Abstract
The repair of DNA damage is a complex process that relies on particular pathways to remedy specific types of damage to DNA. The range of insults to DNA includes small, modest changes in structure including mismatched bases and simple methylation events to oxidized bases, intra- and interstrand DNA crosslinks, DNA double strand breaks and protein-DNA adducts. Pathways required for the repair of these lesions include mismatch repair, base excision repair, nucleotide excision repair, and the homology directed repair/Fanconi anemia pathway. Each of these pathways contributes to genetic stability, and mutations in genes encoding proteins involved in these pathways have been demonstrated to promote genetic instability and cancer. In fact, it has been suggested all cancers display defects in DNA repair. It has also been demonstrated that the ability of cancer cells to repair therapeutically induced DNA damage impacts therapeutic efficacy. This has led to targeting DNA repair pathways and proteins to develop anti-cancer agents that will increase sensitivity to traditional chemotherapeutics. While initial studies languished and were plagued by a lack of specificity and a defined mechanism of action, more recent approaches to exploit synthetic lethal interaction and develop high affinity chemical inhibitors have proven considerably more effective. In this review we will highlight recent advances and discuss previous failures in targeting DNA repair to pave the way for future DNA repair targeted agents and their use in cancer therapy.
Keywords: DNA damage, DNA repair, cancer, nucleotide excision repair, homologous recombination repair, non-homologous end joining, DNA damage response, radiation, PARP, replication protein A
1. DNA damage and repair pathways
DNA damage can arise from many sources, both endogenous and exogenous. Independent of the source, responding to this damage is crucial for the life of the individual cell and also for the life of the organism. In many cases these seemingly similar goals do not coincide and the cell is “sacrificed,” presumably for the greater good of the life of the organism. Interestingly, the transformation to a cancer cell imparts certain advantages to the cells at the expense of the organism but also results in vulnerabilities that can be exploited for treatment. In addition to the acquired changes in cancer cells that drive transformation and progression, the inherent biology of the underlying cell of origin also can impact the response to therapy. In lung epithelial cells for example, one could envision a robust network of DNA repair pathways to accommodate the vast array of genotoxic agents to which cells are exposed in the normal course of breathing. In the decision between cell death or repair and cell maintenance, DNA repair prevails to maintain organ function. Cancers derived from lung epithelial or alveolar cells are endowed with that same innate DNA repair capacity which can limit the effectiveness of agents used to treat the cancer. Germ cells, on the other hand, could be envisioned to be much less tolerant of DNA damage and to maintain the propagation of intact, non-mutated, genetic information, cell death predominates the response to genotoxic stress. Thus germ cell tumors display hypersensitivity to chemotherapeutics that act via the induction of DNA damage.
For those cell types where repairing the damage is advantageous, intricate multiprotein pathways have evolved to handle the array of damage that is encountered. For the purpose of this review we will focus on DNA damage, both endogenous and exogenous and the response and repair of this damage that impacts cancer treatment (Fig. 1). Early classification of DNA repair pathways focused on direct reversal versus excision repair, the former employing damage-specific chemical reversal of the lesion. This is apparent in DNA photolyase and methyl guanine methyl transferase which reverse ultraviolet-induced cyclopyrimidine dimers and O6-methyl guanine modifications respectively. One could also classify the non-homologous end joining (NHEJ) double strand break repair pathway as direct reversal in that the break can be repaired without the addition or loss of nucleotides, though in the context of more complex damage, excision and synthesis steps are needed to ensure repair activity. The excision repair pathways, nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR) each can respond to and repair a variety of lesions. NER is specific for repair of bulky adduct DNA damage while BER is responsible for removing base damage consisting of small oxidative and alkyl adducts. The MMR pathway also has a wide reach and is capable of repairing single base mismatches and a variety of small insertions and deletions to the genome. Homologous recombination (HR) repair or homology directed repair (HDR) is a major player in the response to DNA damage and participates in the tolerance of many types of lesions where the damage is not necessarily removed, but tolerated by using homologous template sequences to synthesize the sequence complementary to the damaged area of the genome. Homology directed repair (HDR) can also remove DNA damage depending on the specific adduct(s) encountered and the cadre of proteins assembled. In addition, both NHEJ and HDR play a crucial role in DSB repair required for CRISPR-Cas9 mediated genome engineering. The repair of DNA interstrand adducts that crosslink the two strands of DNA double helix are particularly problematic and employ the Fanconi Anemia (FA) pathway in collaboration with HDR. These lesions often present absolute blocks to traditional DNA replication machinery and an intricate assembly of proteins in the FA pathway initiate the response and repair to this type of damage.
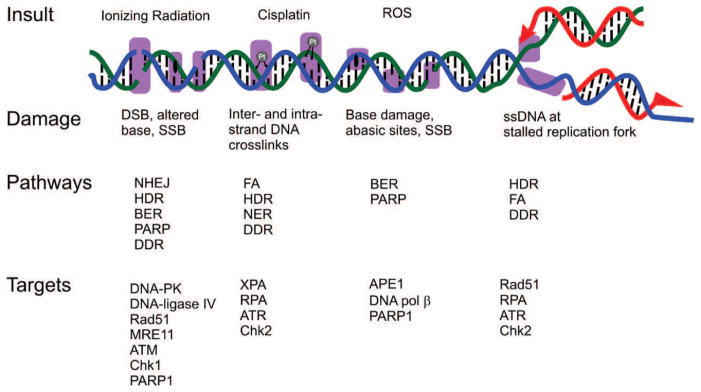
Fig. 1.
DNA damage response and repair. DNA damaged induced by the indicated insults are indicated by the magenta boxes. Pathways involved in responding to and repairing the damage along with current proteins being targeted are indicated.
These pathways responsible for the actual repair of the damaged DNA are all impacted by the larger DNA damage response (DDR) that regulates a number of cellular processes that position the cell to either initiate a repair response or to initiate a cell death response. These however are not mutually exclusive and represent a dynamic process that includes a series of decision points that can impact the fate of the cell and ultimately the organism. With DNA repair playing crucial roles in the development, progression and response to therapy for a wide array of cancers it is not surprising that there are increased efforts to validate DNA damage response and repair proteins as therapeutic targets and develop agents against these targets.
Synthetic lethal approaches to cancer therapy have provided novel mechanisms to specifically target cancer cells while sparing non-cancer cells and thereby reducing toxicity associated with treatment. More recently, the synthetic lethality approach has been exploited with the use of PARP inhibitors for the treatment of BRCA deficient ovarian cancer. Cancer cells are characterized by genetic instability driven by deficiencies in DNA repair and/or DNA recombination genes. This deficiency renders them vulnerable to agents whose effect is part of the synthetic lethal interaction. Synthetically lethal therapeutics known to target the specific gene product that resembles the phenotype caused by a mutation and also exploits inherent differences between cancer cells and normal cells, which is often not feasible with conventional chemotherapeutic drugs. For example, the PARP1 enzyme is involved in repairing single-strand breaks while BRCA is important for repairing double-strand breaks in DNA. The inhibition of PARP1 is selectively lethal to BRCA deficient cancers due to accumulation of single-strand breaks which are converted to double strand breaks during DNA replication which are not repairable in BRCA mutant or deficient cells. In this review we summarize recent development in targeting DNA repair with a focus on events of the past and how they may influence the future of DNA repair targeted therapy.
2: Development and deployment of PARP inhibitors
Poly (ADP-ribose) polymerases (PARPs) and its inhibition
PARPs are a family of nuclear proteins whose actions include DNA damage recognition via binding to single strand breaks (SSB) and synthesis of poly(ADP-ribose) (pADPr) chains on glutamate, aspartate and lysine residues of target proteins. The PARP family consists of 17 isoforms that are classified on the basis of their functional multi-domain architecture and cellular localization (Krishnakumar & Kraus, 2010). PARP1 is the most well studied and dominant protein within this family, although PARP2 may play a similar but smaller and partially redundant role (Yelamos, Schreiber, & Dantzer, 2008). PARP1 has been shown to either directly or indirectly influence the repair of DNA via multiple pathways including BER, HR, NER, NHEJ and MMR. With nicotinamide adenine dinucleotide (NAD+) as a substrate, PARP1 synthesizes pADPr chains, thereby modifying its target protein. Target proteins include: PARP itself (automodification), numerous DNA repair proteins, histone H1, and a series of other transcription factors (Javle & Curtin, 2011).
While not directly participating in DNA repair reactions, both PARP1 and pADPr chains can recruit repair proteins that ultimately act on and repair the damaged DNA. pADPr chain formation also allows for the release of PARP from where it is bound to damaged DNA. Such release of PARP from damaged DNA has also been reported to influence repair, thereby allowing already recruited repair proteins to access the damaged site. pADPr modification of histones also facilitates repair protein access by relaxing chromatin structure. Interestingly, under conditions of excessive DNA damage, extensive PARP activity results in cellular NAD+ depletion which has been reported to induce cell death, though the exact mechanism of cell death remains to be determined (Chiarugi, 2002; Del, Xiao, Rangell, Reichelt, & O’Brien, 2014; Andrabi et al., 2014). This phenomenon also appears to be cell type specific and the impact of PARP-induced NAD+ depletion and energy metabolism in cancer cells treated with PARP inhibitors (PARPi) is likely considerably more complex than originally thought (Rouleau, Patel, Hendzel, Kaufmann, & Poirier, 2010; Bixel & Hays, 2015)
Inhibition of PARP reduces the synthesis of pADPr chains and prevents the recruitment of additional repair factors. Additionally, inhibiting PARP would prevent release from the site of DNA damage, thus trapping PARP on damaged DNA and preventing access for other repair proteins. In fact, the ability to trap PARP on DNA and, not necessarily, the inhibition of catalytic activity, has been correlated with in vitro cytotoxicity and likely clinical efficacy of PARP inhibitors (Murai et al., 2012). The prevention of pADPr chain synthesis and release of PARP from damaged DNA lead to accumulation of SSB that could be converted to single-sided DSB during DNA replication, (Bixel & Hays, 2015; Liu, Konstantinopoulos, & Matulonis, 2014; Murai et al., 2012). The ability to survive this insult relies on an active, intact HRR pathway. However, in the absence of intact DSB repair pathway, the persistent DSB is toxic and eventually leads to cell death (Fig. 2). Although this is the most dominant theory applied to the mechanism of PARPi’s in the setting of a nonfunctional DSB repair pathway (e.g. germline BRCA1/2 mutation (BRCAmut) associated malignancy), there are other models proposing alternative mechanisms, as it is known that PARPs play important roles in other biological processes like transcriptional regulation, chromatin modification, mitotic-spindle formation, intracellular trafficking and energy metabolism (Li & Yu, 2013; Murai et al., 2012; Patel, Sarkaria, & Kaufmann, 2011; Sonnenblick, de, Azim, Jr., & Piccart, 2015).
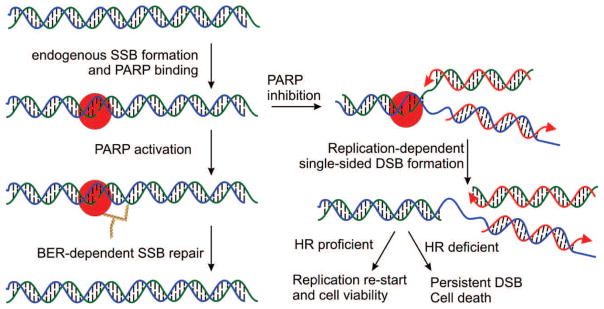
Fig. 2.
Role of PARP inhibitors in DNA repair and synthetic lethality. PARP1 (red) binding to an endogenously induced SSB results in activation and auto ADP-ribosylation (gold triangles) which promotes efficient BER. In the presence of PARPi’s DNA replication (red arrows) generates a DSB with in HR proficient cells can be repaired to allow replication re-start and maintain cell viability while in HR deficient cells, the DSB persists resulting in cell death.
PARP Inhibitors (PARPi)
PARPi are an emerging class of therapeutics with promising activity in ovarian, breast and other cancers. PARPi’s work by competing with the substrate NAD+ at PARP’s enzymatically active site, limiting PARP’s ability to catalyze the formation of pADPr chains (Zaremba & Curtin, 2007). In December 2014, olaparib (AZD-2281) was the first PARPi to be granted FDA approval, specifically as monotherapy for women with ovarian cancer who have had at least three line opf prior chemotherapy and who carry a BRCAmut. This U.S. decision followed The European Medicines Agency’s approval of olaparib as maintenance monotherapy after platinum treatment in the setting of recurrent platinum sensitive epithelial ovarian cancer in women who carry a BRCAmut. Additionally, olaparib is being investigated in several clinical trials as a monotherapy or in combination for the treatment of several solid tumors breast, pancreatic, prostate, and colorectal cancer in addition to other solid cancers.
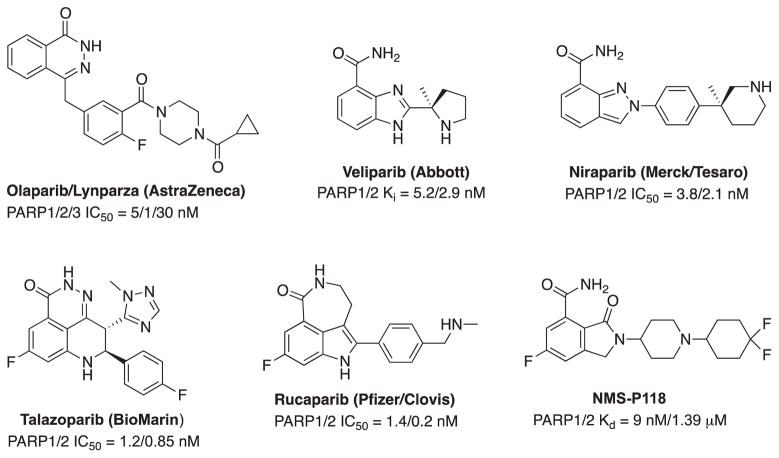
The well-established involvement of PARP1 in the recognition and repair of DNA damage, as well as the success of olaparib has triggered a number of drug discovery programs aimed at the development of additional PARPi. PARPi are adopted two nicotinamide-mimic motifs to compete with nicotinamide portion of NAD+ either as a conventional embedded primary amide (veliparib (ABT-888), niraparib (MK-4827) and NMS-P118) or as an amide enclosed in a cyclic ring (olaparib (AZD-2281), talazoparib (BMN 673) and rucaparib (AG-014699)) (Fig. 3).
Fig. 3.
Structures and activities of several PARP inhibitors.
