Summary
Understanding cardiomyocyte cell cycle regulation after birth is key to optimizing regenerative strategies for the heart post-injury, yet poses multiple technical challenges, as evidenced by recent studies that have arrived at divergent conclusions. In a recent publication in Cell, Alkass et al undertook multiple approaches to examine cardiomyocyte cell cycle regulation in the first three weeks after birth. Here, we summarize results of Alkass et al and three other groups in examining preadolescent cardiomyocyte cell cycle regulation, highlighting the distinct approaches and incumbent caveats.
Understanding cardiomyocyte cell cycle activity during the perinatal and preadolescent period is extremely challenging and is a subject of intense debate. From a classical viewpoint, throughout embryonic development, cardiomyocytes progressively lose their ability to divide and proliferate. Following the period between postnatal day 5 (P5) and 10 (P10), the second and final wave of non-replicative DNA synthesis ends with binucleation of existing cardiomyocytes 1. Recently, Naqvi and colleagues proposed that cardiomyocytes undergo an additional burst of synchronized proliferation on postnatal day 15 (P15) that results in a 40% increase in the number of cardiomyocytes 2. However, in a recent study Alkass et al observed that an increase in cardiomyocyte number after birth is largely restricted to the first postnatal week, with no significant increase in number after postnatal day 11 3. Consistent with Alkass et al, two other groups were not able to substantiate a proliferative burst of cardiomyocytes during preadolescence between the second and third postnatal weeks 4, 5. Interestingly, Alkass et al observed a peak of polyploidization of binucleated cardiomyocytes between the second and third postnatal weeks, introducing further complexity to the model of cardiomyocyte cell cycle behavior.
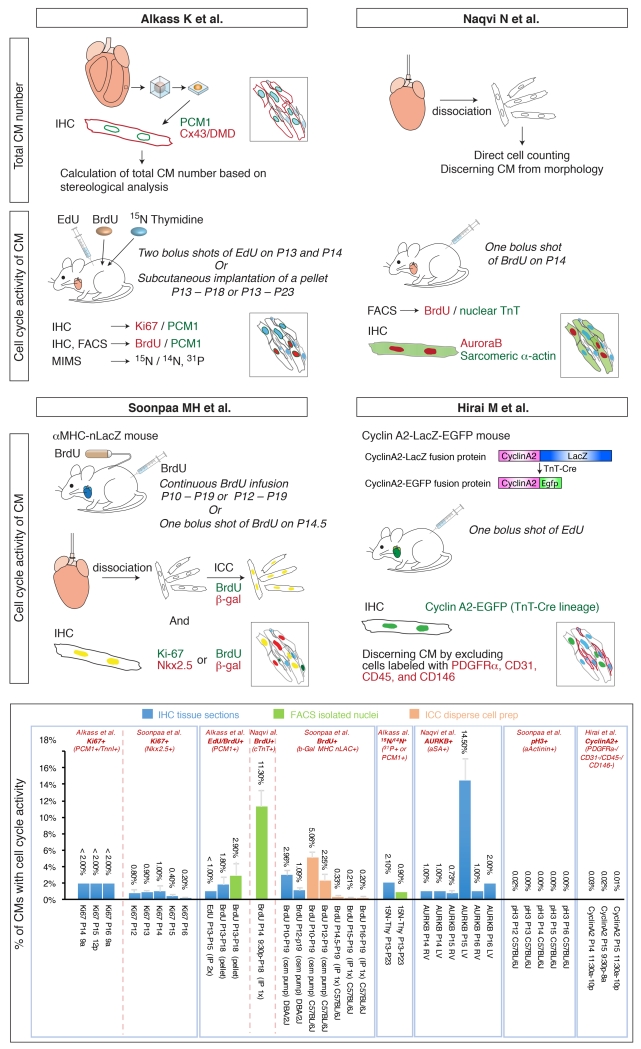
Defining the number, proliferation, multinucleation and ploidy of postnatal cardiomyocytes is technically challenging owing to the complex cellular architecture of the heart, and to the lack of reliable cardiomyocyte-specific nuclear markers 6. Cardiomyocytes are surrounded by multiple smaller cell types, such as fibroblasts, endothelial cells, smooth muscle cells, pericytes, and tissue-resident immune cells, which each have distinct proliferative potentials in both physiological growth and pathological conditions. The key to overcoming these technical challenges depends on the tools and approaches used to identify cardiomyocytes, to quantify their numbers, and to measure their cell cycle activity. As shown in the accompanying Figure, recently several distinct approaches have been undertaken to examine cell cycle behaviors of postnatal cardiomyocytes, resulting in somewhat different conclusions.
Figure.
An illustration of methods undertaken by four independent groups to examine cell cycle activity of cardiomyocytes during preadolescence, and a summary of their results.
