Abstract
Inositol pyrophosphates are high energy signaling molecules involved in cellular processes, such as energetic metabolism, telomere maintenance, stress responses, and vesicle trafficking, and can mediate protein phosphorylation. Although the inositol kinases underlying inositol pyrophosphate biosynthesis are well characterized, the phosphatases that selectively regulate their cellular pools are not fully described. The diphosphoinositol phosphate phosphohydrolase enzymes of the Nudix protein family have been demonstrated to dephosphorylate inositol pyrophosphates; however, the Saccharomyces cerevisiae homolog Ddp1 prefers inorganic polyphosphate over inositol pyrophosphates. We identified a novel phosphatase of the recently discovered atypical dual specificity phosphatase family as a physiological inositol pyrophosphate phosphatase. Purified recombinant Siw14 hydrolyzes the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5 or IP7) in vitro. In vivo, siw14Δ yeast mutants possess increased IP7 levels, whereas heterologous SIW14 overexpression eliminates IP7 from cells. IP7 levels increased proportionately when siw14Δ was combined with ddp1Δ or vip1Δ, indicating independent activity by the enzymes encoded by these genes. We conclude that Siw14 is a physiological phosphatase that modulates inositol pyrophosphate metabolism by dephosphorylating the IP7 isoform 5PP-IP5 to IP6.
Keywords: dual-specificity phosphoprotein phosphatase, inositol phosphate, phosphatase, phosphatidylinositol, Saccharomyces cerevisiae, second messenger, IP7, Siw14, Vip1
Introduction
Inositol pyrophosphates are a novel class of signaling molecules that carry energy-rich diphosphate bonds. In wild-type yeast, the most abundant isoform, diphosphoinositol pentakisphosphate (PP-IP5 or IP7),5 consists of a fully phosphorylated six carbon myo-inositol ring further pyrophosphorylated at one of the carbons. Two physiologically relevant isomers of IP7 include 1PP-IP5 and 5PP-IP5 in which the pyrophosphate group is found at the 1-position or the 5-position, respectively. Sequential phosphorylation of IP7 results in bisdiphosphoinositol tetrakisphosphate (1,5PP-IP4 or IP8) that is pyrophosphorylated at both the 1st and 5th positions (1, 2). Since their discovery more than 20 years ago, inositol pyrophosphates have been implicated in an array of processes, including cellular energetic metabolism, telomere maintenance, oxidative stress response, and vesicle trafficking in animals and fungi (1). Recent work in Saccharomyces cerevisiae found that production of inositol pyrophosphates is essential for mounting environmental stress responses (3). Inositol pyrophosphates are thought to be metabolic regulators displaying expanded cellular roles beyond classic second messengers (1).
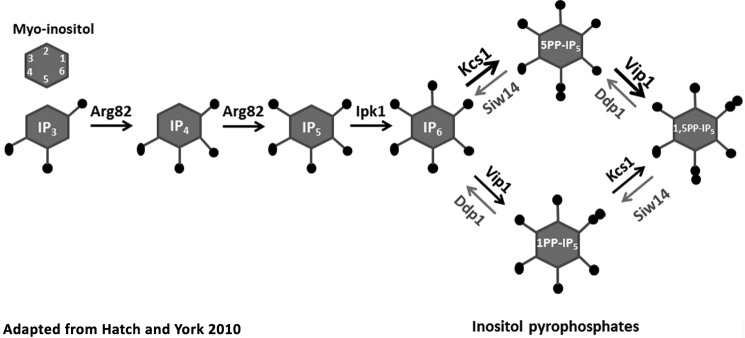
The enzymes that regulate the pools of inositol pyrophosphates are not fully described. The inositol hexakisphosphate kinases synthesize IP7, the most abundant inositol pyrophosphate in the cell; they add the β-phosphate to IP6 at the 5th position generating 5PP-IP5 (2). Yeast possess a single inositol hexakisphosphate kinase gene, KCS1, whereas mammals possess three homologous genes encoding inositol hexakisphosphate kinase (Fig. 1, adapted from Ref. 4). IP8 is synthesized by a PP-IP5 kinase that adds the β-phosphate to IP7 at the 1-position to make 1,5PP-IP4; this enzyme can also use IP6 as a substrate, generating 1PP-IP5. Mammalian genomes have two genes for PPIP5K, and the yeast genome has a single gene, VIP1, that encodes this enzyme. To date, phosphatases described to target the β-phosphate of IP7 and IP8 include members of the Nudix family of hydrolases (2) named DIPP for diphosphoinositol phosphate phosphohydrolase. In mammals, the DIPP enzymes display a preference for the β-phosphate at the 1-position and are encoded by four genes (5). One ortholog is found in yeast, encoded by DDP1; this phosphatase was recently described to prefer polyphosphate (poly-P) over inositol pyrophosphate, although it is able to cleave the β-phosphate at the 1-position on both IP7 and IP8 in vitro (6).
FIGURE 1.
Inositol pyrophosphate synthesis and degradation in yeast. Soluble inositol polyphosphates and inositol pyrophosphates are generated by phosphorylation of myo-inositol-1,4,5-trisphosphate by the inositol phosphate multikinase Arg-82 that produces inositol-1,3,4,5,6-pentakisphosphate after two phosphorylation steps, and inositol phosphate kinase Ipk1 phosphorylates the 2-position to produce IP6. The inositol pyrophosphates are synthesized by adding a β-phosphate to the 5-position preferentially by the IP6 kinase Kcs1 (indicated by the heavier arrows; see Ref. 5), and then at the 1-position by the PP-IP5 kinase Vip1. Our data show that Siw14 opposes the action of Ksc1 by preferentially hydrolyzing the β-phosphate at the 5-position; Ddp1 preferentially hydrolyzes the β-phosphate at the 1-position (6). This schematic was modified from Hatch and York (4).
Siw14 of Saccharomyces is a member of the recently discovered atypical dual specificity phosphatase family, with members in fungi, plants, and protists (7). The SIW14 gene encodes a phosphatase with a basic shallow active site that was previously shown to have low activity with PI(3,5)P2 and no activity with phosphotyrosine (8). Here we describe Siw14 to be a physiological inositol phosphatase dephosphorylating soluble inositol pyrophosphates. Using in vitro assays and in vivo characterization, we show that Siw14 can remove the β-phosphate at the 5-position on IP7 (5PP-IP5). Our results define Siw14 as an important regulator of inositol pyrophosphates and establish its role as an IP7 phosphatase.
Experimental Procedures
Yeast Strains and Plasmids
Yeast strains were purchased from Open Biosystems and Invitrogen, including parental strain BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ) and isogenic mutant strains siw14Δ::KanMX, vip1Δ::KanMX, and ddp1Δ::KanMX. The double mutants vip1Δ::KanMX siw14Δ::URA3 (siw14Δ vip1Δ) and ddp1Δ::KanMX siw14Δ::URA3 (siw14Δ ddp1Δ) were constructed by replacing SIW14 with siw14Δ::URA3 using homologous recombination after amplification of the allele using the primers 5′-AGTTCGGCTTTTTATCTACTCTCTTCTGGATCAATTTTTCTTTTTCATCTAAAGTTTAAAAGGAGTCAATTCATCATTTT-3′ and 5′-TATCAATAAACATCATTTTCGAAGAGACTAGTTACGTAAAGGTAATCACTGTCTACATATTACCATTAGTTTTGCTGGCC-3′. Transformation of Saccharomyces used the protocol of Ito et al. (9).
Plasmid pR492 carries the SIW14 gene with its native promoter in the centromeric pRS316 vector (10) for expression in Saccharomyces. It was cloned after amplification using primers SIW14 forward (5′-ACTCTTCAAGCTTGCTATAGATGGAGCT3′) and SIW14 reverse (5′-AGCTCTCAGCCTATGCATTTAGACTGG-3′), BY4741 genomic DNA as template, and PrimeStar HS polymerase (Clontech). PCR fragments were purified (Qiagen clean-up kit), digested with HindIII and XhoI (New England Biolabs), and ligated into pRS316. The catalytically dead allele, SIW14-C214S (11), was made by fusion PCR (12). Initial synthesis of fragments was completed using PrimeSTAR® HS DNA polymerase (Clontech), following the manufacturer's instructions, pR492 as template, and primer pairs SIW14 forward with C214S reverse (5′-CAACCGATACTGATACATAGTAATAGAGGCAAACATAGAACG-3′) andC214S forward (5′-CGTTCTATGTTTGCCTCTATTACTATGTATCAGTATCGGTTG-3′) with SIW14 reverse. Fragments were purified and combined as templates for the fusion PCR using the SIW14 forward and reverse primers. The fusion product was purified, digested, and ligated as above, and its DNA sequence was determined, generating plasmid pR496.