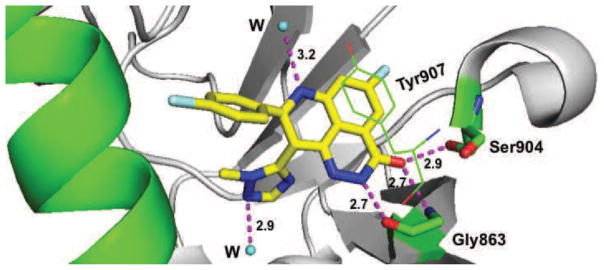
Talazoparib (BMN 673) is a highly potent PARP1/2 inhibitor with demonstrated in vitro selectivity for cells with BRCA1/2 or PTEN mutations (Cardnell et al., 2013), and it has been shown to be nearly a hundred fold more active at trapping PARP-DNA complexes than olaparib and rucaparib, the latter two exhibiting similar trapping potencies (Murai et al., 2014). Furthermore, talazoparib is enjoying early clinical successes, appearing more selectively cytotoxic with a longer half-life and better bioavailability as compared with other PARPi under development. The X-ray co-crystal structure of talazoparib with hPARP1 revealed an extensive hydrogen bonding network between the carboxamide moiety of the inhibitor with the backbone CO and NH of Gly863 and the hydroxyl group of Ser904 (Fig. 4). The tricyclic scaffold of talazoparib forms π-π stacking interactions with Tyr907 in PARP1, and the basic nitrogen of the triazole and tricyclic ring form a tight hydrogen bonding network to structural water molecules. These additional interactions contribute to the increased potency while also indirectly impacting the pharmacokinetic parameters.
Fig. 4.
Molecular interactions of talazoparib (BMN673) (yellow) with hPARP1 (green and gray) (PDB code: 4UND). Interaction with amino acid side chains and structured water molecules are indicated with the dashed magenta lines and distances indicated in Å.
Many current PARPi clinical candidates are nonselective inhibitors of multiple isoforms within the PARP family (Krishnakumar & Kraus, 2010; Yelamos, Farres, Llacuna, Ampurdanes, & Martin-Caballero, 2011). The finding of variable cellular and clinical activity profiles of different, non-selective PARPi may be explained by their polypharmacological profiles. For example, olaparib, rucaparib and veliparib each have wide ranging kinase inhibitory activities (Antolin & Mestres, 2014; Passeri et al., 2015). More recently, Papeo et al. developed a potent PARP1 selective inhibitor, NMS-P118 (Fig. 3) (Papeo et al., 2015). NMS-P118 has an excellent absorption, distribution, metabolism, and excretion profile and high in vivo efficacy as a single agent in triple negative breast cancer with a BRCA1 mutation and also in combination with temozolomide in BRCA2 deficient pancreatic cancer xenograft models. Interestingly, the discovery of a naturally occurring (−)-gossypol as a protein-protein interaction inhibitor of the PARP1 BRCA1 C Terminus (BRCT) domain affords new opportunities for the development of highly potent and specific PARP1 inhibitors available (Na et al., 2015).
Preclinical and Clinical Successes of PARPi for Ovarian and Other Cancers
BRCAmut are associated with increased risks of breast, ovarian, and other cancers. In fact, while the baseline population risk of ovarian cancer is 1.3%, this lifetime risk increases to 15–40% in women with a BRCAmut (Chen & Parmigiani, 2007; Siegel, Ma, Zou, & Jemal, 2014). If a cell is deficient in or has nonfunctional BRCA1/2 protein products, it is unable to localize RAD51 to double strand DNA breaks, and without the localization of RAD51, HR cannot occur effectively. The alternative DNA repair pathways a HR deficient cell is forced to use can be error prone, leading to DNA damage accumulation and malignancy (Venkitaraman, 2014). It is thought that at least 30% of epithelial ovarian and related cancers are associated with defects in the HR pathway (the most common of which are BRCAmut related), and with almost 22,000 cases diagnosed and more than 14,000 deaths in the United States in 2014, therapies that exploit HR deficiencies have great potential to be impactful in the treatment of this largely lethal malignancy (Pennington et al., 2014; Siegel et al., 2014). In unaffected cells, BRCA1/2 proteins are essential for the repair double-strand breaks (DSB) and stalled replication forks by the HRR pathway. However, cells with deficient or non-functional BRCA1/2 treated with PARPi’s are forced to rely on error-prone repair pathways (eg. non-conservative mechanism such as NHEJ or single-strand annealing) to process DNA lesions. This results in high levels of mutations and genetic instability, leads to the cell-cycle arrest and apoptosis.
Applying the concept of synthetic lethality, preclinical PARPi studies demonstrated that PARPi were able to selectively target HRR deficient cells (specifically BRCA mutated cells) (Farmer et al., 2005; Lord, Tutt, & Ashworth, 2015). Phase I data from various PARPi (olaparib, niraparib and rucaparib) as monotherapy in patients with BRCAmut demonstrated impressive response rates, at least 28% using Response Evaluation Criteria in Solid Tumors (RECIST) (Fong et al., 2010; Sandhu et al., 2013). Data from Phase II trials of women with BRCAmut ovarian cancer using olaparib at a dose of 400 mg orally twice daily demonstrated RECIST response rates of 31–41% (Audeh et al., 2010; Gelmon et al., 2011; Kaufman et al., 2015). The Phase II trial by Kaufman et al. demonstrated a 31% response rate (RECIST) and an additional 40% stable disease rate (at more than 8 weeks) in their study subset of 193 women with platinum-resistant ovarian cancer (Kaufman et al., 2015). This trial was fundamental in the FDA approval of olaparib (Bixel & Hays, 2015). Phase I and II data also indicates a greater likelihood of response in women with platinum-sensitive disease (Fong et al., 2010; Gelmon et al., 2011). Additionally, olaparib has been studied in combination with various targeted agents and cytotoxic chemotherapies including cediranib, pegylated liposomal doxorubicin (PLD), carboplatin and paclitaxel, and tolerability and improvements in progression free survival (PFS) have been demonstrated (Del et al., 2014; Liu et al., 2013; Oza et al., 2015). The authors await results of several studies that will further describe the effectiveness of various PARPi, potentially identify the subgroup(s) of women that will receive maximal benefit from PARPi, and determine when to give PARPi in the sequence of ovarian cancer treatment (concomitantly with primary cytotoxic chemotherapy versus maintenance therapy versus recurrent disease). These include ARIEL2/3 (NCT01891344, NCT01968213), SOLO-1,-2,-3 (NCT01844986, NCT01874353, NCT02282020), NOVA (NCT01847274) and GOG 9923 (NCT00989651).
PARPi are generally well-tolerated oral agents. Typically reported treatment related adverse events (mainly grade 1 and 2) include nausea, vomiting, anorexia and fatigue. Myelosuppression is also a relatively common toxicity, appearing to be dose dependent and successfully managed with dose reduction (Audeh et al., 2010; Fong et al., 2010; Gelmon et al., 2011; Kaufman et al., 2015; Sandhu et al., 2013). Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) are rare but serious adverse events that have been reported in the setting of olaparib treatment. Overall, MDS/AML have been reported in less than 1% of patients treated with olaparib, and it is difficult to determine causality as this data only includes a heavily pretreated population (AstraZeneca, 2015). More recently, a phase II study of olaparib in patients with metastatic, hormone resistant prostate cancer showed promising results. Next-generation sequencing was used to identify homologous deletions, mutations or both in a series of DNA repair genes in tumor samples. Patients whose tumors harbor these genetic alterations had a response rate of 33%, which is much higher than expected for this patient population (Mateo et al., 2015). This study, in addition to confirming a potential role for PARPi outside of ovarian cancer highlight the importance of assessing individual DNA repair pathways to identify patient populations most likely to response to therapy.
In summary, PARPi are a new class of therapeutics with well-demonstrated effectiveness against ovarian cancers with BRCAmut associated deficiencies in the HRR pathway. They are generally well tolerated oral agents, and olaparib is FDA approved for treatment after at least 3 prior chemotherapies for women with ovarian cancer who carry a BRCAmut. Research is ongoing to further advance our understanding of these agents, develop more specific and active agents and to define the population(s) of most benefit, with the ultimate goal of improving treatment and survival for patients with ovarian, breast and other cancers.
3. NER inhibitors and combination Pt-therapy
The NER pathway is an incredibly versatile pathway and remedies DNA bulky adduct damage. There are over 30 polypeptides that participate in the repair pathway and extensive genetic and biochemical analyses have led to thorough understanding of the necessary steps and the proteins that carry out those steps (Woods & Turchi, 2013). The pathway can be broken down into 4 essential steps; recognition, incision/excision, resynthesis and ligation. There are two modes employed to identify bulky adduct damage. Either sensor proteins are employed to scan the genomic DNA and recognize distortions and chemical modifications in the DNA, termed global genomic NER (GG-NER) (Maillard, Camenisch, Clement, Blagoev, & Naegeli, 2007), or recognition is coupled to transcription, transcription–coupled NER (TC-NER) (Sancar & Reardon, 2004). The two recognition pathways converge for verification of the damage which involves additional components including TFIIH, RPA and XPA, each of them essential for NER. The incision/excision step relies on two structure specific endonucleases, XPG and ERCC1/XPF, which hydrolyze phosphodiester bonds of the damaged strand of the DNA 3′ and 5′ to the DNA adduct, respectively. Their activity also relies on the formation of the single-strand and double-strand junctions carried out by the XPD and XPB helicases, which are part of the TFIIH component. Resynthesis and ligation conclude the process resulting in an accurately repaired duplex DNA.
As many cancer therapeutics including the widely prescribed platinum-based agents cisplatin, carboplatin, and oxaliplatin impart their clinical efficacy via the induction of DNA damage, and the NER pathway plays a considerable role in modulating the activity of these therapies. With the realization of the importance of NER in modulation therapy, targeting the NER pathway has been pursued. Early agents shown to augment the activity of platinum-based therapeutics included DNA replication inhibitors that were argued to block the resynthesis step, though there is no convincing demonstration of this mechanism. Additivity and synergy observed with these agents were likely multi-factorial (Katz, Andrews, & Howell, 1990; Frankfurt, Seckinger, & Sugarbaker, 1993). The failures of many of these agents to progress in clinical evaluation to either reverse resistance to platinum therapy or increase sensitivity to platinum therapy cast doubt clinically on the utility of inhibiting NER for cancer therapy.
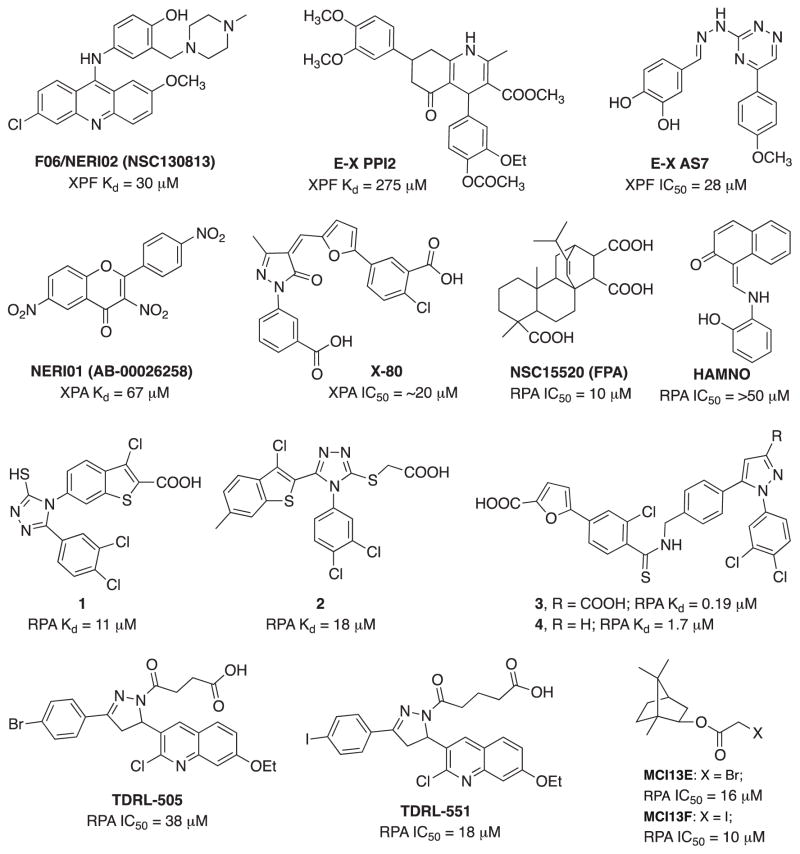
Recently however, with the concept of synthetic lethality and utility of PARP inhibitors there has been renewed interest in targeting DNA repair. In NER, the focus has been on the DNA verification proteins RPA and XPA, and the structure specific nuclease ERCC1-XPF interaction. The potential utility of targeting ERCC1/XPF stems from a series of studies that inversely correlated ERCC1 expression with survival in a series of lung cancer studies. While the specificity of ERCC1 detection is in question, the biologic rationale remains solid (Bhagwat et al., 2009; Niedernhofer, Bhagwat, & Wood, 2007). More recently, it has been demonstrated that genetic manipulation of ERCC1 and XPF levels can influence sensitivity of cisplatin (Arora, Kothandapani, Tillison, Kalman-Maltese, & Patrick, 2010). The identification of F06/NERI02 (NSC130813) in an in silico screen that blocked the ERCC1-XPF interaction further solidifies this target (Jordheim et al. 2013). F06 displays modest in vitro affinity for XPF (30 μM) but has the expected biologic activities and can reduce the XPF-ERCC1 interaction in cells. The synergy observed with cisplatin and mitomycin C is interesting, though the single agent activity is likely a result of an off-target effect as ERCC1 and XPF deficient cells are viable. The alternative is that the presence of F06 induces the accumulation of ERCC1 and XPF monomers and these monomers are toxic to cells. However, this is unlikely to occur as coordinate regulation of proteins levels has not been observed. Despite the unknown mechanism for single agent activity of F06, the increase in sensitivity to cisplatin and MMC are consistent with inhibition of the ERCC1-XPF interaction. In addition, F06 showed a synergistic interaction with olaparib in BRCA1-deficient breast cancer cells; however, the role of ERCC1-XPF in this synergy needs further validation. More recently, several catechol and hydroxyl-imide/pyridine/pyrimidinones have been identified as ERCC1-XPF inhibitors from both in silico and high throughput screenings (Fig. 5) (Chapman et al., 2015; McNeil et al., 2015). Sub or low micromolar potency was observed in a biochemical endonuclease assay against XPF-ERCC1 with limited activity against FEN-1 and DNase I. E-X PPI2 significantly reduced the level of ERCC1-XPF heterodimers in ovarian cancer cells, inhibited NER with an IC50 of 20 μM and enhances melanoma cell sensitivity to cisplatin. Catechol scaffold containing E-X AS7 binds to ERCC1-XPF through metal-based interaction, inhibits NER in low micromolar concentrations (IC50 = 2 μM) and specifically increases the cisplatin sensitivity of NER-proficient human and mouse cells without having any effect on NER-deficient cell lines. Clearly more potent and specific inhibitors are needed to advance this class of molecules into the clinic as bona fide NER inhibitors. Another essential role of NER is in the interaction between ERCC1 and XPA. Barakat et. al. performed an in silico screen targeting the ERCC1-XPA interface and identified NERI01 as an inhibitor of this interaction. NERI01 significantly sensitized human colon cancer cells to UV irradiation but displayed a weaker effect on cisplatin sensitivity (Barakat et al., 2012). A similar modest potency >50 uM, undetermined specificity and cisplatin sensitivity suggest further development is required to obtain lead molecules from in these in silico hits.
