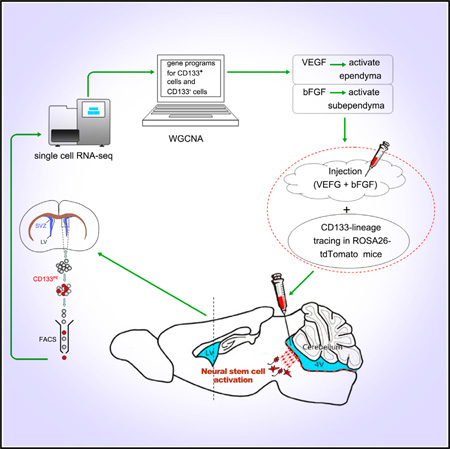
SUMMARY
The scarcity of tissue-specific stem cells and the complexity of their surrounding environment have made molecular characterization of these cells particularly challenging. Through single-cell transcriptome and weighted gene co-expression network analysis (WGCNA), we uncovered molecular properties of CD133+/GFAP− ependymal (E) cells in the adult mouse forebrain neurogenic zone. Surprisingly, prominent hub genes of the gene network unique to ependymal CD133+/GFAP− quiescent cells were enriched for immune-responsive genes, as well as genes encoding receptors for angiogenic factors. Administration of vascular endothelial growth factor (VEGF) activated CD133+ ependymal neural stem cells (NSCs), lining not only the lateral but also the fourth ventricles and, together with basic fibroblast growth factor (bFGF), elicited subsequent neural lineage differentiation and migration. This study revealed the existence of dormant ependymal NSCs throughout the ventricular surface of the CNS, as well as signals abundant after injury for their activation.
Graphical Abstract
INTRODUCTION
Tissue-specific stem cells reside in highly complex cellular environments (Doetsch, 2003; Beckervordersandforth et al., 2010; Coskun et al., 2008; Ming and Song, 2011; Morrison and Spradling, 2008; Merkle et al., 2007) and are in close contact with stem cell niches and progenies to maintain homeostasis, balancing between quiescent and activated states (Lugert et al., 2010; Li and Clevers, 2010). While a cell can be, for the most part, defined by the pattern of genes it expresses, a major challenge for genome-wide transcriptome analyses of tissues with heterogeneous cellular composition is that the readout is the sum or average of all of the different cells in that particular tissue. Unfortunately, the transcriptome of such an “averaged cell” does not truly reflect any particular cells in the tissue, and when it comes to tissue-specific quiescent stem cells or any rare but important cell types in the tissue, population-based transcriptome analyses become nearly impractical and may provide unintentional misleading results (Shapiro et al., 2013; Shalek et al., 2013; Nolan et al., 2013; Meacham and Morrison, 2013; Wu and Tzanakakis, 2013; Snippert and Clevers, 2011). To circumvent such a problem, single-cell-based transcriptome analyses become imperative.
The field of single-cell transcriptome analyses has developed quickly in recent years (Shalek et al., 2013; Xue et al., 2013; Yan et al., 2013; Tang et al., 2009, 2010). Major challenges for single-cell transcriptome analyses lie in technicality. One of the bottlenecks is to maintain the authenticity of gene expression levels during cDNA conversion and amplification. Single-cell RNA sequencing (RNA-seq) analyses are different from single-cell DNA sequencing analyses. The focus of the latter is the exact nucleotide sequence, while transcriptome deals with gene expression levels (mRNA levels); therefore, efficient reverse transcription (cDNA conversion) and linear amplification to keep the relative abundance of different transcripts constant are very important for transcriptome analysis but not for genomic sequencing. Another bottleneck is related to bioinformatics analyses (i.e., big-data processing). Since no detection system is perfect, one must know the nuts and bolts of the single-cell transcriptome analyses, including the detection limit and system-generated variations, which is different from true biological variations, in order to apply the technology well into solving the aforementioned heterogeneity-related difficult biological problems.
The ependymal/subependymal regions of the adult mouse forebrain have been reported to harbor neural stem cells (NSCs), which give rise to olfactory bulb interneurons throughout life. This region contains the previously described four cell types related to adult NSC activities: (1) ependymal E cells; (2) subependymal GFAP+ B cells, some of which are also referred to as mono-ciliated, CD133 (encoded by the prominin1 gene), and GFAP double-positive NSCs (Beckervordersandforth et al., 2010); (3) transit-amplifying C cells; and (4) neuroblast A cells (Figure S1). It has been postulated and widely accepted that GFAP+ B cells contain NSC activity. During NSC activation, B cells produce transit-amplifying C cells, and C cells give rise to large numbers of PSA-NCAM (polysialylated neural cell adhesion molecule)-positive neuroblast A cells, which repopulate the olfactory bulb (Doetsch, 2003). A contentious issue in the field lies in the understanding of the ependyma. While several studies demonstrated that ependymal multi-ciliated cells (E cells) contain stem cell activities (Johansson et al., 1999; Coskun et al., 2008; Nakafuku et al., 2008), others suggested that E cells were structural cells, which did not divide and, therefore, could not serve as NSCs. Previously, we have demonstrated that during embryonic cortical development, immunoreactivity for CD133 labeled almost all cells in the germinal ventricular zone (VZ) lining the ventricular surface, which were considered NSCs (Coskun et al., 2008). Postnatally, CD133 still labeled the layer of cells lining the ventricular surface, which is referred to as the ependyma. It is likely that embryonic VZ cells turn into ependymal cells postnatally. However, whether they still maintain NSC activity has been debated. Using prominin1 gene-based lineage-tracing studies, we have shown that CD133+ cells give rise to neuroblast A cells in the rostral migratory stream (RMS) and interneurons in the olfactory bulb, suggesting that CD133+ ependymal cells still maintain NSC activity (Coskun et al., 2008). An alternative interpretation of the study, however, is that while the prominin1 promoter labeled all CD133-expressing cells, not all, but only the mono-ciliated CD133 and GFAP double-positive cells manifested NSC activities (Codega et al., 2014; Beckervordersandforth et al., 2010). There is no evidence, however, to prove or disprove such an interpretation. Clearly, the CD133 population is heterogeneous in the ependymal and/or subependymal regions. The lack of comprehensive knowledge of the molecular makeups/signatures of the ependymal and subependymal cells hampered the understanding of lineage relationships among the complex cell populations in those areas.
In this study, we carried out single-cell transcriptome analyses on adult mouse CD133+ ependymal and some CD133− subependymal cells. Using weighted gene coexpression network analysis (WGCNA) (Zhang and Horvath, 2005; Langfelder and Horvath, 2008), we uncovered specific gene-regulatory modules correlated with what appeared to be critically staged cells along stem cell activation and subsequent neural lineage development, including CD133+ E cells, CD133 and GFAP double-positive B cells, and DLX2+ and doublecortin+ (DCX+) neuroblast A cells (Coskun et al., 2008; Doetsch et al., 1999; Bonaguidi et al., 2011; Zhao et al., 2008). Moreover, we revealed and validated a tight link between CD133+ adult NSC activation and vasculature development/angiogenesis-related signals.
