Abstract
Statins (HMGCR/HMG-CoA reductase [3-hydroxy-3-methylglutaryl-CoA reductase] inhibitors) are widely used to lower blood cholesterol levels but have been shown to increase the risk of type 2 diabetes mellitus. However, the molecular mechanism underlying diabetogenic effects remains to be elucidated. Here we show that statins significantly increase the expression of key gluconeogenic enzymes (such as G6PC [glucose-6-phosphatase] and PCK1 (phosphoenolpyruvate carboxykinase 1 [soluble]) in vitro and in vivo and promote hepatic glucose output. Statin treatment activates autophagic flux in HepG2 cells. Acute suppression of autophagy with lysosome inhibitors in statin treated HepG2 cells reduced gluconeogenic enzymes expression and glucose output. Importantly, the ability of statins to increase gluconeogenesis was impaired when ATG7 was deficient and BECN1 was absent, suggesting that autophagy plays a critical role in the diabetogenic effects of statins. Moreover autophagic vacuoles and gluconeogenic genes expression in the liver of diet-induced obese mice were increased by statins, ultimately leading to elevated hepatic glucose production, hyperglycemia, and insulin resistance. Together, these data demonstrate that chronic statin therapy results in insulin resistance through the activation of hepatic gluconeogenesis, which is tightly coupled to hepatic autophagy. These data further contribute to a better understanding of the diabetogenic effects of stains in the context of insulin resistance.
Keywords: autophagy, diabetes, gluconeogenesis, HMG-CoA reductase inhibitor, statin
Abbreviations
- ACTB
actin beta
- AKT1
v-akt murine thymoma viral oncogene homolog 1
- ATG7
autophagy-related 7
- Baf A1
bafilomycin A1
- BECN1
Beclin 1 autophagy related
- CQ
chloroquine
- FOXO1
forkhead box O1
- G6PC
glucose-6-phosphatase catalytic subunit
- GCK
glucokinase (hexokinase 4)
- GFP
green fluorescent protein
- HMGCR/HMG-CoA reductase
3-hydroxy-3-methylglutaryl-CoA reductase
- MAP1LC3A/LC3A
microtubule-associated protein 1 light chain 3 alpha
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- O-GluNAc
O-linked β-N-acetyl glucosamine
- PCK1
phosphoenolpyruvate carboxykinase 1 (soluble)
- PIK3C3
phosphatidylinositol 3-kinase catalytic subunit type 3
- PKLR
pyruvate kinase liver and RBC
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- RFP
red fluorescent protein
- RPS6KB1
ribosomal protein S6 kinase
- 70kDa
polypeptide 1
- shRNA
short hairpin RNA
- T2DM
type 2 diabetes mellitus
- XBP1
X-box binding protein 1.
Introduction
Statins (HMGCR inhibitors) are the most widely prescribed drugs for the primary and secondary prevention of cardiovascular diseases. Although these drugs effectively reduce circulating cholesterol levels, numerous studies have demonstrated that statin therapy is linked to the development of type 2 diabetes mellitus (T2DM). Increased incidence of diabetes was observed in clinical trials evaluating pravastatin (Pravastatin or Atorvastatin Evaluation and Infection Therapy),1 simvastatin (Heart Protection Study),2 atorvastatin (Anglo-Scandinavian Cardiac Outcomes Trial—Lipid-Lowering Arm),3 and rosuvastatin (Controlled Rosuvastatin Multinational Trial in Heart Failure; Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin).4,5 Recent metaanalyses of major statin trials also demonstrated that the risk for T2DM is higher in statin users than in nonusers.6,7 This effect is dose-dependent8: the risk of developing diabetes is 12% higher in patients on intensive-dose statin therapy than those on moderate-dose therapy, and this effect is likely to be class-dependent rather than drug-specific. Given that numerous well-designed clinical studies have reported the diabetogenic effect of statins, elucidating the underlying mechanism is of great importance.
The major site of therapeutic action for statins is the liver. Studies in rats have demonstrated that statins are selectively taken up by the liver9 and transported into hepatocytes by a high-affinity process.10 The hepatic specificity of statins may be due to efficient first-pass metabolism.11 The liver plays a critical role in regulating blood glucose levels, especially under a fasting state, maintaining glucose homeostasis through glycogenolysis and gluconeogenesis. Statins have been shown to increase fasting plasma glucose levels in individuals with or without diabetes12 and induce gluconeogenic gene expression in primary cultured human hepatocytes.13 These results suggest that statins raise fasting blood glucose levels in vivo by stimulating gluconeogenesis in the liver.
Autophagy is the catabolic mechanism by which cells regulate the turnover of cellular organelles and proteins. This process also supplies various substrates for energy generation, leading to alterations in cell metabolism.14 In carbohydrate metabolism, autophagy contributes to glycogen breakdown in lysosomes15 and the maintenance of pancreatic β-cell mass and function.16 In the liver, autophagy appears to play an important role in glucose homeostasis by promoting the conversion of amino acids to glucose.17 Statins induce autophagy in various cell types (e.g., macrophages, cancer cells, coronary arterial myocytes),18-21 therefore, we postulate that statins induce autophagy in the liver, thereby stimulating hepatic gluconeogenesis, which manifests clinically as diabetes. In this study we investigated this potential molecular mechanism underlying the diabetogenic effect of statins. Our results show that induction of liver autophagy is integral to statin-induced upregulation of hepatic gluconeogenesis, leading to dysglycemia in mice.
Results
Statins increase gluconeogenic enzyme expression in HepG2 cells
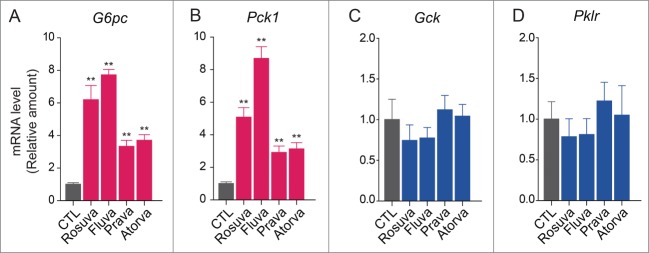
To evaluate whether hepatic gluconeogenesis is involved in the diabetogenic effects of statins, we tested the effects of rosuvastatin, fluvastatin, pravastatin, and atorvastatin on the expression of key enzymes involved in gluconeogenesis and glycolysis in HepG2 hepatocellular carcinoma cells (Fig. 1A). To validate the system, we treated HepG2 cells with insulin to confirm that insulin promotes glycolysis in these cells. Results of real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) confirmed that insulin decreased expression of genes encoding the gluconeogenic enzymes G6PC (glucose-6-phosphatase, catalytic subunit) and PCK1 (phosphoenolpyruvate carboxykinase 1 [soluble]) and increased expression of genes encoding the glycolytic enzymes GCK (glucokinase [hexokinase 4]) and PKLR (pyruvate kinase, liver and RBC) (Fig. 1B). After 24-h treatment with rosuvastatin, fluvastatin, pravastatin, or atorvastatin (20 µM), mRNA levels of G6PC and PCK1 increased in response to each statin (Fig. 1C and Fig. 1D) and protein levels of these enzymes also increased (Fig. 1E), suggesting increased gluconeogenesis. In contrast, statins had little or no effect on mRNA levels of GCK and PKLR (Fig. 1F and Fig. 1G). These data indicate that statins specifically affect the expression of gluconeogenic enzymes in HepG2 cells.
