Abstract
Melanoma, the most aggressive forms of skin cancer, is often fatal if not treated early. Therefore, novel target-based strategies are required to combat this neoplasm. The objective of this study was to determine the role and functional significance of the mitochondrial sirtuin SIRT3 in melanoma. We found that compared to normal primary and immortalized human melanocytes, SIRT3 is significantly overexpressed in multiple human melanoma cells at mRNA and protein levels. Further, employing human tissue microarray, we found that SIRT3 is significantly upregulated in clinical melanoma tissues, compared to melanocytic nevi tissues. Furthermore, a short hairpin RNA (shRNA)-mediated knockdown of SIRT3 in human melanoma cells resulted in 1) decrease in cellular proliferation, colony formation and cellular migration, 2) induction of senescence as shown by increase in SA-β-Gal activity and formation of SAHF as well as increase in mRNA and protein levels of p16INK4a and p21Waf1, 3) G1-phase arrest of the cell cycle, and 4) decreases in mRNA and protein levels of Cyclins (D1, E1) and Cdks (2, 4, 6). Conversely, forced exogenous overexpression of SIRT3 promoted increase in proliferative potential of Hs294T melanoma cells and normal immortalized Mel-ST melanocytes. Finally, we found that SIRT3 knockdown significantly inhibited tumorigenesis in a xenograft model in vivo. To our knowledge, this is the first study supporting the pro-proliferative function of SIRT3 in melanoma.
INTRODUCTION
Melanoma is one of the deadliest cancers of skin and its incidence have been increasing in the USA at a constant pace in the past few decades. Approximately 73,870 new melanoma cases and 9,940 melanoma-related deaths are predicted in the USA this year (Siegel et al., 2015). The existing preventive and therapeutic approaches have not been able to effectively manage melanoma incidences or fatalities, making it a significant clinical problem. Therefore, further research is needed to understand the mechanism and biology of melanocytic transformation and melanoma progression, in order to identify novel targets and strategies for the management of this neoplasm.
The role of sirtuins (SIRTs) in cancer has been an area of intense investigation, recently. The mammalian SIRTs constitute a family of seven members (SIRT1 – SIRT7) with nicotinamide adenine dinucleotide (NAD+) -dependent protein deacetylase and/or adenosine diphosphate (ADP)-ribosyltransferase activities (North and Verdin, 2004). In parallel, they are also known for regulating post-translational acyl modifications in organisms ranging from bacteria to humans (Chen et al., 2007; Guan and Xiong, 2011; Thao and Escalante-Semerena, 2011). SIRTs play critical roles in a variety of cellular processes, including cellular metabolism, cell cycle, cell division and transcriptional regulation. In addition, SIRTs are involved in the pathogenesis of a variety of diseases, including metabolic diseases, cardiovascular diseases, neurodegenerative disorders, aging and cancer (Sebastian et al., 2012). The role of SIRTs in cancer is extremely complex as they appears to have dichotomous functions depending on cell contexts (Bosch-Presegue and Vaquero, 2011). The mitochondrial sirtuin SIRT3 is a 44 kDa nuclear protein that is activated by cleavage of a 16 kDa fragment when translocated to mitochondria under cellular stress (Scher et al., 2007). When localized to mitochondria, SIRT3 coordinates global shifts in mitochondrial activity by deacetylating proteins involved in diverse mitochondrial functions including energy metabolism, ATP-production, redox-balance and mitochondrial biogenesis (Giralt and Villarroya, 2012; Wang et al., 2010). It also plays important roles in the regulation of a variety of cellular processes, including transcription, insulin secretion, and apoptosis (North and Verdin, 2004). The fact that SIRT3 can regulate several cellular processes which are critical in cancer cell proliferation, makes it a potential therapeutic target for cancer management (Alhazzazi et al., 2013). However, the role of SIRT3 in cancer, like other SIRTs, appears to be complex; with evidences suggesting both tumor promoter and suppressor functions. While the research on the role of SIRT3 in cancer is still in its infancy, studies have suggested tumor suppressor (Desouki et al., 2014; Wang et al., 2014) as well as tumor promoter (Huang et al., 2014; Yan et al., 2014) function for this sirtuin. However, the role of SIRT3 in melanoma is not known. This study was designed to define the role and functional significance of SIRT3 in melanoma.
