Abstract
TDP-43 is the major disease-associated protein involved in the pathogenesis and progression of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions linked to TDP-43 pathology (FTLD-TDP). Abnormal phosphorylation, truncation and cytoplasmic mis-localization are known to be the characteristics for the aggregated forms of TDP-43, and gain of toxic abnormal TDP-43 or loss of function of physiological TDP-43 have been suggested as the cause of neurodegeneration. However, most of the post-translational modifications or truncation sites in the abnormal TDP-43 in brains of patients remain to be identified by protein chemical analysis. In this study, we carried out a highly sensitive liquid chromatography-mass spectrometry analysis of Sarkosyl-insoluble pathological TDP-43 from brains of ALS patients and identified several novel phosphorylation sites, deamidation sites, and cleavage sites. Almost all modifications were localized in the Gly-rich C-terminal half. Most of the cleavage sites identified in this study are novel and are located in N-terminal half, suggesting that these sites may be more accessible to proteolytic enzymes. The data obtained in this study provide a foundation for the molecular mechanisms of TDP-43 aggregation and ALS pathogenesis.
Transactivation response (TAR) DNA-binding protein 43 (TDP-43), encoded by the TARDBP gene, is a highly conserved, ubiquitously expressed nuclear protein. It has conserved RNA recognition motifs (RRM1/RRM2) flanked on either side by N-terminal and glycine-rich C-terminal domains that mediate protein-protein interactions1,2. Recent studies show that TDP-43 is a nuclear ribonucleoprotein implicated in exon splicing, gene transcription, regulation of mRNA stability, mRNA biosynthesis, and formation of nuclear bodies3,4,5,6,7,8,9,10,11. Furthermore, TDP-43 is thought to be essential for early development, because homozygous disruption of the TARDBP gene causes early embryonic lethality12,13.
TDP-43 is the major disease-associated protein involved in the pathogenesis of amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD) linked to TDP-43 pathology (FTLD-TDP)14,15, and other neurodegenerative disorders16,17,18,19,20. Mutations of TDP-43 in familial and sporadic ALS and FTLD cases have been linked to the development of TDP-43 pathology21,22,23,24. These mutations are mostly located in the C-terminal glycine-rich region25, suggesting that conformational change of this region is closely related to the pathology.
TDP-43 is predominantly localized in the nucleus, but under pathological conditions it is translocated to the cytosol14,15,26,27,28,29,30. Thus, loss of normal function of nuclear TDP-43 due to cytoplasmic mislocalization, and toxic gain of function due to TDP-43 aggregation are potential disease mechanisms14,15.
Immunoblotting of the Sarkosyl-insoluble fractions from FTLD and ALS cases using phosphospecific antibodies clearly demonstrated that hyperphosphorylated full-length TDP-43 of 45 kDa, smearing substances, and fragments at 18–25 and 35kDa are the major species of TDP-43 26,31,32. We identified at least three C-terminal banding patterns that distinguish diseases with TDP-43 proteinopathy and reported that the banding pattern in different brain regions and spinal cord of individual patients is indistinguishable. Corresponding patterns of protease-resistant phosphorylated TDP-43 are also seen among the pathological phenotypes26,27,32. Recently, we showed that insoluble TDP-43 aggregates in brains of ALS and FTLD-TDP patients have prion-like properties33.
TDP-43 phosphorylation, cleavage and cytoplasmic mislocalization are all associated with TDP-43 aggregation. However it remains to be clarified whether these modifications are the cause of the disease. Recent reports on post-translational modifications, including fragmentation, were based on studies of cellular or animal models overexpressing TDP-43 or its derivatives34,35,36,37. However, TDP-43 aggregate formation in these artificial models may be different from that in the brains of patients. Therefore, it is important and essential for elucidation of the pathogenesis to identify the pathological events in TDP-43 truly occurred in the brain of patients. In this study, we carried out detailed molecular analysis of pathological TDP-43 aggregates in ALS brains.