Figure 1.
Statins upregulate the expression of gluconeogenic enzymes but not glycolytic enzymes in HepG2 cells. (A) Glycolysis and gluconeogenesis pathways. (B) Insulin inhibits gluconeogenesis and enhances glycolysis in this system. Statins (20 µM) upregulated not only mRNA levels of G6PC (C) and PCK1 (D), but also protein levels of G6PC and PCK1 (E). However statins had little effect on mRNA levels of GCK (F) and PKLR (G) in HepG2 cells. *, P <0.05; **, P<0.01 compared with control (CTL).
Statins induce autophagic flux leading to enhanced expression of gluconeogenic enzymes
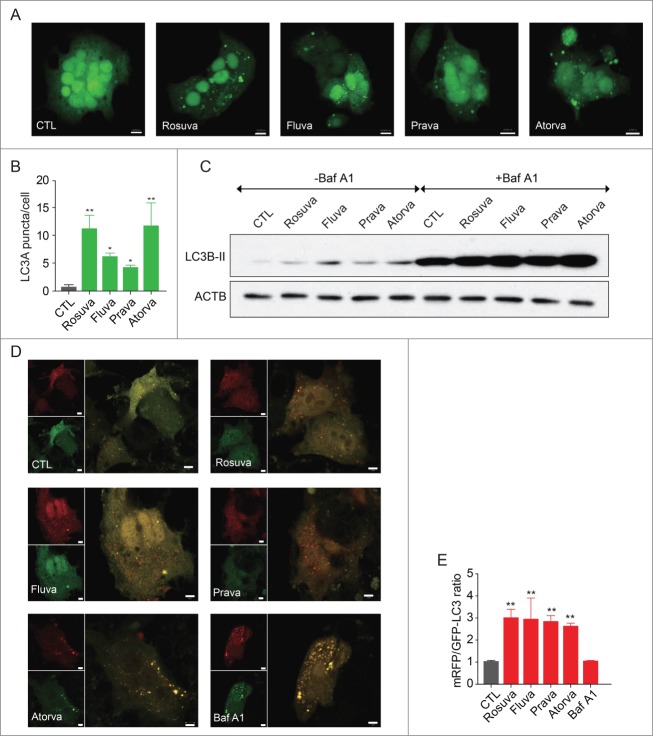
Because autophagy promotes gluconeogenesis in the liver,17 we tested whether statins increase autophagy in hepatocytes by transfecting HepG2 cells with a vector expressing the autophagy marker MAP1LC3A/LC3A (microtubule-associated protein 1 light chain 3 α) fused to green fluorescent protein (GFP). We found that statin treatment increased the number of GFP-LC3A fluorescent puncta representing autophagosomes in the cytosol (Fig. 2A and Fig. 2B). In contrast, puncta were barely discernible in control cells (Fig. 2A and Fig. 2B). To confirm this result, we evaluated LC3B-II expression by western blot analysis, which showed increased LC3B-II levels in statin-treated cells compared with controls, suggesting that statins promote autophagy in HepG2 cells (Fig. 2C). We treated cells with bafilomycin A1 (Baf A1), a lysosomal blocker, to block autophagy and treated with statins to reveal whether increased LC3B with statin is due to autophagy induction or autophagy flow blockade. Statins additionally increased LC3B-II in cells pretreated with Baf A1, which suggests that statins induce autophagy rather than blocking autophagy flow (Fig. 2C). Then we transfected mRFP (monomeric red fluorescent protein)-GFP-LC3 tandem construct encoding LC3 fused to mRFP and GFP to HepG2 cells to evaluate autophagic flux.
Figure 2.
Statins induce autophagy in HepG2 cells. (A) HepG2 cells were transfected with the autophagy sensor GFP-LC3A then treated with rosuvastatin, fluvastatin, pravastatin, or atorvastatin (20 µM) for 24 h. Fluorescent images were obtained by confocal microscopy. (B) Columns in the histogram represent the number of LC3A puncta per cell. At least 6 random fields were chosen from each sample. (C) Results of western blot analysis showed increased LC3B-II levels in statin-treated cells. The Baf A1-treated group showed that statins induced autophagy flux. (D) After transfecting HepG2 cells with mRFP-GFP-LC3B, statins were added for 24 h. Fluorescent images were obtained by confocal microscopy. The GFP protein is unstable in low pH inside of the lysosome and thereby degraded. In contrast, RFP is more stable in acidic conditions and thereby it could maintain red fluorescence. (E) Columns in the histogram represent the ratio of mRFP and GFP LC3B puncta. *, P < 0.01 compared with control.
The GFP protein is degraded in acidic conditions inside the lysosome, leading to loss of the green fluorescent signal whereas RFP is more stable in acidic condition, maintaining the red fluorescent signal. Therefore autophagosomes show the yellow fluorescent signal (merged signal of mRFP and GFP) and autolysosomes show only red signals (mRFP). The number of red and yellow puncta increased in HepG2 cells treated with statin, indicating that stains indeed induce autophagosome and autolysosome formation, representing an increase in autophagic flux (Fig. 2D and Fig. 2E). The possibility that the statins' effect is due to mere inhibition of lysosomal degradation is ruled out based on the result that Baf A1 increased number of yellow puncta without an increase in red puncta (Fig. 2D and Fig. 2E).
Blockage of autophagic flux attenuates the effect of statins on gluconeogenesis.
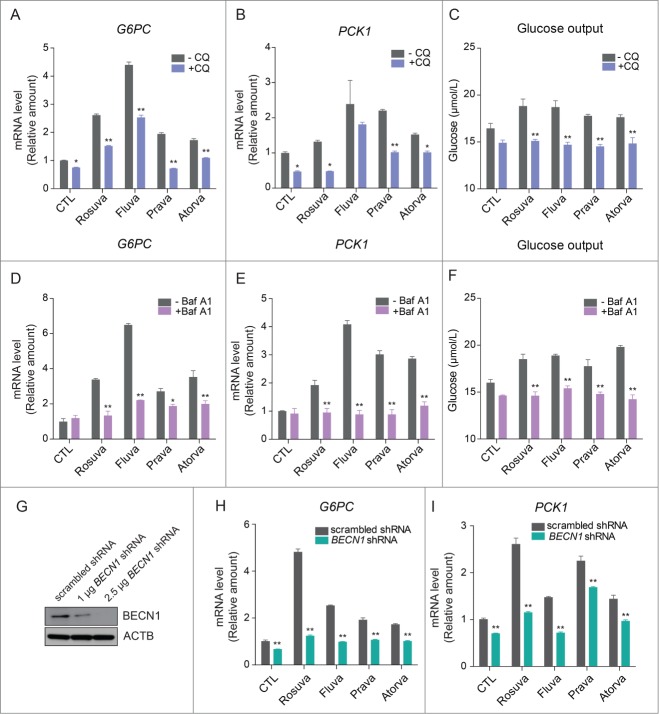
To see whether the gluconeogenic effect of statins is mediated by the autophagic process, we utilized a lysosomal inhibitor such as chloroquine (CQ) or Baf A1, with a statin treatment. Increased G6PC and PCK1 expression with statin treatment were attenuated by CQ (Fig. 3A and Fig. 3B) and glucose production was also decreased (Fig. 3C). Inhibition of the autophagic process with Baf A1 treatment resulted in attenuation of increased G6PC and PCK1 expression with statins (Fig. 3D and Fig. 3E). In addition glucose production was also decreased with Baf A1 (Fig. 3F).