For expression of proteins in Escherichia coli, plasmid vectors pGEX-4T-1 and pGEX-4T-SIW14 were obtained (7). The mutant allele of SIW14 (SIW14-C214S) was constructed using fusion PCR (12), as described above, using primers pGEX forward (5′-GCAGGGCTGGCAAGCCAC-3′) and reverse (5′-CGAAACGCGCGAGGCAGATC-3′) in place of the SIW14 primers. Fusion products were purified, digested with BamHI and XhoI (New England Biolabs), ligated into pGEX-4T-1, and confirmed by DNA sequencing.
For expression in mammalian cells, the SIW14 gene was cloned into the donor vector plasmid pDONR221 using BP Clonase II and sequentially transferred to a Gateway compatible N-terminally tagged pCMV-Myc destination vector using LR Clonase II, following the manufacturer's directions in the Gateway Cloning Technology Instruction Manual (Life Technologies, Inc.). The SIW14-C214S allele was constructed in the pCMV-Myc plasmid via site-directed mutagenesis using the following primer pairs: forward 5′-CTATTACTATGTATCAGTATCGGTTGATTCGCAGG-3′ and reverse 5′-GATACATAGTAATAGAGGCAAACATAGAACGGGG-3′. Thesequence of all plasmid inserts was determined and confirmed to be correct.
Expression and Purification of Recombinant Proteins
Plasmids pGEX-4T-1, pGEX-4T-SIW14, and pGEX-4T-SIW14-C214S were transformed into the E. coli strain RosettaTM(DE3)pLysS (EMD Millipore). Overnight bacterial cultures were inoculated 1:20 into fresh LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) with 100 μg/μl ampicillin and 34 μg/μl chloramphenicol. Cells were grown at 37 °C with shaking for 2 h (∼0.3 OD600), and protein expression was induced at room temperature for 3 h with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. Transformed cells were lysed as described (7), substituting Tris-buffered saline (19.98 mm Tris-HCl, pH 8.0, 135 mm NaCl) containing 0.1% Triton X-100, 5 mm dithiothreitol (DTT), and protease inhibitors (Halt®, ThermoScientific) as the lysis buffer. Proteins were purified using glutathione-agarose (Pierce) resin following the manufacturer's instructions with the following change: resin with bound protein was incubated in wash buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl) containing 28 mm reduced glutathione (Sigma) for 5 min at room temperature to elute. Three elutions were combined, and proteins were concentrated using Amicon ultracentrifugal 30-kDa filters for 15 min at 4000 × g at 4 °C (Sorvall RC6+ centrifuge). Proteins were examined using SDS-PAGE (Any kDTM Mini-PROTEAN® TGXTM Gel, Bio-Rad) followed by Coomassie Blue staining or immunoblot analysis with rabbit α-GST-HRP antibody (Abcam) at a 1:5000 dilution in phosphate-buffered saline (10 mm sodium phosphate, pH 7.4, 137 mm NaCl, 2.7 mm KCl), 2% milk, and 0.01% Tween 20 and were compared with Kaleidoscope Prestained Standards (Bio-Rad).
Phosphatase Activity Assays
Substrates
p-Nitrophenol phosphate (pNPP) was purchased from Sigma. The phosphatidylinositol phosphates phosphatidylinositol 3-phosphate, phosphatidylinositol 4-phosphate, phosphatidylinositol 5-phosphate, phosphatidylinositol 3,4-bisphosphate, phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), and phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3), each with caprylic acid (8:0) as the two acyl chains, were purchased from Echelon Biosciences. IP6 was purchased from Sigma. Four isoforms of IP7 (5-diphosphoinositol pentakisphosphate (5PP-IP5), non-hydrolyzable 5-methylene-bisphosphonate inositol pentakisphosphate (5PCP-IP5), 1PP-IP5, and non-hydrolyzable 1-methylene-bisphosphonate inositol pentakisphosphate (1PCP-IP5)) and IP8 (3,5-bisdiphosphoinositol tetrakisphosphate (3,5PP-IP4)) were synthesized following the methods described previously (13–15).
[32P]Polyphosphate (poly-32P) was prepared as described previously (16) using purified recombinant E. coli polyphosphate kinase (gift of E. Crooke) and 5 μCi of [δ-32P]ATP (3000 Ci/mmol; PerkinElmer Life Sciences). Substrate preparations were analyzed by scintillation counting to calculate the concentration of the poly-P synthesized.
Detection of Orthophosphate
Orthophosphates cleaved from the phosphatidylinositol phosphates were detected with the EnzChek phosphate assay kit (Life Technologies. Inc.) following the manufacturer's instructions, except reaction volumes were scaled down to 100 μl, and the absorbance at 360 nm was measured using a NanoDrop ND-1000. The orthophosphates cleaved from the inositol pyrophosphates were measured using the Malachite Green Phosphate Assay kit (Cayman Chemical Co.) following the manufacturer's guidelines.
Assay Conditions with pNPP and Phosphatidylinositol Phosphates (PIPn)
Purified recombinant Siw14 (10 μg) was incubated with 10 mm of pNPP in 25 mm HEPES, pH 6, 50 mm NaCl, 10 mm MgSO4, and 1 mm DTT for 90 min at 37 °C. pNPP assays were analyzed by the colorimetric change at 420 nm using the GloMax Multi Detection System (Promega).
Purified recombinant Siw14 (50 μg) was incubated with 100 μm of the mono-, di-, and triphosphorylated phosphatidylinositol phosphates PIPn in reaction buffer (0.1 m Tris-HCl, pH 6.0, 10 mm DTT, 40 mm NaCl) for 90 min at 37 °C (7). PTEN (0.08 μg; Echelon Biosciences) was used with PI(3,4,5)P3. The specific activity of both Siw14 and PTEN with PIPn was normalized to the activities with pNPP.
Assay Conditions with Soluble Inositol Pyrophosphates (IPn)
Purified recombinant Siw14 (10 μg) was incubated with each inositol polyphosphate substrate for 90 min at 37 °C in 25 mm HEPES, pH 6.8, 50 mm NaCl, 10 mm MgSO4, and 1 mm DTT. Human DIPP1 (hDIPP1) enzyme (1 μg) (gift of A. Saiardi) was incubated for 15 min under the same conditions. Reaction products were separated by electrophoresis in 35% acrylamide gels in TBE buffer (89 mm Tris, pH 8.0, 89 mm boric acid, 2 mm EDTA) and stained with toluidine blue (20% methanol, 0.1% toluidine blue, and 3% glycerol) as described (6, 17).
Assay Conditions with Polyphosphate
Recombinant Siw14 (20 μg) was incubated with 600 μm 32P in the form of poly-32P for 90 min at 37 °C in 50 mm HEPES-KOH, pH 7.0, 5 mm MgCl2, 0.4% β-mercaptoethanol, and 2% glycerol. As a positive control, 2.5 μg of E. coli polyphosphate phosphatase (a gift from E. Crooke) was incubated for 15 min under the same conditions. Reaction products were spotted onto thin layer chromatography plates and assayed as described (16). Plates were exposed to phosphorimaging screens, and the density of each spot on the TLC plate was measured using ImageQuant software on a STORM 840 instrument (Molecular Dynamics).