Fig. 5.
Structures and activity of NER inhibitors.
The identification of XPA inhibitors has also been pursued focusing not on the protein-protein interactions required for NER, but the necessary and important protein-DNA interactions. An in silico screen identified three hits that were validated using a series of in vitro DNA binding assays. The X-80 compound (Fig. 5) identified in the original screen displayed reasonable potency in fluorescence polarization DNA binding assays (Neher, Shuck, Liu, Zhang, & Turchi, 2010). More recent development of this class of compounds has identified a series of analogs that display a >50-fold increase in potency and excellent specificity (data not shown). The ultimate utility of XPA-DNA binding inhibitors will however require an extensive cellular and in vivo analysis in combination with clinically approved agents that act via the induction of DNA damage.
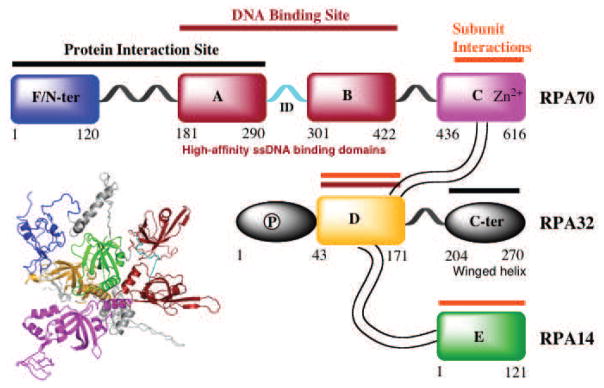
The identification of molecules capable of inhibiting protein-protein interaction has also focused on the major single stranded DNA binding protein in humans, RPA (Replication Protein A). RPA plays an integral role in both NER and HR pathways apart from its essential role in DNA replication, DNA damage checkpoint activation and DNA repair (Haring, Mason, Binz, & Wold, 2008). RPA is overexpressed in a number of cancers including lung, ovarian, breast, colon and esophageal (Jekimovs et al., 2014). RPA is a heterotrimeric complex consisting of 70 kDa (RPA70), 32 kDa (RPA32) and 14 kDa (RPA14) subunits (Fig. 6). The 70 kDa subunit contains the two major high affinity ssDNA binding domains A and B, as well as C and F. Domains D and E are located in the 32-kDa and 14-kDa subunit, respectively (Fan & Pavletich, 2012). RPA consists of a series of oligonucleotide/oligosaccharide binding folds (OB-folds) (domains A–F) many of which are engaged with DNA during the course of its participation in DNA metabolism. The F-domain located on the N-terminus of the 70 kDa subunit (RPA70N) of RPA does not bind with high affinity to ssDNA; however, it participates in a series of protein-protein interactions. The interaction of RPA70N with proteins involved in the DNA damage response pathway such as ATRIP, Rad 9 of the 9-1-1 damage recognition complex, p53 and MRE11 has generated considerable interest in targeting this domain. The first compound identified as an inhibitor of the F-domain of RPA70, NSC15520 (Fumaropimaric acid, FPA), was the discovered in an in vitro high throughput screening (HTS). The compound displays reasonable potency at 10 μM and has been shown to disrupt both RPA70N-Rad9 and RPA70N-p53 interactions (Glanzer, Liu, & Oakley, 2011; Glanzer et al., 2013). NSC15520 did not inhibit the DNA binding activity of RPA, suggesting some degree of specificity for the N-terminal F domain and not the main DNA binding domains A and B which are located in the central region of RPA70. No cellular data was presented possibly as a function of the highly charged nature of the compound limiting its membrane permeability and cellular uptake. A second compound, HAMNO [(1Z)-1-[(2-hydroxyanilino)methylidene]naphthalen-2-one], that selectively blocks the activity of DBD-F was also identified by Oakley et. al. through HTS (Glanzer et al., 2014). HAMNO shows a synergistic interaction with etoposide, enhances DNA replication stress in cancer cells and slows down tumor growth in a xenograft tumor model. The in vitro activity of HAMNO in blocking DBD-F function is weak, with concentrations greater than 50 μM required to observe significant inhibition. However, cellular activity, while consistent with inhibition of RPA activity occurs at considerably lower concentrations. These interesting findings suggest a few possibilities, including cellular metabolism of HAMNO results in the generation of a more potent metabolite or inconsistency in cellular effect and RPA inhibition might be due to off-target effects of HAMNO. A more detailed study of the specificity and cellular activity of HAMNO are needed to ultimately determine the target and activity of this interesting compound. Recently, Stephen Fesik’s group from Vanderbilt University exploited a fragment based NMR screening approach in the discovery of sub-micromolar or low micromolar stapled helix peptides and small molecules as inhibitors of the RPA70-N-terminal domain (Frank et al., 2014; Waterson et al., 2015). The increased potency will undoubtedly facilitate development of cellular inhibitors, though cellular activity or specificity is not documented to date.
Fig. 6.
Schematic representation of the RPA heterotrimeric subunits (70, 32 and 14 kDa) and OB-fold domains (A–F). The DNA interactions sites are indicated by the red bars, subunit interaction sites by gold bars and domains associated with binding other proteins by the black bars.
While much of the effort has been placed on identification of protein-protein interaction inhibitors, many of the proteins required for NER have essential interactions with DNA. Development of inhibitors of protein-DNA interactions presents its own hurdles to overcome, but successful development of agents capable of blocking these essential macromolecular interaction hold considerable promise and can open up an entirely new class of ‘druggable’ targets for therapeutic intervention.
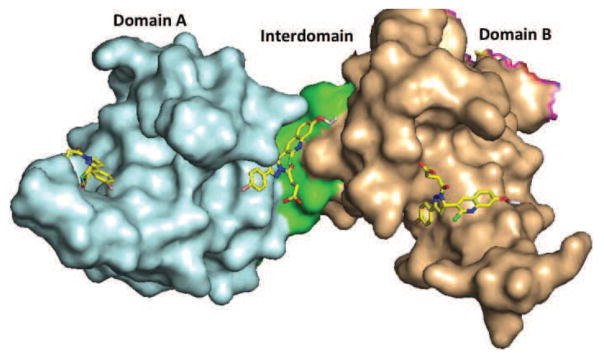
The RPA protein also is involved in essential interaction with DNA to support NER catalyzed repair of bulky adduct DNA damage. The OB-fold in DNA binding domains A and B are responsible for high affinity binding of RPA to single stranded DNA and duplex damaged DNA. Towards identifying inhibitors of the RPA-DNA interactions we established HTS approach (Andrews & Turchi, 2004) and screened the NCI diversity set of chemical compounds. This proof-of-concept study identified inhibitors capable of blocking the RPA-DNA interaction and inhibition of NER catalyzed repair of cisplatin damaged DNA. Further development of the HTS and analysis of a larger, more diverse library resulted in the identification of TDRL-505 (Shuck & Turchi, 2010). TDRL-505 prevents cell cycle progression, induces cytotoxicity, and increases the efficacy of the chemotherapeutic DNA damaging agents cisplatin and etoposide in vitro. In vitro studies and molecular docking delineated the TDRL-505 binding in the high-affinity ssDNA binding domains A and B of RPA70 (Fig. 7). Further synthetic structural modification of TDRL-505 generated several analogs and TDRL-551 was identified as the most potent compound of the series (Mishra, Dormi, Turchi, Woods, & Turchi, 2014). TDRL-551 blocks the RPA-DNA interaction, displays modest single agent activity in lung and ovarian cancer cell lines and also synergizes with cisplatin and etoposide. The mechanism of RPA inhibition we determined to be the direct, reversible interaction with the central OB-folds in domain A and B of RPA70. In vivo analysis of TDRL-551 revealed minimal toxicity alone and in combination with cisplatin and robust anticancer activity in a NSCLC human xenograft model. Favorable pharmacokinetics and a defined mechanism of action position this class of compounds for continued therapeutic development. Currently, we are performing systematic structural modification of TDRL-551 in our laboratory to develop highly potent RPA inhibitors for preclinical settings.
Fig. 7.
Surface representation of RPA70 A and B domains (PDB code: 1FGU) with TDRL-505. TDRL-505 docked in domain A (cyan), domain B (wheat), and the interdomain region (green). ΔG suggests higher affinity of TDRL-505 for domain B and the interdomain region, whereas modest affinity for domain A.
The initial screen of the NCI diversity set also identified NSC73101 as an RPA inhibitor. The structural analog analysis identified the tetrachloro-trimethylbicycloheptane-2-carboxylic acid as the active moiety and further structure activity relationship (SAR) and biochemical analysis revealed an irreversible mechanism of inhibition involving covalent modification of RPA. More reactive isoborneol haloacetate MCI13E and MCI13F exhibited potent single agent activity consistent with blocking RPA’s role in DNA replication (Neher, Bodenmiller, Fitch, Jalal, & Turchi, 2011). MCI13E decreased cell viability, induced apoptosis and showed a synergistic interaction with cisplatin in lung cancer cells. Further SAR is needed to increase the drug-like properties of this class of compounds and in light of recent single agent anticancer activity portend a bright future for these agents (data not shown).
Trabectedin (Ecteinascidin 743 or ET-743) showed high efficacy against the TC-NER sub-pathway but it does not bind to any of the proteins of NER pathway. ET-743 uniquely binds to DNA in the minor groove by forming a covalent bond at the exocyclic N2 position of guanine, which probably recruits NER proteins. ET-743 was recently approved for the treatment of soft tissue sarcomas, ovarian cancer and is currently in clinical trials for the treatment of breast, prostate, and pediatric sarcomas (Goldstein et al., 2014). Another compound, F11782, is a topoisomerase I and II inhibitor but is also found to inhibit NER’s helicase or incision steps, with more preference given to the incision step (Kruczynski et al., 2004).
The complexity of the NER pathway, while daunting at the molecular level, presents a plethora of opportunity to impact therapeutic response in the context of cancer treatment. Exploiting the structural analysis of many of these proteins and their interactions with DNA and other proteins holds the potential to fine tune the pathway and its response to chemotherapy induced DNA damage toward maximizing efficacy, overcoming resistance and reducing the toxicities associated with chemotherapy.
4: Targeting DNA Double Strand Break repair and radiation therapy
Radiation therapy continues to be a mainstay in the treatment of a variety of cancers. Exposure to ionizing radiation (IR) results in a variety of DNA lesions that include DNA termini modifications, SSBs, DSBs and base damage (Pastwa, Neumann, Mezhevaya, & Winters, 2003). DSBs are thought to be the most deleterious to cell survival and the main mechanism driving therapeutic efficacy (Povirk, 2006). The inability or reduced ability to repair DNA DSBs results in increased sensitivity to IR (Jeggo & Lavin, 2009). Furthermore, cells that have increased DSB repair activity, as observed in many cancers, display resistance to standardized radiation therapy and in many cases, resistance to radiation therapy is an adaptive response linked to hyperactive DSB repair mechanisms (Begg, Stewart, & Vens, 2011). IR therapy is often given in combination with chemotherapy, where many of the agents in the combination also induce DNA damage. Synergistic interactions between IR and DNA damaging agents have been observed and the mechanism linked to an inability to repair the IR-induced DSBs. For example, it has been shown that combination platinum-IR therapy results in the inability to repair DSBs by the NHEJ pathway (Boeckman, Trego, & Turchi, 2005; Sears & Turchi, 2012). Developing drugs aimed at modulating DNA DSB repair activity are likely to have a profound impact on the efficacy of radiation therapy. These observations have made targeting proteins in the DNA DSB repair pathways a popular approach for potential cancer treatments.
Repair of DNA DSBs
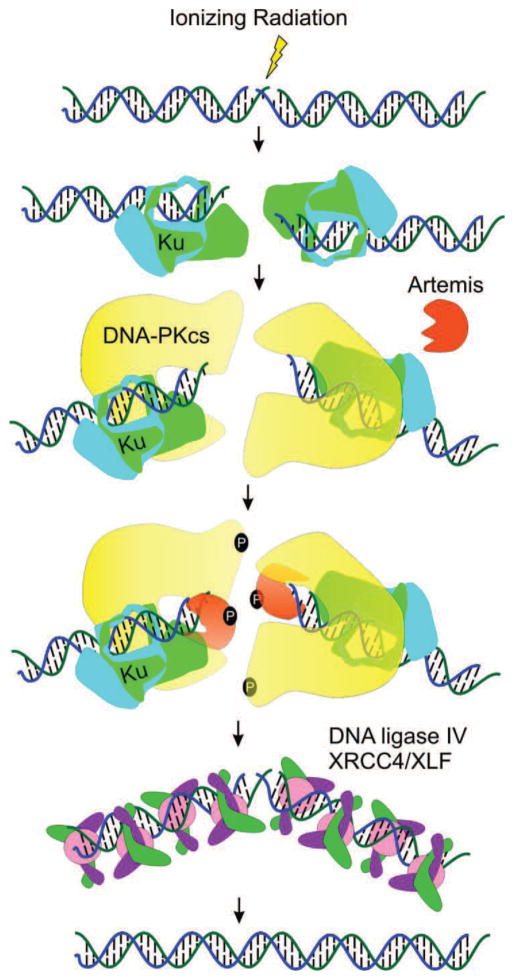
Human cells have two major pathways responsible for the repair of DNA DSBs: HDR and NHEJ. HDR is involved in DSB repair exclusively during S and G2 phases of the cell cycle due to the requirement for a homologous DNA sequence or sister chromatid as a template for the repair process (Chapman, Taylor, & Boulton, 2012). While HDR is restricted to a subset of cell cycle phases, NHEJ is active throughout the cell cycle and is the predominant pathway for the repair of IR induced DSB in higher eukaryotes, particularly those induced by ionizing radiation (Jeggo & Lavin, 2009). There are four specific steps in NHEJ: DNA termini recognition, bridging of the DNA ends (also known as formation of the synaptic complex), DNA end processing and DNA ligation. The specifics of the pathway are outlined in Fig. 8. An IR-induced DNA DSB recruits the heterodimeric Ku (Ku70 and Ku80) complex to the DNA terminus, which upon binding to DNA recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). DNA-PKcs forms a heterotrimeric complex with Ku and its serine/threonine protein kinase activity is activated once bound to the DNA terminus. DNA-PK coordinates NHEJ through autophosphorylation and phosphorylation of other target proteins, leading to DNA end processing by nucleases (Artemis), polymerases (Pol X family), PNK, and TDP. Following end processing, the Ligase IV/XRCC4/XLF complex is recruited to DNA termini and catalyzes ligation of the DNA DSB.
Fig. 8.
NHEJ Pathway. Following a IR-induced DSB, the Ku dimer (green/magenta) engage the DNA terminus. Association and activation of DNA-PKcs (yellow) results in target protein phosphorylation of Artemis (orange) and autophosphorylation. Termini processing by Artemis and pol X family polymerases (not shown) is followed by ligation via DNA ligase IV/XRCC4 and XLF complex.