Figure 3.
Statins increase autophagy-dependent gluconeogenesis in HepG2 cells. After treatment with 20 µM statins for 22 h, 50 µM chloroquine was added to HepG2 cells for 2 h, with statins. HepG2 cells treated with statins and CQ (A) G6PC and (B) PCK1 were analyzed with qRT-PCR. (C) Glucose production by HepG2 cells treated with statins and CQ. HepG2 cells treated with statins and 20 nM Baf A1. (D) G6PC and (E) PCK1 were analyzed with qRT-PCR. (F) Glucose production was measured. (G) After transfecting HepG2 cells with BECN1 shRNA, knockdown of BECN1 was confirmed by western blot analysis. Inhibition of autophagy by BECN1 knockdown decreased expression of G6PC (H) and PCK1 (I). *, P < 0.05, **, P < 0.01 compared with control cells or control cells transfected with scrambled shRNA.
To better understand the effect of statins on autophagy, we transfected HepG2 cells with short hairpin RNAs (shRNAs) against the gene encoding BECN1, which plays an important role in autophagy induction.22 After confirming the knockdown of BECN1 (Fig. 3G), we observed statin-dependent increases in G6PC and PCK1 mRNA levels in transfected cells (Fig. 3H and Fig. 3I). These data support the role of autophagy in statin-induced gluconeogenesis.
ATG7 is necessary for statin-induced gluconeogenesis in the liver
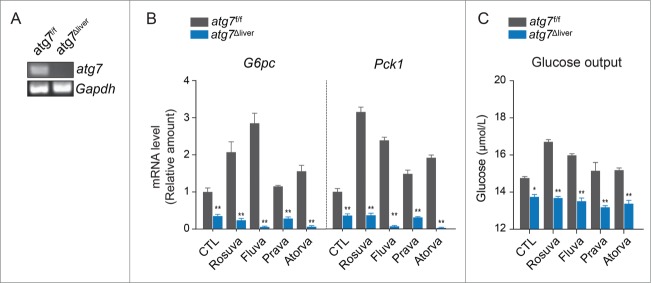
To further evaluate the role of autophagy in regulating the expression of gluconeogenic enzymes, ex vivo studies were performed with primary hepatocytes isolated from liver-specific Atg7 (autophagy-related 7)-deficient mice. Deficiency of Atg7 in the liver was confirmed by qRT-PCR (Fig. 4A), the cultured hepatocytes were treated with statins for 24 h. Our results showed that the statin-induced increase in expression of the genes encoding gluconeogenic enzymes (G6pc and Pck1) was blocked in Atg7-deficient hepatocytes (Fig. 4B). In addition, glucose output did not increase in response to statins in Atg7-deficient hepatocytes (Fig. 4C). Collectively, these data suggest that ATG7, the core autophagy regulator, is required for statin-induced gluconeogenesis in the liver, supporting the role of autophagy in statin-induced hepatic gluconeogenesis.
Figure 4.
The gluconeogenic effect of statins is attenuated in primary hepatocytes derived from liver-specific atg7-deficient mice. (A) Loss of Atg7 in liver tissue was confirmed by qRT-PCR. (B) Primary hepatocytes derived from wild-type and liver specific atg7-deficient mice were treated with statins (20 µM) for 24 h. Increased expression of G6pc and Pck1 was observed in wild-type hepatocytes but not in atg7-deficient hepatocytes. (C) Glucose output by cultured hepatocytes from wild-type and liver specific atg7-deficient mice after 24-h statin treatment. *, P < 0.05; **, P < 0.01 compared with wild-type hepatocytes.
Statins increase hepatic gluconeogenesis, leading to hepatic insulin resistance in high-fat diet-fed mice
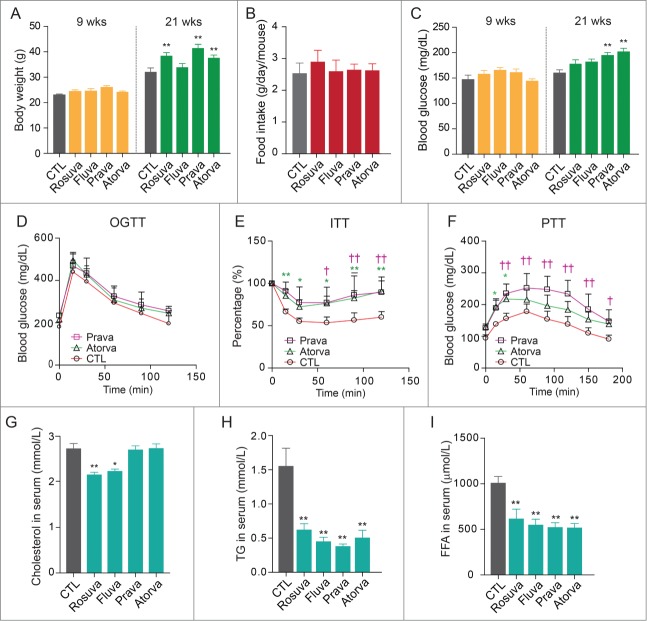
To evaluate the in vivo effects of statins on glucose homeostasis, beginning at 5 wk of age mice were fed a high-fat diet supplemented with rosuvastatin, fluvastatin, pravastatin, or atorvastatin for 16 wk. Mean body weight significantly increased in statin-treated mice compared with untreated mice (Fig. 5A), independent of food intake (Fig. 5B). We measured fasting blood glucose levels and performed the oral glucose tolerance test and insulin tolerance test at wk 15 and pyruvate tolerance test at wk 16. Our results showed that fasting blood glucose levels were higher in mice treated with pravastatin or atorvastatin compared with mice treated with rosuvastatin or fluvastatin (Fig. 5C). Results of the oral glucose tolerance test did not differ between pravastatin- and atorvastatin-treated mice and control mice (Fig. 5D), indicating that pancreatic β-cell function was not impaired at 15 wk. However, blood glucose levels failed to decrease upon insulin treatment in statin-treated mice, indicating insulin resistance (Fig. 5E), and area under the curve for the insulin tolerance test differed significantly between the control group and the statin-treated groups (Fig. S1). To evaluate the possibility that statins induce hepatic insulin resistance by increasing hepatic glucose production, we performed the pyruvate tolerance test, which showed significantly elevated blood glucose levels in pravastatin- and atorvastatin-treated mice over 3 h (Fig. 5F and Fig. S1: area under the curve). These data suggest that pravastatin and atorvastatin increase blood glucose levels in vivo, at least in part, by stimulating hepatic gluconeogenesis.
Figure 5.