Single-cell transcriptome analysis indicated that CD133+/ GFAP− cells in the ependyma expressed vascular endothelial growth factor (VEGF) receptors (VEGFRs) but not basic fibroblast growth factor (bFGF) receptors (bFGFRs); in contrast, CD133+/GFAP+ subependymal B cells appeared to express bFGFRs but not VEGFRs. When bFGF was introduced into the lateral ventricles, only subependymal cells were mitotically further activated. On the contrary, when VEGF was added, mitotic activation of the ependymal cells took place. The epen-dymal/ventricular surface of the fourth ventricle has not been reported to be neurogenic in vivo in postnatal mice (Martens et al., 2002). When exposed to VEGF and bFGF, ependymal quiescent NSCs lining the fourth ventricle were also mitotically activated. Using ROSA26-tdTomato reporter mice with electroporation of prominin1 promoter-driven Cre, we found that CD133+/GFAP− ependymal cells of the fourth ventricle not only became mitotically activated but also created a lineage by differentiating into MAP2+ neurons and GFAP+ glia, indicative of endogenous NSC activity.
We predict that the single-cell transcriptome analysis established here would be a powerful tool for identification of molecular signatures of tissue stem cells and cancer stem cells, as well as revealing molecular features of complex biological systems, including various neural circuits in the brain.
RESULTS
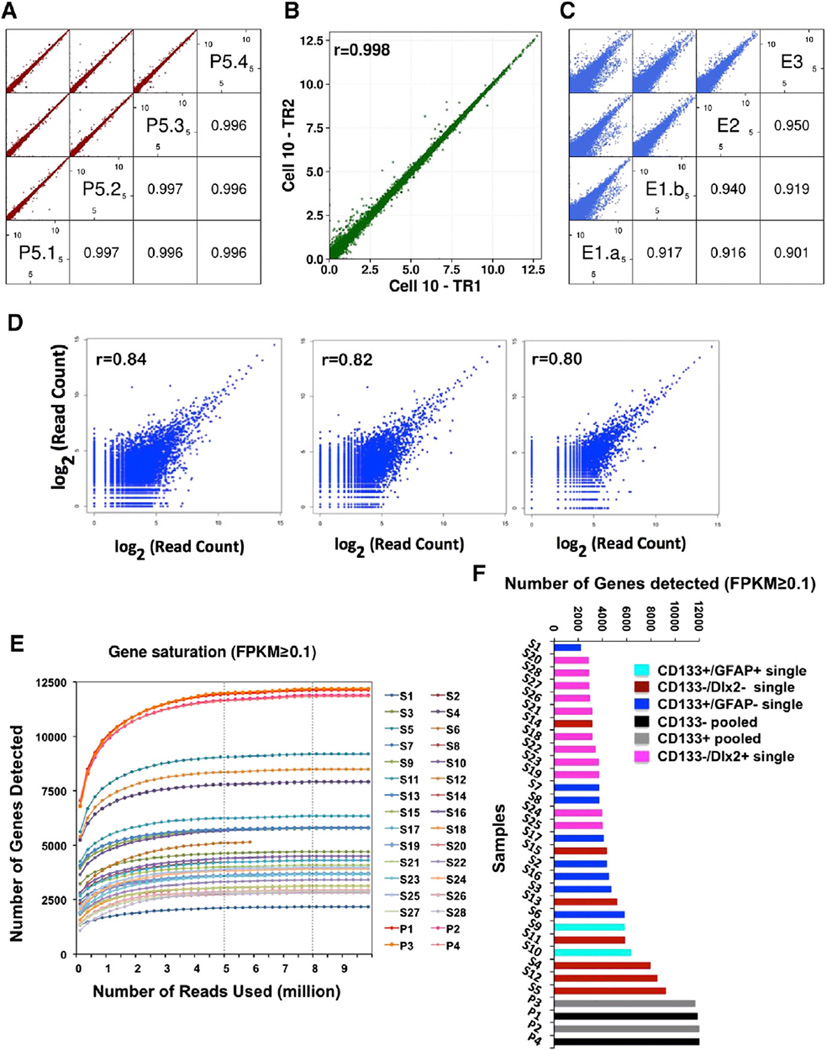
Quality Control of Single-Cell RNA-Seq
To establish single mouse CD133+ and CD133− adult NSCs and/ or progenitor cell transcriptome profiles, we used fluorescence-activated cell sorting (FACS) to isolate CD133+ and CD133− cells, followed by manual picking of single cells under the microscope (Figure S1). Single cells were subjected to RNA-seq sample preparation using previously published protocols (Kurimoto et al., 2007; Tang et al., 2009, 2010; Ramsköld et al., 2012; Xue et al., 2013), with modifications at several steps (for details, see Supplemental Information). We used single-cell qRT-PCR to detect a few well-known markers, including CD133 (prominin1), GFAP, MBP, CNP, DCX, and DLX2, and based on the presence or absence of their expression to represent a few different types of cells in the ependymal/subependymal regions (Figure S1). From a total of more than 200 single cells analyzed by qRT-PCR, we picked 28 cells representing CD133+ and different CD133− populations for subsequent RNA deep sequencing using both SOLiD-3 and Illumina HiSeq 2500 sequencing platforms. On average, about 15–20 million reads mapped to the reference genome (Ensembl genome browser, Mus_musculus, GRCm38.74) (Table S1) were obtained per sample, and FPKM (fragments per kilobase of transcript per million mapped reads) conversion was performed to represent levels of expression. Acquisition of good-quality single-cell RNA-seq data is presumably important to the downstream extrapolation of useful biological information. Therefore, we performed the following assays to assess the validity/quality of our single-cell RNA-seq data: (1) sequencing reproducibility controls and (2) control for batch effect. For sequencing reproducibility controls, we evaluated the reproducibility of our RNA-seq platform by independently sequencing quadruplicate cDNA samples aliquoted from one cDNA library and found Pearson correlation coefficients of the sequencing results to be 0.996 or higher based on log-transformed FPKM, indicative of good sequencing reproducibility and that even sampling of the cDNA library for sequencing could be achieved with our protocol (Figure 1A). For control for batch effect, to assess sequencing batch effect of our system, we sequenced a single-cell cDNA library twice, 6 months apart, and found the Pearson correlation coefficient to be 0.998 of the two batches, demonstrating minimal sequencing batch effect (Figure 1B). To assess potential technical and biological variations of single-cell RNA-seq results, we compared two-cell-stage mouse developing embryos, where the two sister cells are presumably very “similar.” RNA-seq results indicated that Pearson correlation coefficients of similar cells were within the 0.901–0.950 range (Figure 1C). Assuming these samples are the “same,” the maximal technological variations of our single-cell RNA-seq system should be less than or within the range of 0.05 (i.e., 1–0.950 = 0.05) to 0.1 (i.e., 1–0.90 = 0.1) at most. Since early developing mouse embryos, such as two-cell-stage embryos, carry relatively large amounts of RNA (about 0.47 ng per cell) (Olszańska and Borgul, 1993), including stored maternal RNA, it is still unknown whether such an assessment of technical variations may apply to adult somatic NSCs, which contain less amount of RNA (about 10–20 pg per cell) (Tietjen et al., 2003; Beckervordersandforth et al., 2010). Unfortunately, there are no two somatic cells in the adult system known to be truly identical; therefore, we decided to sequence the two halves of one NSC using our single-cell RNA-seq system instead. Presumably, materials from the two halves of one NSC should give the same sequencing result. The challenge was that the total amount of RNA materials in this situation were even less than that in one single NSC. Moreover, we found that when we split the 5-µl single-cell lysate into two halves (2.5 µl), the cell-membranous debris prevented a homogeneous even split of the material. In the end, Pearson correlation coefficients of the three pairs of half-cell RNA-seq were 0.84, 0.82, and 0.80 (Figure 1D). Moreover, we also used the External RNA Control Consortium (ERCC) spike-in method and found that when about 5 pg (similar to the total estimated mRNAs in half of a somatic cell) of ERCC standard poly(A) RNAs were spiked into the half-cell cDNA RNA-seq system, the Pearson correlation coefficient between ERCC concentrations and detected ERCC read counts was 0.93 (Figure S2A). All of these observations demonstrated the validity and reasonable technical variability of our single-cell RNA-seq system. To examine whether the sequencing depths of our data were sufficient, we carried out saturation analyses and found that with a sequencing depth of 15–20 million total reads, when we chose an FPKM cutoff at 0.1, all samples reached saturation in mRNA detection with about 8 million reads used (Figure 1E). The vast majority of single-cell samples saturated with only 5 million reads used (Figure 1E). If we chose an FPKM cutoff at 1, the number of reads required for saturation was even less, about 2–3 million (Figure S2B). Additional quality control results are shown in the Supplemental Information (Figures S2C–S2F).