Results
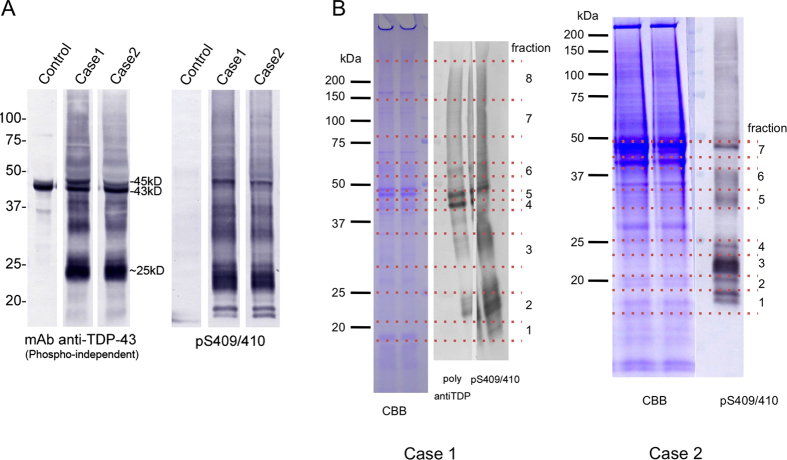
Immunoblot analysis revealed that abundant abnormal TDP-43 was recovered in the Sarkosyl-insoluble fractions of brains from both ALS cases as shown in Fig. 1A. A phosphorylation-independent anti-TDP-43 detected normal TDP-43 of 43 kD in the fractions of both ALS and control brains, but also detected abnormal TDP-43 bands, including full-length phosphorylated TDP-43 of 45 kD, ~25 kD fragments and smears in ALS brains (Fig. 1A). The pS409/410 antibody, which specifically recognizes the abnormal phosphorylation of Ser409 and 410 strongly reacted with these pathological TDP-43 bands and additional fragments of 18 ~ 24 kD (Fig. 1A). No such bands were detected in the same fraction of control brain. Intensities of pS409/410 immunoreactive bands in both ALS cases were 5 ~ 10 times higher compared with those from the other cases previously analyzed in our lab (data not shown). Therefore, we thought these two cases are good for the protein chemical analysis of pathological TDP-43.
Figure 1. SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting of Sarkosyl-insoluble fraction from ALS case 1 brain and ALS case 2 brain.
(A) A phosphorylation-independent anti-TDP-43 detected normal TDP-43 of 43 kD in the fractions of both ALS and control brains, but also detected abnormal TDP-43 bands, including full-length phosphorylated TDP-43 of 45 kD, ~25 kD fragments and smears in ALS brains. (B) TDP-43-positive bands were excised as indicated.
After SDS-PAGE, the gel portions corresponding to these bands were cut out as shown in Fig. 1B and digested with trypsin and chymotrypsin. Digests were analyzed with nano-flow LC-MS/MS. As shown in Tables 1 and 2, we identified 229 peptides from brain of case 1 and 92 peptides from brain of case 2; this difference may reflect the differences in total amounts of pathological TDP-43 in these brains and the numbers of fractions. These peptides covered about 90% of the intact molecule (Tables 1 and 2). Identified peptides (representative), modifications and cleavage sites are shown and summarized in Figs 2 and 3. Further cleavage site peptides are shown in supplemental Fig. S1. Other proteins contaminating the Sarkosyl-insoluble fractions are listed in supplemental Table S1.
Table 1. List of identifid peptides derived from accumulated TDP-43 in ALS patient case 1.
| Query | Start | End | Observed | Mr(expt) | Mr(calc) | Delta | Score | Expect | Peptide |
|---|---|---|---|---|---|---|---|---|---|
| fraction1 | chymotrypsin | ||||||||
| 9373 | 212 | 226 | 871.6568 | 1741.299 | 1741.8386 | −0.5395 | 91 | 5.40E-09 | F.SQYGDVMDVFIPKPF.R |
| 9389 | 212 | 226 | 872.741 | 1743.4674 | 1741.8386 | 1.6288 | 41 | 0.00021 | F.SQYGDVMDVFIPKPF.R |
| 9442 | 212 | 226 | 880.2186 | 1758.4227 | 1757.8335 | 0.5892 | 60 | 2.70E-05 | F.SQYGDVMoDVFIPKPF.R |