RESULTS
SIRT3 is overexpressed in human melanoma cell lines and clinical tissues
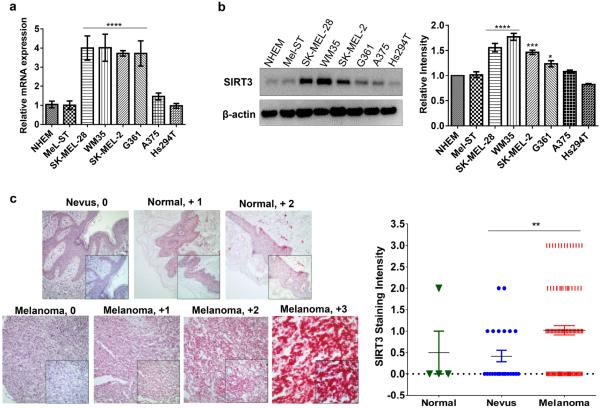
As the first step of our effort to understand the role of SIRT3 in melanoma, we determined the expression profile of SIRT3 in normal human epidermal melanocytes (NHEM), immortalized melanocytes (Mel-ST) and a panel of human melanoma cell lines (SK-MEL-28, WM35, SKMEL-2, G361, A375 and Hs294T), differing in their mutational status of BRAFV600E, p53 and NRAS (Table S1). Compared to the normal NHEM and Mel-ST, several melanoma cell lines showed significantly higher expression of SIRT3 mRNA and protein as assessed by qRT-PCR and immunoblot analyses, respectively (Figure 1a and 1b). In view of these data, for future mechanistic studies, we chose to focus on SK-MEL-2, G361 and SK-MEL-28 (highly SIRT3 expressing) and Hs294T (low SIRT3 expressing) melanoma cells as well as Mel-ST (immortalized normal melanocytes; low SIRT3 expressing).
Figure 1. SIRT3 is overexpressed in human melanoma cell lines and clinical tissues.
(a) qRT-PCR, and (b) Western blot analysis for SIRT3 expression in normal human melanocytes (NHEM), non-transformed but immortalized human melanocyte line (Mel-ST) and six human melanoma cell lines (SK-MEL-2, WM35, SK-MEL-28, G361, Hs294T, A375). β-actin and Gapdh were used as controls, respectively. (c) Representative images for SIRT3 immunohistochemical analysis in two melanoma tissue microarray containing a total of 4 normal skin, 24 nevi and 96 malignant melanoma (stage II, III and IV) tissues. Positive SIRT3 staining is indicated by the intensity of the red color from the Vector Red chromagen used in staining. Individual tissue cores were blindly scored for SIRT3 staining intensity as negative (0), trace, weak (+1), moderate (+2), or strong (+3) staining in 50% of the cells examined. Representative cores for normal, nevus and melanoma samples are shown. The magnification of large panels is 10× and inset windows are 40×. The qRT-PCR and Western blot data represent experiments done in triplicates and the results are presented as means ± SEM. Statistical significance are indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Next, we determined the expression pattern of SIRT3 in clinical human melanoma tissues using two different tissue microarrays (TMAs), containing a total of 4 normal skin tissue, 24 nevus tissues and 96 malignant melanoma tissues of stage II, III and IV (Supplementary Figure S1). Detailed information on the TMAs analyzed, as well as the score of each tissue core is provided in Supplementary Table S2. For tissue score analyses, the normal-, nevus-, and melanomas- tissues from both the TMAs were grouped together in three groups. As shown in Figure 1c, compared to melanocytic nevi, we found significantly higher expression (P<0.01) of SIRT3 in melanoma tissues. This encouraged us to further explore the role of SIRT3 in melanoma survival.
SIRT3 knockdown inhibits growth, colony formation and migration in multiple human melanoma cell line
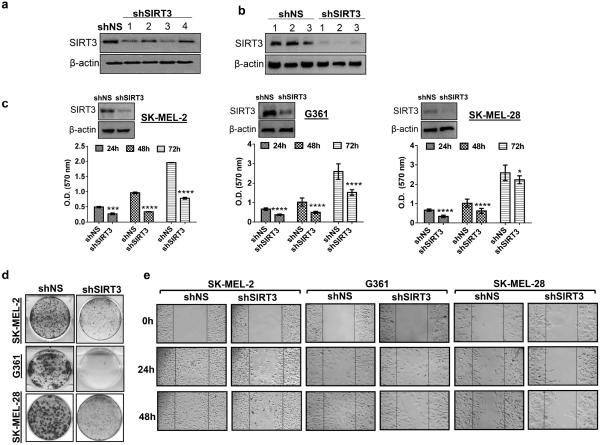
As a starting point, to understand the functional significance of SIRT3 in melanoma cells, employing lentiviral shRNA we generated SIRT3 knocked-down stable SK-MEL-2 cell line. We tested four different shRNAs (#1- #4) which showed a considerable SIRT3-knockdown, compared to non-specific shRNA (shNS) in SK-MEL-2 cells (Figure 2a). Of these, TRCN0000038889 and TRCN0000038892 (#1 and #3) showed the greatest suppression of SIRT3. The most effective shRNA construct (#1) or the empty vector control (pLKO.1) were used further and stable SK-MEL-2 clones were selected with puromycin (2 g/ml) as a selection marker. SIRT3 protein levels showed a marked decrease in shSIRT3 cells compared to the shNS cells (Figure 2b), demonstrating an effective silencing of SIRT3 in puromycin selected clones. Among the clones obtained, a substantial reduction in SIRT3 level was observed in shSIRT3 clone#2 (Figure 2b), which was selected for further experiments. Similarly, the shRNA construct (#1) was also used to generate stable shSIRT3-G361 and shSIRT3-SK-MEL-28 cell lines following a puromycin selection of best clones.
Figure 2. SIRT3 knockdown inhibits proliferation, colony formation and migration of human melanoma cells.