Figure 1. Quality Controls of Single-Cell RNA-Seq Analysis.
(A) Quadruplicate independent sequencing of the same cDNA sample demonstrating good reproducibility as shown by the Pearson correlation coefficient being 0.996–0.997 among all four sequencing results. P, pooled sample.
(B) The same single-cell cDNA sample (S10) was subjected to two independent sequencings in two batches, with the exact sequencing time being 6 months apart. High Pearson correlation coefficient (0.998) indicates minimum sequencing batch effect and, therefore, reliable sequencing technology. TR, technical repetition.
(C) Estimation of technological and biological variations of single-cell sequencing analysis. Two-cell-stage mouse embryos were sequenced either as a whole or separately as individual cells. Pearson correlation coefficients among the four datasets are between 0.901 and 0.95. Assuming these cells are identical, our technical variation is ranging between 0.05 and 0.1. E, embryo.
(D) Pearson correlation coefficient of the three pairs of half-cell RNA-seq.
(E) Saturation curves of RNA-seq data with FPKM ≥ 0.1.
(F) Number of genes detected in pooled samples and single-cell samples (FPKM ≥ 0.1).
See also Figures S1, S2, S3, and S4 and Table S1.
To assess the addition and/or averaging effect when sequencing heterogeneous population samples, from approximately 200 single-cell cDNA libraries confirmed for CD133 positivity via RT-PCR, we collected and mixed ten CD133+ and ten CD133− single-cell libraries, respectively, and sequenced the mixed/pooled samples (Figure 1F, samples P1, P2, P3, and P4). The pooling process increased the cDNA complexity of the samples for subsequent sequencing. Since random sampling and sequencing of one such cDNA mix four times gave highly correlated sequencing results (Pearson correlation coefficient being 0.996 among the quadruplicate samples; Figure 1A), we were confident that sequencing of the pooled sample was saturated. The number of genes detected in single-cell sequencing samples as well as after the pooling of ten single-cell libraries as CD133+ population and CD133− population samples using an FPKM cutoff at 0.1 (Figure 1F) demonstrated that pooled samples contained more expressed genes than any of the single-cell samples. Obviously, population-based analyses did not represent any of the single cells and would likely give false information on gene co-expression. For example, genes co-detected in the same population-based samples— therefore, “seemingly co-expressed”—could, in fact, be expressed in different cells in that population. On the other hand, if we pooled sequencing data of all four pooled samples (P1–P4), with an FPKM cutoff at 0.1, we detected 12,701 genes expressed in 40 CD133+ and CD133− cells. However, if we pooled sequencing data of all 28 single cells together (S1–S28), we detected a total of 13,611 genes expressed in 28 CD133+ and CD133− cells, suggesting that single-cell detection was more sensitive, as more genes could be detected. To carry out a more precise comparison, we designed an experimental paradigm so that the exact ten CD133+ and seven CD133− single cells were both sequenced as individual cells and as pooled population samples after mixing individual cDNAs as described earlier. With detection saturation at FPKM > 0.1, we found that there were more than 1,000 genes, which were detected when the single-cell transcriptome was analyzed individually followed by pooling of the sequencing data but were undetectable in pooled-then-sequenced samples (Figure S3). This is because pooling the cDNAs before sequencing would end up mixing/averaging all cDNA species so that genes that are highly expressed in rare cells (e.g., one out of ten cells) will be diluted or averaged out and, therefore, could fall below the established detection threshold. It is worth noting that, here, we only demonstrated how mixing and averaging cDNA samples would lead to reduced sequencing detection sensitivity. The inevitable stochastic loss of transcripts associated with single-cell RNA-to-cDNA conversion and PCR amplification was not addressed here, which likely contributed to the fact that our half-cell RNA-seq samples only correlated with each other by about 80%–84%, not 99.6%. Nevertheless, with regard to a heterogeneous population, single-cell-resolution, RNA-seq data provide more detailed and abundant information of the molecular properties and cellular compositions of the population compared to population-based analyses. Additionally, with this approach, we detected more than 600 novel multi-exonic long noncoding RNAs (lncRNAs) (Figure S4) that failed to be detected in previously reported lncRNA expression profiles using microdissected adult subventricular zone (SVZ) tissues (Ramos et al., 2013). This revealed the potential discovery power of single-cell RNA-seq. Taken together, these studies demonstrated that single-cell transcriptome profiling provides new information that could be difficult to acquire using population-based analyses.
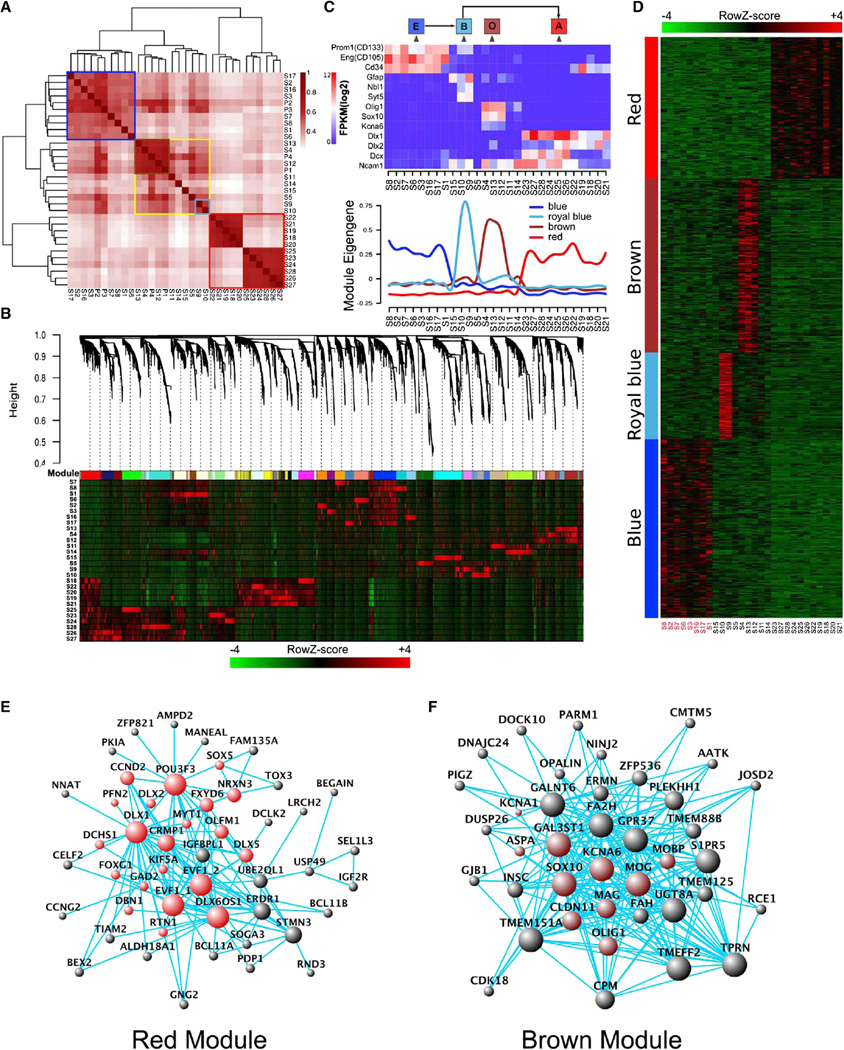
Gene-Network Modules Underlying NSC Activation Identified by WGCNAs
Unsupervised hierarchical clustering of 28 single-cell samples and 4 pooled-cell samples (FPKM ≥ 0.1) demonstrated that data from all eight CD133+/GFAP− single and two pooled CD133+/GFAP− cell samples (ten cells per sample) clustered together naturally (Figure 2A, blue frame). In addition, CD133− and DLX2+ neuronal lineage samples clustered together (Figure 2A, red frame). The rest of the samples formed a loose cluster (Figure 2A, yellow frame), within which there were two more closely correlated sub-clusters (Figure 2A, brown and royal blue frames). S9 and S10 in the royal blue subgroup expressed both CD133 and GFAP.