Extraction of [3H]Inositol Phosphates (3H-IPn) and HPLC Analysis
myo-Inositol pools in yeast cells were uniformly radiolabeled by culturing strains overnight in YPD (1% yeast extract, 2% peptone, and 2% dextrose), and 0.005 OD600 cells were inoculated into 10 ml of synthetic complete (SC) medium lacking inositol (0.17% yeast nitrogen base-inositol (MP Biomedicals), 0.5% ammonium sulfate, 0.069% complete supplement mixture (CSM) −Leu amino acid mix (MP Biomedicals), 0.76 mm leucine, and 2% dextrose) and containing 75 μCi of myo-[3H]inositol (>60 Ci/mmol, PerkinElmer Life Sciences). Cultures were grown for 20–24 h until cells were in mid-log phase (∼0.6 OD600). Tritiated soluble inositol phosphates ([3H]-IPn) were extracted as described (18), except that cells were broken via a vortex in the presence of glass beads for four cycles of 2 min with cooling in-between for 2 min on ice. The inositol phosphates in extracts were separated using HPLC using a Hichrom 110 × 4.6-mm Partisphere strong anion exchange column and collecting 1-ml fractions per min for 90 min. The counts/min of each fraction was measured by scintillation counting (Beckman Coulter) in Ultima-Flo AP (PerkinElmer Life Sciences) scintillation fluid. All inositol phosphate profiles were corrected for baseline counts/min before analysis.
Extraction of [3H]Inositol Phosphates (3H-IPn) from Mammalian Cells and HPLC Analysis
One ml of HEK-293T cells was seeded in 6-cm well plates at 0.2 × 106 cells/ml confluency in medium (1× DMEM (Corning CellGro), 10% FBS (HyClone), 1% l-alanyl-l-glutamine (Life Technologies, Inc.)) containing 1% penicillin/streptomycin (Lonza) and 0.1 mg/ml Normocin (InvivoGen). The following day, the medium was exchanged with 1 ml of inositol-free medium supplemented with myo-[3H]inositol (1× DMEM without l-glutamine and without inositol (MP Biomedicals), 10% FBS (HyClone), 1% l-alanyl-l-glutamine (Life Technologies, Inc.), 1% penicillin/streptomycin (Lonza), 0.1 mg/ml Normocin (InvivoGen), and 20 μCi/ml myo-[3H]inositol (PerkinElmer Life Sciences)) and labeled for 72 h. Cells were transfected in duplicate with mammalian expression plasmids that express Myc alone (pCMV-Myc), Myc-tagged Siw14 (Myc-Siw14), or Myc-tagged Siw14-C214S; transfection occurred for 48 h in 2 ml of the myo-[3H]inositol labeling medium with Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions. Soluble [3H]inositol phosphates were extracted as described (18) with the modifications that 250 μl of extraction buffer was added to each well, and duplicate transfected wells were combined and neutralization occurred for 2 h on ice.
Extraction of Poly-P and Quantification
Poly-P was extracted as described (6) and ∼150,000 cpm of poly-32P was added to the lysis buffer per sample to measure extraction efficiency; recovery was measured by scintillation counting. Cells were lysed for 8 min in 2-min cycles. To measure the total poly-P pools, poly-P was converted to ATP using 3.4 μg of polyphosphate kinase, 10 mm ADP in 50 mm Tris-HCl, pH 7.0, 4 mm MgCl2, 40 mm (NH4)2SO4 for 2 h. Reactions were stopped with 4 mm EDTA and 80 mm Tris-HCl, pH 8.0. ATP was measured using an ATP luciferase assay kit (Invitrogen) on a Victor3V 1420 MultiLabel counter (PerkinElmer Life Sciences) plate reader. Twelve biological replicates were analyzed over four experiments; averages were calculated after samples had been normalized to extraction efficiency.
Extraction of Phosphatidyl[3H]inositol Phosphates (P[3H]IPn) and HPLC Analysis
Cells were grown overnight in YPD, and 0.005 OD600 cells were inoculated into SC-inositol medium containing 75 μCi of [3H]inositol. Cultures were grown for 20–24 h until cells were in log phase (∼0.6 OD600). Tritiated phosphatidylinositides (P[3H]IPn) were extracted and deacylated using (19, 20) as guidelines. Briefly, cells were killed by mixing them with 2 volumes of methanol/HCl (100:1) for 5 min on ice and harvested by centrifugation at 6000 × g for 10 min at 4 °C. Lysis was accomplished by 16 rounds of vortexing in 300 μl of H2O/methanol/HCl (66:100:1) with glass beads for 30 s and resting on ice for 1 min between rounds. Phosphatidylinositol phosphates were isolated by extracting the samples with 1.33 ml of ice-cold chloroform and 470 μl of methanol/HCl (100:1) on ice for 10 min. Samples were mixed with 20 μg of nonradioactive bovine brain phosphatidylinositol phosphate as a carrier (Sigma) and extracted with 0.8 m HCl, 5 mm tetrabutylammonium hydrogen sulfate, and 1 mm EDTA. The lower layer was dried in a vacuum dryer and resuspended in 750 μl of methanol. To deacylate the PIPn, samples were incubated with methylamine reagent (Sigma) at 53 °C for 50 min, cooled to room temperature, and dried under vacuum. Samples were washed with water, and then the lower aqueous phase was removed using n-butanol/petroleum ether/ethyl formate (20:4:1) two times and dried again. Once deacylated, the IPn were separated using a Waters 1525 Binary HPLC pump with a Hichrom 125 × 4.6-mm Partisphere strong anion exchange column as described above except that 120 fractions of 1 ml each were collected. The [3H]inositol was quantified for each elution as described above.
Statistical Analysis of Data
Enzyme assays were performed in triplicate except for the PIPn, which were performed in duplicate; this is noted in the figure legends. Experiments with radiolabeled cells and extraction of soluble or lipid inositol phosphates were performed between four and nine times, as indicated in the figure legends, using an overnight culture that had been inoculated with a freshly grown colony of S. cerevisiae with the indicated genotype. For the mammalian transfection experiments, three to five samples containing HEK-293T cells were transfected with the indicated constructs and were analyzed for inositol polyphosphates. The mean and standard error of the mean are reported graphically. The paired Student's t test was used to determine significance, and p values are reported using the following scheme: *, p value ≤0.05; **, p value ≤0.01; ***, p value ≤0.001; and ****, p value ≤0.0001.
Results
Biochemical Characterization of Siw14
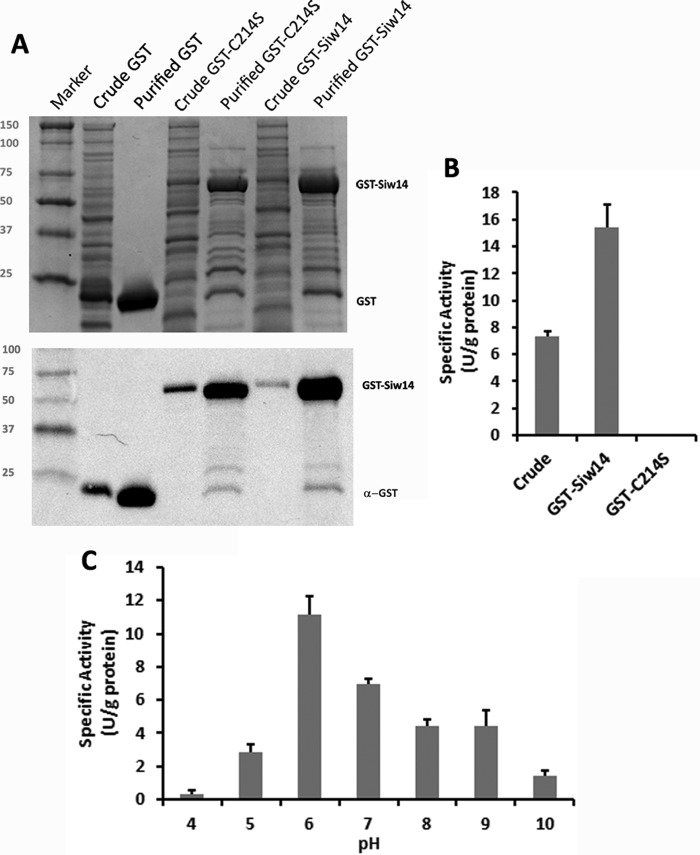
The SIW14 gene of S. cerevisiae encodes a phosphatase with a basic and shallow active site (7, 21). To characterize substrates of the yeast SIW14-encoded phosphatase, GST-tagged Siw14 was expressed in E. coli and purified by affinity chromatography. The predicted mass of the recombinant fusion protein was 52 kDa, and it migrated at ∼50 kDa by SDS-PAGE (Fig. 2A). The purified recombinant protein displayed activity using the generic phosphatase substrate p-nitrophenyl phosphate (pNPP, Fig. 2B). We performed site-directed mutagenesis to generate a phosphatase-inactive mutant of Siw14 by mutating the conserved cysteine within the putative active site (position 214) to serine (11). The activity of the purified GST-Siw14-C214S mutant protein was undetectable (Fig. 2B), confirming that the phosphatase activity detected in the purification of GST-Siw14 was due to Siw14 itself. We determined that the optimal conditions for the Siw14 phosphatase reaction are 37 °C in a buffer of pH 6.0 (Fig. 2C, and data not shown). Treatment with thrombin to remove the GST moiety had no effect on activity toward pNPP (data not shown). The activity of Siw14 for pNPP was low, with a Km of 3 mm, a kcat of 4.4 × 10−7 s−1, and a specificity constant of 1.5 × 10−4 m−1 s−1 (Fig. 3, A and B; Table 1).