DNA-PK and Ku70/80 Inhibitors: Inhibition of termini recognition and end bridging
Many attempts have been made to target DNA-PK, a crucial component of the NHEJ pathway, with molecules showing varying degrees of success via targeting the ATP binding site of the kinase domain. Early attempts to inhibit DNA-PK kinase activity with molecules like Wortmannin and LY294002 (Fig. 9) resulted in potent radiosensitization. Wortmannin, a furanosteroid metabolite of the fungi Penicillium funiculosum, is a potent, non-specific and non-competitive inhibitor of PI3 kinases, which also exhibits equipotent activity for DNA-PK, ATM and polo-like kinases. Wortmannin inhibits DNA-PK irreversibly by covalent modification of Lys802, an amino acid residue in the active site of DNA-PK critical for the phosphate transfer reaction. LY294002, a morpholine derivative of the flavonoid, is also a non-specific, competitive inhibitor that binds reversibly to the kinase domain of DNA-PK. Rapid metabolic clearance, instability, in vivo toxicity and off-target effects of wortmannin and LY294002 made further preclinical and clinical evaluation impractical (Collis, DeWeese, Jeggo, & Parker, 2005; Davidson, Amrein, Panasci, & Aloyz, 2013). However, LY294002 has been extensively utilized by KuDOS Pharmaceuticals Ltd. (UK) and Ihmaid et al. as a lead compound for further modifications to improve specificity and efficacy to reduce off-target toxicities (Leahy et al., 2004; Clapham et al., 2011; Munck et al., 2012; Ihmaid, Al-Rawi, Bradley, Angove, & Robertson, 2012; Andrs et al., 2015).
Fig. 9.
Structures and activity of DNA-PK, PNKP and Ligase IV inhibitors.
NU7026 and NU7441 (KU-57788) were developed from the LY294002 scaffold. Both compounds display high selectivity for DNA-PK compared to PI3K (~55 and ~355-fold, respectively) and are inactive against both ATM and ATR (>100 μM). NU7026 showed potent radiosensitization activity and potentiation of cell death with various chemotherapeutic agents (Peddi et al., 2010; Willmore et al., 2004). Despite promising in vitro and in vivo results, pharmacokinetic studies revealed that NU7026 has a poor aqueous solubility and exhibits rapid clearance due to multiple oxidations and glucuronidation, predominantly at C-2 position of the morpholine ring (Nutley et al., 2005). NU7441 enhanced the cytotoxicity of IR and etoposide in SW620, LoVo, and V3-YAC colon cancer cells but not in DNA-PK deficient V3 cells, assuring that potentiation of DNA damage and cell death was primarily due to DNA-PK inhibition (Zhao et al., 2006). However, the limited aqueous solubility, poor oral bioavailability and marginal potentiation of radiation and chemotherapy sensitivity restricted further clinical evaluation of NU7441. KU-0060648 emerged by further SAR analysis of NU7441 as a highly potent cell-permeable dual DNA-PK and PI3K inhibitor with better oral bioavailability and pharmacokinetic profiles (Munck et al., 2012). Although NHEJ is considered to be the main pathway for repair of IR-induced DSBs, relatively little success has been observed with inhibitors targeting the main proteins in this pathway. However, future development of highly specific DNA-PK inhibitors with good ADME profiles will likely be achieved by developing novel chemical entities based on the recently reported X-ray crystal structure of DNA-PK (Sibanda et al., 2010). Three DNA-PK related inhibitors MSC2490484A, CC-122 and CC-115 (DNA-PK and mTOR dual inhibitor) are currently being investigated in phase I clinical trial either for solid tumors, non-Hodgkin lymphoma, multiple myeloma or hematologic malignancies (Goodwin & Knudsen, 2014).
While considerable effort has been placed on inhibition of the kinase active site, it has not escaped our notice that inhibition of the regulatory Ku subunits could also result in reduced DNA-PK activity and NHEJ. This is consistent with the existing data demonstrating that shRNA depletion of Ku70 or Ku80 showed cytotoxicity and radiosensitization in pancreatic cancer cells (Li et al., 2012b). Despite the crucial role of Ku early in the NHEJ pathway (DNA termini recognition and end bridging), no small molecule inhibitors have been published to date. However, our group has developed a class of compounds that abolish the Ku-DNA end binding activity in vitro. In vivo studies confirm decreased Ku foci formation in the presence of the inhibitors as well as decreased NHEJ mediated re-circularization of a plasmid molecule (unpublished data). Further development of Ku and DNA-PK inhibitors hold considerable potential to impact cancer therapy.
Inhibition of DNA-end processing
The processing step in NHEJ continues to be an active area of study as the list of nucleic acid enzymes involved in the pathway expands. However, there has not been a concerted effort to validate these proteins as potential targets. Radiation therapy is known to produce DSBs with various compound lesions at the termini that must be processed before repair can be completed. Targeting NHEJ processing enzymes in an effort to increase radiosensitivity seems a worthwhile endeavor. Artemis, the DNA-PK dependent endonuclease, has confirmed activity in the NHEJ pathway and interestingly, Artemis deficient cells show extreme radiosensitization (Rooney et al., 2003). However, no inhibitors have been identified, likely a result of the difficulty in purifying the proteins and the complex assay requirements (Pawelczak & Turchi, 2010). Interestingly, strides have been made to target human polynucleotide kinase/phosphatase (hPNKP), the enzyme responsible for phosphorylation of 5′-hydroxyl termini and dephosphorylation of 3′-phosphate termini, which are frequently altered at DNA termini from IR treatment and repair of which is required for NHEJ (Karimi-Busheri, Rasouli-Nia, lalunis-Turner, & Weinfeld, 2007). A12B4C3 (Fig. 9) was found to be a noncompetitive inhibitor that regulates the phosphatase activity of hPNKP by disrupting the secondary structure, resulting in radiosensitization in lung and breast cell culture models (Freschauf et al., 2009). While not a particularly potent inhibitor with no assessment of clinical efficacy, this work raises interesting possibilities for hPNKP as a target for developing useful radiosensitizer molecules.
DNA Ligase Inhibitors: Inhibition of DNA ligation
The ligation step of NHEJ is an attractive target for inhibition, as DNA Ligase IV has an essential function in the pathway. Early attempts to inhibit the ligase resulted in the development of compound L189 (Fig. 9), a molecule with poor specificity that showed equipotent inhibitory activity against Ligase I, III and IV (Chen et al., 2008). Recently the inhibitor SCR7 (an L189 derivative) was identified as a more specific inhibitor of NHEJ, potentially in a Ligase IV dependent manner, leading to the accumulation of DSBs and subsequent cytotoxicity (Srivastava et al., 2012). However, a higher concentration (IC50 = 20–150 μM) is required to observe a significant effect in radio- and chemo-sensitization assays, making this less likely to be therapeutically useful. Furthermore, SCR7 has been used at lower concentrations (1 μM) to increase HDR efficiencies (through direct inhibition of NHEJ) for CRISPR mediated genome engineering techniques, but cellular toxicity was observed with concentrations above 1 μM, suggesting that cell-dependent toxicity or off-target effects may be occurring with the inhibitor (Maruyama et al., 2015; Chu et al., 2015). Further optimization continues to be necessary to pursue a potential therapeutically relevant inhibitor of Ligase IV, which remain in the pre-clinical stage.
5: DNA Damage Response (DDR) Kinases: MRN, ATM, ATR and Chk Inhibitors
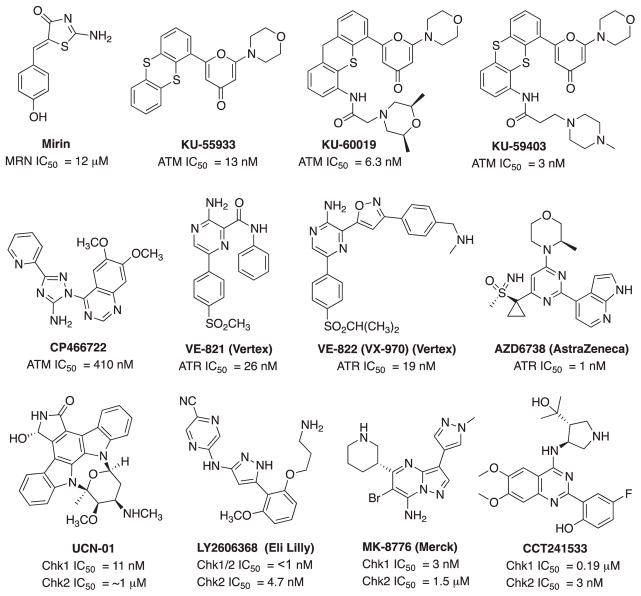
The DNA damage response is initiated by recognition of specific DNA damage and determines the cellular response to DNA damaging therapies. The initial response to DNA damage includes activation by phosphorylation of one of three PIKKs ATM, ATR or DNA-PKcs. While there is significant overlap between the pathways, the specific PIKK and downstream pathways activated are largely determined by the type of DNA damage, with DNA DSBs able to activate all three (Woods & Turchi, 2013). Additionally, the MRN complex (Mre11-Rad50-Nbs1) plays a role in detection of DNA DSBs through activation of ATM. A forward chemical genetic screen identified Mirin (Fig. 10), a molecule that inhibits MRN dependent activation of ATM and blocks the Mre11 exonuclease activity (Dupre et al., 2008). Expansion studies on this molecule resulted in a class of inhibitors that selectively block the differential nuclease activities of Mre11, resulting in a deeper understanding of the mechanisms of Mre11 (Shibata et al., 2014). This research revealed that inhibition of endonuclease activity pushes the cell to NHEJ rather than HDR, and blocking the exonuclease activity results in a repair defect. These studies demonstrate the potential impact, both mechanistically and therapeutically, of targeting the damage response pathway.
Fig. 10.
Structures and activity of MRN, ATM, ATR and Chk1/2 inhibitors.
Targeting of the early DDR kinases has focused largely on the three PIKKs. DNA-PK phosphorylation, while able to activate the DDR, particularly in ATM-deficient cell lines, largely results in activation of DSB repair through NHEJ and the impact of DNA-PK inhibition is discussed above. We will discuss in some detail small molecular inhibitors of ATM, ATR and the major downstream damage signaling protein of ATR, Chk1. While there has been some interest in targeting Chk2, the major downstream DDR effector of ATM, development has largely been hampered by a lack of specificity and limited combination cytotoxicity of Chk2 targeting compounds (Anderson et al., 2011; Duong et al., 2013).
ATM is activated following DSBs and plays a major role in the DDR to DSBs caused by IR. It impacts cell cycle regulation and cytotoxicity by phosphorylation of its major downstream factors, Chk2 and p53. Cells deficient in ATM are hypersensitive to radiation and radiomimetic drugs, making this an ideal target for radiosensitization (Li et al., 2001). Caffeine and wortmannin were two of the earliest compounds shown to inhibit ATM and resulted in increased sensitivity to IR and other chemotherapeutic agents. However, lack of specificity with caffeine inhibiting both ATM and ATR, and wortmannin inhibiting ATM, PI3K and DNA-PKcs, in addition to low potency and high in vivo toxicity has restricted further development of these compounds. KuDOS Pharmaceuticals (acquired by AstraZeneca, 2005) developed the first potent and selective ATM inhibitor, KU-55933, identified during screening of small molecule library based on the LY294002 (non-specific DNA-PK inhibitor) scaffold (Fig. 10). KU-55933 is a highly selective inhibitor over the related PIKKs (DNA-PK, PI3K/PI4K, ATR and mTOR; IC50 in the sub-micromolar range) and sensitizes tumor cells to ionizing radiation and radiomimetic drugs, including camptothecin, doxorubicin and etoposide. Importantly, patients without functional ATM expression showed no radiosensitization, confirming that the ATM kinase is specifically targeted by KU-55933 and related compounds (Hickson et al., 2004; Weber & Ryan, 2015). Further SAR analysis of KU-55933 to improve the pharmacokinetic properties led to the development of the more potent KU-60019. KU-60019 is 10-fold more effective than KU-55933 at blocking the radiation-induced DDR, highly radiosensitizes the human glioma cells and might be useful in adjuvant therapy for patients with mutant p53 brain cancers (Biddlestone-Thorpe et al., 2013). Furthermore, KU-60019 alone (without radiation) inhibits glioma cell migration, invasion and growth suggesting that ATM might be controlling growth and motility via pro-survival signaling pathways, such as AKT and MEK/ERK. Despite the impressive in vitro results of KU-60019, poor bioavailability limited utility for in vivo studies (Golding et al., 2009; Biddlestone-Thorpe et al., 2013). Another class of compound, CP466722, was identified as a highly selective and rapidly reversible ATM inhibitor by Rainey et al. in collaboration with Pfizer. Although CP466722 showed a distinct radiosensitizing effect, no further in vivo data of this compound has been reported. The most recent ATM inhibitor, KU-59403, showed improved potency and significantly enhanced in vitro cytotoxicity as a sensitizer to radiation as well as to camptothecin, etoposide and doxorubicin. Colon cancer xenograft models revealed increased cytotoxicity of camptothecin and irinotecan in combination with KU-59403 as well as improved pharmacokinetics and bioavailability as compared to other ATM inhibitors (Batey et al., 2013). These recent developments are encouraging and support the future clinical development of ATM inhibitors; however, at this time no clinical trials are underway.
ATR is a DDR kinase activated early after treatment with a number of DNA damaging agents, including chemotherapeutic drugs, ultraviolet light and ionizing radiation. This signaling molecule is primarily activated by SSBs and responds to DNA replication stress and is therefore active in the S and G2 phases of the cell cycle. Although the role of ATR and Chk1 in chemotherapeutic and UV-induced DNA damage is better studied, a number of recent publications have investigated their roles in toxicity to single and double-strand breaks, such as those caused by ionizing radiation. Pharmacologic inhibition of both ATR and its downstream effector, Chk1, has demonstrated differential effects of radiosensitization in certain cancer types and mutational profiles. For instance, treatment with the ATR inhibitors VE-821 and VE-822 in vitro, results in radiosensitization to ionizing radiation in p53 mutant pancreatic cells (Prevo et al., 2012; Fokas et al., 2014). In vitro studies support a role for ATR in promoting HDR, and radiosensitization to ATR and Chk1 inhibitors is felt to be due to two mechanisms. First, inhibition of ATR and Chk1 leads to cell cycle arrest and increases replication stress, leading to an increase in DNA DSBs at the sites of DNA damage, such as those caused by gemcitabine. Secondly, ATR and Chk1 inhibition leads to a decrease in HDR, which is particularly cytotoxic in combination with ionizing radiation, in those cells already deficient in ATM or p53, both of which are commonly mutated in solid-organ tumors. (Wang, Wang, Powell, Iliakis, & Wang, 2004; Fokas et al., 2014; Prevo et al., 2012). VE-821, developed by Vertex Pharmaceuticals is a potent, selective ATR inhibitor that also exhibits increased single-agent toxicity against radiotherapy resistant hypoxic tumor cells (Fig. 10). VE-822 (VX-970), an optimized analog of VE-821 with improved potency, solubility, safety and pharmacokinetic properties is currently under phase I clinical trials for the treatment of advanced solid tumor and relapsed or refractory small cell lung cancer in combination with several DNA damaging agents (Weber & Ryan, 2015). Another selective ATR inhibitor in phase I clinical trial is AZD6738 (an analog of AZ20) to access the safety, tolerability and dosing profile as a monotherapy and in combination with chemoradiation therapy in patients with solid tumors. AZD6738 induces ATM kinase-dependent DNA damage signaling and remarkably enhances the cytotoxicity of cisplatin in ATM-deficient lung cancer xenograft models. AZD6738 is an orally active and bioavailable single agent anti-tumor agent which showed a constant increase in γH2AX pan-nuclear staining in tumor tissue but only a temporary increase in bone marrow and gut tissues suggesting a favorable therapeutic index for ATR inhibitors (Foote et al., 2013; Vendetti et al., 2015).