Figure 2. WGCNA Revealed Gene-Network Modules Enriched in CD133+ E Cells, GFAP+/CD133+ B Cells, DLX2+ A Cells, and SOX10+/OLIG1+ O Cells.
(A) Unsupervised hierarchical clustering of 28 single-cell samples and four pooled-cell datasets with FPKM cutoff of 0.1.
(B) WGCNA dendrogram indicating expression of different gene modules in all 28 single-cell samples.
(C) Eigengene expression of four selected modules across all 28 single-cell samples. Color code of the modules is preserved. Expressions of a few known markers for E, B, O, and A cells were also shown in 28 samples. Based on these two sets of information, it is obvious that the blue module has enriched expression in 8 CD133 single-positive cells, the brown module is expressed in 3 O cells, the red module is highly expressed in 11 DLX2+ A cells, and the royal blue module is expressed in 2 GFAP+ and CD133+ B cells.
(D) Expanded view of expression of all genes in each of the four modules across all 28 single-cell samples.
(E) Hub-gene network of the red module (the A module). Size of the dots represents hubness. Red highlights the genes known for neurogenesis.
(F) Hub-gene network of the brown module (the O module). Size of the dots represents hubness. Brown highlights the genes known for oligodendrocyte differentiation.
See also Figure S5.
When single-cell RNA-seq data (FPKM ≥ 0.1) were subjected to WGCNAs, multiple gene-network modules were obtained. We decided to focus initially on modules that were shared by more than one sample so that shared properties of cell subpopulations could be revealed. Four modules of interest were identified: blue, red, brown, and royal blue (Figures 2B–2D). Module Eigengene (i.e., the first principal component of a given module, which can be considered a representative of the gene expression profiles in a module) expression across all single cells is presented in Figure 2C. Based on limited marker gene expression-PROM1 (CD133) as an E-cell marker, GFAP as a B-cell marker, and DLX2 and DCX as A-cell markers—we assigned CD133+/GFAP− cells as E cells, and the blue module turned out to be an E-cell-specific module. Similarly, S9 and S10 appeared to belong to B cells due to GFAP and prominin1 double positivity, and the royal blue module seemed to be B cell specific, although it is based on only two cells captured and sequenced in these analyses. While it is possible that these two B cells only represent a subset of the B-cell population, this is still the first exploration of the molecular signatures of some B cells. Hub-gene-network analysis of the modules revealed a hierarchical organization of highly connected genes in each module, through which key controlling (hub) genes in the modular network can be identified (Figures 2E, 2F, and 3A). The red module contains many neuronal differentiation hub genes, including, for example, Dlx1, Dlx2, and Sox5, which resemble A cells (Figure 2E). On the other hand, the brown module hub genes contain Sox10, Olig1, Mog, and Mag, which are all oligodendrocyte differentiation genes; therefore, this module is likely oligodendrocyte specific and, hence, referred to as the “O-cell” module (Figure 2F). Although only 8 E cells, 11 A cells, 3 O cells, and 2 B cells were sequenced, being able to easily assign different modules to different cells (i.e., E, B, A, and O cells) (Figures 2D–2F and 3A) and to extrapolate the hub-gene networks indicated that WGCNA was powerful in providing useful information related to cell-type specificity. Although, based on previous knowledge, B and A cells are likely derived from E cells, we could not yet establish a link between E cells and O cells. While more sampling of cells will be needed— specifically with regard to B and O cell populations to assess whether the identified modules represent the entire B and O cell populations or only a subset of those cells—the discovery of these modules would certainly help with better understanding of the neurogenic zone cells as well as the molecular features of different cell types along the process of stem cell activation and differentiation in future studies.
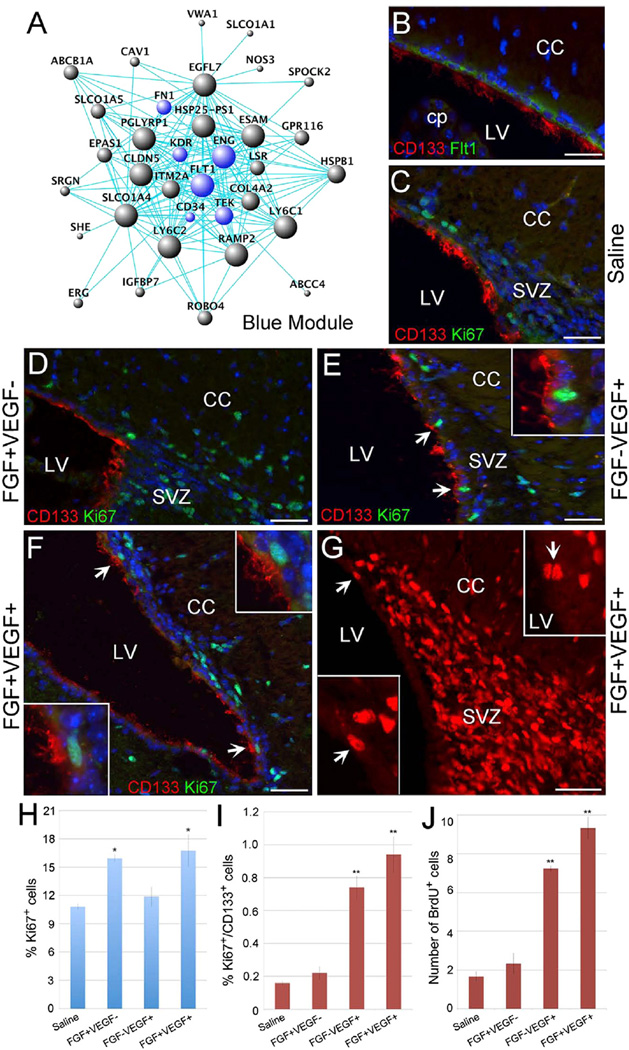
Figure 3. Mitotic Activation of Ependymal CD133+ Cells by VEGF.