FIGURE 2.
Enzymatic activity of purified GST-Siw14 with pNPP. A, purification of GST-Siw14 from E. coli. A representative Coomassie-stained gel (top panel) and anti-GST immunoblot (bottom panel) of crude extracts (crude) and purified empty vector, GST-Siw14, and GST-Siw14 carrying the substitution C214S (GST-C214S) are shown. Ten μg of each protein sample was used for both panels; marker, Kaleidoscope Prestained Standards. B, activity of the crude extract before purification and after purification, GST-Siw14, and a catalytically dead version of GST-Siw14 (GST-C214S). C, GST-Siw14 activity with pNPP while varying the pH. All enzymes assays were completed in triplicate; error bars represent the standard error of the mean.
FIGURE 3.
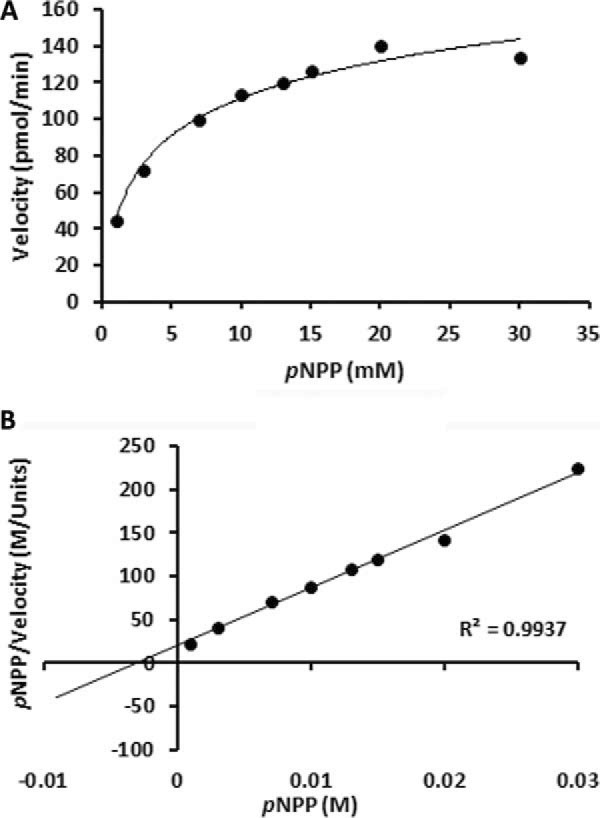
Kinetic characterization of Siw14 with pNPP. A, Michaelis-Menten, B, Hanes-Woolf plots showing the rate of GST-Siw14 activity over a range of substrate concentrations. Each point represents an average of triplicate reactions using 10 μg of protein in buffer (25 mm HEPES, pH 6, 1 mm DTT, 50 mm NaCl, and 10 mm MgSO4) unless otherwise noted.
TABLE 1.
Kinetic measurements for Siw14
| Measurement | pNPP | 5PP-IP5 |
|---|---|---|
| Km | 3 mm | 35 μm |
| kcat | 4.4 × 10−7 s−1 | 2.5 × 10−3 s−1 |
| Specificity Constant | 1.5 × 10−4 m−1 s−1 | 74 m−1 s−1 |
Siw14 Has a Low Activity toward Poly-P and Phosphatidylinositol Phosphates (PIPn)
To identify the substrate(s) of Siw14, we tested polyphosphate and lipid inositol phosphates (7, 21). Siw14 is homologous with the Arabidopsis thaliana protein AtPFA-DSP1 that is able to cleave short chain poly-P molecules (21). Siw14 cleaved orthophosphate from radiolabeled poly-P but only with the use of more protein and longer reaction times as compared with pNPP and with a control phosphatase, E. coli polyphosphate phosphatase (data not shown). Thus, we concluded that poly-P is a poor substrate for Siw14 and is not its specific substrate.
We tested the ability of Siw14 to dephosphorylate phosphatidylinositol phosphates (PIPn) because the purified enzyme was previously described to act upon PI(3,5)P2 (7). We used each of the mono-, di-, and triphosphorylated forms of phosphatidylinositol phosphates with two caprylic acid (8:0) acyl chains as potential substrates and measured the release of orthophosphate. As shown in Fig. 4A, we found that the recombinant Siw14 protein had only minimal activity with each of the PIPn, showing no clear preference for any of these substrates; the greatest activity occurred with PI(3,5)P2 as substrate. We compared Siw14 with PTEN, a well characterized PI(3,4,5)P3 phosphatase, after normalizing their activities to pNPP as a common substrate. The specific activity of Siw14 for PI(3,5)P2 was nearly 20-fold lower than that of PTEN for PI(3,4,5)P3 (Fig. 4A). Although we could replicate the results of Romá-Mateo et al. (7), these data suggested to us that none of the PIPn are the authentic physiological substrates for Siw14.
FIGURE 4.
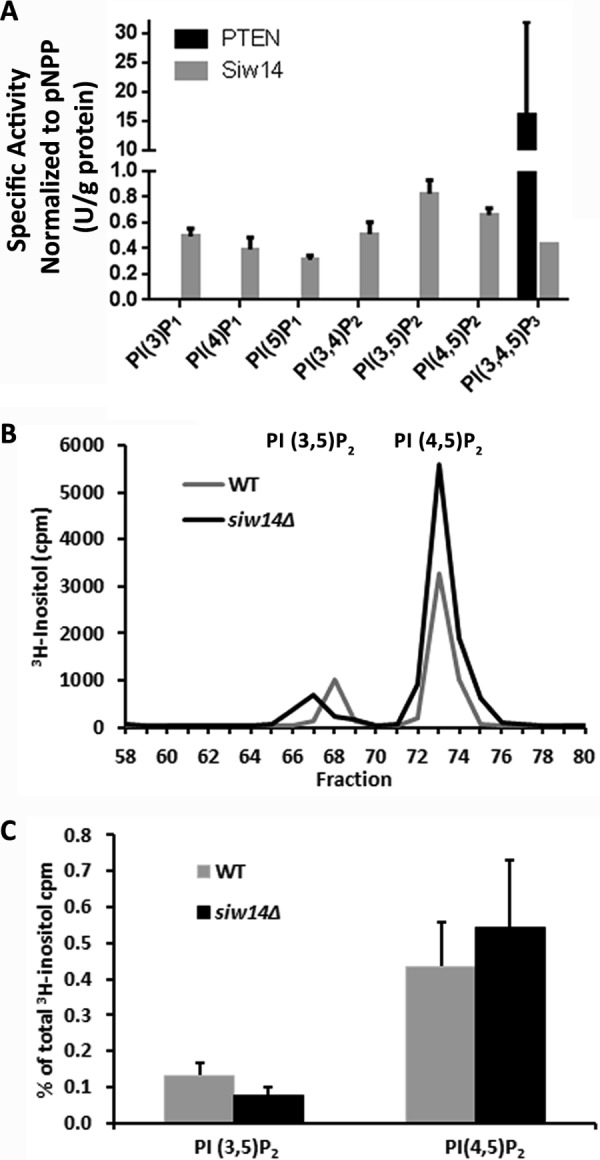
Siw14 has low activity with phosphatidylinositol phosphates. A, specific activity of duplicate samples of Siw14 (50 μg) with each of the seven isoforms of PIPn and PTEN (0.08 μg) with PI(3,4,5)P2, normalized to their specific activity with pNPP. Error bars represent the standard error of the mean. B, representative PIPn profiles for WT and siw14Δ mutant; this experiment was repeated four times. C, average PI(3,5)P2 and PI(4,5)P2 expressed as a percentage of the total [3H]inositol counted in four biological replicates; error bars represent the standard error of the mean. The differences were not significantly different using the Student's t test; p value = 0.22 for PI(3,5)P2 and p value = 0.63 for PI(4,5)P2.