(a) Western blot analysis for confirming SIRT3 knockdown in SK-MEL-2 cells. Four independent shRNAs construct (1: TRCN0000038889, 2: TRCN0000038890, 3: TRCN0000038892, 4: TRCN0000038893) were used for SIRT3 knockdown (shSIRT3) as described in Materials and Methods. Control cells were transduced with pLKO.1 empty vector (shNS). β-actin was used as a loading control. (b) Western blot analysis of stable SIRT3 knockdown (shSIRT3 #1 - #3) and control vector (shNS #1 - #3) clones selected with puromycin (2 μg/ml) was performed to confirm the knockdown. β-actin was used as a loading control. (c) Western blot analysis of stable SIRT3 knockdown (shSIRT3) in SK-MEL-2, G361 and SK-MEL-28 melanoma cells. β-actin was used as a loading control. Proliferative potential was assessed at 24, 48 and 72 h in stable cells using MTT assay. (d) Clonogenic cell survival of stable cells was assessed by colony formation assay. Representative images after 14 days (SK-MEL-2 cells) and 11 days (G361 and SK-MEL-28 cells) in culture are shown. (e) Cell migration was determined using an Ibidi Culture Insert. Representative images shown at 0, 24 and 48 h were analyzed under microscope. Scale bars indicate 100 μm. All experiments were done in triplicates and the results are presented as means ± SEM. Statistical significance are indicated as *P < 0.05, ***P<0.001, ****P<0.0001.
Using the stable transfectants, generated, we determined the effect of SIRT3 knockdown on growth and proliferative potential of melanoma cells. We sub-cultured the cells (shSIRT3-SK-MEL-2, shSIRT3-G361 and shSIRT3-SK-MEL-28) and conducted MTT, colony formation and cell migration assays, to determine the anti-proliferative response of SIRT3 knockdown in these cells. As shown in figure 2, SIRT3 knockdown resulted in a significant decrease in cell proliferation (Figure 2c) and clonogenic survival (Figure 2d) in all the three melanoma cell lines. Western blot analysis confirmed SIRT3 knockdown in stable shSIRT3-SK-MEL-2, shSIRT3-G361 and shSIRT3-SK-MEL-28 cell lines (Figure 2c). Further, SIRT3 depletion also resulted in a marked decreased migration capacity of these cells in a time-dependent fashion (Figure 2e). These results suggested SIRT3 has a pro-proliferative function in human melanoma that is possibly not cell type specific.
SIRT3 knockdown induces senescence-like phenotype in human melanoma cells
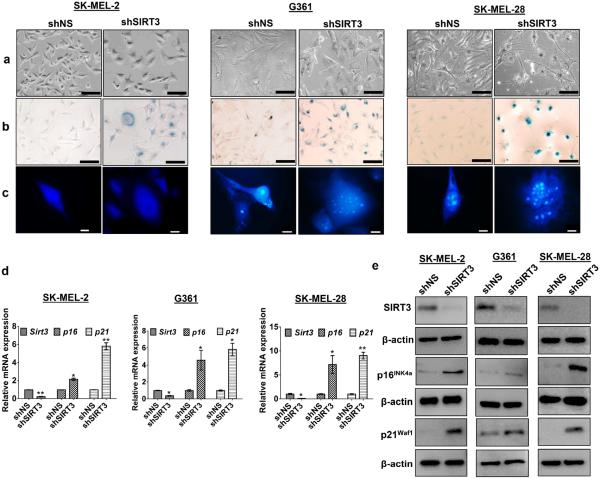
Interestingly, we found that SIRT3-knockdown resulted in an alteration in cell morphology, showing somewhat irregular, enlarged, flat and multinucleated phenotype that is consistent with senescent (Figure 3a). In order to validate this, we determined the effect of SIRT3 knockdown on senescence-associated beta-galactosidase (SA-β-Gal) staining and senescence-associated heterochromatin foci (SAHF) in all the three cell lines (SK-MEL-2, G361, SK-MEL-28). As shown in Figure 3b and 3c, when compared to nonsense cells, SIRT3 knockdown resulted in a marked increase in SA-β-Gal activity and SAHF formation, indicative of senescence induction as a result of SIRT3 depletion. Further, we determined the effect of SIRT3 knockdown on senescence-associated markers viz. p16INK4a and p21Waf1. We observed a striking upregulation in levels of p16INK4a and p21Waf1 in SIRT3 knockdown cells (Figure 3d and e). These results suggested that SIRT3 knockdown causes anti-proliferative effects in human melanoma cells, possibly via p53-independent senescence, since the SK-MEL-2 cells lack functional p53 (Kato et al., 2003).
Figure 3. SIRT3 knockdown induces senescence-like phenotypes in human melanoma cells.