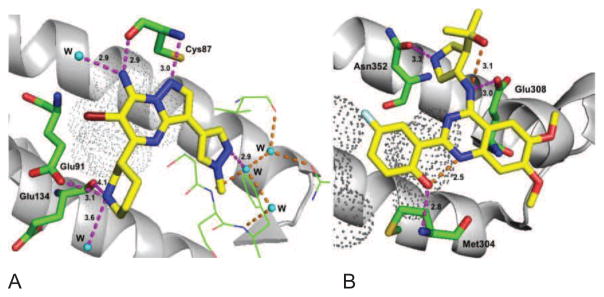
The checkpoint kinase 1 (Chk1) plays a critical role in DNA replication, the regulation of cell cycle progression and survival following the induction of DSBs and acts as one of the guardians to maintain genomic integrity of cells. Chk1 inhibitors are used in vitro in combination with chemotherapeutic and radiation treatment and a number of these are currently being investigated in clinical trials. Early compounds included UCN-01, a derivative of staurosporine, a serine/threonine kinase inhibitor which targets a number of cyclin-dependent kinases that regulate G1 checkpoint control (Fig. 10). Although well tolerated, a further clinical development was terminated in the first stage of the study due to lack of response, pharmacokinetic and toxicity issues (Li et al., 2012a). AZD7762, a Chk1/2 inhibitor revealed a promising pre-clinical profile and has been evaluated in two separate phase I clinical trials but further studies were halted due to cardiac toxicity. A clinical trial in combination with gemcitabine was recently completed which excluded patients with heart failure or significant cardiac disease (Seto et al., 2013). LY2603618 is a highly potent and selective Chk1 inhibitor, developed by Array biopharma in collaboration with ICOS (ICOS acquired by Eli Lilly in 2007). LY2603618 in combination with the chemotherapeutic drugs pemetrexed and cisplatin, showed increased antitumor effects in non-small cell lung culture models in vitro and in mouse xenograft models. Clinical trials of LY2603618 in combination with pemetrexed or cisplatin + pemetrexed in solid organ tumors showed favorable pharmacologic profiles, with a number of patients achieving stable disease or a partial response to therapy. However cardiac toxicity, myelosuppression and low tolerance were observed in phase I trials for the treatment of solid tumors in combination with pemetrexed (Weiss et al., 2013; Calvo et al., 2014; Doi et al., 2015) and Lilly has stopped further development of LY2603618. A derivative, LY2606368, is an ATP-competitive inhibitor with sub-nanomolar IC50 in vitro and in vivo data revealed DNA damage response modulation and xenograft tumor growth inhibition as a single agent and in combination (Lainchbury et al., 2012). This agent is currently under investigation in a number of phase I trials is also being evaluated in a phase II trial for the treatment of certain ovarian, breast and prostate cancers (King et al., 2015). However, LY2606368 is only administered intravenously, likely a result of its poor oral bioavailability. The highly selective Chk1 (over Chk2) inhibitor, MK-8776 (SCH-900776), has been studied in vitro in leukemias, NSCLC, ovarian and pancreatic cells and results in enhanced sensitivity of chemotherapeutic drugs and to chemoradiation therapy. MK-8776 has been well-tolerated in phase I trials in the treatment of solid tumors and leukemias, with a phase II study currently underway for the treatment of acute myeloid leukemia (Grabauskiene et al., 2013; Engelke et al., 2013; Daud et al., 2015). The role of Chk2 inhibitors as anticancer agents remains controversial because the highly selective Chk2 inhibitor, VRX0466617, has failed to potentiate cisplatin and doxorubicin cytotoxic effects in tumor cells (Carlessi, Buscemi, Fontanella, & Delia, 2010; Carlessi et al., 2007). Other selective agents including PV1019 and CCT241533 were found to potentiate the cytotoxicity of genotoxic agents and PARP inhibitors, respectively. Both compounds exhibited mild antitumor activity and radioprotective effects in mouse thymocytes (Garrett & Collins, 2011). GDC-0425 and GDC-0575 developed by Array Biopharma-Genentech as highly selective and orally bioavailable Chk1 inhibitors are currently under phase I clinical trials in combination with gemcitabine in patients with refractory solid tumors or lymphoma. Specific molecular interactions of selective Chk1 (MK-8776) and Chk2 (CCT241533) inhibitors are outlined in the Fig. 11 and the differences in the binding region of Chk1 in comparison to Chk2 can be exploited for further selectivity (Labroli et al., 2011; Caldwell et al., 2011).
Fig. 11.
Molecular interaction of Chk inhibitors A) Molecular interactions of MK-8776 with hChk1: MK-8776 is docked using predecessor X-ray crystal structure (PDB code: 3OT3) to delineate the key interactions. The N-1 moiety and exocyclic NH2 of the pyrazolo[1,5-a]pyrimidine binds to the Cys87 and water molecule in the hinge region, pyrazole moiety interacts with an array of ordered water molecules in the Chk1 specificity domain, piperidine amine makes several key interactions with acidic residues (Glu91 and Glu134) and a conserved water molecule and pyrazolo[1,5-a]pyrimidine moiety is surrounded by hydrophobic residues Leu136 and Leu137 (black dots).; B) Molecular interactions of CCT241533 with hChk2 (PDB code: 2XBJ): A planar and pseudotetracyclic structure formed due to intramolecular hydrogen bonding in between the phenolic hydroxyl and the quinazoline N-1, the phenolic hydroxyl binds to Met304 in the hinge region, quinazoline exocyclic amine makes contact with Glu308, pyrrolidine amine forms a charge-assisted hydrogen interaction with Asn352, two methoxy substituents occupying the solvent exposed region and quinazoline moiety is surrounded by hydrophobic residues Val234, Leu354, Ala247, Leu301 and Leu303 (some of them shown in black dots). Intermolecular and Intramolecular hydrogen bonds showed in magenta and orange dotted lines, respectively and distances indicated in Å.
Clinical studies of the highly potent Chk1/2 inhibitors XL844 (Exelixis) and PF-00477736 (Pfizer) have been prematurely discontinued without stating specific reason (McNeely, Beckmann, & Bence Lin, 2014). In addition, although outside the scope of this article, a number of agents not directly involved in DNA damage repair or DDR have been demonstrated to sensitize to radiation therapy indirectly by inhibition of HDR. Many of these are currently being evaluated in clinical trials, including inhibitors of histone deacetylase (CHR-3996, CHF-2845, 4SC-202, JNJ-26481585, ITF2357 and AR-42) and HSP90 (including IPI-504, AUY922, STA-9090, AT13387, SNX-5422 and 17-AAG).
The link between DNA repair and ionizing radiation has been firmly established and work to date suggests that targeting the DNA DSB repair pathways has considerable potential for increasing radio- and chemosensitization in the clinic. Clearly, the search for potent and specific inhibitors to target the cellular response to DNA DSBs is still required. It is encouraging that studies continue to evolve to identify new radiosensitizer agents like mibefradil dihydrochloride that was recently identified as a DSB repair inhibitor and a radiosensitizer (Goglia et al., 2015). Additionally, the synthetic lethality approach to target inhibition of two DNA repair pathways simultaneously is expanding. This is evidenced by the molecule YU238259 that shows synergy to ionizing radiation and potentiates chemotoxicity through inhibition of HDR and shows increased synergy in a BRCA-2 null background (Stachelek et al., 2015). However, in spite of strong pre-clinical data showing highly specific and potent inhibitors, relatively few have been further assessed in clinical trials, many of which have been closed due to toxicities or lack of clinical efficacy.
6: Targeting homology directed repair and Rad51 in cancer
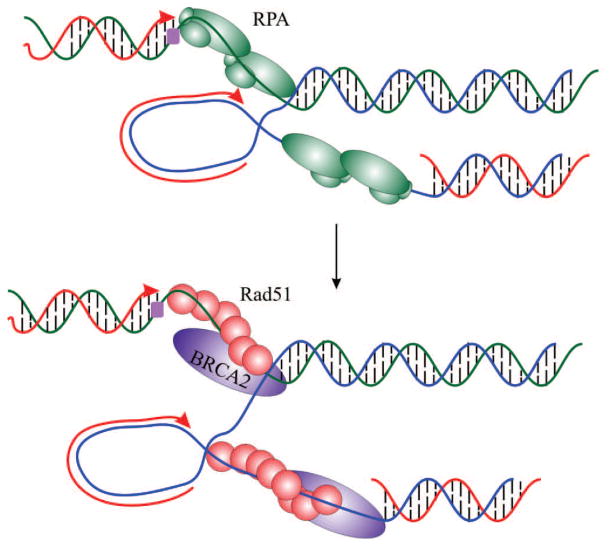
HDR is a major mechanism to ensure the accurate repair of a variety of DNA lesions including double strand breaks, single strand DNA gaps and intra- and inter-strand crosslinks (San Filippo, Sung, & Klein, 2008). In addition HDR is critical for reestablishing replication forks at the sites of damage and proper chromosome segregation during meiosis. The canonical HDR pathway requires numerous factors and many of their biochemical functions have been elucidated. Central to HDR is Rad51, the eukaryotic ortholog to E.coli RecA, and its function as a DNA strand exchange protein. The role of Rad51 involves replacing RPA bound to ssDNA and the formation of a RAD51-ssDNA filament termed a presynaptic filament (Fig. 12). The binding of Rad51 stretches the DNA facilitating a fast and efficient homology search. The invasion between the Rad51-ssDNA and homologous duplex DNA leads to a heteroduplex DNA and formation of a Holliday junction. Dissociation of Rad51 accompanies the synthesis step and ultimately allows the final ligation.
Fig. 12.
Rad51 catalyzed homologous recombination repair. A leading strand replication block (purple box) stalls fork progression leading to the generation of single stranded DNA and binding of RPA (green trimer). Rad51 (red) replacement of RPA and association of BRCA2 (magenta) precedes homology driven recombination to by-pass the lesion and fork restart.
Consistent with the central role in HDR there is considerable evidence that Rad51 plays a significant role in cancer development and response to therapy. Rad51 binding to ssDNA is facilitated by a series of proteins including the Rad51paralogs Rad51C and Rad51D. Germline mutations in either Rad51C or D were reported to increase a woman’s susceptibility for the development of epithelial ovarian cancer, suggesting that accurate regulation of RAD51 activity is critical to maintaining genetic stability (Song et al., 2015). While Rad51 mutations have not been identified in cancer, increased expression of Rad51 has been reported in nearly every cancer assessed (Raderschall et al., 2002; Klein, 2008). Elevated Rad51 mRNA expression by at least 2 fold as compared to normal tissue samples has been described in invasive ductal carcinoma of the breast, non-small cell lung cancer, bladder cancer, glioblastoma multiforme, sarcomas, prostate and pancreatic cancers (Oncomine database search). In addition, numerous cancer cell lines display increased Rad51 foci independent of exogenous DNA damage further supporting evidence of abnormalities of Rad51 regulation in cancer (Mathews et al., 2011; Raderschall et al., 2002). The increase in expression and foci formation could be the result of the increased DNA replication stress experienced by the rapidly dividing cancer cells. Rad51 expression is associated with worse prognosis in multiple malignancies (Mitra et al., 2009; Qiao et al., 2005). Colon cancer patients with strong Rad51 tumor expression have a median survival of 11 months as compared to 76 months in patients with weak expression (Tennstedt et al., 2013). Of note, increased Rad51 expression in cancer is not a consequence of gene amplification but rather results from increased transcription of the Rad51 gene.
The negative correlation between Rad51 expression and prognosis is paradoxical but clearly suggests that “more” is not necessarily “better”. One could envision increased expression would correlate with increased repair and maintenance of genetic stability. Current evidence however does not support these assumptions and suggests tight regulation of DNA repair proteins including Rad51 is needed for HDR to maintain its role in preserving genomic integrity with either too much or too little protein and activity being detrimental. Recent in vivo data incriminates Rad51 in promoting the formation of RNA-DNA hybrids which represent a potent source of altering genome structure and inducing chromosome instability (Wahba, Gore, & Koshland, 2013). In addition, increased Rad51 in multiple myeloma and esophageal adenocarcinoma was associated with elevated HR activity while knockdown of Rad51 prevented the acquisition of genomic changes (Shammas et al., 2009; Pal et al., 2011). Rad51 inhibition was shown to reduce breast cancer migration suggesting it contributes to metastases as well (Wiegmans et al., 2014). These data were quite surprising demonstrating that a protein that has been long recognized as a DNA repair protein can in fact have genotoxic effects and be implicated in both DNA damage and promotion of genomic alterations.
Rad51 has also been shown to contribute to a therapy resistant phenotype in cancer specifically with therapeutics that induce DNA damage (Takenaka et al., 2007). Fewer Rad51 foci in breast cancer patients correlated with better responses to anthracycline based chemotherapy (Graeser et al., 2010). Lower Rad51 expression or Rad51 inhibition have been shown to sensitize cancer cells to cisplatin, doxorubicin and ionizing radiation (Hannay et al., 2007; Liu et al., 2011). In addition to its role in HDR, Rad51 seems to play an independent role in the repair of DNA inter-strand crosslinks and a separation of function mutation has been identified. Cells expressing a Rad51 T131P mutant were found to be HDR proficient but inter-strand crosslink repair defective. This suggests that a unique Rad51 activity or interaction is required for FA-dependent interstrand crosslink repair that is not required for canonical HDR (Wang et al., 2015).