To investigate this further, we radiolabeled cells with myo-[3H]inositol, extracted phosphatidylinositol phosphates from wild-type and siw14Δ mutant cells, and examined the deacylated products by HPLC; using this method, we can detect the inositol headgroups from PI(3,5)P2 and PI(4,5)P2. We found no difference in the levels of tritiated PI(3,5)P2 or PI(4,5)P2 in the siw14Δ mutant as compared with the isogenic wild-type strain, consistent with the in vitro experiments (Fig. 4, B and C).
Soluble Inositol Pyrophosphates Are the Physiological Substrates for Siw14
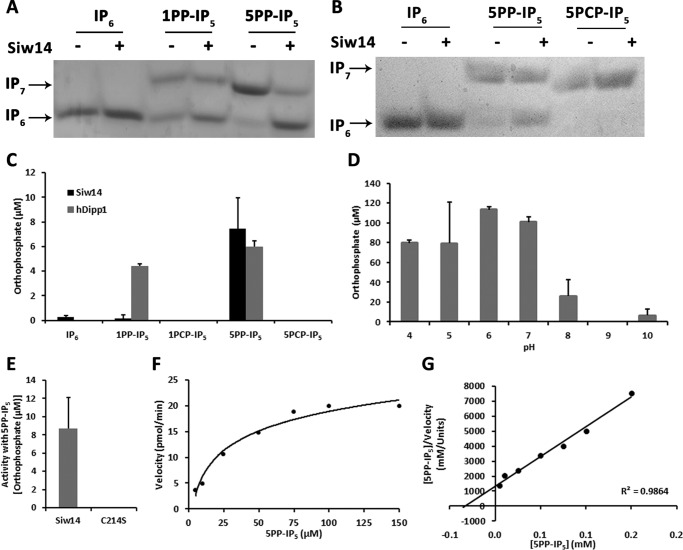
We considered whether soluble inositol polyphosphates might be the natural substrates for Siw14 because of their structural similarity to phosphatidylinositol phosphates for which Siw14 had low preference and the shallow and basic active site of Siw14 that might accommodate them (7). Purified recombinant GST-Siw14 was tested with commercially prepared IP6, two different isoforms of chemically synthesized IP7 (5-diphosphoinositol pentakisphosphate (5PP-IP5) and 1-diphosphoinositol pentakisphosphate (1PP-IP5)). The human DIPP1 (hDIPP1) enzyme was used as a positive control during quantitation because it is a known inositol pyrophosphate phosphatase (5, 6).
As shown in Fig. 5, A and B, Siw14 exhibited no detectable activity with IP6 as a substrate and very low activity with 1PP-IP5 as a substrate. However, Siw14 dephosphorylates 5PP-IP5 to IP6 as shown by the change in mobility of bands on a 35% acrylamide gel (Fig. 5, A and B). We quantified these reactions using a malachite green assay (Fig. 5C), and found that Siw14 exhibited greater activity with 5PP-IP5 than with 1PP-IP5 or with IP6. We also assessed activity from purified recombinant GST-Siw14 and GST-Siw14-C214S proteins with 5PP-IP5 as substrate; we detected no orthophosphate product formed from the C214S mutant protein (Fig. 5E). These data showed that 5PP-IP5 is a preferred substrate for Siw14.
FIGURE 5.
Siw14 dephosphorylates IP7. A and B, reactions containing 10 μg of GST-Siw14 (+) or water (−) were incubated with 400 μm IP6, 1PP-IP5, 5PP-IP5, or the non-hydrolyzable analog 5PCP-IP5 (as indicated) and then separated on a 35% acrylamide gel. The mobility of IP6, IP7, and IP8 is indicated. Representative gels are shown; experiments with 1PP-IP5 and 5PP-IP5 were repeated three times; experiments with 5PCP-IP5 were repeated twice. C, reactions containing 10 μg of GST-Siw14 or 1 μg of hDIPP1 were incubated with 50 μm IP6, 5PP-IP5, 1PP-IP5, or the non-hydrolyzable analogs 5PCP-IP5 or 1PCP-IP5. The cleaved orthophosphate was quantified using a malachite green assay. Reactions were performed in triplicate and were performed with three separate enzyme preparations (data shown are from one of the preparations). D, GST-Siw14 activity with 5PP-IP5 in buffers with the pH varying from pH 4 to pH 10. The liberated orthophosphate was quantified using malachite green. E, activity of purified recombinant GST-Siw14 or GST-Siw14-C214S with 5PP-IP5; F, Michaelis-Menten; G, Hanes-Woolf plots assessing the affinity and kinetics of Siw14 with 5PP-IP5. Bars or points represent the average of triplicate reactions normalized to the no enzyme control for each substrate. Error bars for graphs in C–E represent the standard error of the mean.
To confirm that we were detecting cleavage of the pyrophosphate of 5PP-IP5, we tested the ability of Siw14 to utilize analogs of IP7 that contain non-hydrolyzable methylene-bisphosphonate moieties (13, 14). In these analogs, carbon replaces the oxygen in the anhydride linkage to the β-phosphate. In semi-quantitative gel assays, GST-Siw14 hydrolyzed 5PP-IP5 to IP6 but was unable to use 5PCP-IP5 as a substrate (Fig. 5B). This finding is consistent with Siw14 hydrolyzing the β-phosphate at the 5-position. To quantify this, malachite green assays were performed with GST-Siw14 using 5PP- and 1PP-isomers of IP7 as well as the bisphosphonate analogs; hDIPP1 was used as a positive control. As can be seen in Fig. 5C, we detected no product formed when 5PCP-IP5 and 1PCP-IP5 were used as substrates for GST-Siw14, indicating that Siw14 is unable to cleave any of the phosphates from these molecules. Together with the above data presented in Fig. 4, our results provide support that Siw14 is capable of dephosphorylating the β-phosphate of inositol pyrophosphates as the physiological substrate, preferring 5PP-IP5 over 1PP-IP5, to produce IP6.
We characterized the biochemical properties of GST-Siw14 using 5PP-IP5. We examined the activity of Siw14 under different pH conditions and found that it has highest activity at a pH of 6.0 (Fig. 5D), similar to pNPP. We determined the affinity Siw14 for 5PP-IP5 and assayed its kinetic parameters under these conditions (Fig. 5, E and F). We found a Km of 34 μm, a kcat of 2.5 × 10−3 s−1, and a specificity constant of 74 m−1 s−1 using Hanes-Woolf plot analysis. We note that the Km for 5PP-IP5 is ∼100 times lower than for pNPP, and the kcat for 5PP-IP5 is ∼6000 times faster, supporting an enhanced kinetic efficiency of 5 orders of magnitude with 5PP-IP5 (compare the specificity constants of 74 m−1 s−1 for 5PP-IP5 versus 1.5 × 10−4 m−1 s−1 for pNPP) (Table 1).