(A) Hub-gene network of the blue module (the CD133-specific E module). The size of the dots represents hubness. Blue highlights the genes being discussed in the text.
(B) A representative sagittal section from the adult SVZ region stained with CD133 (red) and Flt1 (green).
(C–F) Fluorescent photomicrographs of representative sagittal sections from the adult SVZ region stained with CD133 (red) and the proliferation marker Ki67 (green). The animals were injected with saline (C), bFGF (D), VEGF (E), or bFGF + VEGF (F). Arrows and the insets in (E) and (F) indicate CD133+/Ki67+ ependymal cells from the same panels.
(G) A representative sagittal section from the SVZ region of adult mouse that was injected with bFGF + VEGF followed by 7-day oral BrdU administration. BrdU+ nuclei (red) are present within the SVZ. The arrows and the lower inset show BrdU+ nuclei within the ependymal layer from the same panel. The upper inset is taken from a different section and shows additional BrdU+ cells in the ependymal layer adjacent to the lateral ventricle.
(H) The percentage of Ki67+ cells in the subependymal region under saline, bFGF+VEGF−, bFGF−VEGF+, and bFGF+VEGF+ conditions.
(I) The percentage of Ki67+/CD133+ ependymal cells under saline, bFGF+VEGF−, bFGF−VEGF+, and bFGF+VEGF+ conditions.
(J) Quantification of BrdU+ cells in the ependymal layer under saline, bFGF+VEGF−, bFGF-VEGF+, and bFGF+VEGF+ conditions.
Scale bars, 100 µm in (B)-(G). Error bars indicate the SD from three different experiments. *p < 0.05; **p < 0.01. CC, corpus callosum; cp, choroid plexus; LV, lateral ventricle.
E-Cell Module Is Enriched in Other Stem Cell Markers and in Immune-Responsive and Angiogenesis-Related Genes
When the CD133 ependymal-cell-specific blue module was examined in combination with Gene Ontology (GO) analysis, major GO terms associated with the blue (E) module were related to vasculature development/angiogenesis and injury immune responses. Some of the big “hubs” in this module were FLT1, KDR, and TEK, which are VEGF receptors and an angiopoietin receptor, respectively (Figure 3A). Immunocytochemical analyses indicated that CD133+ ependymal cells did express the VEGF receptor FLT1, suggesting that this module indeed marked ependymal cells but not other lineage cells such as endothelial cells (Figure 3B). To our surprise, a hematopoietic stem cell (HSC) marker CD34 and a mesenchymal stem cell (MSC) marker ENG (CD105) were also present in this module. Since CD133 labels the VZ in the developing cortex (Coskun et al., 2008; Fischer et al., 2011; Uchida et al., 2000), it will be interesting to know whether these angiogenesis-, hematopoiesis-, and mesenchyme-related features are present early on or are acquired later during postnatal development. CD133+ ependymal cells also express extracellular matrix proteins such as fibronectin (FN1) and collagen (COL4A2), which are features of the mesenchymal lineage. It remains to be determined whether epithelial-to-mesenchymal transition (EMT) is involved in the development of adult tissue stem cells.
We tried to address whether the E (blue) module, which was extrapolated from transcriptomes of eight CD133+ GFAP− cells, actually represented features shared by ependymal NSCs. We analyzed five additional single-cell and two 10-cell pools of CD133+ GFAP− ependymal cells via RNA-seq. We then plotted the expression levels of 30 hub genes of the E (blue) module in eight initially sequenced single cells, as well as the “5 + 20” additionally sequenced samples, and found that the expression data clustered considerably well, suggesting that these 33 cells share common properties regarding E-module hub-gene expression (Figure S5). Based on these data, it appears that, from the hub-gene expression perspective, a large number of CD133+ GFAP− ependymal cells are similar. Therefore, without knowing whether all or only a fraction of ependymal CD133+ cells are quiescent NSCs, we speculated that the E module probably represented common features of most, if not all, CD133+ ependymal cells, including CD133+ ependymal NSCs.
Based on the transcriptome data indicating the expression of VEGF receptors (FLT1 and KDR) as hub genes in the CD133+ ependymal cell-specific gene regulatory network, we confirmed that FLT1 and CD133 were indeed co-expressed in the ependyma (Figure 3B), which is a very quiescent zone, because under normal conditions, Ki67+ cells in the ependymal layer are extremely rare (Coskun et al., 2008; Ihrie and Alvarez-Buylla, 2011). Subsequently, we explored the function of VEGF signaling in regulating the proliferation and differentiation of adult ependymal NSCs (Figures 3D–3J). In orderto determine whether ependymal cells were responsive to mitogen stimulation, we injected bFGF, VEGF, or bFGF + VEGF into the lateral ventricles of adult mice and examined the proliferation of CD133+ cells. Compared to saline injections, bFGF treatment mainly increased Ki67 labeling in the subependymal region (i.e., 10.81% ± 0.31% versus 15.94% ± 0.40%), with minimal effects on the ependyma (i.e., 0.16% ± 0.01 % versus 0.22% ± 0.04%) (Figures 3C, 3D, 3H, and 3I). In contrast, when VEGF alone was injected, the ependyma (i.e., 0.16% ± 0.01% versus 0.74% ± 0.07%) was mitotically activated, but not so much the subependyma (i.e., 10.81% ± 0.31% versus 11.88% ± 1.04%) (Figures 3C, 3E, 3H, and 3I). When bFGF and VEGF were applied together, we detected an increased number of Ki67+ ependymal cells (i.e., 0.16% ± 0.01% versus 0.94% ± 0.11%) and subependymal cells (Figures 3F, 3H, and 3I). Injected animals were also administered with bromodeoxyuridine (BrdU) orally for 7 days following surgery. In agreement with the Ki67 data, animals that were injected with bFGF and VEGF together showed numerous BrdU+ ependymal cells (Figures 3G and 3J). These results indicated that while subependymal NSCs (B cells) were mitotically responsive to bFGF, ependymal CD133+ quiescent cells were more responsive to VEGF. Since the vascular system has been postulated to be one of the major components of the NSC niche (Ihrie and Alvarez-Buylla, 2011; Mirzadeh et al., 2008) and also vascular reconstruction is a necessary step via VEGF/an-giopoietin signaling during injury, the VEGF responsiveness of CD133+ cells might serve to coordinate the activation of quiescent NSCs and angiogenesis processes to facilitate neural repair. Since quiescent CD133+ ependymal cells can give rise to activated NSCs (B cells) (Coskun et al., 2008), CD133+/GFAP+ cells might represent E-to-B transitioning cells. B cells express VEGF according to single-cell RNA-seq results, which could potentially serve as an endogenous mitogen to activate ependymal cells even though, after nerve injury, VEGF levels in the CNS would be much higher. Since CD133+ cells exist throughout the ependyma of the lateral, third, and fourth ventricles (Figure 5A), as well as the central canal of the spinal cord (Coskun et al., 2008), but VEGF-expressing B cells (i.e., subependymal NSCs) are only known to exist in the forebrain, it is possible that the lack of B-cell-secreted VEGF is part of the reason for CD133+ stem cells not to manifest stem cell activity in the CNS regions other than the forebrain, under normal conditions.