Statin treatment increases body weight and fasting blood glucose levels and impairs pyruvate tolerance in high-fat diet-fed mice. (A) Mice were fed a high-fat diet with or without a statin (0.01%, w/w) for 16 wk. Mean body weight gain was significantly higher in statin-treated mice compared with untreated control mice. (B) Food intake did not differ among the groups. (C) Fasting blood glucose levels were elevated in statin-treated mice. (D) Results of the oral glucose tolerance test performed at 20 wk of age showed no differences among groups (pravastatin-treated, atorvastatin-treated, and untreated control mice). (E) Results of the insulin tolerance test showed attenuated insulin responses in statin-treated mice. (F) Results of the pyruvate tolerance test performed at 21 wk of age showed elevated blood glucose levels in pravastatin- and atorvastatin-treated mice. Statin treatments significantly decreased serum cholesterol (G), triglyceride (TG) (H), and free fatty acid (FFA) (I) levels. *, P < 0.05; **, P < 0.01 compared with untreated mice (control, n = 9 ; rosuvastatin, n = 7 ; fluvastatin, n = 7 ; pravastatin, n = 11 and atorvastatin, n = 11 ).
To determine whether statins function as HMGCR inhibitors under our experimental conditions, we measured serum levels of cholesterol, triglycerides, and free fatty acids in the high-fat diet-fed mice. Serum cholesterol levels were only marginally decreased by rosuvastatin and fluvastatin (Fig. 5G); however, serum triglyceride and free fatty acid levels were significantly decreased in the statin-treated groups compared with the control group (Fig. 5H and Fig. 5I). These data suggest that under conditions in which statins increase hepatic gluconeogenesis, they function as HMGCR inhibitors within hepatocytes, and these statin-induced metabolic changes may be related to the inhibition of endogenous cholesterol synthesis.
Statins increase hepatic gluconeogenesis and autophagy in high-fat diet-fed mice
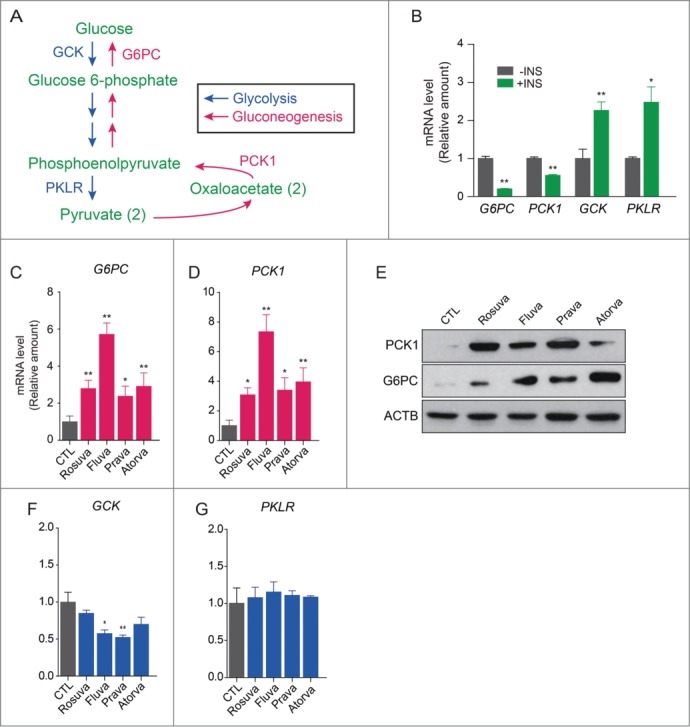
To determine whether statins increase hepatic gluconeogenesis and autophagy in vivo, expression of key gluconeogenic enzymes in the livers of high-fat diet-fed mice was evaluated by qRT-PCR. Consistent with in vitro results, statin treatment caused a significant increase in the expression of hepatic gluconeogenic genes (G6pc and Pck1) in mice (Fig. 6A and Fig. 6B). However, expression of glycolytic genes (Gck and Pklr) was not affected by statins (Fig. 6C and Fig. 6D).
Figure 6.
Statin treatment elevates expression of gluconeogenic enzymes but not glycolytic enzymes in the livers of statin-treated mice. Results of qRT-PCR showed that statins increase expression of G6pc (A) and Pck1 (B), which encode gluconeogenic enzymes. In contrast, expression of Gck (C) and Pklr (D), which encode glycolytic enzymes, did not differ between statin-treated and untreated control mice. *, P < 0.05; **, P < 0.01 compared with control.
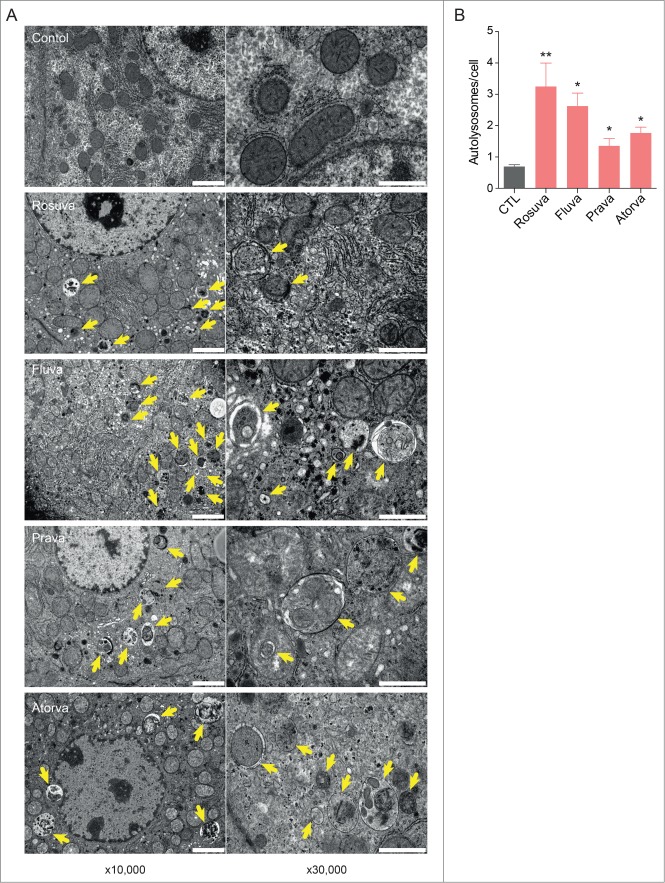
To determine whether statins induce autophagy in mouse livers in vivo, electron microscopy analysis was performed. Transmission electron microscopy analysis revealed prominent vacuolization and autophagosomes in the hepatocytes of statin-treated mice (Fig. 7A). Autophagic vacuoles are increased in statin treated mouse livers (Fig. 7B). Collectively, these data demonstrate that statin treatment leads to insulin resistance by increasing gluconeogenesis, which is tightly coupled to autophagy.
Figure 7.
Electron microscopy analysis of autophagosomes in the livers of statin-treated mice. (A) Hepatocytes of statin-treated mice showed prominent vacuolization and autophagosomes, as assessed by transmission electron microscopy. Arrows indicate double membranes of autophagosomes; scale bars: 2 µm at ×10,000 and 1 µm at ×30,000 magnification. (B) All statins significantly increased autophagic vacuole formation in mouse livers. *, P < 0.05; **, P < 0.01 compared with control.
Discussion
Although numerous clinical trials and epidemiologic studies have demonstrated that statin therapy increases the risk of T2DM,1-7 the molecular mechanism underlying this unexpected drug action has not been elucidated. In this study, we showed that statin treatment leads to insulin resistance by activating hepatic gluconeogenesis, which is tightly coupled to hepatic autophagy.