| 6387 | 215 | 226 | 683.1282 | 1364.2418 | 1363.6846 | 0.5572 | 23 | 0.0077 | Y.GDVMDVFIPKPF.R |
| 3616 | 235 | 243 | 520.0455 | 1038.0765 | 1039.4223 | −1.3458 | 24 | 0.025 | F.ADDQIAQSpL.C |
| 4502 | 277 | 289 | 583.8335 | 1165.6524 | 1164.4949 | 1.1576 | 38 | 0.00028 | F.GGNPGGFGNQGGF.G |
| 12543 | 290 | 316 | 829.3413 | 2485.0021 | 2486.0452 | −1.0431 | 45 | 0.0016 | F.GNSRGGGAGLGN*NQGSNMGGGMNFGAF.S |
| trypsin | |||||||||
| 7855 | 84 | 95 | 641.5232 | 1281.0318 | 1280.5918 | 0.44 | 48 | 3.10E-05 | R.KMDETDASSAVK.V |
| 8748 | 103 | 114 | 671.5735 | 1341.1324 | 1340.7704 | 0.362 | 21 | 0.052 | K.TSDLIVLGLPWK.T |
| 3193 | 115 | 121 | 417.6076 | 833.2006 | 833.4131 | −0.2124 | 26 | 0.0036 | K.TTEQDLK.E |
| 3198 | 115 | 121 | 417.6429 | 833.2713 | 833.4131 | −0.1417 | 23 | 0.0071 | K.TTEQDLK.E |
| 6245 | 152 | 160 | 572.4623 | 1142.9101 | 1143.5448 | −0.6347 | 20 | 0.014 | R.FTEYETQVK.V |
| 6249 | 152 | 160 | 572.5684 | 1143.1223 | 1143.5448 | −0.4225 | 24 | 0.0057 | R.FTEYETQVK.V |
| 10549 | 215 | 227 | 513.2426 | 1536.7058 | 1535.7807 | 0.9252 | 26 | 0.025 | Y.GDVMDVFIPKPFR |
| 7515 | 252 | 263 | 626.6138 | 1251.2131 | 1250.6619 | 0.5512 | 36 | 0.0004 | K.GISVHISNAEPK.H |
| 7516 | 252 | 263 | 626.6459 | 1251.2772 | 1251.6459 | −0.3687 | 33 | 0.0037 | K.GISVHISN*AEPK.H |
| 10016 | 276 | 291 | 742.3951 | 1482.7757 | 1482.6277 | 0.148 | 42 | 0.00011 | R.FGGNPGGFGNQGGFGN.S |
| 12266 | 276 | 293 | 863.7729 | 1725.5312 | 1725.7608 | −0.2296 | 105 | 2.30E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 12269 | 276 | 293 | 864.0314 | 1726.0482 | 1725.7608 | 0.2874 | 103 | 2.30E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 12274 | 276 | 293 | 864.0852 | 1726.1559 | 1725.7608 | 0.3951 | 98 | 2.10E-09 | R.FGGNPGGFGNQGGFGNSR.G |
| 12276 | 276 | 293 | 864.1173 | 1726.2201 | 1725.7608 | 0.4592 | 97 | 2.30E-09 | R.FGGNPGGFGNQGGFGNSR.G |
| 12278 | 276 | 293 | 864.1233 | 1726.232 | 1725.7608 | 0.4712 | 101 | 3.40E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 12279 | 276 | 293 | 864.1337 | 1726.2528 | 1725.7608 | 0.492 | 111 | 8.90E-11 | R.FGGNPGGFGNQGGFGNSR.G |
| 12280 | 276 | 293 | 576.432 | 1726.274 | 1725.7608 | 0.5132 | 26 | 0.0082 | R.FGGNPGGFGNQGGFGNSR.G |
| 12290 | 276 | 293 | 576.6025 | 1726.7856 | 1725.7608 | 1.0248 | 36 | 0.0006 | R.FGGNPGGFGNQGGFGNSR.G |
| 12268 | 276 | 293 | 576.265 | 1725.7732 | 1726.7448 | −0.9716 | 37 | 0.0007 | R.FGGNPGGFGNQGGFGN*SR.G |
| 12271 | 276 | 293 | 864.0555 | 1726.0964 | 1726.7448 | −0.6484 | 90 | 1.30E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| fraction2 | chymotrypsin | ||||||||
| 5089 | 176 | 188 | 742.6311 | 1483.2476 | 1483.7518 | −0.5042 | 43 | 0.00092 | C.KLPN*SKQSQDEPL.R |
| 5096 | 176 | 188 | 743.321 | 1484.6275 | 1483.7518 | 0.8757 | 46 | 0.00041 | C.KLPN*SKQSQDEPL.R |