(a) Following stable SIRT3 knockdown in melanoma cells, the morphological changes were analyzed under microscope (Nikon Digital Sight DS-Fi1 camera) and images were captured using NIS Elements AR 3.1 software. Representative images of control (shNS) and SIRT3 knockdown (shSIRT3) cells are shown. Scale bars indicate 100 μm. (b) SA-β-Gal staining in cells, showing accumulation of blue color in senescent cells. Representative images were analyzed under microscope. Scale bar=100 μm. (c) SAHF formation in cells was analyzed by DAPI staining of heterochromatin foci. Representative images are analyzed under microscope. Scale bar=10 μm. (d) qRT-PCR and (e) Western blot analysis for SIRT3 and senescence associated proteins, p16INK4a and p21Waf1 levels in stable cells. β-actin and Gapdh were used as controls, respectively. All experiments were done in triplicates and the results are presented as means ± SEM. Statistical significance are indicated as *P < 0.05, **P<0.01.
SIRT3 knockdown caused a perturbation in cell cycle in human melanoma cells
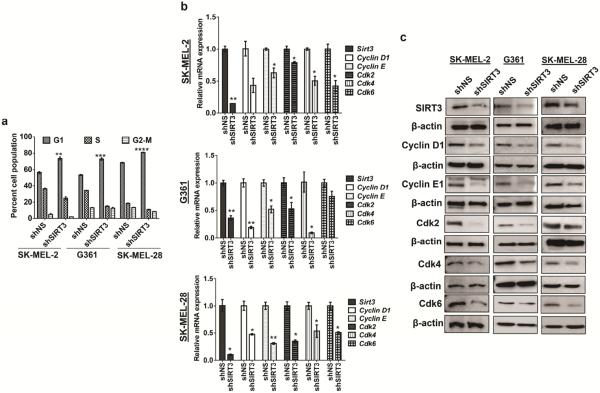
As described above, SIRT3 knockdown melanoma cells showed decreased cell proliferation when compared to the respective controls (Figure 2c). To determine if this decrease in cell proliferation was due to a dysregulation in cell cycle, we performed a cell cycle distribution analysis by flow cytometry. We found that SIRT3 knockdown, in all the three cell line, resulted in a significantly enhanced accumulation of cells in G0/G1 phase. This increase in G1 population was accompanied with a concomitant decrease of cells in S phase and G2-M phase (Figure 4a). However, we did not observe a sub-G1 cell population. To corroborate the observed G1 phase arrest, we examined the effect of SIRT3 knockdown on the cell cycle regulatory molecules involved in the G1 phase (Cyclin D1, Cyclin E1, Cdk2, Cdk4, Cdk6), at both mRNA and protein levels. As shown in Figure 4b and 4c, we observed a marked reduction in the levels of these cell cycle regulatory molecules as a result of SIRT3 knockdown in all three melanoma cells, both at mRNA and protein levels.
Figure 4. SIRT3 knockdown induces G1-phase arrest in human melanoma cells.
(a) Following SIRT3 knockdown in melanoma cells, they were sub-cultured, collected and analyzed for cell cycle distribution using FACS analysis. (b) qRT-PCR and (c) Western blot analysis for SIRT3, Cyclin D1, Cyclin E1, Cdk2, Cdk4 and Cdk6 expression levels in stable cells. β-actin and Gapdh were used as controls, respectively. All experiments were done in triplicates and the results are presented as means ± SEM. Statistical significance are indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Forced SIRT3 overexpression enhances cell proliferation in Hs294T melanoma cells
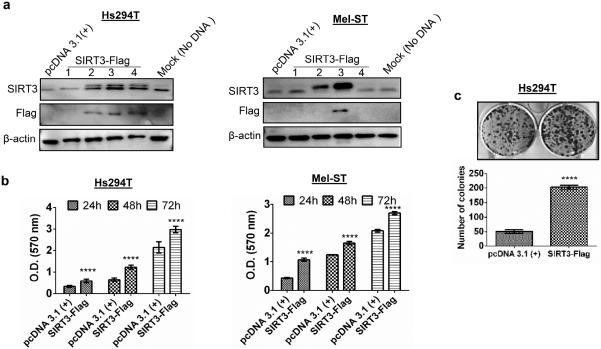
To further substantiate our finding that SIRT3 has a possible tumor promoter function in melanoma, in an additional strategy, we overexpressed SIRT3 in human melanoma Hs294T cells and normal immortalized melanocytic line Mel-ST, which showed very low expression of endogenous SIRT3 (Figure 1a and b). We achieved successful enhanced expression of SIRT3 using SIRT3-Flag plasmid, as shown by increase in the levels of SIRT3 protein, in both cell lines (Figure 5a). Next, we determined the effect of forced SIRT3 overexpression on cell growth and proliferation, using MTT and colony formation assays. As expected, forced overexpression of SIRT3 resulted in a significantly increased proliferation and colony formation in Hs294T cells as compared to control cells (Figure 5b and c). Similarly, SIRT3 overexpression in Mel-ST cells also showed significantly enhanced proliferation (Figure 5). However, SIRT3 overexpression did not result in formation of colonies in the normal Mel-ST melanocytes. Overall, these data further substantiated that SIRT3 is crucial for melanoma growth and survival.
Figure 5. Exogenous SIRT3 overexpression increases growth and proliferation of Hs294T melanoma cells and normal immortalized Mel-ST melanocytes.