Total Number of Cardiomyocytes
To assess the total number of cardiomyocytes in left ventricle from P2 to P100, Alkass et al utilized “design-based stereology”. Tissue segments were isolated from left ventricle, made into spherical isectors and utilized to generate thick frozen sections. Myocardial nuclei and cells were labeled by immunostaining for Pericentriolar material 1 (PCM-1), Connexin43 (Cx43) and Dystrophin (DMD), and counted to deduce myocardial cell density. The total number of cardiomyocytes was calculated by multiplying the myocardial cell density within the sample by the estimated volume of the entire left ventricle, calculated from actual left ventricular weight. With this approach Alkass et al concluded that the number of left ventricular cardiomyocytes increases 40% from P2 (1.7 × 106 ± 0.2 × 106) to P5 (2.3 × 106 ± 0.2 × 106), but that the number of left ventricular cardiomyocytes does not increase during preadolescence from P14 to P18.
With this method, actual myocardial cell numbers could be different if there are regional differences in myocardial cell density within the left ventricle. In addition, as PCM-1 is a centrosomal matrix protein expressed in multiple cell types, it is not clear how specifically PCM-1 marks cardiomyocyte nuclei, although two groups have previously used PCM-1 as a cardiomyocyte specific marker 7, 8.
On the other hand, Naqvi et al performed enzymatic dissociation of both ventricles and calculated the total cardiomyocyte number by direct cell counting of small sample aliquots with a hemocytometer, defining cardiomyocytes by their morphology. In this manner, Naqvi et al found a similar 40% increase in the number of cardiomyocytes in the early perinatal period (P1 to P4), although the number of cardiomyocytes summed for both ventricles was about two-fold lower than the number of cardiomyocytes calculated by Alkass et al for left ventricle alone. However, in contrast to Alkass et al, Naqvi et al found an additional 40% increase in cardiomyocyte numbers during preadolescence (P14 to P18). As indicated by Alkass et al, a potential issue with this approach is that the efficiency of dissociation can be variable.
Cardiomyocyte Cell Cycle Activity
One of the key issues in determining cardiomyocyte cell cycle activity is unequivocal identification of cardiomyocytes. BrdU, EdU, Ki67, and phosphorylated Histone 3 are intranuclear markers generally used to label cells undergoing proliferation or DNA synthesis. Optimally, these markers should be analyzed in conjunction with a marker of cardiomyocyte nuclei 6.
To investigate a potential burst of cardiomyocyte cell cycle activity in preadolescence, Alkass et al used an anti-PCM1 antibody to label cardiomyocyte nuclei for immunohistochemistry or flow cytometry. However, as PCM-1 disassembles in prometaphase, this method might lead to an underestimation of the actual number of cardiomyocytes. However, this possibility was taken into consideration by Alkass et al, who addressed this issue by co-immunostaining for Troponin I.
Recent studies by other groups investigating cell cycle behavior in postnatal cardiomyocytes utilized distinct nuclear markers for cardiomyocytes. Soonpaa et al used an anti-Nkx2-5 antibody or an αMHC-nLacZ reporter mouse 5. Hirai et al generated and used a CyclinA2-LacZ-EGFP reporter mouse. In this reporter, a CyclinA2-lacZ fusion reporter is expressed under control of the CyclinA2 locus within BAC sequences, and upon Troponin T-Cre mediated excision, is converted to a CyclinA2-EGFP fusion reporter, expressed under the control of the CyclinA2 locus only in cycling cardiomyocytes 4. In contrast to the nuclear markers mentioned above, the original report suggesting a preadolescent burst of cardiomyocyte proliferation largely relied on immunostaining for α-Sarcomeric Actin to identify cardiomycytes in tissue sections 2. Although this marker has been used broadly to identify cardiomyocytes, it is challenging to unequivocally determine whether a nucleus belongs to an α-Sarcomeric Actin positive cardiomyocyte, or to closely juxtaposed non-cardiomyocytes.
Alkass et al utilized three distinct assays to determine cardiomyocyte cell cycle activity, and did not observe a burst of cardiomyocyte proliferation during preadolescence with any of their approaches. The first approach was section immunostaining with antibodies to Ki67 in conjunction with antibodies to PCM-1 to mark cardiomyocyte nuclei. Results demonstrated that fewer than 2% of cardiomyocytes were positive for Ki67 from P14 through P16. In agreement with this, Soonpaa et al reported that fewer than 1% of cardiomyocytes were Ki67 positive from P12 through P16. In contrast, Naqvi et al utilized antibodies to Aurora B kinase, and found that 14% of cardiomyocytes were Aurora B positive at P15. Since Ki67 is expressed from G1 to anaphase, in theory Ki67 should label a broader range of cell cycle activity than Aurora B, which is expressed from G2 to M.