Siw14 Modulates IP7 Levels in Vivo
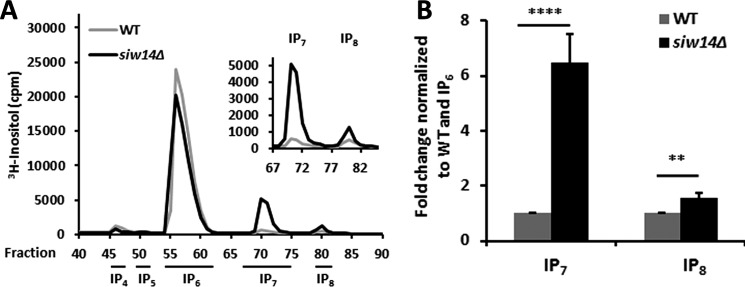
Our in vitro studies demonstrated Siw14 to be a 5PP-IP5 phosphatase. To evaluate the in vivo relevance of these results, we examined the intracellular inositol polyphosphate levels in siw14Δ mutant strains. The relative inositol phosphate levels were quantified by radiolabeling cells with myo-[3H]inositol, extracting soluble inositol phosphates, separating fractions by HPLC, and quantifying tritium by scintillation counting. The IP7 levels were determined relative to IP6 and then expressed as normalized to the WT strain. In the siw14Δ mutant, levels of IP7 increased by an average of 6.5 ± 1.0-fold (Fig. 6A), and the levels of IP8 also increased by about 1.6 ± 0.2-fold (Fig. 6B). Alternatively, when these data are expressed as a ratio of IP6 to IP7 to IP8 in each cell type, the WT cells have a ratio of 96.4:2.1:1.5, whereas the ratio in the siw14Δ mutant changed to 85.5:12.8:1.7. Thus, we saw substantial changes in the pools of both IP6 and IP7.
FIGURE 6.
In vivo IP7 and IP8 levels increase in the SIW14 yeast deletion mutant. A, representative inositol polyphosphate profiles of WT and siw14Δ. B, ratios of IP7 or IP8 to IP6 in yeast siw14Δ, normalized to WT. Average values are from nine replicates; error bars represent the standard error of the mean. Significance was determined by Student's t test; **, p ≤ 0.01; ****, p ≤ 0.0001.
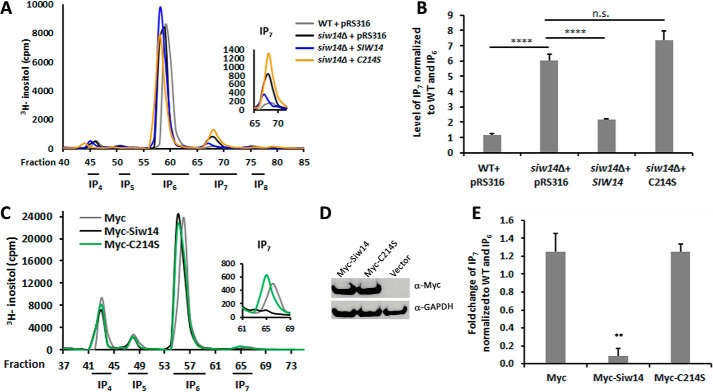
We complemented the siw14Δ mutant with the wild-type and C214S mutant alleles of SIW14, and we fractionated the radiolabeled soluble inositol polyphosphates. As shown in Fig. 7, the wild-type allele of SIW14 restored native levels of IP7 to the strain, but the catalytically dead allele of SIW14 (C214S) was unable to complement and the IP7 levels were not significantly different from the null mutant (Fig. 7, A and B).
FIGURE 7.
Catalytic activity of Siw14 is necessary to modulate IP7 levels. A, representative inositol polyphosphate profiles of WT and the siw14Δ mutant complemented with empty vector, wild-type gene SIW14, and an allele expressing the catalytically dead Siw14-C214S. B, ratio of IP7 to IP6 normalized to WT, performed in triplicate. C, representative inositol phosphate profiles from empty vector, Myc-SIW14, and Myc-SIW14-C214S transfected HEK-293T cells. D, Western blot showing successful expression of the two alleles of Myc-Siw14 relative to α-GAPDH. E, ratios showing a significant fold decrease of IP7 in Myc-Siw14-transfected cells compared with empty vector. The control and Myc-SIW14 experiment was performed on five samples of transfected cells; the Myc-SIW14-C214S experiment was performed in triplicate. For graphs in B and E, the bars represent the average of the replicates with standard error. Significance was determined using the Student t test, **, p values ≤0.01; ****, p values ≤0.0001. n.s., not significant.
We extended this analysis by testing whether Siw14 is sufficient for in vivo regulation of IP7 by overexpression in mammalian cells. Vector (pCMV-Myc) or myc-tagged SIW14 expression constructs were transfected into HEK-293T cells radiolabeled with myo-[3H]inositol. In the presence of Siw14, there was a reduction in endogenous IP7 levels compared with the cells transfected with the empty vector control pCMV-Myc. Furthermore, expression of the catalytically dead protein (myc-tagged SIW14-C214) did not lead to a change in the endogenous IP7 pools (Fig. 7, C and E). Together, these in vivo findings are consistent with our in vitro data supporting a role for Siw14 as an IP7 phosphatase.
Higher Levels of Inositol Pyrophosphates Do Not Affect Poly-P pools
Yeast cells that are unable to synthesize inositol pyrophosphates have undetectable levels of poly-P (6). We wondered whether the opposite might be true, i.e. whether increased levels of inositol pyrophosphates might result in higher levels of poly-P. To examine this, the relative poly-P levels were measured by extracting poly-P, enzymatically converting it to ATP, and quantifying ATP via the release of light using a luciferase assay (6, 22). We found no difference in poly-P pools in the siw14Δ mutant as compared with the parental strain (non-significant decrease, p value of 0.09) over four separate experiments (data not shown). We conclude that higher levels of inositol pyrophosphates do not result in higher levels of poly-P.
Siw14 and Other Enzymes That Metabolize IP7 Act Independently
A number of mutant yeast strains have been shown to accumulate IP7 (Fig. 1) (6, 23, 24). Vip1 is the kinase that adds the β-phosphate at the 1-position to convert IP7 (5PP-IP5) to IP8 (1,5PP-IP4) (23). DDP1 encodes the Saccharomyces homolog of hDIPP1. In vitro, the Ddp1 enzyme is able to dephosphorylate the β-phosphate from IP7 only at prolonged time points or at non-physiological concentrations; however, in vivo IP7 levels increase in the ddp1 mutant (6). We measured the levels of IP7 in the ddp1Δ and the vip1Δ mutants and compared them with the siw14Δ mutant. We found that the increase in IP7 levels for the siw14Δ mutant (6.5 ± 1.0-fold) was of the same magnitude as the accumulation in the ddp1Δ (7.0 ± 1.1-fold) and vip1Δ (4.0 ± 1.7-fold) mutants (Fig. 8, A–C), as had been seen previously (6).
FIGURE 8.
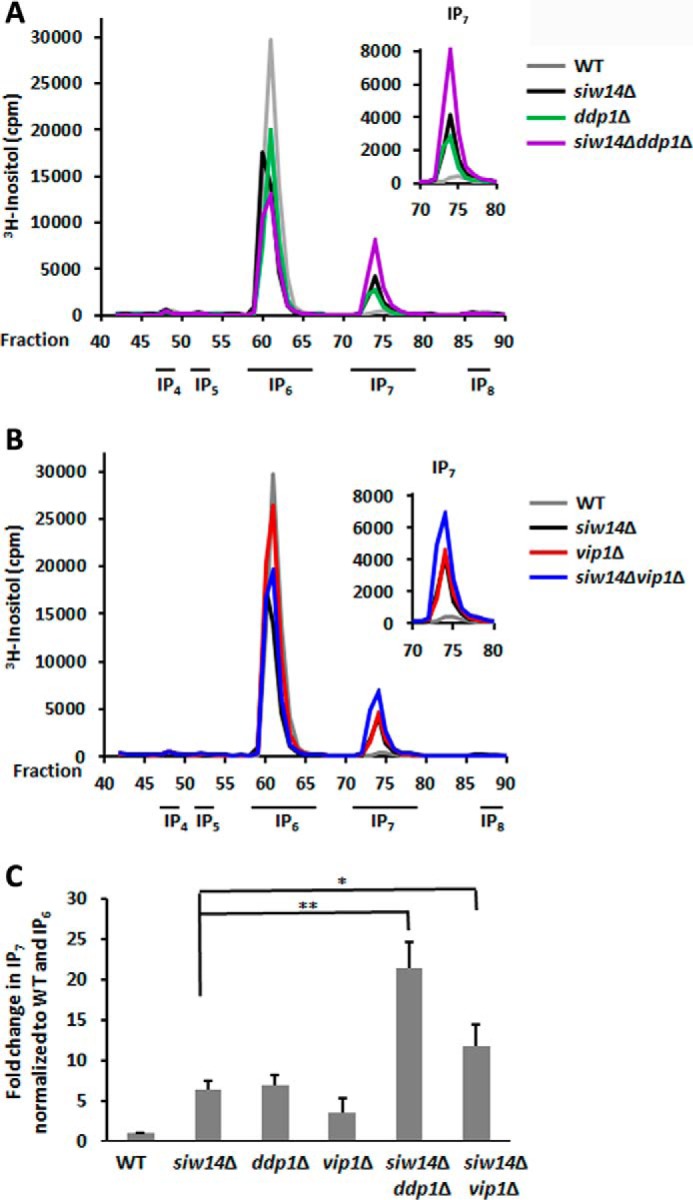
Levels of IP7 increase in single and double kinase and phosphatase mutants. A, representative inositol polyphosphate profiles of WT, and single and double mutants of siw14Δ and ddp1Δ. B, representative inositol polyphosphate profiles of WT, and single and double mutants of siw14Δ and vip1Δ. C, levels of IP7 in each mutant, normalized to WT and IP6, over four separate experiments. Error bars represent standard error of the mean, and significance was determined by Student's t test. Each mutant was significantly different than the WT, p = ≤ 0.0001; *, p ≤ 0.05; **, p ≤ 0.01.