(a) Western blot analysis of stable SIRT3 overexpression in Hs294T and Mel-ST cells transfected with control empty vector pcDNA 3.1(+), SIRT3-Flag (#1- #4) clones and mock control (no DNA) as described in Materials and Methods. β-actin was used as a loading control. Stable cells were selected with G-418 (2 mg/ml) for 14 days for Hs294T and 12 days for Mel-ST. (b) Proliferative potential was assessed at 24, 48 and 72 h in stable cells using MTT assay. (c) Clonogenic cell survival of stable cells was assessed by colony formation assay. Representative images after 10 days in culture are shown. The number of colonies were counted and plotted for quantitation. All experiments were done in triplicates and the results are presented as means ± SEM. Statistical significance are indicated as ****P<0.0001.
SIRT3 knockdown causes a decrease in tumor growth in a xenograft nude mouse model
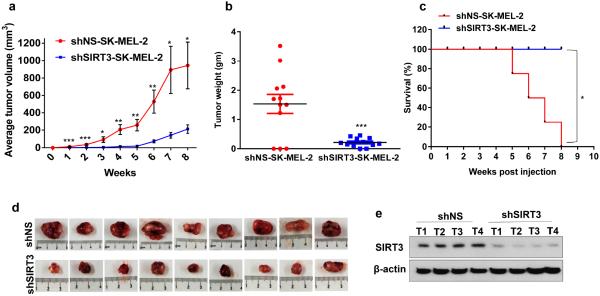
To determine the relevance of our in vitro finding to in vivo situation, we determined the tumorigenic potential of SIRT3-kockdown cells in NU/NU Nude Mouse. The mice were subcutaneously implanted with shNS-SK-MEL-2 (control) and shSIRT3-SK-MEL-2 (SIRT3-knockdown) cells, followed by assessing tumorigenesis. In our experimental plan, the animals were individually followed for tumor growth and the mice reaching with a tumor at 20 mm in the largest dimension were withdrawn and sacrificed. All the mice were euthanized at 8 weeks following tumor implantation. As shown in figure 6, SIRT3 knockdown resulted in a significant decrease in average tumor volume, measured on a weekly basis. Further, at termination of the study, SIRT3 knockdown was found to result in a significant decrease in average tumor weight (~86%) (Figure 6b) in mice. The Kaplan-Meier analysis showed that SIRT3 knockdown conferred a significant survival advantage, in terms of reaching to the cutoff tumor size (Figure 6c). SIRT3 downregulation was confirmed by Western blotting in tumor tissues (Figure 6e). Overall, our data clearly suggested that SIRT3 inhibition imparts a significant decrease in melanoma tumorigenesis.
Figure 6. SIRT3 knockdown causes a decrease in tumor growth in Nu/Nu xenograft mouse model.
The melanoma cells (shNS and shSIRT3-SK-MEL-2 cells) were implanted and allowed to grow in Nu/Nu mice and tumorigenesis was assessed as described in `Materials and Methods'. (a) Average tumor volume (on a weekly basis); (b) Tumor weight (at termination of experiment); and (c) Kaplan–Meier curve for target tumor size (on a weekly basis) are shown. (d) Pictures of resected tumors (at termination of experiment) were captured using a digital camera. (e) Western blot analysis of SIRT3 expression in tumor tissues lysates was conducted, β-actin was used as loading control. Statistical significance are indicated as *P<0.05, ***P<0.001,****P<0.0001.
DISCUSSION
Melanoma is an aggressive form of skin cancer that grows rapidly and exhibits resistance to most currently available therapeutic strategies. Therefore, identifying novel mechanism-based targets are required for developing newer therapeutics to interfere with this disease, alone or in combination with current drugs. Similarly, novel mechanism-based biomarkers are needed for melanoma diagnosis and prognosis. This study was designed to decipher the role and functional significance of SIRT3 in melanoma. SIRT3 is a major mitochondrial deacetylase that is currently being investigated as a potential modulator of tumorigenesis (Finley and Haigis, 2012). However, the role of SIRT3 in cancer seems to be complex and somewhat controversial, with evidence for its tumor promoter as well as tumor suppressor functions. Studies have shown that SIRT3 is downregulated in gastric cancer (Yang et al., 2014a) and hepatocellular carcinoma tissues (Zhang et al., 2012). Interestingly, low expression of SIRT3 was found to significantly inhibit mitotic entry, growth, proliferation and promoted apoptosis of lung cancer cell lines through deacetylation of nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) (Li et al., 2013). Another study has suggested that SIRT3 functions as a tumor suppressor in pancreatic cancer, via modulating cellular iron metabolism (Jeong et al., 2014). In contrast, higher expression of SIRT3 was reported in esophageal and breast cancer (Ashraf et al., 2006; Zhao et al., 2013). Also, downregulation of SIRT3 in oral cancer cells inhibits cell growth and proliferation as well as enhances radiotherapeutic and chemotherapeutic drug cytotoxicity (Alhazzazi et al., 2011). Similarly, silencing of SIRT3 was shown to inhibit the proliferation, invasion, migration and increased the apoptosis in the cultured colon cancer cell lines (Liu et al., 2014). Thus, based on available data, it appears that SIRT3 can act either as a tumor promoter or as a tumor suppressor (Chen et al., 2014). Indeed, this dual role of SIRT3 in cancer emphasizes the importance of intense research in this area. We focused our study to determine the role of SIRT3 in melanoma, which has not been yet investigated.