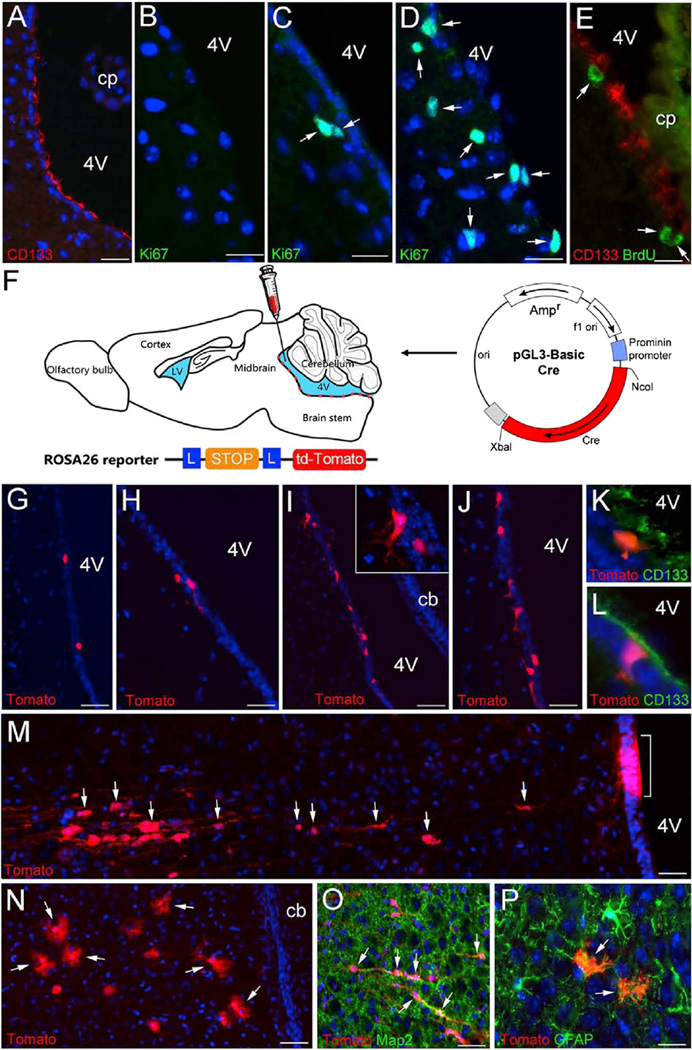
Figure 5. Activation of Dormant CD133+ NSCs in the Fourth Ventricle by Treatment of FGF and VEGF.
(A) A sagittal section of adult fourth ventricular region stained with CD133 (red).
(B–D) Immunostained sections of the postnatal fourth ventricular region from control (B, saline) and experimental (C, bFGF, and D, bFGF + VEGF administration) groups showing numerous proliferating Ki67+ cells (green, arrows).
(E) A fluorescent image showing BrdU+ cells (green) double-labeled with CD133 (red) lining the fourth ventricle following the local administration of bFGF and VEGF.
(F) Diagram illustrating the experimental paradigm of CD133+ ependymal cell lineage-tracing studies. The prominin 1 minimum promoter-driven Cre construct used for electroporation is shown. (G–L) Sagittal sections of P14 ROSA26-tdTomato reporter mice showing tdTomato+ cells (red) around the fourth ventricle; the animals were injected with mP2 + saline (G), mP2 + bFGF (H), or mP2 + FGF + VEGF (I–L) followed by electroporation at P7. The inset in (I) shows the higher magnification of tdTomato+ cells. (K and L) Fluorescent images of ROSA26-tdTomato mice stained with CD133 showing tdTomato+ (red) and CD133+ (green) cells lining the fourth ventricle following the injection of FGF + VEGF and electroporation of mP2.
(M–P) tdTomato+ cells (red) in the parenchyma of ROSA26-tdTomato reporter mice at P21; the animals were injected with mP2 + FGF + VEGF followed by electroporation at P7. The bracket in (M) indicates a group of cells at the ependymal layer expressing tdTomato, while arrows point to some of the apparently migrating tdTomato+ cells. The arrows in (N) denote tdTomato+ cells with bushy processes resembling astrocytes. (O) and (P) show tdTomato+ cells expressing MAP2 or GFAP, respectively.
Scale bars, 100 µm in (A) and (G)–(J); 40 µm in (B)–(E); and 75 µm in (M)–(P). 4V, fourth ventricle; cb, cerebellum; cp, choroid plexus.
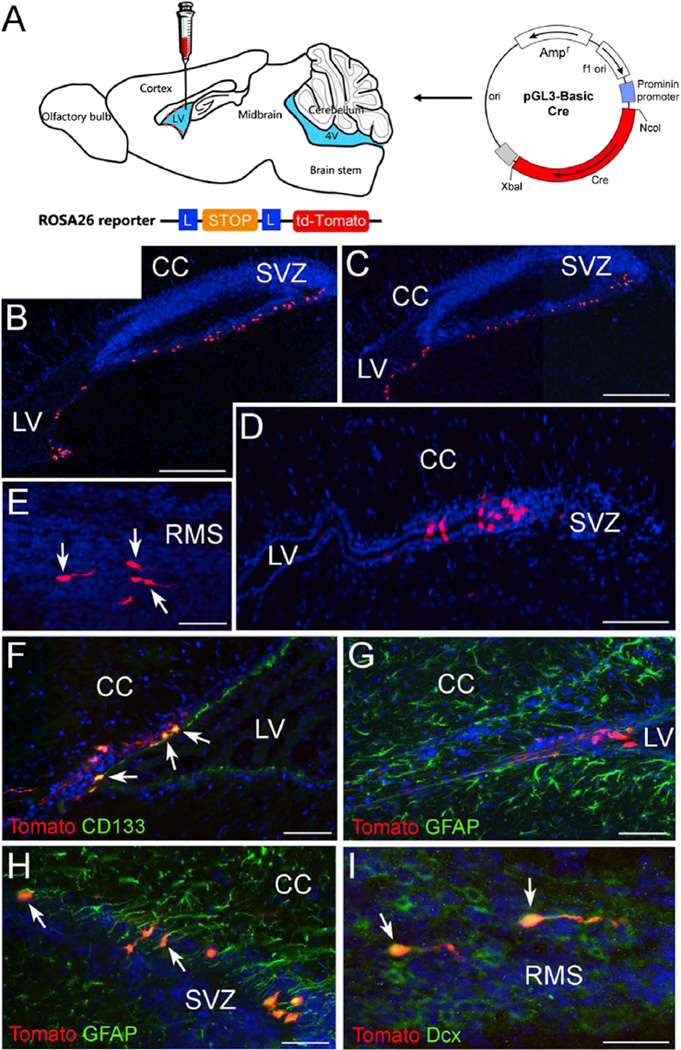
To establish a CD133+ ependymal cell downstream-lineage-tracing paradigm, we used ROSA26-tdTomato reporter mice (Figure 4A). When a plasmid containing CD133 (prominin1) minimum promoter-driven Cre recombinase was electroporated into postnatal mouse lateral ventricle walls, tdTomato-expressing CD133-lineage cells were clearly labeled (Figure 4B–4I). In this approach, only CD133+ cells could acquire Cre recombinase activity, which expels the stop cassette in the ROSA26 locus, allowing tdTomato to be permanently expressed not only in the CD133+ cells but also in their downstream lineage. With this approach, the lineage included CD133+ ependymal cells, GFAP+ astrocytes or B cells, and DCX+ neuroblasts (Figure 4B–4I), consistent with what we previously reported using ROSA26-LacZ reporter mice (Coskun et al., 2008).