| 4168 | 211 | 221 | 654.5313 | 1307.0479 | 1306.554 | 0.4939 | 39 | 0.00027 | F.FSQYGDVMDVF.I |
| 6859 | 211 | 226 | 945.4351 | 1888.8556 | 1888.907 | −0.0514 | 48 | 3.70E-05 | F.FSQYGDVMDVFIPKPF.R |
| 6861 | 211 | 226 | 946.2177 | 1890.4207 | 1889.891 | 0.5298 | 30 | 0.0016 | F.FSQ*YGDVMDVFIPKPF.R |
| 6880 | 211 | 226 | 953.7546 | 1905.4947 | 1904.9019 | 0.5928 | 52 | 4.10E-05 | F.FSQYGDVMoDVFIPKPF.R |
| 3531 | 212 | 221 | 580.9187 | 1159.8228 | 1159.4856 | 0.3373 | 35 | 5.70E-04 | F.SQYGDVMDVF.I |
| 6412 | 212 | 226 | 872.3367 | 1742.6589 | 1741.8386 | 0.8203 | 73 | 1.50E-07 | F.SQYGDVMDVFIPKPF.R |
| 6461 | 212 | 226 | 880.2487 | 1758.4828 | 1757.8335 | 0.6493 | 52 | 9.50E-05 | F.SQYGDVMoDVFIPKPF.R |
| 4522 | 215 | 226 | 682.6916 | 1363.3687 | 1363.6846 | −0.3159 | 38 | 3.10E-04 | Y.GDVMDVFIPKPF.R |
| 4524 | 215 | 226 | 683.6007 | 1365.1869 | 1363.6846 | 1.5022 | 21 | 2.20E-02 | Y.GDVMDVFIPKPF.R |
| 6869 | 271 | 289 | 633.079 | 1896.2151 | 1897.8456 | −1.6305 | 42 | 0.00049 | L.ERSGRFGGNPGGFGN*QGGF.G |
| 3544 | 277 | 289 | 583.3915 | 1164.7685 | 1164.4949 | 0.2737 | 28 | 0.0025 | F.GGNPGGFGNQGGF.G |
| 3546 | 277 | 289 | 583.4573 | 1164.9001 | 1164.4949 | 0.4053 | 27 | 0.0051 | F.GGNPGGFGNQGGF.G |
| 3548 | 277 | 289 | 583.528 | 1165.0413 | 1164.4949 | 0.5465 | 30 | 0.0014 | F.GGNPGGFGNQGGF.G |
| 7741 | 290 | 313 | 1106.3008 | 2210.587 | 2210.9182 | −0.3312 | 95 | 1.50E-08 | F.GNSRGGGAGLGNNQGSN*MGGGMNF.G |
| 7743 | 290 | 313 | 737.9193 | 2210.7361 | 2210.9182 | −0.1821 | 50 | 0.00044 | F.GNSRGGGAGLGNNQGSN*MGGGMNF.G |
| 7770 | 290 | 313 | 743.0682 | 2226.1827 | 2226.9131 | −0.7304 | 61 | 2.20E-05 | F.GNSRGGGAGLGN*NQGSNMoGGGMNF.G |
| 7772 | 290 | 313 | 743.2908 | 2226.8505 | 2226.9131 | −0.0626 | 38 | 0.0067 | F.GNSRGGGAGLGN*NQGSNMGGGMoNF.G |
| 7775 | 290 | 313 | 743.557 | 2227.6492 | 2226.9131 | 0.7361 | 48 | 0.00017 | F.GNSRGGGAGLGNNQGSN*MGGGMoNF.G |
| 7791 | 290 | 313 | 1121.8962 | 2241.7779 | 2241.924 | −0.1461 | 65 | 7.30E-07 | F.GNSRGGGAGLGNNQGSNMoGGGMoNF.G |
| 7792 | 290 | 313 | 748.4241 | 2242.2504 | 2242.908 | −0.6576 | 66 | 1.00E-05 | F.GNSRGGGAGLGN*NQGSNMoGGGMoNF.G |
| 7795 | 290 | 313 | 748.7297 | 2243.1672 | 2243.892 | −0.7248 | 36 | 0.01 | F.GN*SRGGGAGLGNNQ*GSNMoGGGMoNF.G |
| 7796 | 290 | 313 | 748.7909 | 2243.3508 | 2243.892 | −0.5412 | 47 | 0.00068 | F.GNSRGGGAGLGN*NQGSN*MoGGGMoNF.G |
| 7799 | 290 | 313 | 1123.2252 | 2244.4359 | 2243.892 | 0.5439 | 69 | 3.10E-07 | F.GNSRGGGAGLGN*NQGSN*MoGGGMoNF.G |
| 8563 | 290 | 316 | 829.6099 | 2485.8078 | 2486.0452 | −0.2374 | 58 | 8.60E-05 | F.GN*SRGGGAGLGNNQGSNMGGGMNFGAF.S |
| 8567 | 290 | 316 | 830.2059 | 2487.5958 | 2487.0292 | 0.5666 | 67 | 1.20E-05 | F.GNSRGGGAGLGN*NQGSN*MGGGMNFGAF.S |
| 8603 | 290 | 316 | 834.7759 | 2501.3058 | 2503.0241 | −1.7183 | 56 | 0.00012 | F.GN*SRGGGAGLGNNQGSNMGGGMoN*FGAF.S |