As described above, our data demonstrated that SIRT3 is upregulated, both at mRNA and protein levels, in a range of human melanoma cell lines with different gene mutational status (Table S1), compared to normal primary melanocytes and immortalized melanocytes. To further explore the clinical relevance of SIRT3, we used commercially available TMAs containing normal/nevus and different stages of human melanoma tissues. SIRT3 was shown to be overexpressed in melanoma tissues compared to nevi. However, due to limited number of specimens in the TMAs used, we were unable to find a stage specific correlation data on SIRT3 expression in melanomas. Altogether, our cell culture and TMA data strongly implicated SIRT3 overexpression in melanoma progression.
In next series of experiments, in order to determine whether SIRT3-overexpression in melanoma cells promoted proliferation, we stably knocked-down SIRT3 in melanoma cells and showed that it resulted in significantly reduced cellular growth, migration and colony-formation, suggesting a possible pro-proliferative function of SIRT3 in human melanoma. Further, our data also demonstrated that SIRT3 knockdown causes senescence-like phenotype in melanoma cells, as evident from morphological alterations characterized by flat cell phenotype (Ben-Porath and Weinberg, 2004) as well as increased senescence-associated SA-β-Gal staining and accumulation of SAHF, which are typical markers to identify senescence in growth-arrested cells (Dimri et al., 1995; Narita et al., 2003). The senescent phenotype is also characterized by increases in cyclin kinase inhibitors p16INK4a and p21Waf1 (Alcorta et al., 1996; Chang et al., 1999). Consistent with this, we observed an increase in p16INK4a and p21Waf1 following SIRT3 depletion in melanoma cells harboring mutant and wild type p53. Therefore, this appears to be a p53-independent mechanism (Fang et al., 1999; Jacobs and de Lange, 2004; Phalke et al., 2012). Although p21 is a principal p53 target gene and a central component in a variety of p53-mediated stress responses, its expression can be positively regulated through several p53-independent mechanisms (Zeng and el-Deiry, 1996). Indeed p21Waf1 has recently been reported to be increased upon SIRT3 depletion in esophageal squamous cell carcinoma cells (Yang et al., 2014b). Interestingly, we found that G361 cells which possess wild-type p53 also showed decreased cell proliferation and colony formation upon SIRT3 depletion. Future, in-depth studies are required to dissect the interaction of SIRT3 with p53 in melanoma cells with wild type p53 versus mutant p53. It is possible that SIRT3 may modulate p53 (in cells with active p53) to affect the proliferation of melanoma cells. This will be important because melanoma is one of the few cancers that rarely possess p53 mutations and SIRT3 may provide an alternative route for altered p53 regulation without actual mutations. At present, our data suggest that SIRT3 inhibition imparts anti-proliferative response in melanoma cells, irrespective of their p53 status.
Since p21 is known to control cell cycle by inhibiting cyclin-cyclin dependent kinase (cdk) network (Dobashi et al., 1998; Fotedar et al., 1996; Sherr and Roberts, 1999), we determine the effect of SIRT3 knockdown on cell cycle distribution. Our data demonstrated that SIRT3 knockdown triggered a marked accumulation of cells in G1 phase at the expense of G2/M and S phases. Interestingly, we did not see any change in sub-G1 population that excludes an induction of apoptosis following SIRT3 knockdown in melanoma cells. However, it remains to be seen if p53 status plays a role in induction of apoptosis versus senescence in melanoma cells.
We next determined the effect of SIRT3 knockdown on cyclin-cdk network operative in G1 phase (Cyclin D1, Cdk4, Cdk6) and G1/S transition (Cyclin E1, Cdk2) because the progression and transition from G1 to S phase requires the formation and action of Cyclin D1-Cdk4 or Cyclin D1-Cdk6 and Cyclin E1-Cdk2 complexes. As expected, we found a down-regulation of Cdk4, Cdk6, Cdk2 and its associated cyclins, Cyclin D1, Cyclin E1 in SIRT3-depleted cells. Interestingly, these targets (p16, p21, Cyclin D1, Cyclin E1, Cdk4, Cdk6 and Cdk2), evaluated in our study have not been reported as the direct targets of SIRT3. However, detailed studies are needed to further dissect the cause-and-effect mechanisms.
Next, we demonstrated that exogenous overexpression of SIRT3 in Hs294T cell line also resulted in a marked increase in cell growth and colonies formation. In addition, immortalized normal melanocytes Mel-ST also demonstrated an enhanced proliferative potential following SIRT3 overexpression.
