain, phosphoinositide interacting
Supplementary Material
Funding Statement
This work was supported by grants from the Danish Cancer Society, the Danish National Research Foundation, the European Research Council (LYSOSOME), the Danish Medical Research Council, the Alfred Benzon Foundation, the Lundbeck Foundation, the Association for International Cancer Research and the Novo Nordisk Foundation (M.J.), and Deutsche Forschungsgemeinschaft SFB 877 (D.A.) as well as postdoctoral fellowships from the Federation of European Biochemical Societies (FEBS) and the Danish Medical Research Council (E.F.).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank D. Skousborg Larsen, K. Grøn Henriksen, M. Mari, S. Mathieu and S. Iversen for excellent technical assistance, H. Leffler, Y. Luo, P. Ross-Macdonald, K. Sandhoff and T. Yoshimori for valuable reagents, M. Høyer-Hansen and J. Nylandsted for helpful discussions, and the Developmental Studies Hybridoma Bank for the LAMP2 antibody. We acknowledge L. Plantard and the Core Facility for Integrated Microscopy, University of Copenhagen for the help in 3D-SIM analyses.
References
- [1].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Weidberg H, Shvets E, Elazar Z. Biogenesis and Cargo Selectivity of Autophagosomes. Annu Rev Biochem 2011; 80:125-56; PMID:21548784; http://dx.doi.org/ 10.1146/annurev-biochem-052709-094552 [DOI] [PubMed] [Google Scholar]
- [3].Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280-93; PMID:20965422; http://dx.doi.org/ 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 2013; 14:759-74; PMID:24201109; http://dx.doi.org/ 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- [5].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology 2011; 27:107-32; PMID:21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- [6].Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/ 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 2014; 55:238-52; PMID:24954904; http://dx.doi.org/ 10.1016/j.molcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659-75; PMID:22613832; http://dx.doi.org/ 10.1083/jcb.201111079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013; 154:1285-99; PMID:24034251; http://dx.doi.org/ 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Molecular biology of the cell 2012; 23:896-909; PMID:22219374; http://dx.doi.org/ 10.1091/mbc.E11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res 2013; 52:424-37; PMID:23684760; http://dx.doi.org/ 10.1016/j.plipres.2013.05.001 [DOI] [PubMed] [Google Scholar]
- [12].Simons K, Ikonen E. Functional rafts in cell membranes. Nature 1997; 387:569-72; PMID:9177342; http://dx.doi.org/ 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- [13].Shakor AB, Taniguchi M, Kitatani K, Hashimoto M, Asano S, Hayashi A, Nomura K, Bielawski J, Bielawska A, Watanabe K, et al.. Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. The Journal of biological chemistry 2011; 286:36053-62; PMID:21856749; http://dx.doi.org/ 10.1074/jbc.M111.228593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Duran JM, Campelo F, van Galen J, Sachsenheimer T, Sot J, Egorov MV, Rentero C, Enrich C, Polishchuk RS, Goni FM, et al.. Sphingomyelin organization is required for vesicle biogenesis at the Golgi complex. EMBO J 2012; 31:4535-46; PMID:23178595; http://dx.doi.org/ 10.1038/emboj.2012.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Grimmer S, Spilsberg B, Hanada K, Sandvig K. Depletion of sphingolipids facilitates endosome to Golgi transport of ricin. Traffic 2006; 7:1243-53; PMID:16919154; http://dx.doi.org/ 10.1111/j.1600-0854.2006.00456.x [DOI] [PubMed] [Google Scholar]