| 8606 | 290 | 316 | 835.1727 | 2502.4962 | 2503.0241 | −0.5279 | 73 | 2.50E-06 | F.GN*SRGGGAGLGNNQGSNMoGGGMN*FGAF.S |
| 8611 | 290 | 316 | 835.5923 | 2503.755 | 2503.0241 | 0.7309 | 59 | 6.60E-05 | F.GN*SRGGGAGLGNNQGSNMGGGMoN*FGAF.S |
| 8667 | 290 | 316 | 840.1269 | 2517.3588 | 2518.035 | −0.6762 | 61 | 4.90E-05 | F.GNSRGGGAGLGNNQ*GSNMoGGGMoNFGAF.S |
| 8671 | 290 | 316 | 840.6654 | 2518.9744 | 2519.019 | −0.0446 | 64 | 2.20E-05 | F.GN*SRGGGAGLGNNQGSNMoGGGMoN*FGAF.S |
| 1915 | 317 | 323 | 779.2965 | 778.2892 | 778.3353 | −0.0462 | 37 | 0.00039 | F.SINPAMoM.A |
| 1916 | 317 | 323 | 779.4086 | 778.4013 | 778.3353 | 0.066 | 37 | 0.00038 | F.SINPAMoM.A |
| 4592 | 317 | 330 | 688.1877 | 1374.3609 | 1374.6635 | −0.3026 | 28 | 0.0041 | F.SINPAMMoAAAQAAL.Q |
| 4704 | 317 | 330 | 696.6204 | 1391.2262 | 1391.6424 | −0.4163 | 48 | 0.00028 | F.SINPAMoMoAAAQ*AAL.Q |
| 5282 | 317 | 331 | 760.6963 | 1519.378 | 1518.717 | 0.661 | 62 | 1.30E-05 | F.SINPAMoMoAAAQAALQ.S |
| 6767 | 317 | 334 | 616.6206 | 1846.84 | 1846.8705 | −0.0305 | 21 | 0.019 | F.SINPAMMAAAQAALQSSW.G |
| 6768 | 317 | 334 | 924.6777 | 1847.3409 | 1846.8705 | 0.4704 | 43 | 9.10E-05 | F.SINPAMMAAAQAALQSSW.G |
| 6835 | 317 | 334 | 941.1243 | 1880.234 | 1879.8444 | 0.3896 | 33 | 0.014 | F.SINPAMoMoAAAQ*AALQSSW.G |
| 3280 | 324 | 334 | 552.4872 | 1102.9598 | 1102.5407 | 0.4191 | 68 | 7.20E-07 | M.AAAQAALQSSW.G |
| 7465 | 340 | 359 | 1037.6927 | 2073.3709 | 2072.893 | 0.478 | 38 | 0.00028 | M.LASQQNQSGPSGNNQNQGNM.Q |
| 10098 | 340 | 367 | 1015.7645 | 3044.2718 | 3044.3391 | −0.0673 | 68 | 1.60E-05 | M.LASQQNQSGPSGNNQNQGN*MQREPNQAF.G |
| 10096 | 340 | 367 | 1015.7145 | 3044.1216 | 3045.3231 | −1.2014 | 60 | 8.90E-05 | M.LASQQNQSGPSGNNQN*Q*GNMQREPNQAF.G |
| 10136 | 340 | 367 | 1020.859 | 3059.5552 | 3060.334 | −0.7788 | 75 | 9.60E-07 | M.LASQQNQSGPSGNNQNQ*GNMoQREPNQAF.G |
| 10145 | 340 | 367 | 1021.6365 | 3061.8876 | 3061.318 | 0.5696 | 57 | 0.00017 | M.LASQQNQSGPSGNNQ*N*QGNMoQREPNQAF.G |
| 10150 | 340 | 367 | 1021.8416 | 3062.5028 | 3062.302 | 0.2008 | 45 | 0.0027 | M.LASQQNQSGPSGN*N*Q*NQGNMoQREPNQAF.G |
| 9721 | 341 | 367 | 978.223 | 2931.6472 | 2931.255 | 0.3922 | 50 | 0.00076 | L.ASQQNQSGPSGNNQNQGN*MQREPNQAF.G |
| 9722 | 341 | 367 | 978.352 | 2932.0343 | 2931.255 | 0.7793 | 48 | 0.0014 | L.ASQQNQSGPSGNNQN*QGNMQREPNQAF.G |
| 9763 | 341 | 367 | 983.592 | 2947.7543 | 2947.2499 | 0.5044 | 34 | 0.012 | L.ASQQNQSGPSGNNQNQGNMoQREPNQ*AF.G |
| 9764 | 341 | 367 | 983.6382 | 2947.8927 | 2947.2499 | 0.6428 | 40 | 0.0091 | L.ASQQNQSGPSGNNQNQGNMoQREPNQ*AF.G |
| 9765 | 341 | 367 | 983.6984 | 2948.0733 | 2947.2499 | 0.8233 | 50 | 9.70E-05 | L.ASQQNQSGPSGNNQNQGNMoQREPN*QAF.G |
| 9772 | 341 | 367 | 983.9515 | 2948.8328 | 2948.2339 | 0.5989 | 37 | 0.018 | L.ASQQNQ*SGPSGN*NQNQGNMoQREPNQAF.G |