We wondered whether these enzymes acted independently. We assessed accumulation of IP7 in the double siw14Δ ddp1Δ and siw14Δ vip1Δ mutants. Both double mutants accumulate higher levels of IP7 than either single mutant (Fig. 8, A–C); we found a 21.3 ± 3.2-fold increase for the siw14Δ ddp1Δ mutant that was slightly more than additive, and an 11.8 ± 2.6-fold increase for the siw14Δ vip1Δ mutant that was additive.
Discussion
In this study we have discovered Siw14 to be a physiological regulator of the soluble inositol pyrophosphate 5PP-IP5 (see Fig. 1). Siw14 displays pyrophosphate-specific phosphatase activity both in vitro and in vivo. The Siw14 enzyme has a strong preference for inositol pyrophosphates over other previously described putative substrates (such as poly-P and PI(3,5)P2) (7, 21). Siw14 is a member of the plant and fungi atypical-dual specificity phosphatase (PFA-DSP) subfamily (7). This subfamily has members in plants, fungi, kinetoplastids, and slime molds (7), and there is only limited homology with the broader dual specificity phosphatase family at the signature motif C(X)5R in the catalytic center (21). The proteins within the PFA-DSP subfamily have not been well studied, and the evolutionary conservation of their inositol pyrophosphate phosphatase activity remains unknown. However, DSP enzymes are distinct from other characterized inositol pyrophosphate phosphatases that are of the Nudix hydrolase family.
The A. thaliana genome encodes five isoforms of this protein, and S. cerevisiae has three isoforms (7, 8). The structure of the A. thaliana AtPFA-DSP1 was determined by x-ray crystallography (21), and the yeast Siw14 ortholog was modeled based on this structure (7). The active site is predicted to be basic and shallow, consistent with our finding that the substrate is large and negatively charged (7, 21). Siw14 and AtPFA-DSP1 are highly conserved throughout the catalytic core and share 61% identity and 76% similarity (7). Although the AtPFA-DSP1 protein was shown to dephosphorylate phosphotyrosine, phosphatidylinositol mono-, di-, and triphosphates, and short chain inorganic polyphosphate (poly-P) (7, 21), it was not tested with soluble inositol pyrophosphates, and thus it is unclear what substrates it would prefer. A potentially important role in plants for these PFA-DSP enzymes is implied given the genomic expansion of this phosphatase subfamily.
Despite apparently similar roles in vivo, we found that recombinant Siw14 had a lower specific activity toward 5PP-IP5 when compared with hDIPP1, the previously characterized inositol pyrophosphate phosphatase from humans. The specificity constant for 5PP-IP5 from our Siw14 data (74 m−1 s−1) is significantly lower than the previously reported data for hDIPP1, which ranges from 2.5 × 106 to 4.76 × 107 m−1 s−1 (5, 25). There are several possible reasons to explain this difference. The first is that the enzymes belong to different classes as follows: Siw14 is a member of the atypical dual specificity phosphatase family (EC 3.1.3, phosphoric monoester hydrolase), and hDIPP1 is a member of the Nudix family (EC 3.6.1, acting on phosphorus-containing anhydrides). Second, this difference may be specific to the recombinant form of Siw14 enzyme. It is possible that the activity of Siw14 is regulated by a post-translational modification; it has two putative phosphorylation sites (26) and E. coli is unable to make these post-translational modifications. Alternatively, the purification process may have yielded substantial inactive protein or that Siw14 may require additional partner proteins for full activity. Using two-hybrid and affinity purification approaches, several high throughput studies found that Siw14 is in a complex with Oca1 and Oca2 (27–30). It is possible that full activity from Siw14 may require these (or other) partner proteins. Ongoing experiments are being performed to address the role(s) for these proteins in their interactions with Siw14.
Consistent with the in vitro biochemical assays, IP7 levels rise in the siw14Δ mutant. The increase in IP7 in the siw14Δ mutant (Fig. 6) was similar to that observed in the vip1Δ and ddp1Δ deletion mutants (6, 23, 24). The IP7 that we detected is likely to be the 5PP-IP5 isoform given the presence of Kcs1 (the kinase specific for the 5-position). We see a small but significant increase in the accumulation of IP8 in the siw14Δ mutant, likely due to the increased amount of substrate for the Vip1 kinase (23); we can also detect a 2.8-fold increase in IP8 in the siw14Δ ddp1Δ double mutant (data not shown). Our data are consistent with a model that indicates that the primary pathway from IP6 to IP8 is through the addition of a β-phosphate to the 5-position of IP6 by Kcs1 prior to the addition at the 1-position by Vip1 (Fig. 1). Because the siw14Δ vip1Δ deletion mutant has twice the IP7 levels as either of the single mutants, this finding suggests an additive effect on the IP7 pools due to independent activity by these enzymes. We cannot rule out additional pathway regulation in the siw14Δ ddp1Δ mutant because the IP7 increase was slightly more than additive in that strain.
Inositol pyrophosphates have been linked to cellular stress response, vesicle trafficking, ribosome biogenesis, telomere length, energy charge, and starvation (3, 31–34). The Sudbery laboratory showed that the siw14Δ mutant does not respond appropriately to nutrient deprivation in stationary phase and is defective in fluid phase endocytosis (11). Our laboratory noted an up-regulation of the environmental stress response in unstressed siw14Δ mutants compared with the wild-type, consistent with the results of Worley et al. (3).6 The siw14Δ mutant of Saccharomyces also has actin clumps and cytoskeletal defects during stationary phase (11); Schizosaccharomyces pombe strains with a mutation in the asp1+ gene, a member of the VIP1 family, also exhibit cytoskeleton structure defects (35). We note that yeast strains with mutations in SIW14, VIP1, and KCS1 genes share similar phenotypes such as resistance to H2O2 and other stresses (3, 24).6 The intrinsic levels of or the cycling through of inositol pyrophosphates might be important for the regulation of the stress response in yeast, although it has yet to be determined whether the accumulation of the different isoforms of IP7 have the same or overlapping phenotypes. IP7 levels are highly dynamic in mammalian cells (36); whether and how Siw14-like functionality is conserved in higher organisms remain to be defined. Our identification of a novel inositol pyrophosphate phosphatase provides additional support for what is likely to emerge as a highly regulated signaling network characterized by this novel high energy molecule class whose cellular pools are combinatorially altered by phosphotransfer to proteins, kinase-mediated synthesis, and dephosphorylation.