- [16].Barcelo-Coblijn G, Martin ML, de Almeida RF, Noguera-Salva MA, Marcilla-Etxenike A, Guardiola-Serrano F, Luth A, Kleuser B, Halver JE, Escriba PV. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc Natl Acad Sci U S A 2011; 108:19569-74; PMID:22106271; http://dx.doi.org/ 10.1073/pnas.1115484108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kirkegaard T, Roth AG, Petersen NH, Mahalka AK, Olsen OD, Moilanen I, Zylicz A, Knudsen J, Sandhoff K, Arenz C, et al.. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 2010; 463:549-53; PMID:20111001; http://dx.doi.org/ 10.1038/nature08710 [DOI] [PubMed] [Google Scholar]
- [18].Petersen NH, Olsen OD, Groth-Pedersen L, Ellegaard AM, Bilgin M, Redmer S, Ostenfeld MS, Ulanet D, Dovmark TH, Lonborg A, et al.. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell 2013; 24:379-93; PMID:24029234; http://dx.doi.org/ 10.1016/j.ccr.2013.08.003 [DOI] [PubMed] [Google Scholar]
- [19].Schuchman EH. The pathogenesis and treatment of acid sphingomyelinase-deficient Niemann-Pick disease. Int J Clin Pharmacol Ther 2009; 47 Suppl 1:S48-57; PMID:20040312 [DOI] [PubMed] [Google Scholar]
- [20].Li X, Xu M, Pitzer AL, Xia M, Boini KM, Li PL, Zhang Y. Control of autophagy maturation by acid sphingomyelinase in mouse coronary arterial smooth muscle cells: protective role in atherosclerosis. J Mol Med (Berl) 2014; 92:473-85; PMID:24463558; http://dx.doi.org/ 10.1007/s00109-014-1120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gabande-Rodriguez E, Boya P, Labrador V, Dotti CG, Ledesma MD. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ 2014; 21:864-75; PMID:24488099; http://dx.doi.org/ 10.1038/cdd.2014.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ostenfeld MS, Hoyer-Hansen M, Bastholm L, Fehrenbacher N, Olsen OD, Groth-Pedersen L, Puustinen P, Kirkegaard-Sorensen T, Nylandsted J, Farkas T, et al.. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy 2008; 4:487-99; PMID:18305408; http://dx.doi.org/ 10.4161/auto.5774 [DOI] [PubMed] [Google Scholar]
- [23].Lozano J, Morales A, Cremesti A, Fuks Z, Tilly JL, Schuchman E, Gulbins E, Kolesnick R. Niemann-Pick Disease versus acid sphingomyelinase deficiency. Cell Death Differ 2001; 8:100-3; PMID:11313707; http://dx.doi.org/ 10.1038/sj.cdd.4400775 [DOI] [PubMed] [Google Scholar]
- [24].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; PMID:17534139; http://dx.doi.org/ 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- [25].May WS, Tyler G. Phosphorylation of the surface transferrin receptor stimulates receptor internalization in HL60 leukemic cells. J Biol Chem 1987; 262:16710-8; PMID:3479431 [PubMed] [Google Scholar]
- [26].Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL. Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol 2014; 34:4379-88; PMID:25266655; http://dx.doi.org/ 10.1128/MCB.00740-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jiang M, Liu K, Luo J, Dong Z. Autophagy is a renoprotective mechanism during in vitro hypoxia and in vivo ischemia-reperfusion injury. Am J Pathol 2010; 176:1181-92; PMID:20075199; http://dx.doi.org/ 10.2353/ajpath.2010.090594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, Kitamura H, Niimura F, Matsusaka T, Soga T, et al.. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 2011; 22:902-13; PMID:21493778; http://dx.doi.org/ 10.1681/ASN.2010070705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature 2014; 510:48-57; PMID:24899304; http://dx.doi.org/ 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- [30].Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 2012; 23:1860-73; PMID:22456507; http://dx.doi.org/ 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG. Exploring the human genome with functional maps. Gen Res 2009; 19:1093-106; PMID:19246570; http://dx.doi.org/ 10.1101/gr.082214.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Haobam B, Nozawa T, Minowa-Nozawa A, Tanaka M, Oda S, Watanabe T, Aikawa C, Maruyama F, Nakagawa I. Rab17-mediated recycling endosomes contribute to autophagosome formation in response to Group A Streptococcus invasion. Cell Microbiol 2014; 16:1806-21; PMID:25052408; http://dx.doi.org/ 10.1111/cmi.12329 [DOI] [PubMed] [Google Scholar]