| 6163 | 389 | 405 | 561.7853 | 1682.334 | 1680.5164 | 1.8176 | 20 | 0.034 | A.SpNAGSpGSGFN*GGFGSSM.D |
| 2063 | 398 | 405 | 426.5261 | 851.0376 | 851.2521 | −0.2145 | 13 | 0.063 | F.NGGFGSpSMo.D |
| 2064 | 398 | 405 | 426.5549 | 851.0953 | 851.2521 | −0.1568 | 13 | 0.057 | F.NGGFGSpSMo.D |
| trypsin | |||||||||
| 4541 | 252 | 263 | 626.1067 | 1250.1988 | 1250.6619 | −0.4631 | 47 | 3.70E-05 | K.GISVHISNAEPK.H |
| 4554 | 252 | 263 | 626.5655 | 1251.1164 | 1251.6459 | −0.5295 | 30 | 0.0069 | K.GISVHISN*AEPK.H |
| 10298 | 273 | 293 | 676.4501 | 2026.3284 | 2026.8994 | −0.571 | 49 | 3.20E-05 | R.SGRFGGN*PGGFGNQGGFGNSR.G |
| 8412 | 276 | 293 | 863.5454 | 1725.0763 | 1725.7608 | −0.6845 | 68 | 4.20E-07 | R.FGGNPGGFGNQGGFGNSR.G |
| 8413 | 276 | 293 | 863.6377 | 1725.2608 | 1725.7608 | −0.5 | 74 | 1.00E-07 | R.FGGNPGGFGNQGGFGNSR.G |
| 8414 | 276 | 293 | 863.6544 | 1725.2943 | 1725.7608 | −0.4665 | 81 | 2.80E-08 | R.FGGNPGGFGNQGGFGNSR.G |
| 8415 | 276 | 293 | 863.7958 | 1725.577 | 1725.7608 | −0.1838 | 96 | 4.00E-09 | R.FGGNPGGFGNQGGFGNSR.G |
| 8421 | 276 | 293 | 864.0331 | 1726.0516 | 1725.7608 | 0.2908 | 90 | 1.30E-08 | R.FGGNPGGFGNQGGFGNSR.G |
| 8424 | 276 | 293 | 864.0687 | 1726.1228 | 1725.7608 | 0.3619 | 82 | 2.10E-08 | R.FGGNPGGFGNQGGFGNSR.G |
| 8425 | 276 | 293 | 864.0773 | 1726.14 | 1725.7608 | 0.3792 | 102 | 8.60E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 8426 | 276 | 293 | 864.0829 | 1726.1512 | 1725.7608 | 0.3904 | 112 | 8.80E-11 | R.FGGNPGGFGNQGGFGNSR.G |
| 8427 | 276 | 293 | 864.0865 | 1726.1585 | 1725.7608 | 0.3977 | 83 | 1.60E-08 | R.FGGNPGGFGNQGGFGNSR.G |
| 8428 | 276 | 293 | 864.1036 | 1726.1926 | 1725.7608 | 0.4318 | 74 | 1.10E-07 | R.FGGNPGGFGNQGGFGNSR.G |
| 8429 | 276 | 293 | 864.1146 | 1726.2146 | 1725.7608 | 0.4537 | 103 | 5.60E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 8416 | 276 | 293 | 863.8204 | 1725.6263 | 1726.7448 | −1.1185 | 87 | 2.10E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8417 | 276 | 293 | 863.9819 | 1725.9493 | 1726.7448 | −0.7955 | 88 | 2.50E-08 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8419 | 276 | 293 | 863.9979 | 1725.9813 | 1726.7448 | −0.7635 | 90 | 1.30E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8420 | 276 | 293 | 864.0292 | 1726.0438 | 1726.7448 | −0.701 | 81 | 2.40E-08 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8423 | 276 | 293 | 864.066 | 1726.1175 | 1726.7448 | −0.6273 | 69 | 1.70E-06 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8430 | 276 | 293 | 864.1177 | 1726.2209 | 1726.7448 | −0.5239 | 90 | 1.20E-08 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8434 | 276 | 293 | 864.2111 | 1726.4077 | 1726.7448 | −0.3371 | 96 | 3.30E-09 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8435 | 276 | 293 | 864.2555 | 1726.4964 | 1726.7448 | −0.2484 | 95 | 4.30E-09 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8436 | 276 | 293 | 576.5455 | 1726.6148 | 1726.7448 | −0.1301 | 31 | 0.012 | R.FGGNPGGFGNQGGFGN*SR.G |