Targeting Rad51: prior failure and future successes
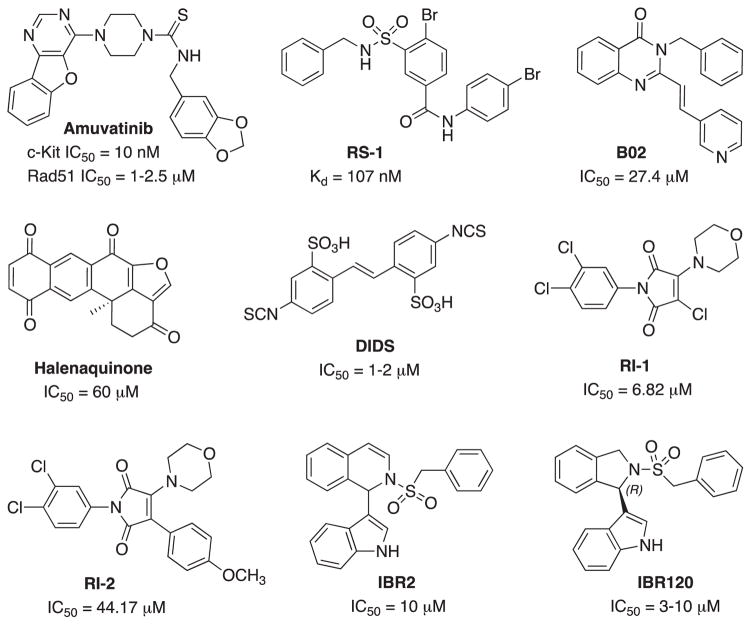
The role of Rad51 in the repair and tolerance of chemotherapy and radiation induced DNA damage provides an excellent rationale for Rad51 as a therapeutic target in cancer treatment. This is further supported by the increased expression of Rad51 in many cancers and the reliance of higher Rad51 activity to combat the replication stress observed in cancers. Inhibition of Rad51 in this context could provide a therapeutic window where inhibition of Rad51 in non-cancerous cells would have less influence and therefore reduced toxicity. Earlier attempts at targeting Rad51 were unsuccessful mainly due to the fact the drugs used were not specific Rad51 inhibitors. For example, amuvatinib (MP-470) was labeled as a Rad51 inhibitor when in fact it was a multi-targeted tyrosine kinase inhibitor that happened to reduce Rad51 expression in tumor cell lines by an unknown mechanism (Zhao, Luoto, Meng, & Bristow, 2011). The clinical development of amuvatinib was halted when phase 2 data indicated a failure to achieve its primary endpoint. The lack of efficacy of amuvatinib was a reflection of its unclear mechanism of action rather than evidence against targeting Rad51.
A number of more specific Rad51 inhibitors are currently in the early phases of drug development; however, further optimization is likely to be needed prior to their progress into clinical trials. Two different strategies are currently being employed in targeting Rad51 with promising pre-clinical results. The first strategy exploits the overexpression of Rad51 in cancer by further stimulating the formation of toxic Rad51 complexes on undamaged chromatin using chemical agents such as RS-1 (Fig. 13). This strategy could potentially prove selective to cancer cells with increased baseline Rad51 expression thereby sparing normal cells. RS-1 acts as an allosteric effector of active hRad51 filament formation on ssDNA that stimulates DNA binding and recombination activities of hRad51 by locking its active conformation without affecting active site ATP hydrolysis. RS-1 has so far been demonstrated to have single agent activity in tumor cell lines that display increased Rad51 expression and its activity is dependent on Rad51 and the Rad54B/Rad54L translocase (Mason et al., 2015). In addition, RS-1 has in vivo anti-cancer activity in a xenograft animal model (Mason et al., 2014). The second strategy for targeting Rad51 is inhibition of DNA strand exchange activity via disrupting Rad51’s ability to bind ssDNA. B02 is one of the compounds identified that specifically inhibit human Rad51 binding to DNA. Targeting Rad51 using B02 has been shown to inhibit HDR and increase cancer sensitivity to DNA damaging agents including ionizing radiation, cisplatin, mitomycin C, doxorubicin, and etoposide (Alagpulinsa, Ayyadevara, & Shmookler Reis, 2014). Halenaquinone inhibits Rad51-dsDNA binding specifically, but not Rad51-ssDNA binding (Takaku et al., 2011) (Fig. 13). In human cells, halenaquinone significantly decreased the IR induced Rad51 foci formation probably by inhibiting the ternary complex formation which promotes the DNA homologous pairing step during the HR process. DIDS is another potent competitive Rad51 inhibitor that directly binds to Rad51 and disassembles Rad51 from ssDNA (Ishida et al., 2009). However, further effect of DIDS on the DNA repair and HR was not evaluated due to high human cell toxicity. Chloromaleimide derivative RI-1 irreversibly binds to the thiol group on the C319 residue of human Rad51. In order to overcome off-target covalent interactions and improve compound stability in the cells, RI-2 was developed. RI-2 reversibly binds to the same site on Rad51 as does RI-1 but with a 6-fold decrease in potency.
Fig. 13.
Structures and activity of Rad51 stimulator (RS-1) and inhibitors.
An alternative to inhibition of the protein-DNA interaction site is to block the protein-protein interaction sites (Budke et al., 2012; Budke et al., 2013). The inhibitor IBR2 disrupts the Rad51-BRC interaction, Rad51 multimerization, and enhances proteasome-mediated RAD51 protein degradation. The cellular effect of IBR2 included an increase in the sensitivity of MCF7 breast cancer cells to IR (Zhu et al., 2013). Further SAR analysis of IBR2 generated the stereo selective inhibitor IBR20 (Fig. 13). This compound has been demonstrated to disrupt Rad51 multimer formation, inhibit HDR and display cytotoxic activity in a variety of cancer cell lines with low micromolar IC50s (Zhu et al., 2015). While clearly in the developmental stages, each of the strategies currently under investigation could be an effective addition to cancer therapy.
7: BER
BER represents an attractive target for cancer therapy as many of the proteins involved in the BER have been shown to be dysregulated in a wide variety of cancers (Wallace, 2014). The relatively small number of proteins required and the convergence of the pathway after the creation of the AP-site represents a limited number of proteins to target. Similar to other excision repair pathways, BER is initiated by a recognition event where the modified base is recognized by a DNA glycosylase. There is a variety of different glycosylases that recognize the array of possible base modifications. Following recognition and hydrolysis of the glycosidic bond, the resulting abasic site is recognized by APE1 and a nick made in the phosphodiester bond 5′ of the abasic site resulting in 3′ OH and 5′ deoxyribose-phosphate (dRP) termini. Herein, the pathway can diverge and repair can be completed via long-patch or short patch repair. Long-patch BER (LP-BER) occurs by the excision of at least two nucleotides and DNA synthesis catalyzed by pol β or by pol δ/ε. In long patch repair, DNA pol β performs displacement synthesis and the resulting flap is cut by FEN1 prior to ligation. Alternatively, short patch BER (SP-BER) occurs by the excision of one nucleotide. In short patch repair, the 5′ lyase activity of DNA pol β cleaves the 5′ dRP and adds a single base prior to ligation. However, if the dRP cannot be effectively removed the BER pathway proceeds via the long-patch mechanism. The reconstitution of LP-BER has demonstrated an absolute requirement for the endonuclease activity of FEN1 but for SP-BER it is not required at all. Ligation is accomplished by DNA ligase I or DNA ligase III in complex with XRCC1 (Svilar, Goellner, Almeida, & Sobol, 2010). Beyond the role of PARP in BER, efforts to target BER have focused largely on APE1, the human AP- endonuclease and DNA polymerase β.
APE1 inhibitors
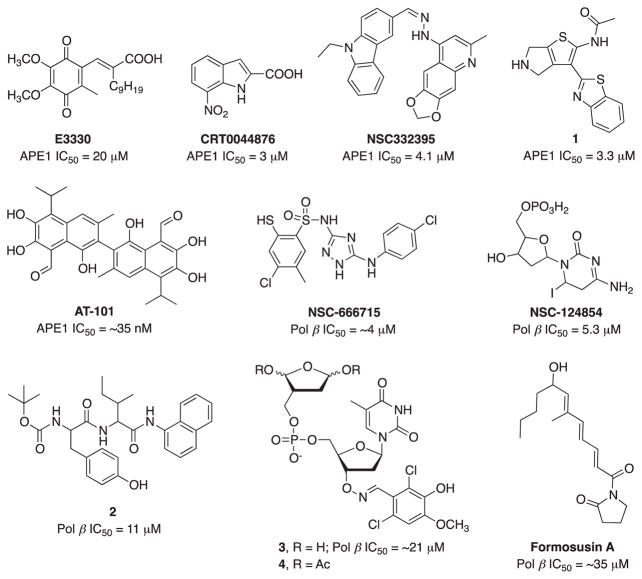
Overexpression or altered level of AP-endonuclease, APE1 in several cancers has been shown to increase the resistance of tumor cells to treatment with various chemotherapeutic agents emphasizing APE1 as an important target for cancer therapy (Kaur, Cholia, Mantha, & Kumar, 2014). APE1 is arguably the most studied, and a large number of APE1 inhibitors have been reported and expertly reviewed in 2012 (Al-Safi, Odde, Shabaik, & Neamati, 2012). E3330 and its analogs hold clinical therapeutic potential as a specific inhibitor of APE1 redox activity but do not affect APE1 DNA repair activity (Kelley, Georgiadis, & Fishel, 2012) (Fig. 14). While the redox function of APE1 is also an active area of investigation as a therapeutic target, we will focus mainly on the repair-specific agents identified since 2012. CRT0044876 is the first potent and selective APE1 inhibitor and enhanced the cytotoxicity of several DNA damaging agents in vitro but failed to have further success due to poor cellular activity and toxicity issues (Madhusudan et al., 2005). To identify novel agents capable of inhibiting APE1, multiple approaches have been employed. An in silico screening was used to focus on the endonuclease region of APE1 and resulted in the discovery of a series of APE1 inhibitors with varying potency (Mohammed et al., 2011). Similarly, a HTS approach was also used and resulted in the identification of a large number of hits that was subsequently culled to identify cellular activity (Abbotts et al., 2014; Dorjsuren et al., 2012). Srinivasan et. al found several unique nanomolar to sub-micromolar APE1 inhibitors based upon molecular modeling and virtual screening strategy (Srinivasan, et. al. 2012). NSC332395 is the most active compound of the quinoline series and potentiated the toxicity of a DNA damaging agent, though in vivo activity remains to be determined. A systematic medicinal chemistry approach driven by the NIH generated N-(3-(benzo[d]thiazol-2-yl)-6-isopropyl-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)acetamide and N-(3-(benzo[d]thiazol-2-yl)-5,6-dihydro-4H-thieno[2,3-c]pyrrol-2-yl)acetamide (1) as potent APE1 inhibitors with favorable in vitro ADME profile and also showed good plasma and brain exposure in mice (Fig. 14). Both compounds also potentiated the cytotoxicity of methylmethane sulfonate (MMS) and temozolomide (TMZ) (Rai et al., 2012).
Fig. 14.
Structures and activity of BER inhibitors.
Furthermore, gossypol is a natural product BH3 mimetic that was shown to directly interact with the BH domains in the Bcl-2 family of proteins (Kitada et al., 2003). APE1 also contains a BH domain and it was subsequently demonstrated that gossypol could bind directly to APE1 and inhibit both repair and redox functions (Qian et al., 2014). The inhibitor is potent with an IC50 of ~35 nM though a high degree of specificity is unlikely. The AT-101 isomer of gossypol was subsequently found to potentiate the cytotoxicity of cisplatin and etoposide in lung cancer models (Ren et al., 2015). The ability to attribute any clinical effect of APE inhibition is questionable considering long history of this agent and the lack of specificity.
POL β inhibitors
Expanding beyond APE1, numerous recent reports have identified putative inhibitors of DNA pol β (Barakat, Gajewski, & Tuszynski, 2012). Pol β plays a significant role in chemotherapeutic agent resistance, because its expression and altered level reduces the efficacy of DNA damaging drugs. An in silico screening identified NSC-666715 as a putative pol β inhibitor and subsequent follow up work demonstrated direct inhibition of pol β (Jaiswal et al., 2009; Jaiswal et al., 2015) (Fig. 14). Inhibition of both long patch and short patch BER was demonstrated but NSC-666715 did not inhibit other proteins in the BER pathways suggesting that the effect of BER was exclusively through pol β However, there is no specificity analyses reported against other DNA polymerases. The demonstration that NSC-666715 sensitized cells to TMZ is also consistent with inhibition of BER and polβ. A similar approach was utilized to identify the specific pol β inhibitor NSC-124854. NSC-124854 effectively increases the sensitivity of TMZ to colon cancer cells both in vitro and in vivo models (Jaiswal et al., 2011) (Fig. 14). A series of peptides based on TT-232, a somatostatin structural analogue were assessed as pol β inhibitors. Peptide 2 was identified to be the most effective and displayed minimal inhibition against a panel of mammalian DNA polymerases (Kuriyama, Miyazaki, Tsuda, Yoshida, & Mizushina, 2013). Modest activity (IC50 ~11 μM) was observed in vitro however, 200 μM concentration was used in many experiments suggesting that cellular effects observed including synergy with MMS may be due to off-target effect.
Very recent studies have identified derivative of the irreversible pol β inhibitor 2-phosphato-1,4-dioxobutane (DOB) (Arian et al., 2014). Compound 3 and pro-inhibitor 4 exhibits irreversible inactivation of DNA pol β inhibiting both lyase and displacement synthesis activity. These data suggests that both long patch and short patch BER would be targeted by these agents. Finally, a focus on natural product and derivatives has identified formosusin A [(6Z,8E,10E)-variotin] as a pol β inhibitor where three closely related isomers displayed no inhibitory activity (Mizushina et al., 2014). Importantly, formosusin A was shown to be specific for pol β and despite a relatively weak affinity (IC50 = ~35 μM), displayed no detectable inhibitory activity against a panel of other DNA polymerases. Cellular activity was not reported and clearly SAR analysis will be needed to increase potency and efficacy.
8: Conclusions and future directions
The 2015 Nobel Prize in Chemistry was awarded to three DNA repair researchers Paul Modrich, Aziz Sancar and Thomas Lindahl, who detailed the molecular mechanisms of MMR, NER and BER, respectively. The advances in our understanding of these pathways have been instrumental in developing novel agents to block or, in some cases, enhance repair activity. The cadre of proteins and enzymes that respond to and repair DNA damage holds considerable potential to impact human health. From re-establishing genomic integrity to identifying useful synthetic lethal interactions to enhance the sensitivity to widely prescribed chemotherapeutics, the impact of DNA repair cannot be underscored. The complexity of the cellular response to DNA damage however must be considered at every stage of development, from initial target identification to design of clinical trials and deployment in the clinic. Ignoring the biology behind the development holds dire consequence and can result in significant delays and abandoning potentially useful agents. The high cost and extended times associated with drug development necessitates a detailed understanding of the biology behind the drug development to allow effective pre-clinical and clinical investigations on drug combinations which capitalize on well-defined functions in DNA damage repair and response. We are poised for a rapid expansion of DNA repair targeted agents that move from the lab to the clinic. These will have the potential to treat a variety of conditions and capitalize on the numerous discoveries and ultimately will positively impact on human health.
Acknowledgments
Research in the Turchi lab is supported by NIH grants R01-CA180710 and R41-CA162648 and the Tom and Julie Wood Family Foundation. The authors recognize the vast contributions to the DNA damage and repair pathways by various worldwide groups, and we regret not being able to cite all relevant publications due to limitation and scope of the review.