- [33].Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell 2000; 11:2775-91; PMID:10930469; http://dx.doi.org/ 10.1091/mbc.11.8.2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kornhuber J, Tripal P, Reichel M, Muhle C, Rhein C, Muehlbacher M, Groemer TW, Gulbins E. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological group of drugs with broad clinical applications. Cell Physiol Biochem 2010; 26:9-20; PMID:20502000; http://dx.doi.org/ 10.1159/000315101 [DOI] [PubMed] [Google Scholar]
- [35].Hunt RC, Marshall-Carlson L. Internalization and recycling of transferrin and its receptor. Effect of trifluoperazine on recycling in human erythroleukemic cells. J Biol Chem 1986; 261:3681-6; PMID:3005297 [PubMed] [Google Scholar]
- [36].Aits S, Kricker J, Liu B, Ellegaard AM, Hamalisto S, Tvingsholm S, Corcelle-Termeau E, Hogh S, Farkas T, Holm Jonassen A, et al.. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 2015; 11:1408-24; PMID:26114578; http://dx.doi.org/ 10.1080/15548627.2015.1063871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kuemmel TA, Thiele J, Schroeder R, Stoffel W. Pathology of visceral organs and bone marrow in an acid sphingomyelinase deficient knock-out mouse line, mimicking human Niemann-Pick disease type A. A light and electron microscopic study. Pathol Res Pract 1997; 193:663-71; PMID:9505258; http://dx.doi.org/ 10.1016/S0344-0338(97)80025-8 [DOI] [PubMed] [Google Scholar]
- [39].Linkermann A, Green DR. Necroptosis. The N Engl J Med 2014; 370:455-65; PMID:24476434; http://dx.doi.org/ 10.1056/NEJMra1310050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Llacuna L, Mari M, Garcia-Ruiz C, Fernandez-Checa JC, Morales A. Critical role of acidic sphingomyelinase in murine hepatic ischemia-reperfusion injury. Hepatology 2006; 44:561-72; PMID:16941686; http://dx.doi.org/ 10.1002/hep.21285 [DOI] [PubMed] [Google Scholar]
- [41].Gotoh K, Lu Z, Morita M, Shibata M, Koike M, Waguri S, Dono K, Doki Y, Kominami E, Sugioka A, et al.. Participation of autophagy in the initiation of graft dysfunction after rat liver transplantation. Autophagy 2009; 5:351-60; PMID:19158494; http://dx.doi.org/ 10.4161/auto.5.3.7650 [DOI] [PubMed] [Google Scholar]
- [42].Ding WX, Yin XM. Dissection of the multiple mechanisms of TNF-alpha-induced apoptosis in liver injury. J Cell Mol Med 2004; 8:445-54; PMID:15601573; http://dx.doi.org/ 10.1111/j.1582-4934.2004.tb00469.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lang PA, Schenck M, Nicolay JP, Becker JU, Kempe DS, Lupescu A, Koka S, Eisele K, Klarl BA, Rubben H, et al.. Liver cell death and anemia in Wilson disease involve acid sphingomyelinase and ceramide. Nat Med 2007; 13:164-70; PMID:17259995; http://dx.doi.org/ 10.1038/nm1539 [DOI] [PubMed] [Google Scholar]
- [44].Ullio C, Casas J, Brunk UT, Sala G, Fabrias G, Ghidoni R, Bonelli G, Baccino FM, Autelli R. Sphingosine mediates TNFalpha-induced lysosomal membrane permeabilization and ensuing programmed cell death in hepatoma cells. J Lipid Res 2012; 53:1134-43; PMID:22454477; http://dx.doi.org/ 10.1194/jlr.M022384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Farkas T, Daugaard M, Jäättelä M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem 2011; 286:38904-12; PMID:21930714; http://dx.doi.org/ 10.1074/jbc.M111.269134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jäättelä M, Benedict M, Tewari M, Shayman JA, Dixit VM. Bcl-x and Bcl-2 inhibit TNF and Fas-induced apoptosis and activation of phospholipase A2 in breast carcinoma cells. Oncogene 1995; 10:2297-305; PMID:Can't [PubMed] [Google Scholar]