| 8438 | 276 | 293 | 864.4259 | 1726.8372 | 1726.7448 | 0.0924 | 82 | 1.00E-07 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8439 | 276 | 293 | 864.4338 | 1726.853 | 1726.7448 | 0.1082 | 82 | 1.00E-07 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8440 | 276 | 293 | 864.4481 | 1726.8817 | 1726.7448 | 0.1369 | 77 | 3.30E-07 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8441 | 276 | 293 | 864.4512 | 1726.8878 | 1726.7448 | 0.143 | 72 | 9.10E-07 | R.FGGNPGGFGNQ*GGFGNSR.G |
| 8443 | 276 | 293 | 864.5175 | 1727.0205 | 1726.7448 | 0.2757 | 90 | 3.90E-09 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8444 | 276 | 293 | 576.6903 | 1727.0491 | 1726.7448 | 0.3043 | 40 | 0.00049 | R.FGGNPGGFGNQGGFGN*SR.G |
| 8445 | 276 | 293 | 864.5552 | 1727.0959 | 1726.7448 | 0.3511 | 78 | 5.70E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| 8447 | 276 | 293 | 864.6735 | 1727.3324 | 1726.7448 | 0.5875 | 78 | 1.50E-07 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8448 | 276 | 293 | 864.687 | 1727.3595 | 1726.7448 | 0.6146 | 81 | 2.60E-08 | R.FGGN*PGGFGNQGGFGNSR.G |
| 8446 | 276 | 293 | 864.6155 | 1727.2164 | 1727.7288 | −0.5124 | 79 | 1.50E-07 | R.FGGN*PGGFGNQ*GGFGNSR.G |
| 8458 | 276 | 293 | 865.0176 | 1728.0206 | 1727.7288 | 0.2918 | 53 | 1.20E-05 | R.FGGN*PGGFGNQGGFGN*SR.G |
| 8463 | 276 | 293 | 865.0424 | 1728.0703 | 1727.7288 | 0.3415 | 39 | 0.00023 | R.FGGN*PGGFGNQ*GGFGNSR.G |
| 8470 | 276 | 293 | 865.0927 | 1728.1707 | 1727.7288 | 0.4419 | 50 | 2.10E-05 | R.FGGN*PGGFGNQGGFGN*SR.G |
| 8508 | 276 | 293 | 865.5012 | 1728.9879 | 1728.7128 | 0.275 | 32 | 0.0016 | R.FGGN*PGGFGN*Q*GGFGNSR.G |
| 5555 | 280 | 293 | 676.4501 | 1350.8856 | 1350.6065 | 0.2791 | 75 | 1.00E-07 | N.PGGFGNQGGFGNSR.G |
| fraction3 | chymotrypsin | ||||||||
| 9282 | 211 | 226 | 945.6774 | 1889.3402 | 1888.907 | 0.4332 | 32 | 0.001 | F.FSQYGDVMDVFIPKPF.R |
| 8489 | 212 | 226 | 872.2216 | 1742.4286 | 1741.8386 | 0.59 | 68 | 4.10E-07 | F.SQYGDVMDVFIPKPF.R |
| 8572 | 212 | 226 | 879.9073 | 1757.8001 | 1757.8335 | −0.0333 | 33 | 0.0012 | F.SQYGDVMoDVFIPKPF.R |
| 4043 | 277 | 289 | 583.3818 | 1164.7491 | 1164.4949 | 0.2543 | 35 | 0.00048 | F.GGNPGGFGNQGGF.G |
| 4044 | 277 | 289 | 583.4044 | 1164.7942 | 1164.4949 | 0.2993 | 27 | 0.0032 | F.GGNPGGFGNQGGF.G |
| 4048 | 277 | 289 | 583.4793 | 1164.9441 | 1164.4949 | 0.4492 | 37 | 0.00059 | F.GGNPGGFGNQGGF.G |
| 4057 | 277 | 289 | 583.5714 | 1165.1283 | 1164.4949 | 0.6334 | 20 | 0.014 | F.GGNPGGFGNQGGF.G |
| 10527 | 290 | 313 | 743.5016 | 2227.4829 | 2225.9291 | 1.5539 | 32 | 0.0086 | F.GNSRGGGAGLGNNQGSNMoGGGMNF.G |