Finally, employing Nu/Nu xenograft mice implanted with melanoma cells we found that SIRT3 knocked-down cells had significantly decreased tumorigenic potential in vivo. Overall, our study provides several lines of evidence supporting a pro-proliferative/tumor promoter function of SIRT3 in melanoma. This suggests that SIRT3 needs to be further evaluated as a therapeutic target for anti-melanoma drug development. Detailed mechanistic studies are required to further dissect the downstream targets of SIRT3 in melanoma. For example, it will be interesting to determine the effect of SIRT3 knockdown on metabolic pathways in melanoma cells. Similarly, in vivo studies in additional relevant melanoma models, such as BrafV600E/Pten−/− melanoma mouse model and pharmacological inhibitor(s) of SIRT3, are needed to further validate the target ability of SIRT3 for treatment.
MATERIALS AND METHODS
Cell lines and cell culture
Human melanoma cell lines (SK-MEL-28, WM35, SK-MEL-2, G361, A375, Hs294T), human embryonic kidney cell line (HEK293T) and normal human melanocytes (NHEM) cells were purchased from ATCC (Manassas, VA). Non-transformed immortalized primary human melanocyte cell line (Mel-ST) was generously provided by Dr. Robert Weinberg (Whitehead Institute, Cambridge, MA). Details of cell culture are provided in Supplementary Information online.
Quantitative real time-PCR (qRT-PCR)
qRT-PCR was performed using StepOnePlus Real-Time PCR systems (Life Technologies Corp., Carlsbad, CA) and SYBR Premix Ex Taq II (TaKaRa, WI) with first strand cDNA, forward and reverse primers. Details are provided in Supplementary Information online.
Western blotting and immunohistochemistry (IHC)
Western blotting and IHC was performed as detailed in Supplementary Information online.
Lentiviral SIRT3 knockdown
For viral creation, HEK293T cells were transfected using the calcium phosphate transfection method (Schmit et al., 2012) with control empty vector pLKO.1 (SHC001V) and four different SIRT3 targeting short hairpin RNA (shRNA). Details are provided in Supplementary Information online.
MTT assay
The SIRT3 knockdown stable cells were seeded into 24- well plates at a density of 25 × 103 in each well. After culturing for 24, 48 and 72 h, MTT assay was performed as described previously (Wilking et al., 2014).
Clonogenic cell survival assay
The cells with stable SIRT3 knockdown (3 × 103 cells) or overexpression (2 × 103 cells) were seeded in 6-well plates. After 14 days, colony formation was assessed as described previously (Wilking et al., 2014).
Migration assay
Cell migration was analyzed using an Ibidi Culture-Insert (Ibidi, Munich, Germany) in a 35-mm μ-Dish, according to manufacturer's protocol. Briefly, 70 μl of SIRT3-knockdown stable cells or the control cell suspensions (5 × 105 cells/ml) were transferred into each well and incubated at 37°C and 5% CO2. After appropriate cell attachment was achieved (24 h), the Culture-Insert was removed with sterilized forceps and the μ-Dish was filled with cell-free medium (2 ml of media for 35-mm μ-Dish). Migration was assessed, at various time points and images were captured with a Nikon Digital Sight DS-Fi1 inverted microscope (Nikon Instruments Inc., Melville, NY) using NIS Elements AR 3.1 software.
SA-β-Gal staining and SAHF analysis
SA-β-Gal stained positive cell is one of the cellular senescent characteristics (Dimri et al., 1995). Similarly, SAHF formation is regarded to be another marker of cellular senescence (Narita et al., 2003). To determine the induction of senescence, the cells were cultured and SA-β-Gal staining and SAHF analysis were performed as s described in Supplementary Information.
Cell cycle analysis
Cell cycle analysis in SIRT3 knockdown melanoma cells were done as described in Supplementary Information.
Transfections for forced SIRT3 overexpression
For SIRT3 overexpression, Hs294T and Mel-ST cells were transfected with four isolated plasmid DNA of SIRT3-Flag and empty vector pcDNA 3.1(+) as described in Supplementary Information.
Xenograft experiment
Homozygous female nude mice (NU-Foxn1nu NU/NU) aged 6 weeks were purchased from Charles River Laboratories International, Inc. (Wilmington, MA, USA) and quarantined for 2 weeks in pathogen-free conditions at animal facility. At age 8 weeks each mouse was injected subcutaneously on right flank with 2 × 106 cells (shNS-SK-MEL-2 and shSIRT3-SK-MEL-2) mixed with Matrigel (BD Biosceinces) at the ratio of 1:1. Twelve animals per group were used in this study. Once tumors are palpable, they were measured twice per week using digital calipers and volume was calculated by according to the formula 0.52 (height × length × width). Mice were sacrificed when tumors reached 20 mm in the largest dimension. Western blotting of tumor tissue homogenate was used to examine SIRT3 protein expression as described above. All xenograft experiments were performed under a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee.
Statistical analysis
Statistical analyses were performed with two-tailed unpaired Student's t-test between two experimental groups and One-Way ANOVA for more than two experimental groups followed by the Dunnett's test for multiple comparisons using PRISM version 5.0 software (GraphPad Software, Inc., La Jolla, CA). Data was expressed as mean ± SEM unless otherwise stated. P value less than 0.05 was considered as significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Weinberg (Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, MA) for kindly providing Mel-ST cells, used in this study. We are also thankful to Ms Mary Ndiyae for Technical Assistance in melanoma cell implantation in Nu/Nu mice. This work was partially supported by funding from the NIH (R01AR059130, R01CA176748) and the Department of Veterans Affairs (VA Merit Review Award 1I01BX001008).