Abbreviations
- NER
nucleotide excision repair
- MMR
DNA mismatch repair
- BER
base excision repair
- HDR
homology directed repair
- FA
Fanconi Anemia
- HR
homologous recombination
- DDR
DNA Damage Response
- NHEJ
non-homologous end joining
- SSB
single strand breaks
- pADPr
poly(ADP-ribose)
- PARP
Poly (ADP-ribose) polymerase
- NAD+
nicotinamide adenine dinucleotide
- PARPi
PARP inhibitors
- FDA
Food and Drug administration
- MDS
Myelodysplastic syndrome
- AML
acute myeloid leukemia
- GG-NER
global genomic NER
- TC-NER
transcription–coupled NER
- XP
xeroderma pigmentosum
- ERCC
Excision repair cross-complementing
- Cisplatin
Cis-diamminedichloroplatinum (II)
- UV
Ultra violet
- RPA
Replication Protein A
- OB
Oligonucleotide/oligosaccharide binding
- HTS
High throughput screen
- FPA
Fumaropimaric acid
- IR
ionizing radiation
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- PI3K
phosphatidylinositol-3-kinase-like
- SAR
structure activity relationship
- ATM
Ataxia telangiectasia mutated kinase
- ATR
ATM-Rad3 related kinase
- ATRIP
ATR interacting protein
- hPNKP
human polynucleotide kinase/phosphatase
- Chk
Checkpoint kinase
- Mre11-Rad50-Nbs1
MRN complex
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbotts R, Jewell R, Nsengimana J, Maloney DJ, Simeonov A, Seedhouse C, et al. Targeting human apurinic/apyrimidinic endonuclease 1 (APE1) in phosphatase and tensin homolog (PTEN) deficient melanoma cells for personalized therapy. Oncotarget. 2014;5:3273–3286. doi: 10.18632/oncotarget.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Safi RI, Odde S, Shabaik Y, Neamati N. Small-molecule inhibitors of APE1 DNA repair function: an overview. Curr Mol Pharmacol. 2012;5:14–35. [PubMed] [Google Scholar]
- Alagpulinsa DA, Ayyadevara S, Shmookler Reis RJ. A Small-Molecule Inhibitor of RAD51 Reduces Homologous Recombination and Sensitizes Multiple Myeloma Cells to Doxorubicin. Front Oncol. 2014;4:289. doi: 10.3389/fonc.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson VE, Walton MI, Eve PD, Boxall KJ, Antoni L, Caldwell JJ, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Research. 2011;71:463–472. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci US A. 2014;111:10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews BJ, Turchi JJ. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Molecular Cancer Therapeutics. 2004;3:385–391. [PubMed] [Google Scholar]
- Andrs M, Korabecny J, Jun D, Hodny Z, Bartek J, Kuca K. Phosphatidylinositol 3-Kinase (PI3K) and phosphatidylinositol 3-kinase-related kinase (PIKK) inhibitors: importance of the morpholine ring. Journal of Medicinal Chemistry. 2015;58:41–71. doi: 10.1021/jm501026z. [DOI] [PubMed] [Google Scholar]
- Antolin AA, Mestres J. Linking off-target kinase pharmacology to the differential cellular effects observed among PARP inhibitors. Oncotarget. 2014;5:3023–3028. doi: 10.18632/oncotarget.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arian D, Hedayati M, Zhou H, Bilis Z, Chen K, DeWeese TL, et al. Irreversible inhibition of DNA polymerase beta by small-molecule mimics of a DNA lesion. J Am Chem Soc. 2014;136:3176–3183. doi: 10.1021/ja411733s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Kothandapani A, Tillison K, Kalman-Maltese V, Patrick SM. Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair (Amst) 2010;9:745–753. doi: 10.1016/j.dnarep.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AstraZeneca. Lynparza (olaparib) Capsules, Prescribing Information. 2015. 10-4-2015. Ref Type: Online Source. [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Barakat KH, Gajewski MM, Tuszynski JA. DNA polymerase beta (pol beta) inhibitors: a comprehensive overview. Drug Discov Today. 2012;17:913–920. doi: 10.1016/j.drudis.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Barakat KH, Jordheim LP, Perez-Pineiro R, Wishart D, Dumontet C, Tuszynski JA. Virtual screening and biological evaluation of inhibitors targeting the XPA-ERCC1 interaction. PLoS One. 2012;7:e51329. doi: 10.1371/journal.pone.0051329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey MA, Zhao Y, Kyle S, Richardson C, Slade A, Martin NM, et al. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol Cancer Ther. 2013;12:959–967. doi: 10.1158/1535-7163.MCT-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- Bhagwat NR, Roginskaya VY, Acquafondata MB, Dhir R, Wood RD, Niedernhofer LJ. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer Research. 2009;69:6831–6838. doi: 10.1158/0008-5472.CAN-09-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlestone-Thorpe L, Sajjad M, Rosenberg E, Beckta JM, Valerie NC, Tokarz M, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clinical Cancer Research. 2013;19:3189–3200. doi: 10.1158/1078-0432.CCR-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixel K, Hays JL. Olaparib in the management of ovarian cancer. Pharmgenomics Pers Med. 2015;8:127–135. doi: 10.2147/PGPM.S62809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckman HJ, Trego KS, Turchi JJ. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol Cancer Res. 2005;3:277–285. doi: 10.1158/1541-7786.MCR-04-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, et al. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. Journal of Medicinal Chemistry. 2013;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B, Logan HL, Kalin JH, Zelivianskaia AS, Cameron MW, Miller LL, et al. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Research. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell JJ, Welsh EJ, Matijssen C, Anderson VE, Antoni L, Boxall K, et al. Structure-based design of potent and selective 2-(quinazolin-2-yl)phenol inhibitors of checkpoint kinase 2. Journal of Medicinal Chemistry. 2011;54:580–590. doi: 10.1021/jm101150b. [DOI] [PubMed] [Google Scholar]
- Calvo E, Chen VJ, Marshall M, Ohnmacht U, Hynes SM, Kumm E, et al. Preclinical analyses and phase I evaluation of LY2603618 administered in combination with pemetrexed and cisplatin in patients with advanced cancer. Invest New Drugs. 2014;32:955–968. doi: 10.1007/s10637-014-0114-5. [DOI] [PubMed] [Google Scholar]
- Cardnell RJ, Feng Y, Diao L, Fan YH, Masrorpour F, Wang J, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clinical Cancer Research. 2013;19:6322–6328. doi: 10.1158/1078-0432.CCR-13-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlessi L, Buscemi G, Fontanella E, Delia D. A protein phosphatase feedback mechanism regulates the basal phosphorylation of Chk2 kinase in the absence of DNA damage. Biochim Biophys Acta. 2010;1803:1213–1223. doi: 10.1016/j.bbamcr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6:935–944. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chapman TM, Gillen KJ, Wallace C, Lee MT, Bakrania P, Khurana P, et al. Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg Med Chem Lett. 2015;25:4097–4103. doi: 10.1016/j.bmcl.2015.08.031. [DOI] [PubMed] [Google Scholar]
- Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. Journal of Clinical Oncology. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Research. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi A. Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Clapham KM, Bardos J, Finlay MR, Golding BT, Griffen EJ, Griffin RJ, et al. DNA-dependent protein kinase (DNA-PK) inhibitors: structure-activity relationships for O-alkoxyphenylchromen-4-one probes of the ATP-binding domain. Bioorg Med Chem Lett. 2011;21:966–970. doi: 10.1016/j.bmcl.2010.12.047. [DOI] [PubMed] [Google Scholar]
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- Daud AI, Ashworth MT, Strosberg J, Goldman JW, Mendelson D, Springett G, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. Journal of Clinical Oncology. 2015;33:1060–1066. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- Davidson D, Amrein L, Panasci L, Aloyz R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del CG, Sessa C, von MR, Vigano L, Digena T, Locatelli A, et al. Phase I study of olaparib in combination with liposomal doxorubicin in patients with advanced solid tumours. Br J Cancer. 2014;111:651–659. doi: 10.1038/bjc.2014.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del NC, Xiao Y, Rangell L, Reichelt M, O’Brien T. Depletion of the central metabolite NAD leads to oncosis-mediated cell death. Journal of Biological Chemistry. 2014;289:35182–35192. doi: 10.1074/jbc.M114.580159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi T, Yoshino T, Shitara K, Matsubara N, Fuse N, Naito Y, et al. Phase I study of LY2603618, a CHK1 inhibitor, in combination with gemcitabine in Japanese patients with solid tumors. Anticancer Drugs. 2015;26:1043–1053. doi: 10.1097/CAD.0000000000000278. [DOI] [PubMed] [Google Scholar]
- Dorjsuren D, Kim D, Vyjayanti VN, Maloney DJ, Jadhav A, Wilson DM, III, et al. Diverse small molecule inhibitors of human apurinic/apyrimidinic endonuclease APE1 identified from a screen of a large public collection. PLoS One. 2012;7:e47974. doi: 10.1371/journal.pone.0047974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HQ, Hong YB, Kim JS, Lee HS, Yi YW, Kim YJ, et al. Inhibition of checkpoint kinase 2 (CHK2) enhances sensitivity of pancreatic adenocarcinoma cells to gemcitabine. J Cell Mol Med. 2013;17:1261–1270. doi: 10.1111/jcmm.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke CG, Parsels LA, Qian Y, Zhang Q, Karnak D, Robertson JR, et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clinical Cancer Research. 2013;19:4412–4421. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treatment Reviews. 2014;40:109–117. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. Journal of Clinical Oncology. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. Journal of Medicinal Chemistry. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Frank AO, Vangamudi B, Feldkamp MD, Souza-Fagundes EM, Luzwick JW, Cortez D, et al. Discovery of a potent stapled helix peptide that binds to the 70N domain of replication protein A. Journal of Medicinal Chemistry. 2014;57:2455–2461. doi: 10.1021/jm401730y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfurt OS, Seckinger D, Sugarbaker EV. Inhibition of DNA repair in cells treated with a combination of alkylating agents. Anticancer Res. 1993;13:947–952. [PubMed] [Google Scholar]
- Freschauf GK, Karimi-Busheri F, Ulaczyk-Lesanko A, Mereniuk TR, Ahrens A, Koshy JM, et al. Identification of a small molecule inhibitor of the human DNA repair enzyme polynucleotide kinase/phosphatase. Cancer Research. 2009;69:7739–7746. doi: 10.1158/0008-5472.CAN-09-1805. [DOI] [PubMed] [Google Scholar]
- Garrett MD, Collins I. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol Sci. 2011;32:308–316. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- Glanzer JG, Carnes KA, Soto P, Liu S, Parkhurst LJ, Oakley GG. A small molecule directly inhibits the p53 transactivation domain from binding to replication protein A. Nucleic Acids Research. 2013;41:2047–2059. doi: 10.1093/nar/gks1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer JG, Liu S, Oakley GG. Small molecule inhibitor of the RPA70 N-terminal protein interaction domain discovered using in silico and in vitro methods. Bioorg Med Chem. 2011;19:2589–2595. doi: 10.1016/j.bmc.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer JG, Liu S, Wang L, Mosel A, Peng A, Oakley GG. RPA inhibition increases replication stress and suppresses tumor growth. Cancer Research. 2014;74:5165–5172. doi: 10.1158/0008-5472.CAN-14-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goglia AG, Delsite R, Luz AN, Shahbazian D, Salem AF, Sundaram RK, et al. Identification of novel radiosensitizers in a high-throughput, cell-based screen for DSB repair inhibitors. Mol Cancer Ther. 2015;14:326–342. doi: 10.1158/1535-7163.MCT-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009;8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LJ, Gurtler J, Del Prete SA, Tjulandin S, Semiglazov VF, Bayever E, et al. Trabectedin as a single-agent treatment of advanced breast cancer after anthracycline and taxane treatment: a multicenter, randomized, phase II study comparing 2 administration regimens. Clin Breast Cancer. 2014;14:396–404. doi: 10.1016/j.clbc.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabauskiene S, Bergeron EJ, Chen G, Chang AC, Lin J, Thomas DG, et al. CHK1 levels correlate with sensitization to pemetrexed by CHK1 inhibitors in non-small cell lung cancer cells. Lung Cancer. 2013;82:477–484. doi: 10.1016/j.lungcan.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clinical Cancer Research. 2010;16:6159–6168. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannay JA, Liu J, Zhu QS, Bolshakov SV, Li L, Pisters PW, et al. Rad51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol Cancer Ther. 2007;6:1650–1660. doi: 10.1158/1535-7163.MCT-06-0636. [DOI] [PubMed] [Google Scholar]
- Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. Journal of Biological Chemistry. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Research. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Ihmaid SK, Al-Rawi JM, Bradley CJ, Angove MJ, Robertson MN. Synthesis, DNA-PK inhibition, anti-platelet activity studies of 2-(N-substituted-3-aminopyridine)-substituted-1,3-benzoxazines and DNA-PK and PI3K inhibition, homology modelling studies of 2-morpholino-(7,8-di and 8-substituted)-1,3-benzoxazines. Eur J Med Chem. 2012;57:85–101. doi: 10.1016/j.ejmech.2012.08.035. [DOI] [PubMed] [Google Scholar]
- Ishida T, Takizawa Y, Kainuma T, Inoue J, Mikawa T, Shibata T, et al. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Research. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal AS, Banerjee S, Aneja R, Sarkar FH, Ostrov DA, Narayan S. DNA polymerase beta as a novel target for chemotherapeutic intervention of colorectal cancer. PLoS One. 2011;6:e16691. doi: 10.1371/journal.pone.0016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal AS, Banerjee S, Panda H, Bulkin CD, Izumi T, Sarkar FH, et al. A novel inhibitor of DNA polymerase beta enhances the ability of temozolomide to impair the growth of colon cancer cells. Mol Cancer Res. 2009;7:1973–1983. doi: 10.1158/1541-7786.MCR-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal AS, Panda H, Law BK, Sharma J, Jani J, Hromas R, et al. NSC666715 and Its Analogs Inhibit Strand-Displacement Activity of DNA Polymerase beta and Potentiate Temozolomide-Induced DNA Damage, Senescence and Apoptosis in Colorectal Cancer Cells. PLoS One. 2015;10:e0123808. doi: 10.1371/journal.pone.0123808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? International Journal of Radiation Biology. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- Jekimovs C, Bolderson E, Suraweera A, Adams M, O’Byrne KJ, Richard DJ. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front Oncol. 2014;4:86. doi: 10.3389/fonc.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Busheri F, Rasouli-Nia A, lalunis-Turner J, Weinfeld M. Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Research. 2007;67:6619–6625. doi: 10.1158/0008-5472.CAN-07-0480. [DOI] [PubMed] [Google Scholar]
- Katz EJ, Andrews PA, Howell SB. The effect of DNA polymerase inhibitors on the cytotoxicity of cisplatin in human ovarian carcinoma cells. Cancer Commun. 1990;2:159–164. doi: 10.3727/095535490820874515. [DOI] [PubMed] [Google Scholar]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. Journal of Clinical Oncology. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Cholia RP, Mantha AK, Kumar R. DNA repair and redox activities and inhibitors of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1): a comparative analysis and their scope and limitations toward anticancer drug development. Journal of Medicinal Chemistry. 2014;57:10241–10256. doi: 10.1021/jm500865u. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol. 2012;5:36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C, Diaz HB, McNeely S, Barnard D, Dempsey J, Blosser W, et al. LY2606368 Causes Replication Catastrophe and Antitumor Effects through CHK1-Dependent Mechanisms. Mol Cancer Ther. 2015;14:2004–2013. doi: 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- Kitada S, Leone M, Sareth S, Zhai D, Reed JC, Pellecchia M. Discovery, characterization, and structure-activity relationships studies of proapoptotic polyphenols targeting B-cell lymphocyte/leukemia-2 proteins. Journal of Medicinal Chemistry. 2003;46:4259–4264. doi: 10.1021/jm030190z. [DOI] [PubMed] [Google Scholar]
- Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruczynski A, Barret JM, Van HB, Chansard N, Astruc J, Menon Y, et al. Decreased nucleotide excision repair activity and alterations of topoisomerase IIalpha are associated with the in vivo resistance of a P388 leukemia subline to F11782, a novel catalytic inhibitor of topoisomerases I and II. Clinical Cancer Research. 2004;10:3156–3168. doi: 10.1158/1078-0432.ccr-1305-2. [DOI] [PubMed] [Google Scholar]
- Kuriyama I, Miyazaki A, Tsuda Y, Yoshida H, Mizushina Y. Inhibitory effect of novel somatostatin peptide analogues on human cancer cell growth based on the selective inhibition of DNA polymerase beta. Bioorg Med Chem. 2013;21:403–411. doi: 10.1016/j.bmc.2012.11.024. [DOI] [PubMed] [Google Scholar]
- Labroli M, Paruch K, Dwyer MP, Alvarez C, Keertikar K, Poker C, et al. Discovery of pyrazolo[1,5-a]pyrimidine-based CHK1 inhibitors: a template-based approach--part. Bioorg Med Chem Lett. 2011;21:471–474. doi: 10.1016/j.bmcl.2010.10.114. [DOI] [PubMed] [Google Scholar]
- Lainchbury M, Matthews TP, McHardy T, Boxall KJ, Walton MI, Eve PD, et al. Discovery of 3-alkoxyamino-5-(pyridin-2-ylamino)pyrazine-2-carbonitriles as selective, orally bioavailable CHK1 inhibitors. Journal of Medicinal Chemistry. 2012;55:10229–10240. doi: 10.1021/jm3012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Christensen SD, Frankel PH, Margolin KA, Agarwala SS, Luu T, et al. A phase II study of cell cycle inhibitor UCN-01 in patients with metastatic melanoma: a California Cancer Consortium trial. Invest New Drugs. 2012a;30:741–748. doi: 10.1007/s10637-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Wang X, Pan Y, Lee DH, Chowdhury D, Kimmelman AC. Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PLoS One. 2012b;7:e39588. doi: 10.1371/journal.pone.0039588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YL, Carty MP, Oakley GG, Seidman MM, Medvedovic M, Dixon K. Expression of ATM in ataxia telangiectasia fibroblasts rescues defects in DNA double-strand break repair in nuclear extracts. Environmental and Molecular Mutagenesis. 2001;37:128–140. doi: 10.1002/em.1020. [DOI] [PubMed] [Google Scholar]
- Liu JF, Konstantinopoulos PA, Matulonis UA. PARP inhibitors in ovarian cancer: current status and future promise. Gynecologic Oncology. 2014;133:362–369. doi: 10.1016/j.ygyno.2014.02.039. [DOI] [PubMed] [Google Scholar]
- Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, et al. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 2013;49:2972–2978. doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Jiang H, Liu Z, Wang Y, Zhao M, Hao C, et al. Berberine radiosensitizes human esophageal cancer cells by downregulating homologous recombination repair protein RAD51. PLoS One. 2011;6:e23427. doi: 10.1371/journal.pone.0023427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nuc–eic Acids Research. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard O, Camenisch U, Clement FC, Blagoev KB, Naegeli H. DNA repair triggered by sensors of helical dynamics. Trends in Biochemical Sciences. 2007 doi: 10.1016/j.tibs.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Dusad K, Wright WD, Grubb J, Budke B, Heyer WD, et al. RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells. Nucleic Acids Research. 2015;43:3180–3196. doi: 10.1093/nar/gkv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Logan HL, Budke B, Wu M, Pawlowski M, Weichselbaum RR, et al. The RAD51-stimulatory compound RS-1 can exploit the RAD51 overexpression that exists in cancer cells and tumors. Cancer Research. 2014;74:3546–3555. doi: 10.1158/0008-5472.CAN-13-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews LA, Cabarcas SM, Hurt EM, Zhang X, Jaffee EM, Farrar WL. Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas. 2011;40:730–739. doi: 10.1097/MPA.0b013e31821ae25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacology & Therapeutics. 2014;142:1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- McNeil EM, Astell KR, Ritchie AM, Shave S, Houston DR, Bakrania P, et al. Inhibition of the ERCC1-XPF structure-specific endonuclease to overcome cancer chemoresistance. DNA Repair (Amst) 2015;31:19–28. doi: 10.1016/j.dnarep.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Dormi SS, Turchi AM, Woods DS, Turchi JJ. Chemical inhibitor targeting the replication protein A-DNA interaction increases the efficacy of Pt-based chemotherapy in lung and ovarian cancer. Biochemical Pharmacology. 2014 doi: 10.1016/j.bcp.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Jameson C, Barbachano Y, Sanchez L, Kote-Jarai Z, Peock S, et al. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009;55:696–704. doi: 10.1111/j.1365-2559.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushina Y, Suzuki-Fukudome H, Takeuchi T, Takemoto K, Kuriyama I, Yoshida H, et al. Formosusin A, a novel specific inhibitor of mammalian DNA polymerase beta from the fungus Paecilomyces formosus. Bioorg Med Chem. 2014;22:1070–1076. doi: 10.1016/j.bmc.2013.12.038. [DOI] [PubMed] [Google Scholar]
- Mohammed MZ, Vyjayanti VN, Laughton CA, Dekker LV, Fischer PM, Wilson DM, III, et al. Development and evaluation of human AP endonuclease inhibitors in melanoma and glioma cell lines. Br J Cancer. 2011;104:653–663. doi: 10.1038/sj.bjc.6606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck JM, Batey MA, Zhao Y, Jenkins H, Richardson CJ, Cano C, et al. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol Cancer Ther. 2012;11:1789–1798. doi: 10.1158/1535-7163.MCT-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Research. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Z, Peng B, Ng S, Pan S, Lee JS, Shen HM, et al. A small-molecule protein-protein interaction inhibitor of PARP1 that targets its BRCT domain. Angew Chem Int Ed Engl. 2015;54:2515–2519. doi: 10.1002/anie.201410678. [DOI] [PubMed] [Google Scholar]
- Neher TM, Bodenmiller D, Fitch RW, Jalal SI, Turchi JJ. Novel irreversible small molecule inhibitors of replication protein A display single-agent activity and synergize with cisplatin. Mol Cancer Ther. 2011;10:1796–1806. doi: 10.1158/1535-7163.MCT-11-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher TM, Shuck SC, Liu J, Zhang JT, Turchi JJ. Identification of novel small molecule inhibitors of the XPA protein using in silico based screening. ACS Chem Biol. 2010;5:953–965. doi: 10.1021/cb1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N Engl J Med. 2007;356:2538–2540. doi: 10.1056/NEJMc070742. [DOI] [PubMed] [Google Scholar]
- Nutley BP, Smith NF, Hayes A, Kelland LR, Brunton L, Golding BT, et al. Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. British Journal of Cancer. 2005;93:1011–1018. doi: 10.1038/sj.bjc.6602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza AM, Cibula D, Benzaquen AO, Poole C, Mathijssen RH, Sonke GS, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2015;16:87–97. doi: 10.1016/S1470-2045(14)71135-0. [DOI] [PubMed] [Google Scholar]
- Pal J, Bertheau R, Buon L, Qazi A, Batchu RB, Bandyopadhyay S, et al. Genomic evolution in Barrett’s adenocarcinoma cells: critical roles of elevated hsRAD51, homologous recombination and Alu sequences in the genome. Oncogene. 2011;30:3585–3598. doi: 10.1038/onc.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papeo G, Posteri H, Borghi D, Busel AA, Caprera F, Casale E, et al. Discovery of 2-[1-(4,4-Difluorocyclohexyl)piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoind ole-4-carboxamide (NMS-P118): A Potent, Orally Available, and Highly Selective PARP-1 Inhibitor for Cancer Therapy. Journal of Medicinal Chemistry. 2015;58:6875–6898. doi: 10.1021/acs.jmedchem.5b00680. [DOI] [PubMed] [Google Scholar]
- Passeri D, Camaioni E, Liscio P, Sabbatini P, Ferri M, Carotti A, et al. Concepts and Molecular Aspects in the Polypharmacology of PARP-1 Inhibitors. ChemMedChem. 2015 doi: 10.1002/cmdc.201500391. [DOI] [PubMed] [Google Scholar]
- Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiation Research. 2003;159:251–s261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci US A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak KS, Turchi JJ. Purification and characterization of exonuclease-free Artemis: Implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair (Amst) 2010;9:670–677. doi: 10.1016/j.dnarep.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddi P, Loftin CW, Dickey JS, Hair JM, Burns KJ, Aziz K, et al. DNA-PKcs deficiency leads to persistence of oxidatively induced clustered DNA lesions in human tumor cells. Free Radic Biol Med. 2010;48:1435–1443. doi: 10.1016/j.freeradbiomed.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clinical Cancer Research. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst) 2006;5:1199–1212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Li M, Sui J, Ren T, Li Z, Zhang L, et al. Identification of a novel potential antitumor activity of gossypol as an APE1/Ref-1 inhibitor. Drug Des Devel Ther. 2014;8:485–496. doi: 10.2147/DDDT.S62963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao GB, Wu YL, Yang XN, Zhong WZ, Xie D, Guan XY, et al. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Research. 2002;62:219–225. [PubMed] [Google Scholar]
- Rai G, Vyjayanti VN, Dorjsuren D, Simeonov A, Jadhav A, Wilson DM, III, et al. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. Journal of Medicinal Chemistry. 2012;55:3101–3112. doi: 10.1021/jm201537d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Shan J, Li M, Qing Y, Qian C, Wang G, et al. Small-molecule BH3 mimetic and pan-Bcl-2 inhibitor AT-101 enhances the antitumor efficacy of cisplatin through inhibition of APE1 repair and redox activity in non-small-cell lung cancer. Drug Des Devel Ther. 2015;9:2887–2910. doi: 10.2147/DDDT.S82724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, et al. Defective DNA repair and increased genomic instability in artemis-deficient murine cells. Journal of Experimental Medicine. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sancar A, Reardon JT. Nucleotide excision repair in E-coli and man. Dna Repair and Replication. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14:882–892. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- Sears CR, Turchi JJ. Complex Cisplatin-Double Strand Break (DSB) Lesions Directly Impair Cellular Non-Homologous End-Joining (NHEJ) Independent of Downstream Damage Response (DDR) Pathways. Journal of Biological Chemistry. 2012;287:24263–24272. doi: 10.1074/jbc.M112.344911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto T, Esaki T, Hirai F, Arita S, Nosaki K, Makiyama A, et al. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer Chemotherapy & Pharmacology. 2013;72:619–627. doi: 10.1007/s00280-013-2234-6. [DOI] [PubMed] [Google Scholar]
- Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113:2290–2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuck SC, Turchi JJ. Targeted inhibition of Replication Protein A reveals cytotoxic activity, synergy with chemotherapeutic DNA-damaging agents, and insight into cellular function. Cancer Research. 2010;70:3189–3198. doi: 10.1158/0008-5472.CAN-09-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, et al. Contribution of Germline Mutations in the RAD51B, RAD51C, and RAD51D Genes to Ovarian Cancer in the Population. Journal of Clinical Oncology. 2015;33:2901–2907. doi: 10.1200/JCO.2015.61.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenblick A, de AE, Azim HA, Jr, Piccart M. An update on PARP inhibitors--moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- Stachelek GC, Peterson-Roth E, Liu Y, Fernandez RJ, III, Pike LR, Qian JM, et al. YU238259 Is a Novel Inhibitor of Homology-Dependent DNA Repair That Exhibits Synthetic Lethality and Radiosensitization in Repair-Deficient Tumors. Mol Cancer Res. 2015;13:1389–1397. doi: 10.1158/1541-7786.MCR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW. Base Excision Repair and Lesion-Dependent Subpathways for Repair of Oxidative DNA Damage. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku M, Kainuma T, Ishida-Takaku T, Ishigami S, Suzuki H, Tashiro S, et al. Halenaquinone, a chemical compound that specifically inhibits the secondary DNA binding of RAD51. Genes Cells. 2011;16:427–436. doi: 10.1111/j.1365-2443.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- Takenaka T, Yoshino I, Kouso H, Ohba T, Yohena T, Osoegawa A, et al. Combined evaluation of Rad51 and ERCC1 expressions for sensitivity to platinum agents in non-small cell lung cancer. International Journal of Cancer. 2007 doi: 10.1002/ijc.22738. [DOI] [PubMed] [Google Scholar]
- Tennstedt P, Fresow R, Simon R, Marx A, Terracciano L, Petersen C, et al. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. International Journal of Cancer. 2013;132:2118–2126. doi: 10.1002/ijc.27907. [DOI] [PubMed] [Google Scholar]
- Vendetti FP, Lau A, Schamus S, Conrads TP, O’Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015 doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Research. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- Waterson AG, Kennedy JP, Patrone JD, Pelz NF, Feldkamp MD, Frank AO, et al. Diphenylpyrazoles as replication protein a inhibitors. ACS Med Chem Lett. 2015;6:140–145. doi: 10.1021/ml5003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacology & Therapeutics. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Weiss GJ, Donehower RC, Iyengar T, Ramanathan RK, Lewandowski K, Westin E, et al. Phase I dose-escalation study to examine the safety and tolerability of LY2603618, a checkpoint 1 kinase inhibitor, administered 1 day after pemetrexed 500 mg/m(2) every 21 days in patients with cancer. Invest New Drugs. 2013;31:136–144. doi: 10.1007/s10637-012-9815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmans AP, Al-Ejeh F, Chee N, Yap PY, Gorski JJ, Da SL, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5:3261–3272. doi: 10.18632/oncotarget.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore E, de CS, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103:4659–4665. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- Woods D, Turchi JJ. Chemotherapy induced DNA damage response: Convergence of drugs and pathways. Cancer Biol Ther. 2013;14:379–89. doi: 10.4161/cbt.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1:328–346. [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem. 2007;7:515–523. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- Zhao H, Luoto KR, Meng AX, Bristow RG. The receptor tyrosine kinase inhibitor amuvatinib (MP470) sensitizes tumor cells to radio- and chemo-therapies in part by inhibiting homologous recombination. Radiother Oncol. 2011;101:59–65. doi: 10.1016/j.radonc.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Research. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Guo XE, Qiu XL, Hu CM, Chamberlin AR, et al. Synthesis, molecular modeling, and biological evaluation of novel RAD51 inhibitors. Eur J Med Chem. 2015;96:196–208. doi: 10.1016/j.ejmech.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhou L, Wu G, Konig H, Lin X, Li G, et al. A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol Med. 2013;5:353–365. doi: 10.1002/emmm.201201760. [DOI] [PMC free article] [PubMed] [Google Scholar]