As a second approach, Alkass et al applied thymidine analogs Edu/BrdU, and quantified Edu/BrdU positive cardiomyocytes utilizing section immunostaining or flow cytometry of nuclei isolated from left ventricular tissue. Following two days of sequential intraperitoneal injections of EdU, fewer than 1% of cardiomyocyte nuclei were EdU positive on P15. Consistent with this result, fewer than 3% of cardiomyocyte nuclei were BrdU positive following 5 days of continuous administration of BrdU via subcutaneously implanted pellets (P13-P18). These results are in agreement with findings reported by Soonpaa et al, who undertook a similar approach (see Figure). The original report by Navqi et al found evidence for a proliferative burst of cardiomyocytes following a single intraperitoneal injection of BrdU the evening of P14, followed by flow cytometry of BrdU+/cTnT+ nuclei on P18, finding that 11% of cardiomyocyte nuclei were positive for BrdU. The presence of cTnT, a sarcomeric protein, in nuclei is somewhat puzzling, although Bergmann and colleagues have previously utilized cTnT as a marker of cardiomyocyte nuclei 9.
To exclude the possibility of toxic or anti-proliferative effects of BrdU/EdU, Alkass et al undertook a third approach, utilizing multi-isotope mass spectrometry (MIMS) analysis of 15N-thymidine incorporation in conjunction with 14N and 31P labeling to identify cardiomyocyte cell borders and their incumbent nuclei. Following 10 days (P13-P23) of continuous administration of 15N-thymidine via subcutaneously implanted pellets, 0.9-2.1% of cardiomyocyte nuclei were found to be 15N positive. These results are in close agreement with their own results with BrdU incorporation, and the study reported by Soonpaa et al. This approach provides another layer of evidence against a proliferative burst of cardiomyocytes during preadolescence.
Using a CyclinA2-EGFP fusion reporter mouse, Hirai et al also did not observe an appreciable increase in cell cycle activity of cardiomyocytes between P14 and P16, but rather a steady decrease in cell cycle activity during this period. At P12, 0.7% of cardiomyocytes were CyclinA2-EGFP positive, very similar to the percentage of Ki-67 positive cardiomyocytes (0.8%) reported by Soonpaa et al. at this stage. However, the percentage of Cyclin A2-EGFP positive cardiomyocytes detected between P14 and P16 was about 0.02%, significantly lower than the percentages of Ki67 positive cardiomyocytes observed by Alkass et al and Soonpaa et al. This discrepancy may result from several factors. CyclinA2-EGFP is expressed from late G1 to prometaphase, not throughout the cell cycle like Ki67, so a potential increase in the length of the cardiomyocyte cell cycle might affect distinct readouts. Additionally, Hirai et al utilized a cocktail of antibodies to mark all non-myocyte cell types in the heart, to negatively define cardiomyocytes. If some non-myocyte cell types were not labeled with this cocktail, the total number of cardiomyocyte nuclei may have been overestimated. As Alkass et al also describe an increase in ploidy following P13 (see below), it is possible that Ki67, but not Cyclin A2, is expressed during polyploidization of myocardial nuclei.
Cardiomyocyte ploidy
Polyploidization occurs in both human and mouse cardiomyocytes during maturation of cardiomyocytes. In humans about 60% of cardiomyocyte nuclei are polyploidy, whereas in mice the majority of cardiomyocyte nuclei are diploid, with ~10% being polyploid 10-12. Alkass et al investigated DNA synthesis in preadolescent mouse heart and found that the 1-3% of cardiomyocytes that are actively engaged in cell cycle activity following P13 are undergoing polyploidization rather than proliferation or multinucleation. They determined a 10% increase in nuclear DNA content during the second and third week after birth using flow cytometry in conjunction with staining for PCM-1 as a marker for cardiomyocyte nuclei. Furthermore, selective gating of Edu+/PCM-1+ or BrdU+/PCM-1+ nuclei revealed a significant increase in the number of tetraploid nuclei not only with maturation but also when compared to BrdU+/PCM-1- nuclei. In contrast to these results, Naqvi et al reported a relative decrease in the percentage of tetraploid nuclei between P10 and P14. The discrepancy between these two studies may result from the distinct methods employed for sample preparation. Naqvi et al assessed DNA content of nuclei from a cardiomyocyte-enriched population obtained by differential gravitational sedimentation while Alkass et al used nuclear preparations from left ventricle. It is possible that Naqvi et al unintentionally excluded a specific subset of cardiomyocytes during this process. Notably, the increase in and the extent of ploidy occurring during preadolescence as described by Alkass et al are in close agreement with previous results of Brodsky et al using cytophotometry of Feulgen-stained DNA in 14C-thymidine-labeled nuclei 10.