Figure 4. Labeling of the Progeny of CD133+ Cells Surrounding the Lateral Ventricles.
(A) A diagram illustrating the injection of mP2 plasmid into the lateral ventricle of ROSA26-tdTomato animals followed by electroporation. (B–E) Sagittal sections of P14 ROSA26-tdTomato reporter mice showing tdTomato+ cells (red) around the lateral ventricle; the animals were injected with mP2 followed by electroporation at P7. Arrows in (E) point out migrating tdTomato+ cells in the RMS.
(F–I) Fluorescent images of ROSA26-tdTomato forebrain sections stained with CD133 (F, green), GFAP (Gand H, green), and doublecortin (I, green). The arrows in (F), (H), and (I) denote tdTomato+ cells that are immunoreactive for CD133, GFAP, and doublecortin, respectively.
Scale bars, 250 µm in (B) and (C); 50 µm in (D)–(H); and 40 µm in (I). CC, corpus callosum, LV, lateral ventricle, cp, choroid plexus.
Activation of CD133+ Ependymal Quiescent NSCs Lining the Fourth Ventricle
To examine whether VEGF and bFGF together can elicit activation of CD133+ ependymal quiescent NSCs in non-neurogenic brain regions, we injected bFGF and/or VEGF into the fourth ventricle and observed mitotic activation of cells in the ependyma and the immediately adjacent parenchymal regions (Figures 5A–5E). To trace the lineage of activated CD133+ ependymal cells lining the fourth ventricle, we electroporated a plasmid carrying prominin1 (CD133) mP2 promoter driving Cre recombinase (Coskun et al., 2008) into the fourth ventricle of ROSA26-tdTomato reporter mice, with or without co-injection of VEGF and bFGF (Figure 5F). We found that injection of VEGF and bFGF was sufficient to mitotically activate the ependymal CD133+ cells as indicated by Ki67 immunoreactivity and BrdU incorporation (Figures 5D and 5E). Following electroporation, tdTomato+ cells only appeared in the ependymal layer of the fourth ventricle in control mice without VEGF or bFGF injections (Figure 5G). Similarly, with bFGF injection alone, only a few tdTomato-labeled cells were observed (Figure 5H). However, the addition of bFGF and VEGF together triggered the CD133+ ependymal NSC activation process (Figures 5I–5L). tdTomato+ cells, indicative of the downstream lineage of CD133+ ependymal NSCs lining the fourth ventricle, were found to migrate into the parenchyma and differentiate into Map2+ neurons and GFAP+ astrocytes (Figures 5M–5P). These data indicate that at least a subset, if not all, of the CD133+ ependymal cells throughout the CNS are dormant NSCs and, upon VEGF and bFGF stimulation, as in the case of injury, can likely be mitotically activated and differentiate into downstream neural-lineage cells.
DISCUSSION
In this study, through single-cell transcriptome analyses, we provide an initial high-resolution picture of the molecular biological characteristics of the CD133+ ependymal quiescent NSCs and other cells in close proximity, some of which could be the progeny of CD133+ cells. While single-cell analyses in combination with WGCNA revealed much information about the cells, modules that were shared by subsets of samples appeared to be quite effective in revealing common features or molecular signatures of specific groups of cells (ependymal CD133+ E cells, A cells, B cells, etc.). Moreover, hub-gene analyses were powerful in revealing important regulatory factors for specific biological features of the subsets of cells. It is rather peculiar that the E-module hubs are enriched for angiogenesis-related and immune factors, as well as MSC and HSC markers CD105 and CD34, blurring the boundaries between neural and immune or epithelial and mesenchymal lineages. Whether this stands for a transitional state during EMT or whether tissue-specific stem cells are vague in their germ-layer specificity is an interesting question and remains to be determined. The hub-gene network analysis was useful for discovering that VEGF was a critical trigger to mitotically activate ependymal CD133+ NSCs in different parts of the ventricular system in the CNS. Given that vasculature disruption and reconstruction are closely linked to CNS injury and repair, our data support the notion that CNS injury is one of the major stimuli that activates CD133+ quiescent NSCs. The fact that we were able to drive CD133+ ependymal cells lining the fourth ventricle into neurogenesis further supports the idea that dormant NSCs exist throughout the CNS, lining the ventricular surface including the central canal of the spinal cord, and upon injury, those cells can be activated through exposure to factors such as VEGF and potentially participate in neural repair.
Although single-cell RNA-seq is set to be used to study cellular diversity, it turned out to be particularly useful for the identification of gene programs shared by subsets of cells, more so than differential gene expression analyses using population-based analyses, because, as discussed before, genes detected to be highly expressed in population-based samples may not be expressed by every single cell in that population. Those genes will surface in population-based differential expression analyses and end up masking genes that are truly unique to cell subpopulations. Therefore, for the discovery of shared common features of cell subpopulations, single-cell transcriptome analyses have proven to be rather effective.
Clearly, when the whole genome-wide transcriptome is considered, there is substantial heterogeneity in gene expression patterns of the CD133+/GFAP− ependymal cell population. Pearson’s correlation coefficients of pairwise samples were about 0.6–0.7. Therefore, even if these cells are all quiescent NSCs, they may reside at different cellular states such as being quiescent or at differently poised states for activation. Alternatively, these cells might be innately distinct in that some of them are structural cells and others are NSCs. Additional single-cell transcriptome analyses will allow us to fully unfold the heterogeneity of these cells as well as the processes underlying quiescent stem cell activation during injury in other regions of the CNS. The analyses might also reveal the ongoing homeostatic stem cell activity in the fore-brain. Moreover, this powerful method could be used in the near future to successfully solve different biological problems that are currently masked by cellular heterogeneity, including the identification of cancer stem cells or molecular characteristics of individual neurons that are functioning in neural circuits.
EXPERIMENTAL PROCEDURES
Isolation of Single Cells from Postnatal SVZ
Three-week-old mice were anesthetized and euthanized in accordance with Tongji University institutional guidelines. The brains were quickly removed from the skull and put into cold PBS. After several washes with cold PBS at 37°C for 30 min, the tissue was dissected under a dissection microscope, and the walls of the lateral ventricles were obtained and enzymatically digested using Papain (Worthington LS003127) at 37°Cfor 30 min. Dissociated cells were labeled with phycoerythrin (PE)-conjugated CD133 (eBiosciences 12-1331) for 1 hr. After several PBS washes, labeled cells were sorted using the Becton Dickinson Fluorescence-Activated Cell Sorter to isolate CD133+ or CD133− cells, followed by manual picking of single cells under the microscope.
Fragment Library Construction and Sequencing
After the generation of cDNA from a single cell, 500 ng of cDNA was used for the SOLiD system’s low-input fragment library preparation. Using the Covaris S2 System (Covaris), cDNA was sheared into 100- to 150-bp short fragments according to the manufacturer’s instructions, and the ends of the target DNA were repaired using the end-polishing enzymes 1 and 2 (Epicentre ER0720). The sheared DNA was then purified with the SOLiD Library Column Purification Kit (QIAGEN 28706) and subsequently ligated to SOLiD P1 and P2 adaptors. The resulting ligated population was resolved on a 3% agarose gel, and the 80- to 300-bp fractions were excised. The adaptor-ligated DNA was subjected to nine cycles of PCR amplification. The amplified PCR products were purified with the SOLiD Library Column Purification Kit (QIAGEN 28106) to yield the SOLiD fragment library ready for emulsion PCR. Emulsion PCR reactions were performed by mixing 500-pg single-cell libraries with 1.6 billion beads (1-mm diameter) with P1 primers covalently attached to their surfaces. The 50-base-pair sequences were obtained using SOLiD sequencing.