One of this study's most important findings is that statins induce autophagy in hepatocytes both in vitro and in vivo. Although a recent study shows that statin treatment blocks autophagy flux in skeletal muscle by inhibiting PRKD (protein kinase D) activity,23 most previous studies have described the effects of statins on autophagy in cultured cells.18-21 In human prostate cancer and rhabdomyosarcoma cells, statins induce autophagy by blocking geranylgeranyl biosynthesis through the inhibition of HMGCR.19,20 In coronary artery myocytes, RAC1 GTPase overexpression, which activates MTOR (mechanistic target of rapamycin [serine/threonine kinase]), blocks simvastatin-induced autophagy.21 Because the role of MTOR in autophagy inhibition is well established,21,24 and simvastatin suppresses MTOR signaling in cardiomyocytes in vitro and in vivo,21 we examined the effect of statins on MTOR complex 1and its downstream target in HepG2 cells. Statins attenuated the MTOR and RPS6KB phosphorylation, which are enhanced with insulin treatment in HepG2 cells (Fig. S2).
G6PC and PCK1 are FOXO1 target genes and they could be regulated by FOXO1. Therefore we examined whether FOXO1 phosphorylation is altered by statins. We found that statin increased FOXO1 phosphorylation in primary mouse hepatocytes (Fig. S3). The canonical insulin signaling pathway for regulation of glucose metabolism involves phosphorylation and export of the transcription factor FOXO1 out of the nucleus. Keeping FOXO1 in the nucleus induces the transcriptional induction of gluconeogenic enzymes. Increased phosphorylation of FOXO1 by statins in primary mouse hepatocytes suggests that statins promote gluconeogenesis enzymes independent of FOXO1 phosphorylation. AKT can induce the phosphorylation of FOXO1, making it accumulate in the cytoplasm. We found that AKT phosphorylation was decreased with statin treatment in primary mouse hepatocyte (Fig. S4). However, paradoxically, statins decreased the phosphorylation of AKT whereas they increased the phosphorylation of FOXO1 in primary mouse hepatocytes. This suggests that the statin-induced increase in FOXO1 phosphorylation is independent of insulin-AKT signaling. Insulin-PIK3C3-AKT-FOXO1 pathway itself is intact in primary cultured hepatocytes because insulin could increase the phosphorylation levels of FOXO1 and AKT, and wortmannin could decrease both the phosphorylation. Mechanisms other than phosphorylation, like FOXO1 acetylation,25-28 XBP1 mediated FOXO1 proteasomal degradation29 and/or O-Glc-NAc glycation30-32 could overcome the effect of FOXO1 phosphorylation and enhance gluconeogenesis. Therefore our data suggest that statin-induced elevation of gluconeogenesis is independent of FOXO1 phosphorylation.
Nevertheless, the mechanism by which statins affect MTOR activity is unknown. A recent study reports a link between MTOR signaling and intracellular cholesterol trafficking.33 In this study, pharmacological depletion of cholesterol from the plasma membrane of endothelial cells inhibits MTOR activity, and this effect is partially reversed by restoration of cholesterol to the membrane, suggesting that MTOR is involved in sensing cellular membrane sterol concentrations.33 Because statins block cholesterol synthesis in the liver,34 these drugs may cause cholesterol depletion from the plasma membrane by suppressing de novo cholesterol synthesis. Indeed, statins have been reported to decrease membrane cholesterol levels in various cell types.35,36 Moreover, mevalonate, the cholesterol intermediate just downstream of HMG-CoA, reverses statin-induced inhibition of MTOR signaling,37,38 indicating that HMGCR inhibition is required for the suppression of MTOR activity. However, additional studies are needed to confirm that statins reduce membrane cholesterol level in hepatocytes and that statin-induced autophagy and gluconeogenesis are mediated through the suppression of MTOR activity.
The mechanism by which autophagy induces gluconeogenesis is also unclear. Autophagy is thought to be a survival mechanism during starvation that supplies amino acids for gluconeogenesis in the liver.17,39 Accordingly, autophagy-deficient atg5 knockout mice generally die from hypoglycemia within 24 h after birth,40 and liver-specific Atg7-deficient mice fed a high-fat diet show improved insulin sensitivity and glucose tolerance compared to wild-type high-fat diet-fed mice.41 Our results showing increased expression of G6pc and Pck1 and elevated glucose output by statin-treated primary hepatocytes, and the blocking of these effects in the hepatocytes of liver-specific Atg7-deficient mice, strongly support the idea that statins stimulate gluconeogenesis through autophagy. A previous study suggests the opposite role of autophagy in gluconeogenesis with the finding that virus-mediated overexpression of Atg7 reduces expression of G6pc and Pck1 in the livers of mice.42 However, the induction of autophagy by Atg7 overexpression in this study is not verified; therefore, it is not clear that this effect is due to autophagy induction.
In addition, our study revealed a potential new mechanism underlying autophagy-induced gluconeogenesis: increased expression of the key gluconeogenic enzymes G6PC and PCK1.
We checked whether glucogenic amino acid, alanine, could enhance glucose output of HepG2 cell treated with statins. We found that treatment of HepG2 cells with alanine significantly increased glucose output in the absence of statin. In addition synergistic effects were seen when alanine and statins were treated simultaneously (Fig. S5). These data suggested that statin-induced gluconeogenesis is working.
Our results showing the attenuated glucose response to exogenous insulin and delayed glucose disposal after pyruvate loading in our mouse model provide additional evidence for elevated hepatic gluconeogenesis with statin treatment. No difference in oral glucose tolerance test results between treatment groups and controls indicates that chronic statin treatment did not impair the ability of pancreatic β-cells to secrete insulin under our experimental conditions. However, the effect of statins on insulin secretion has been controversial.43-45 A previous study shows that pravastatin does not affect insulin secretion in a pancreatic β-cell line,45 which is consistent with our result. In contrast, atorvastatin and simvastatin are reported to inhibit glucose-dependent insulin secretion by blocking calcium signaling in β-cells,43,45 which differs from our result with atorvastatin. This discrepancy may be due to differences in the statin concentrations used, because pravastatin and atorvastatin clearly induced insulin resistance in high-fat diet-fed mice with normal pancreatic β-cell function. Although increased peripheral insulin resistance and impaired insulin secretion are considered the main pathophysiologic features of T2DM, hepatic insulin resistance manifested by elevated gluconeogenesis is another important aspect of diabetic pathophysiology.46 Consistent with this, our results showed that pravastatin and atorvastatin increased fasting glucose level, which is, at least in part, due to enhanced hepatic gluconeogenesis.
Collectively, these data showing statin-induced hepatic insulin resistance and gluconeogenesis in mice suggest that autophagy-induced hepatic gluconeogenesis is a potential mechanism of statin-induced T2DM in humans. In contrast to these results, one study reports that statin can improve insulin sensitivity in liver of obese mice.47 There are some differences between the studies. Their report studies 4-wk-old Wistar rats and uses lovastatin (6 mg/kg/d) whereas we used C57BL/6J mice and the dose of statin was 0.01% of food weight. We administered statins for 16 wk while they included the statin treatment for a wk. Our study represents the chronic administration of statins.
In conclusion, our study found that chronic statin treatment contributes to the development of T2DM in mice. Statin treatment upregulated the gene expression of key enzymes involved in hepatic gluconeogenesis (G6PC and PCK1), increasing glucose production in the liver, and ultimately leading to hepatic insulin resistance. Our results showed that these effects are mediated through autophagy induction in the liver. This work advances our understanding of the mechanism underlying the effects of statins on insulin resistance and T2DM.