The foregoing discussion serves to emphasize the challenges in determining cardiomyocyte cell cycle behavior in postnatal heart. To investigate cell cycle activity, each group utilized distinct combinations of, and distinct methods of utilizing, cell cycle indicators, including BrdU, EdU, 15N-thymidine, Ki-67, Aurora B, and Cyclin A2, each having distinct characteristics and limitations. There was also diversity in methods utilized to identify or isolate cardiomyocytes. Calculating percentages of cardiomyocytes relies on accurate methods of quantifying total cardiomyocyte number, performed either by analyses of sections or isolated cells, each of which has methodological limitations. Mouse strain background might also be considered as contributing to distinct results, however the C57BL/6J strain used by Naqvi et al was also examined by Soonpaa et al, and Alkass et al used the C57BL/6N strain. C57BL/6J and C57BL/6N are both substrains of C57BL/6.
Overall, it appears that the weight of evidence currently would argue against a proliferative burst of cardiomyocytes in the second postnatal week (Figure). However, intriguingly, the work of Alkass et al and previous results from Brodsky et al suggest that the observed low level of cell cycle activity during this period may reflect increased ploidy of cardiomyocytes. This observation introduces further complexity to the dynamics of cardiomyocyte cell cycle activity. This process still remains little understood and requires further investigation. Furthermore, cardiomyocytes undergo DNA synthesis leading to polyploidy also during aging or in pathological settings 1, 13, highlighting the importance of better understanding the role and mechanisms underlying this phenomenon. Why and how does the increase in ploidy occur? Polyploidy is often associated with larger cell size, and may be required to fulfill metabolic and functional demands 14. What are the cell cycle mechanisms employed, endocycling, or endomitosis that result in cardiomyocyte ploidy? Is differential DNA replication occurring, or are all DNA segments replicated equally? How is this polyploidy related to multinucleation? What is the biological function of this process and how does it relate to withdrawal of cardiomyocytes from a canonical mitotic cell cycle?
One of the challenges in regenerating diseased or damaged heart tissue is to replace lost cardiomyocytes, one potential avenue including the prompting of adult cardiomyocytes to proliferate. Cell based therapies to replace cardiomyocytes will also rely on the ability to promote the development and maturation of exogenous sources of cardiomyocytes. Additionally, human models of heart disease will require the ability to produce mature cardiomyocytes in a dish. For all these reasons, it is important for us to have a clear understanding of the regulation of cardiomyocyte cell cycle activity throughout development and during maturation to the adult cardiomyocye phenotype, despite considerable technical hurdles presented.
References
- 1.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis during hypertrophy in adult mice. Am J Physiol. 1994;266:H1439–45. doi: 10.1152/ajpheart.1994.266.4.H1439. [DOI] [PubMed] [Google Scholar]
- 2.Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, Howard WW, Iismaa SE, Chan AY, Crawford BH, Wagner MB, Martin DI, Lefer DJ, Graham RM, Husain A. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell. 2014;157:795–807. doi: 10.1016/j.cell.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, Bergmann O. No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell. 2015;163:1026–36. doi: 10.1016/j.cell.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 4.Hirai M, Chen J, Evans SM. Tissue-Specific Cell Cycle Indicator Reveals Unexpected Findings for Cardiac Myocyte Proliferation. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.115.307697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soonpaa MH, Zebrowski DC, Platt C, Rosenzweig A, Engel FB, Field LJ. Cardiomyocyte Cell-Cycle Activity during Preadolescence. Cell. 2015;163:781–2. doi: 10.1016/j.cell.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Soonpaa MH, Rubart M, Field LJ. Challenges measuring cardiomyocyte renewal. Biochim Biophys Acta. 2013;1833:799–803. doi: 10.1016/j.bbamcr.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, Frisen J. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res. 2011;317:188–94. doi: 10.1016/j.yexcr.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Preissl S, Schwaderer M, Raulf A, Hesse M, Gruning BA, Kobele C, Backofen R, Fleischmann BK, Hein L, Gilsbach R. Deciphering the Epigenetic Code of Cardiac Myocyte Transcription. Circ Res. 2015;117:413–23. doi: 10.1161/CIRCRESAHA.115.306337. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodsky WY, Arefyeva AM, Uryvaeva IV. Mitotic polyploidization of mouse heart myocytes during the first postnatal week. Cell Tissue Res. 1980;210:133–44. doi: 10.1007/BF00232149. [DOI] [PubMed] [Google Scholar]
- 11.Adler CP, Friedburg H, Herget GW, Neuburger M, Schwalb H. Variability of cardiomyocyte DNA content, ploidy level and nuclear number in mammalian hearts. Virchows Arch. 1996;429:159–64. doi: 10.1007/BF00192438. [DOI] [PubMed] [Google Scholar]
- 12.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andra M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H, Frisen J. Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015;161:1566–75. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–6. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenfelder KP, Fox DT. The expanding implications of polyploidy. J Cell Biol. 2015;209:485–91. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]