- [47].Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al.. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 2007; 25:193-205; PMID:17244528; http://dx.doi.org/ 10.1016/j.molcel.2006.12.009 [DOI] [PubMed] [Google Scholar]
- [48].Szyniarowski P, Corcelle-Termeau E, Farkas T, Hoyer-Hansen M, Nylandsted J, Kallunki T, Jaattela M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011; 7:892-903; PMID:21508686; http://dx.doi.org/ 10.4161/auto.7.8.15770 [DOI] [PubMed] [Google Scholar]
- [49].Farkas T, Hoyer-Hansen M, Jaattela M. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy 2009; 5:1018-25; PMID:19652534; http://dx.doi.org/ 10.4161/auto.5.7.9443 [DOI] [PubMed] [Google Scholar]
- [50].Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/ 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Graham JM. Preparation of preformed iodixanol gradients. TheScientificWorldJournal 2002; 2:1351-5; PMID:12805919; http://dx.doi.org/ 10.1100/tsw.2002.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ambjorn M, Ejlerskov P, Liu Y, Lees M, Jäättelä M, Issazadeh-Navikas S. IFNB1/interferon-beta-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy 2013; 9:287-302; PMID:23221969; http://dx.doi.org/ 10.4161/auto.22831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet 2008; 17:2723-37; PMID:18550655; http://dx.doi.org/ 10.1093/hmg/ddn174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jäättelä M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev 2005; 19:570-82; PMID:15741319; http://dx.doi.org/ 10.1101/gad.305405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 2005; 280:18283-90; PMID:15755735; http://dx.doi.org/ 10.1074/jbc.M413957200 [DOI] [PubMed] [Google Scholar]
- [56].Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Hoyer-Hansen M, Weber E, Multhoff G, Rohde M, Jaattela M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med 2004; 200:425-35; PMID:15314073; http://dx.doi.org/ 10.1084/jem.20040531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Changou CA, Wolfson DL, Ahluwalia BS, Bold RJ, Kung HJ, Chuang FY. Quantitative analysis of autophagy using advanced 3D fluorescence microscopy. J Vis Exp 2013:e50047; PMID:23665532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Beneduci A, Chidichimo G, Tripepi S, Perrotta E. Transmission electron microscopy study of the effects produced by wide-band low-power millimeter waves on MCF-7 human breast cancer cells in culture. Anticancer Res 2005; 25:1009-13; PMID:15868940 [PubMed] [Google Scholar]
- [59].Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol 2009; 452:143-64; PMID:19200881; http://dx.doi.org/ 10.1016/S0076-6879(08)03610-0 [DOI] [PubMed] [Google Scholar]
- [60].Horinouchi K, Erlich S, Perl DP, Ferlinz K, Bisgaier CL, Sandhoff K, Desnick RJ, Stewart CL, Schuchman EH. Acid sphingomyelinase deficient mice: a model of types A and B Niemann-Pick disease. Nat Genet 1995; 10:288-93; PMID:7670466; http://dx.doi.org/ 10.1038/ng0795-288 [DOI] [PubMed] [Google Scholar]
- [61].Linkermann A, Brasen JH, Himmerkus N, Liu S, Huber TB, Kunzendorf U, Krautwald S. Rip1 (receptor-interacting protein kinase 1) mediates necroptosis and contributes to renal ischemia/reperfusion injury. Kidney Int 2012; 81:751-61; PMID:22237751; http://dx.doi.org/ 10.1038/ki.2011.450 [DOI] [PubMed] [Google Scholar]
- [62].Linkermann A, Himmerkus N, Rolver L, Keyser KA, Steen P, Brasen JH, Bleich M, Kunzendorf U, Krautwald S. Renal tubular Fas ligand mediates fratricide in cisplatin-induced acute kidney failure. Kidney Int 2011; 79:169-78; PMID:20811331; http://dx.doi.org/ 10.1038/ki.2010.317 [DOI] [PubMed] [Google Scholar]
- [63].Szyniarowski P, Corcelle-Termeau E, Farkas T, Høyer-Hansen M, Nylandsted J, Kallunki T, Jäättelä M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011; 7:892-903; PMID:21508686; http://dx.doi.org/ 10.4161/auto.7.8.15770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.