Materials and Methods
Cell culture and drug treatments
The primary hepatocytes and hepatocellular carcinoma HepG2 cell lines were cultured in Dulbecco's modified Eagle's medium (Thermo Scientific, SH30243.01) containing 10% fetal bovine serum (Thermo Scientific, SH30071.03), 100 U/ml penicillin, and 100 µg/ml streptomycin (Thermo scientific, SV30010) in a 5% CO2 incubator at 37°C. The statin drugs rosuvastatin (Sigma-Aldrich, SML1264), fluvastatin (Sigma-Aldrich, Y0001090), pravastatin (Sigma-Aldrich, P4498), and atorvastatin (Sigma-Aldrich, PZ0001) were dissolved in dimethyl sulfoxide before dilution in the culture medium. In all experiments the final statin concentration was 20 µM, and final dimethyl sulfoxide concentration was ≤ 0.1%. Chloroquine (Sigma-Aldrich, C6628) and bafilomycin A1 (Sigma-Aldrich, B1793) were dissolved in distilled water before treatments. The final concentration was 50 uM and 20 nM each, respectively.
Analysis of autophagy by confocal microscopy
HepG2 cells were transfected with the expression vector GFP-LC3 and mRFP-GFP-LC3 using Lipofectamine 2000 (Invitrogen, 11668–027) for 48 h. The cells were then treated with statins for 24 h and fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 10 min. After fixation, HepG2 cells were washed in phosphate-buffered saline (PBS; Amresco, E404–200TABS) 3 times for 5 min and then observed using LSM 700 and LSM780 confocal microscopes (Zeiss, Gottingen, Germany).
RNA extraction and qRT-PCR
Total RNA was extracted from HepG2 cells using the RNeasy Mini Kit (Qiagen, 74104) and from primary hepatocytes using TRIzol (Invitrogen, 15596–018). Reverse transcription was carried out with 2 μg total RNA using the QuantiTect Reverse Transcription kit (Qiagen, 205311) according to the manufacturer's instruction. Expression of target genes G6PC, PCK1, GCK, and PKLR was analyzed by qPCR using SYBR Premix Ex Taq (Clontech, RR420A) and gene-specific primers designed from sequences submitted to the NCBI nucleotide sequence database. Amplification was carried out using the Takara Thermal Cycler Dice® Real-Time system (Otsu, Shiga, Japan) and the following cycling conditions: 40 cycles of 95°C for 5 sec, 58°C for 10 sec, and 72°C for 20 sec. All reactions were performed in triplicate, and target gene expression was normalized to that of the internal control glyceraldehyde 3-phosphate dehydrogenase.
Immunoblotting
Cells were lysed in buffer consisting of 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate (Sigma-Aldrich, D6750), 1% Nonidet P-40 (Sigma-Aldrich, 74385), 0.1% sodium dodecyl sulfate (Sigma-Aldrich, L3771), 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitor cocktail (Roche, 11 836 153 001). Equivalent amounts of each protein extract were separated on 10% polyacrylamide gels and electrophoretically transferred onto polyvinylidene fluoride membrane (Millipore, IPVH00010). After blocking, the membranes were incubated with primary antibodies against PCK1 (Santa Cruz Biotechnology, sc-32879), G6PC (Santa Cruz Biotechnology, sc-25840), FOXO1 (Cell Signaling Technology, 2880), phospho-FOXO1 (S256; Cell Signaling Technology, 9461), AKT (Cell Signaling Technology, 9272), phospho-AKT (S473; Cell Signaling Technology, #9271), LC3B (Sigma-Aldrich, L7543), ACTB (Sigma-Aldrich, A1978), BECN1 (Santa Cruz Biotechnology, sc-11427), MTOR (Santa Cruz Biotechnology, sc-8319), phospho-MTOR (S2448; Santa Cruz Biotechnology, sc-101738), RPS6KB (Santa Cruz Biotechnology, sc-230), and phospho-RPS6KB (T389; Santa Cruz Biotechnology, sc-11759) followed by horseradish peroxidase-conjugated IgG (Santa Cruz Biotechnology, sc-2371) and anti-rabbit IgG (Santa Cruz Biotechnology, sc-2030). The blots were developed using an enhanced chemiluminescent detection kit.
Glucose output assay
Glucose output from HepG2 cells was quantified using a colorimetric glucose assay kit (BioVision, K686–100) according to the manufacturer's instructions. Briefly, HepG2 cells were treated with or without statin for 24 h. The conditioned medium was then collected and incubated with the reaction mix for 30 min at room temperature. Absorbance at 450 nm was measured in a 96-well plate reader (Molecular Devices, Sunnyvale, CA).
BECN1 RNA interference
Electroporation of shRNA-expressing plasmids in HepG2 cells was performed using the Neon® transfection system (Invitrogen, MPK10096) according to the manufacturer's protocol. Briefly, trypsinized HepG2 cells (1 × 106 cells) were washed in PBS and then resuspended in Neon Resuspension Buffer R. The cell suspension was mixed with 2 µg shRNA against BECN1 (Santa Cruz Biotechnology, sc-29797-SH) or a scrambled shRNA sequence (Santa Cruz Biotechnology, sc-108060) as a negative control and pulsed twice at 1200 V for 50 msec. After electroporation, cells were quickly seeded into 6-well plates and grown in culture medium for further experiments. Successful inhibition of BECN1 expression was verified by western blot analysis.
Animals
Four-wk-old male C57BL/6J mice were housed under controlled conditions (21°C ± 2°C, 60% ± 10% humidity, 12-h light/12-h dark cycle) with ad libitum access to food and water. After 1 wk, the mice were divided into 5 groups according to treatment (untreated control, n = 9 ; rosuvastatin, n = 7 ; fluvastatin, n = 7 ; pravastatin, n = 11 ; atorvastatin, n = 11 ). Beginning at 5 wk of age, all mice were fed a high-fat diet that included 45% lipids (Research Diets, Inc., D12451). The food given to each treatment group was supplemented with 0.01% (w/w) of the appropriate statin. Food intake and body weight of the mice were evaluated 2 times a wk at the same time of day. Fasting blood glucose level was measured weekly in the evening after an 8-h fast. After 16 wk, the mice were anesthetized with zolazepam and tiletamine (Zoletil, 50 mg/kg; Virbac France GTIN: 03597132126045), and blood was collected by cardiac puncture. Primary hepatocytes were isolated from male liver-specific atg7-deficient mice41 and wild-type mice (9 wk old) using a previously described method.48 Atg7f/f mice were crossed with albumin promoter-driven Cre mice to generate liver-specific atg7-deficient mice41 (atg7f/f;alb-Cre mice). The animal protocol was approved by the institutional animal care and use committee at Yonsei University College of Medicine.
Oral glucose tolerance, insulin tolerance, and pyruvate tolerance tests
To perform the oral glucose tolerance test, 40% glucose (2 g/kg body weight) was administered via oral gavage after a 6-h fast. Blood was collected from the tail vein at 0, 30, 60, 90, and 120 min after glucose administration. To assay insulin tolerance, fasting glucose was measured 4 h after fasting, and then mice were intraperitoneally injected with 0.75 U/kg human insulin-R (Sigma-Aldrich, I9278) dissolved in PBS. Blood glucose was measured at 15, 30, 60, 90, and 120 min after injection. To assay pyruvate tolerance, mice were intraperitoneally injected with 2 g/kg sodium pyruvate (Sigma-Aldrich, P2256) dissolved in PBS after an 18-h fast. Blood was collected from the tail vein before pyruvate injection (0 h) and at 15, 30, 60, 90, and 120 min after injection. Glucose levels were determined using an Accu-Chek Performa® glucometer (Boehringer-Mannheim, Indianapolis, IN).