Abbreviations
- ADP
adenosine diphosphate
- HRP
horseradish peroxidase
- IHC
immunohistochemistry
- NAD+
nicotinamide adenine dinucleotide
- NHEM
normal human epidermal melanocytes
- NMNAT2
nicotinamide mononucleotide adenylyltransferase 2
- SAHF
senescence-associated heterochromatin foci
- SA-β-Gal
senescence-associated beta-galactosidase
- SIRTs
sirtuins
- TMAs
tissue microarrays
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST STATEMENT: None
REFERENCES
- Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–7. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–8. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. Sirtuin-3 (SIRT3) and the Hallmarks of Cancer. Genes Cancer. 2013;4:164–71. doi: 10.1177/1947601913486351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, et al. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–61. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Weinberg RA. When cells get stressed: an integrative view of cellular senescence. J Clin Invest. 2004;113:8–13. doi: 10.1172/JCI200420663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2:648–62. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–7. [PubMed] [Google Scholar]
- Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y, et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014;5:e1047. doi: 10.1038/cddis.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45:1071–7. doi: 10.1016/j.humpath.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobashi Y, Shoji M, Jiang SX, Kobayashi M, Kawakubo Y, Kameya T. Active cyclin A-CDK2 complex, a possible critical factor for cell proliferation in human primary lung carcinomas. Am J Pathol. 1998;153:963–72. doi: 10.1016/S0002-9440(10)65638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18:2789–97. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- Finley LW, Haigis MC. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med. 2012;18:516–23. doi: 10.1016/j.molmed.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Doree M, Messier H, et al. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–64. [PubMed] [Google Scholar]
- Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem J. 2012;444:1–10. doi: 10.1042/BJ20120030. [DOI] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–16. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KH, Hsu CC, Fang WL, Chi CW, Sung MT, Kao HL, et al. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J Surg. 2014;38:910–7. doi: 10.1007/s00268-013-2359-0. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, de Lange T. Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr Biol. 2004;14:2302–8. doi: 10.1016/j.cub.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, Haigis MC. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene. 2015;34:2115–24. doi: 10.1038/onc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–9. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Feng Z, Wu W, Li J, Zhang J, Xia T. SIRT3 regulates cell proliferation and apoptosis related to energy metabolism in non-small cell lung cancer cells through deacetylation of NMNAT2. Int J Oncol. 2013;43:1420–30. doi: 10.3892/ijo.2013.2103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu C, Huang Z, Jiang H, Shi F. The sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed Res Int. 2014;2014:871263. doi: 10.1155/2014/871263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, et al. p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534–42. doi: 10.1093/nar/gks858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit TL, Nihal M, Ndiaye M, Setaluri V, Spiegelman VS, Ahmad N. Numb regulates stability and localization of the mitotic kinase PLK1 and is required for transit through mitosis. Cancer Res. 2012;72:3864–72. doi: 10.1158/0008-5472.CAN-12-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: an update. J Biol Chem. 2012;287:42444–52. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Control of protein function by reversible Nvarepsilon-lysine acetylation in bacteria. Curr Opin Microbiol. 2011;14:200–4. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Yi Y, Li YW, Cai XY, He HW, Ni XC, et al. Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer. 2014;14:297. doi: 10.1186/1471-2407-14-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking MJ, Singh C, Nihal M, Zhong W, Ahmad N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch Biochem Biophys. 2014;563:94–100. doi: 10.1016/j.abb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SM, Han X, Han PJ, Chen HM, Huang LY, Li Y. SIRT3 is a novel prognostic biomarker for esophageal squamous cell carcinoma. Med Oncol. 2014;31:103. doi: 10.1007/s12032-014-0103-8. [DOI] [PubMed] [Google Scholar]
- Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun. 2014a;443:156–60. doi: 10.1016/j.bbrc.2013.11.068. [DOI] [PubMed] [Google Scholar]
- Yang M, Yang C, Pei Y. Effects of downregulation of SIRT3 expression on proliferation and apoptosis in esophageal squamous cell carcinoma EC9706 cells and its molecular mechanisms. Biomed Mater Eng. 2014b;24:3883–90. doi: 10.3233/BME-141219. [DOI] [PubMed] [Google Scholar]
- Zeng YX, el-Deiry WS. Regulation of p21WAF1/CIP1 expression by p53-independent pathways. Oncogene. 1996;12:1557–64. [PubMed] [Google Scholar]
- Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y, et al. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One. 2012;7:e51703. doi: 10.1371/journal.pone.0051703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang H, Wang X, Zhang R, Wang C, Guo Z. Sirtuin-3 (SIRT3) expression is associated with overall survival in esophageal cancer. Ann Diagn Pathol. 2013;17:483–5. doi: 10.1016/j.anndiagpath.2013.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.