Author Contributions
E. A. S. purified proteins, performed site-directed mutagenesis, designed, performed, and analyzed enzyme assays, performed poly-P experiments, including synthesizing poly-32P, extracted soluble inositol polyphosphates and poly-P, performed and analyzed the mammalian expression studies, analyzed inositol polyphosphate, PIPn HPLC data, and poly-P data, and wrote the paper. L. S. C. designed, performed, and analyzed the mammalian studies, performed the yeast lipid extractions, performed HPLC analyses and scintillation counting, and analyzed the data. M. W. synthesized all isoforms of IP7. E. C. designed and assessed the poly-P studies. D. F. designed the synthesis of IP7. A. C. R. designed the inositol polyphosphates studies. R. J. R. conceived and coordinated the study. All authors reviewed the results, edited the manuscript, and approved the final version of the manuscript.
Acknowledgments
We thank the Pulido laboratory for sending the pGEX-4T-SIW14 plasmid we used to purify the GST-Siw14 protein. We thank Katherine McIntire in the Crooke laboratory for showing us how to synthesize poly-P and make ATP from poly-P. Purified recombinant polyphosphate kinase and polyphosphate phosphatase enzymes were provided by E. Crooke. We also thank Adolfo Saiardi for supplying the purified hDIPP enzyme.
This work was supported by National Science Foundation CAREER Grant MCB-1253809 (to A. C. R.) and a Georgetown University Pilot Grant (to R. J. R.). The authors declare that they have no conflicts of interest with the contents of this article.
E. A. Steidle, A. C. Resnick, and R. J. Rolfes, unpublished data.
- 5PP-IP5 or IP7
- 5-diphosphoinositol pentakisphosphate
- IP6
- inositol hexakisphosphate
- 1,5PP-IP4
- bis-diphosphoinositol tetrakisphosphate (also known as IP8)
- PCP-IP5
- methylene-bisphosphonate inositol pentakisphosphate
- poly-P
- polyphosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-triphosphate
- Siw14
- phosphatase from SIW14 gene (also called Oca3)
- Kcs1
- IP6 kinase (IP6K)
- Vip1
- PP-IP5 kinase (PPIP5K)
- Ddp1
- diadenosine and diphosphoinositol polyphosphate phosphohydrolase
- DIPP
- diphosphoinositol phosphate phosphohydrolase
- PTEN
- phosphatase and tensin homolog
- Nudix
- nucleoside diphosphate linked to X phosphohydrolase
- PFA-DSP
- plant-fungal atypical dual-specificity phosphatase
- hDIPP1
- human DIPP1
- pNPP
- p-nitrophenol phosphate
- 1PP-IP5
- 1-diphosphoinositol pentakisphosphate
- 5PCP-IP5
- 5-methylene-bisphosphonate inositol pentakisphosphate.
References
- 1. Wilson M. S., Livermore T. M., and Saiardi A. (2013) Inositol pyrophosphates: between signaling and metabolism. Biochem. J. 452, 369–379 [DOI] [PubMed] [Google Scholar]
- 2. Thomas M. P., and Potter B. V. (2014) The enzymes of human diphosphoinositol polyphosphate metabolism. FEBS J. 281, 14–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Worley J., Luo X., and Capaldi A. P. (2013) Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 3, 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hatch A. J., and York J. D. (2010) SnapShot: inositol phosphates. Cell 143, 1030–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kilari R. S., Weaver J. D., Shears S. B., and Safrany S. T. (2013) Understanding inositol pyrophosphate metabolism and function: kinetic characterization of the DIPPs. FEBS Lett. 587, 3464–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., and Saiardi A. (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Romá-Mateo C., Sacristán-Reviriego A., Beresford N. J., Caparrós-Martín J. A., Culiáñez-Macià F. A., Martín H., Molina M., Tabernero L., and Pulido R. (2011) Phylogenetic and genetic linkage between novel atypical dual specificity phosphatases from non-metazoan organisms. Mol. Genet. Genomics 285, 341–354 [DOI] [PubMed] [Google Scholar]
- 8. Romá-Mateo C., Ríos P., Tabernero L., Attwood T. K., and Pulido R. (2007) A novel phosphatase family, structurally related to dual specificity phosphatases, that displays unique amino acid sequence and substrate specificity. J. Mol. Biol. 374, 899–909 [DOI] [PubMed] [Google Scholar]
- 9. Ito H., Fukuda Y., Murata K., and Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., Mannazzu I., and Sudbery P. E. (2004) A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cha-Aim K., Hoshida H., Fukunaga T., and Akada R. (2012) Fusion PCR via novel overlap sequences. Methods Mol. Biol. 852, 97–110 [DOI] [PubMed] [Google Scholar]
- 13. Wu M., Dul B. E., Trevisan A. J., and Fiedler D. (2013) Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem. Sci. 4, 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu M., Chong L. S., Capolicchio S., Jessen H. J., Resnick A. C., and Fiedler D. (2014) Elucidating diphosphoinositol polyphosphate function with nonhydrolyzable analogues. Angew. Chem. Int. Ed. Engl. 53, 7192–7197 [DOI] [PubMed] [Google Scholar]
- 15. Capolicchio S., Wang H., Thakor D. T., Shears S. B., and Jessen H. J. (2014) Synthesis of densely phosphorylated bis-1,5-diphospho-myo-inositol tetrakisphosphate and its enantiomer by bidirectional P-anhydride formation. Angew. Chem. Int. Ed. Engl. 53, 9508–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akiyama M., Crooke E., and Kornberg A. (1993) An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268, 633–639 [PubMed] [Google Scholar]
- 17. Losito O., Szijgyarto Z., Resnick A. C., and Saiardi A. (2009) Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE 4, e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Azevedo C., and Saiardi A. (2006) Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 1, 2416–2422 [DOI] [PubMed] [Google Scholar]
- 19. Dove S. K., and Michell R. H. (2009) Inositol lipid-dependent functions in Saccharomyces cerevisiae: analysis of phosphatidylinositol phosphates. Methods Mol. Biol. 462, 59–74 [DOI] [PubMed] [Google Scholar]
- 20. Auger K. R., Serunain L. A., and Cantley L. C. (1990) in Methods in Inositide Research (Irvine R. F., ed) pp. 159–166, Raven Press, Ltd., New York [Google Scholar]
- 21. Aceti D. J., Bitto E., Yakunin A. F., Proudfoot M., Bingman C. A., Frederick R. O., Sreenath H. K., Vojtik F. C., Wrobel R. L., Fox B. G., Markley J. L., and Phillips G. N. Jr. (2008) Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000. Proteins 73, 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ault-Riché D., Fraley C. D., Tzeng C. M., and Kornberg A. (1998) Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180, 1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., and York J. D. (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 [DOI] [PubMed] [Google Scholar]
- 24. Onnebo S. M., and Saiardi A. (2009) Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 423, 109–118 [DOI] [PubMed] [Google Scholar]
- 25. Safrany S. T., Ingram S. W., Cartwright J. L., Falck J. R., McLennan A. G., Barnes L. D., and Shears S. B. (1999) The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 274, 21735–21740 [DOI] [PubMed] [Google Scholar]
- 26. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 28. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., and Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 30. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 31. Fleischer B., Xie J., Mayrleitner M., Shears S. B., Palmer D. J., and Fleischer S. (1994) Golgi coatomer binds, and forms K+-selective channels gated by, inositol polyphosphates. J. Biol. Chem. 269, 17826–17832 [PubMed] [Google Scholar]
- 32. Ali N., Duden R., Bembenek M. E., and Shears S. B. (1995) The interaction of coatomer with inositol polyphosphates is conserved in Saccharomyces cerevisiae. Biochem. J. 310, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye W., Ali N., Bembenek M. E., Shears S. B., and Lafer E. M. (1995) Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J. Biol. Chem. 270, 1564–1568 [PubMed] [Google Scholar]
- 34. Szijgyarto Z., Garedew A., Azevedo C., and Saiardi A. (2011) Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 [DOI] [PubMed] [Google Scholar]
- 35. Pöhlmann J., Risse C., Seidel C., Pohlmann T., Jakopec V., Walla E., Ramrath P., Takeshita N., Baumann S., Feldbrügge M., Fischer R., and Fleig U. (2014) The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 10, e1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Menniti F. S., Miller R. N., Putney J. W. Jr., and Shears S. B. (1993) Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 268, 3850–3856 [PubMed] [Google Scholar]