Plasma glucose, cholesterol, triglyceride, and free fatty acid measurement
Blood was collected in microcentrifuge tubes and centrifuged to obtain serum, which was divided into aliquots and stored at -80°C for subsequent assays. Serum glucose, cholesterol, triglyceride, and free fatty acid levels were measured with the respective assay kits (BioAssay Systems, EBGL-100 for glucose, ECCH-100 for cholesterol, ETGA-200 for triglyceride and EFFA-100 for free fatty acid) according to the manufacturer's instructions.
Transmission electron microscopy
Autophagic vacuoles in the liver were visualized by transmission electron microscopy. Glutaraldehyde-fixed mouse liver tissues were postfixed in 2% osmium tetroxide, dehydrated in graded alcohol, and flat embedded in Epon 812 (Electron Microscopy Sciences, 100503–876). Ultrathin tissue sections (300 nm) were stained with uranyl acetate and lead citrate and examined with an electron microscope (JEM-1011, JEOL/MegaView III, Olympus, Tokyo, Japan).
Statistical analysis
Data are presented as mean ± standard error of the mean. Groups were compared using the Student t test or one-way analysis of variance followed by the Dunnett multiple comparison test, where appropriate; P < 0.05 was considered significant. Data analysis was carried out using Prism 5.0 software (GraphPad Software, La Jolla, CA).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by the “Kiturami” Faculty Research Assistance Program of Yonsei University College of Medicine (6–2012–0148 and 6–2013–0158); the National Research Foundation of Korea Grant funded by the Korean government (MEST, Basic Research Promotion Fund; NRF-2010–013-E0008 and NRF-2012000891 to E.S. Kang, and NRF-2012R1A12042620 to H Lee); and the National Institute of Health (R01DK083567 to Y.-B.K)
References
- 1.Sabatine MS WS, Morrow DA, McCabe CH, Canon CP High-dose atorvastatin associated with worsening glycemic control: A PROVE-IT TIMI 22 substudy. Circulation 2004; 110(Suppl III):S834. [Google Scholar]
- 2.Collins R, Armitage J, Parish S, Sleigh P, Peto R, Heart Protection Study Collaborative G . MRC/BHF Heart Protection Study of cholesterol-lowering with simvastatin in 5963 people with diabetes: a randomised placebo-controlled trial. Lancet 2003; 361:2005-16; PMID:12814710; http://dx.doi.org/ 10.1016/S0140-6736(03)12475-0 [DOI] [PubMed] [Google Scholar]
- 3.Sever PS, Poulter NR, Dahlof B, Wedel H, Collins R, Beevers G, Caulfield M, Kjeldsen SE, Kristinsson A, McInnes GT, et al. Reduction in cardiovascular events with atorvastatin in 2,532 patients with type 2 diabetes: Anglo-Scandinavian Cardiac Outcomes Trial–lipid-lowering arm (ASCOT-LLA). Diabetes Care 2005; 28:1151-7; PMID:15855581; http://dx.doi.org/ 10.2337/diacare.28.5.1151 [DOI] [PubMed] [Google Scholar]
- 4.Kjekshus J, Apetrei E, Barrios V, Bohm M, Cleland JG, Cornel JH, Dunselman P, Fonseca C, Goudev A, Grande P, et al. Rosuvastatin in older patients with systolic heart failure. N Engl J Med 2007; 357:2248-61; PMID:17984166; http://dx.doi.org/ 10.1056/NEJMoa0706201 [DOI] [PubMed] [Google Scholar]
- 5.Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM Jr., Kastelein JJ, Koenig W, Libby P, Lorenzatti AJ, MacFadyen JG, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 2008; 359:2195-207; PMID:18997196; http://dx.doi.org/ 10.1056/NEJMoa0807646 [DOI] [PubMed] [Google Scholar]
- 6.Sattar N, Preiss D, Murray HM, Welsh P, Buckley BM, de Craen AJ, Seshasai SR, McMurray JJ, Freeman DJ, Jukema JW, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet 2010; 375:735-42; PMID:20167359; http://dx.doi.org/ 10.1016/S0140-6736(09)61965-6 [DOI] [PubMed] [Google Scholar]
- 7.Rajpathak SN, Kumbhani DJ, Crandall J, Barzilai N, Alderman M, Ridker PM. Statin therapy and risk of developing type 2 diabetes: a meta-analysis. Diabetes Care 2009; 32:1924-9; PMID:19794004; http://dx.doi.org/ 10.2337/dc09-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preiss D, Seshasai SR, Welsh P, Murphy SA, Ho JE, Waters DD, DeMicco DA, Barter P, Cannon CP, Sabatine MS, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA 2011; 305:2556-64; PMID:21693744; http://dx.doi.org/ 10.1001/jama.2011.860 [DOI] [PubMed] [Google Scholar]
- 9.McTaggart F, Buckett L, Davidson R, Holdgate G, McCormick A, Schneck D, Smith G, Warwick M. Preclinical and clinical pharmacology of Rosuvastatin, a new 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor. Am J Cardiol 2001; 87:28B-32B; PMID:11256847; http://dx.doi.org/ 10.1016/S0002-9149(01)01454-0 [DOI] [PubMed] [Google Scholar]
- 10.Nezasa K, Higaki K, Takeuchi M, Nakano M, Koike M. Uptake of rosuvastatin by isolated rat hepatocytes: comparison with pravastatin. Xenobiotica 2003; 33:379-88; PMID:12745873; http://dx.doi.org/ 10.1080/0049825031000066259 [DOI] [PubMed] [Google Scholar]
- 11.Parker RA, Clark RW, Sit SY, Lanier TL, Grosso RA, Wright JJ. Selective inhibition of cholesterol synthesis in liver versus extrahepatic tissues by HMG-CoA reductase inhibitors. J Lipid Res 1990; 31:1271-82; PMID:2401858 [PubMed] [Google Scholar]
- 12.Sukhija R, Prayaga S, Marashdeh M, Bursac Z, Kakar P, Bansal D, Sachdeva R, Kesan SH, Mehta JL. Effect of statins on fasting plasma glucose in diabetic and nondiabetic patients. J Investig Med 2009; 57:495-9; PMID:19188844 [DOI] [PubMed] [Google Scholar]
- 13.Hafner M, Juvan P, Rezen T, Monostory K, Pascussi JM, Rozman D. The human primary hepatocyte transcriptome reveals novel insights into atorvastatin and rosuvastatin action. Pharmacogenet Genomics 2011; 21:741-50; PMID:21869732; http://dx.doi.org/ 10.1097/FPC.0b013e32834a5585 [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab 2011; 13:495-504; PMID:21531332; http://dx.doi.org/ 10.1016/j.cmet.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 2013; 11:e1001708; PMID:24265594; http://dx.doi.org/ 10.1371/journal.pbio.1001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 2008; 8:318-24; PMID:18840362; http://dx.doi.org/ 10.1016/j.cmet.2008.08.013 [DOI] [PubMed] [Google Scholar]
- 17.Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 2011; 7:727-36; PMID:21471734; http://dx.doi.org/ 10.4161/auto.7.7.15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, Suzuki H, Marais AD, Brombacher F. Statin therapy reduces the mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J Infect Dis 2014; 209:754-63; PMID:24133190; http://dx.doi.org/ 10.1093/infdis/jit550 [DOI] [PubMed] [Google Scholar]