WGCNA
A signed network was constructed using any gene that was expressed at an FPKM value of 0.1 or higher in at least one of the samples. Soft power parameter was estimated and used to derive a pairwise distance matrix for selected genes using the topological overlap measure, and the dynamic hybrid cut method was used to detect clusters. The node centrality, defined as the sum of within-cluster connectivity measures, was used to rank genes for “hub-ness” within each cluster. For visual analysis of the constructed networks by hard thresholding of edge distances, the closest 150 edges were represented using Cytoscape 3.0.0.
Intraventricular Injections, Electroporation, and BrdU Administration
After postnatal C57B6 mice (wild-type or ROSA26-tdTomato) were fully anesthetized by using isoflurane, a Hamilton syringe was lowered 2.5 mm down to the lateral ventricle from −0.3 mm Bregma and 1.2 mm lateral coordinates using a stereotaxic apparatus. For the fourth-ventricle injections, postnatal C57B6 mice (wild-type or ROSA26-tdTomato), fully anesthetized using isoflurane, received a Hamilton syringe lowered 2 mm down to the fourth ventricle from −6.0 mm Bregma coordinates at the midline using a stereotaxic apparatus. Subsequently, 2 µl of saline, fibroblast growth factor (FGF), VEGF, FGF +VEGF, or mP2 was injected into the right lateral ventricle or the fourth ventricle of postnatal animals. The animals that were injected with mP2 were subjected to electroporation with the BTX ECM 830 electroporator (70 V, 100 ms). The cell proliferation marker BrdU was administered orally from the day of injection for 7 days. Animals were perfused, and their brain tissues were processed as previously described (Coskun et al., 2008) 7 or 14 days after intraventricular injections.
Immunostaining
SuperFrost Plus slides (Fisher Scientific) with tissue sections were dried at room temperature and rinsed a few times with PBS. For the detection of BrdU-incorporated cells, the sections were immersed in 2 N hydrochloric acid (HCl) at 50°C for 30 min to denature the DNA and then rinsed twice with 0.1 M borate buffer for 15 min. After several PBS washes, all slides were immersed in blocking solution (2% normal donkey serum and 0.15% Triton X-100 in PBS) for 1 hr at room temperature. Primary antibody incubation was carried out overnight at 4°C with the following antibodies at specified dilutions in blocking solution: anti-GFAP (Sigma), 1:1,000; anti-BrdU (BD Biosciences), 1:400; anti-CD133 (eBioscience), 1:1,000; anti-Flt-1 (Abcam), 1:500; MAP2 (Sigma), 1:750; doublecortin (Abcam), 1:500; and Ki67 (Vector), 1:500). The following day, slides were incubated with the appropriate Cy2- or Cy3-conjugated secondary antibodies (Jackson Immunoresearch) for 1 hr at room temperature at 1:500 dilution. Hoechst staining was used to label the nuclei. The slides were examined by using Olympus fluorescent microscopes or a confocal system (Zeiss; Axioplan-LSM 510-META). Quantification of Ki67+, Ki67+/CD133+, and BrdU+ cells was performed by counting the labeled cells within the ependyma or subependyma from three separate experiments. Statistical analysis was performed using a one-way ANOVA.
Supplementary Material
Highlights.
Single-cell RNA-seq reveals a signature gene program in ependymal CD133+ cells
VEGF is a mitogen for ependymal CD133+ cells and bFGF for subependymal B cells
CD133+ cells lining the fourth ventricle show NSC activity upon VEGF/bFGF treatment
In Brief.
Using single-cell transcriptome and network analyses, Luo et al. identify a subset of quiescent neural stem cells in non-neurogenic brain regions and show that these cells could be mitotically activated and differentiated into neurons and glia upon stimulation.
Acknowledgments
We thank Gun Woo Byeon for his assistance in data analyses. We thank Xiao Chen from Jiangxi Science & Technology Normal University for making the graphical abstract and some figures. This work was supported by the National Key Basic Research Program of China (grants 2011CBA01106, 2011CB965102, 2010CB945202, 2012CB966300, 2010CB945600, 2011CB966204, and 2014CB964602); the National Natural Foundation of China (grants 91319309, 31271371, 81330030, 31271450, and 31271375); and grants from the NIH (P01 GM081621-01A1), the Transcriptome and Epigenetics Core of Center for Study of Opioid Receptors and Drugs of Abuse (CSORDA center; grant NIH-P50DA005010), and the Intellectual and Developmental Disabilities Research Center (IDDRC center; grant NIH-P30HD004612) at the University of California, Los Angeles. The project is also supported by Yunnan and Shanghai local grants 2012HA013, 2014FC004, 13XD1403600, and ZJ2014-ZD-002.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the RNA-seq data and processed files reported in this paper is GEO: GSE61288.
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.04.001.
AUTHOR CONTRIBUTIONS
Y.L., Y.E.S., and S.L. designed the study. Y.L., V.C., A.L., J.Y., L.C., W.G., Z.S., C. Li, Y.C., D.L., J.W., C. Lin, Z.D., H.Z., J.Z., J.L., H. Liu, J.D.V., S.H., and S.L. carried out experiments or contributed critical reagents and protocols. J.Y., W.G., K.Z., H. Lin, and Y.E.S. analyzed the data and performed statistical analyses. Y.E.S., Y.L., V.C., J.Y., and S.L. wrote the manuscript. All the authors read and approved the manuscript.
REFERENCES
- Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun V, Wu H, Blanchi B, Tsao S, Kim K, Zhao J, Biancotti JC, Hut-nick L, Krueger RC, Jr, Fan G, et al. CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc. Natl. Acad. Sci. USA. 2008;105:1026–1031. doi: 10.1073/pnas.0710000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F. The glial identity of neural stem cells. Nat. Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Fischer J, Beckervordersandforth R, Tripathi P, Steiner-Mezzadri A, Nin-kovic J, Götz M. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat. Protoc. 2011;6:1981–1989. doi: 10.1038/nprot.2011.412. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat. Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Martens DJ, Seaberg RM, van der Kooy D. In vivo infusions of exogenous growth factors into the fourth ventricle of the adult mouse brain increase the proliferation of neural progenitors around the fourth ventricle and the central canal of the spinal cord. Eur. J. Neurosci. 2002;16:1045–1057. doi: 10.1046/j.1460-9568.2002.02181.x. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakafuku M, Nagao M, Grande A, Cancelliere A. Revisiting neural stem cell identity. Proc. Natl. Acad. Sci. USA. 2008;105:829–830. doi: 10.1073/pnas.0711637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding BS, Schachterle W, Liu Y, Rosenwaks Z, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszańska B, Borgul A. Maternal RNA content in oocytes of several mammalian and avian species. J. Exp. Zool. 1993;265:317–320. doi: 10.1002/jez.1402650313. [DOI] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Ray-chowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113–122. doi: 10.1038/embor.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc. Natl. Acad. Sci. USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tzanakakis ES. Deconstructing stem cell population heterogeneity: single-cell analysis and modeling approaches. Biotechnol. Adv. 2013;31:1047–1062. doi: 10.1016/j.biotechadv.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. Genetic programsinhuman and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;12:2005. doi: 10.2202/1544-6115.1128. Published online August 12, 2005. http://dx.doi.org/10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.