- 19.Parikh A, Childress C, Deitrick K, Lin Q, Rukstalis D, Yang W. Statin-induced autophagy by inhibition of geranylgeranyl biosynthesis in prostate cancer PC3 cells. Prostate 2010; 70:971-81; PMID:20135644; http://dx.doi.org/ 10.1002/pros.21131 [DOI] [PubMed] [Google Scholar]
- 20.Araki M, Maeda M, Motojima K. Hydrophobic statins induce autophagy and cell death in human rhabdomyosarcoma cells by depleting geranylgeranyl diphosphate. Eur J Pharmacol 2012; 674:95-103; PMID:22094060; http://dx.doi.org/ 10.1016/j.ejphar.2011.10.044 [DOI] [PubMed] [Google Scholar]
- 21.Wei YM, Li X, Xu M, Abais JM, Chen Y, Riebling CR, Boini KM, Li PL, Zhang Y. Enhancement of autophagy by simvastatin through inhibition of Rac1-mTOR signaling pathway in coronary arterial myocytes. Cell Physiol Biochem 2013; 31:925-37; PMID:23817226; http://dx.doi.org/ 10.1159/000350111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, Vandenabeele P. Beclin1: a role in membrane dynamics and beyond. Autophagy 2012; 8:6-17; PMID:22170155; http://dx.doi.org/ 10.4161/auto.8.1.16645 [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Verity MA, Reue K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab 2014; 20:267-79; PMID:24930972; http://dx.doi.org/ 10.1016/j.cmet.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942-6; PMID:20526321; http://dx.doi.org/ 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A 2005; 102:11278-83; PMID:16076959; http://dx.doi.org/ 10.1073/pnas.0502738102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem 2005; 280:20589-95; PMID:15788402; http://dx.doi.org/ 10.1074/jbc.M412357200 [DOI] [PubMed] [Google Scholar]
- 27.Singh BK, Sinha RA, Zhou J, Xie SY, You SH, Gauthier K, Yen PM. FoxO1 deacetylation regulates thyroid hormone-induced transcription of key hepatic gluconeogenic genes. J Biol Chem 2013; 288:30365-72; PMID:23995837; http://dx.doi.org/ 10.1074/jbc.M113.504845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011; 145:607-21; PMID:21565617; http://dx.doi.org/ 10.1016/j.cell.2011.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Lee J, Reno CM, Sun C, Park SW, Chung J, Lee J, Fisher SJ, White MF, Biddinger SB, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med 2011; 17:356-65; PMID:21317886; http://dx.doi.org/ 10.1038/nm.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem 2008; 283:16283-92; PMID:18420577; http://dx.doi.org/ 10.1074/jbc.M802240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem 2009; 284:5148-57; PMID:19103600; http://dx.doi.org/ 10.1074/jbc.M808890200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS letters 2008; 582:829-34; PMID:18280254; http://dx.doi.org/ 10.1016/j.febslet.2008.02.010 [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Dang Y, Ren YR, Liu JO. Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc Natl Acad Sci U S A 2010; 107:4764-9; PMID:20176935; http://dx.doi.org/ 10.1073/pnas.0910872107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001; 292:1160-4; PMID:11349148; http://dx.doi.org/ 10.1126/science.1059344 [DOI] [PubMed] [Google Scholar]
- 35.Kirsch C, Eckert GP, Mueller WE. Statin effects on cholesterol micro-domains in brain plasma membranes. Biochem Pharmacol 2003; 65:843-56; PMID:12628479; http://dx.doi.org/ 10.1016/S0006-2952(02)01654-4 [DOI] [PubMed] [Google Scholar]
- 36.Wei YM, Li X, Xiong J, Abais JM, Xia M, Boini KM, Zhang Y, Li PL. Attenuation by statins of membrane raft-redox signaling in coronary arterial endothelium. J Pharmacol Exp Ther 2013; 345:170-9; PMID:23435541; http://dx.doi.org/ 10.1124/jpet.112.201442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andres AM, Hernandez G, Lee P, Huang C, Ratliff EP, Sin J, Thornton CA, Damasco MV, Gottlieb RA . Mitophagy is Required for Acute Cardioprotection by Simvastatin. Antioxid Redox Signal 2014; 10:21:1960-73. PMID:23901824; http://dx.doi.org/ 10.1089/ars.2013.5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kou R, Shiroto T, Sartoretto JL, Michel T. Suppression of Galphas synthesis by simvastatin treatment of vascular endothelial cells. J Biol Chem 2012; 287:2643-51; PMID:22144680; http://dx.doi.org/ 10.1074/jbc.M111.303594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A 1983; 80:2179-83; PMID:6340116; http://dx.doi.org/ 10.1073/pnas.80.8.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032-6; PMID:15525940; http://dx.doi.org/ 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- 41.Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim Y-N, Kim SS, Kim H, Hur KY, Kim HK, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 2013; 19:83-92; PMID:23202295; http://dx.doi.org/ 10.1038/nm.3014 [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010; 11:467-78; PMID:20519119; http://dx.doi.org/ 10.1016/j.cmet.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yada T, Nakata M, Shiraishi T, Kakei M. Inhibition by simvastatin, but not pravastatin, of glucose-induced cytosolic Ca2+ signalling and insulin secretion due to blockade of L-type Ca2+ channels in rat islet beta-cells. Br J Pharmacol 1999; 126:1205-13; PMID:10205010; http://dx.doi.org/ 10.1038/sj.bjp.0702397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mita T, Watada H, Nakayama S, Abe M, Ogihara T, Shimizu T, Uchino H, Hirose T, Kawamori R. Preferable effect of pravastatin compared to atorvastatin on beta cell function in Japanese early-state type 2 diabetes with hypercholesterolemia. Endocr J 2007; 54:441-7; PMID:17457013; http://dx.doi.org/ 10.1507/endocrj.K06-198 [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa M, Okajima F, Inoue N, Motomura K, Kato T, Takahashi A, Oikawa S, Yamada N, Shimano H. Distinct effects of pravastatin, atorvastatin, and simvastatin on insulin secretion from a beta-cell line, MIN6 cells. J Atheroscler Thromb 2006; 13:329-35; PMID:17192698; http://dx.doi.org/ 10.5551/jat.13.329 [DOI] [PubMed] [Google Scholar]
- 46.Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014; 510:542-6; PMID:24847880; http://dx.doi.org/ 10.1038/nature13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalli CA, Pauli JR, Prada PO, Cintra DE, Ropelle ER, Velloso LA, Saad MJ. Statin modulates insulin signaling and insulin resistance in liver and muscle of rats fed a high-fat diet. Metabolism 2008; 57:57-65; PMID:18078859; http://dx.doi.org/ 10.1016/j.metabol.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 48.Shen L, Hillebrand A, Wang DQ, Liu M Isolation and primary culture of rat hepatic cells. J Vis Exp 2012;29:pii: 3917; PMID: 22781923; http://dx.doi.org/ 10.3791/3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.