| 10566 | 290 | 313 | 1122.9318 | 2243.849 | 2243.892 | −0.043 | 27 | 0.021 | F.GNSRGGGAGLGN*NQ*GSNMoGGGMoNF.G |
| 11297 | 290 | 316 | 840.3088 | 2517.9047 | 2518.035 | −0.1303 | 38 | 0.0081 | F.GNSRGGGAGLGN*NQGSNMoGGGMoNFGAF.S |
| 11299 | 290 | 316 | 840.5651 | 2518.6734 | 2518.035 | 0.6383 | 30 | 0.0058 | F.GN*SRGGGAGLGNNQGSNMoGGGMoNFGAF.S |
| 7796 | 298 | 313 | 546.0648 | 1635.1726 | 1636.5535 | −1.3808 | 24 | 0.082 | A.GLGN*NQ*GSpN*MGGGMNF.G |
| 5837 | 317 | 330 | 688.5671 | 1375.1196 | 1374.6635 | 0.4561 | 25 | 0.0073 | F.SINPAMMAAAQAAL.Q |
| 3589 | 324 | 334 | 552.6029 | 1103.1912 | 1102.5407 | 0.6506 | 36 | 0.0017 | M.AAAQAALQSSW.G |
| 1956 | 398 | 405 | 426.5643 | 851.1141 | 852.2361 | −1.122 | 23 | 0.038 | F.N*GGFGSpSMo.D |
| trypsin | |||||||||
| 6817 | 103 | 114 | 671.6575 | 1341.3004 | 1340.7704 | 0.53 | 61 | 2.90E-06 | K.TSDLIVLGLPWK |
| 4663 | 152 | 160 | 572.6335 | 1143.2524 | 1143.5448 | −0.2924 | 41 | 0.00021 | R.FTEYETQVK.V |
| 4677 | 152 | 160 | 573.5289 | 1145.0432 | 1143.5448 | 1.4984 | 33 | 0.00079 | R.FTEYETQVK.V |
| 5751 | 252 | 263 | 626.5578 | 1251.101 | 1251.6459 | −0.5448 | 33 | 0.0017 | K.GISVHISN*AEPK.H |
| 11145 | 276 | 293 | 864.0863 | 1726.158 | 1725.7608 | 0.3972 | 104 | 9.80E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 11147 | 276 | 293 | 576.6158 | 1726.8257 | 1725.7608 | 1.0649 | 43 | 0.00028 | R.FGGNPGGFGNQGGFGNSR.G |
| 11138 | 276 | 293 | 864.0644 | 1726.1142 | 1726.7448 | −0.6306 | 99 | 3.00E-09 | R.FGGN*PGGFGNQGGFGNSR.G |
| 11140 | 276 | 293 | 864.1228 | 1726.231 | 1726.7448 | −0.5138 | 92 | 1.60E-08 | R.FGGNPGGFGNQGGFGN*SR.G |
| 11142 | 276 | 293 | 864.1336 | 1726.2527 | 1726.7448 | −0.4922 | 85 | 1.60E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| 11144 | 276 | 293 | 864.2585 | 1726.5024 | 1726.7448 | −0.2424 | 25 | 0.052 | R.FGGN*PGGFGNQGGFGNSR.G |
| 11157 | 276 | 293 | 576.9172 | 1727.7297 | 1726.7448 | 0.9849 | 59 | 3.00E-05 | R.FGGNPGGFGNQGGFGN*SR.G |
| fraction4 | chymotrypsin | ||||||||
| 4202 | 153 | 162 | 622.3894 | 1242.7642 | 1242.5802 | 0.184 | 40 | 0.00024 | F.TEYETQVKVMo.S |
| 9557 | 290 | 313 | 748.4747 | 2242.4024 | 2242.908 | −0.5056 | 65 | 1.20E-05 | F.GN*SRGGGAGLGNNQGSNMoGGGMoNF.G |
| trypsin | |||||||||
| 15817 | 56 | 79 | 875.6911 | 2624.0515 | 2623.3799 | 0.6716 | 30 | 0.0048 | R.LVEGILHAPDAGWGNLVYVVNYPK.D |
| 15818 | 56 | 79 | 875.9263 | 2624.757 | 2623.3799 | 1.3771 | 48 | 0.00018 | R.LVEGILHAPDAGWGNLVYVVNYPK.D |
| 15822 | 56 | 79 | 876.7198 | 2627.1377 | 2625.3479 | 1.7899 | 40 | 0.003 | R.LVEGILHAPDAGWGN*LVYVVN*YPK.D |
| 5505 | 84 | 95 | 641.2124 | 1280.4102 | 1280.5918 | −0.1816 | 35 | 0.0015 | R.KMDETDASSAVK.V |
| 5513 | 84 | 95 | 641.6223 | 1281.2301 | 1280.5918 | 0.6383 | 64 | 1.20E-06 | R.KMDETDASSAVK.V |
| 5516 | 84 | 95 | 428.1922 | 1281.5549 | 1280.5918 | 0.963 | 30 | 0.0056 | R.KMDETDASSAVK.V |
| 5728 | 84 | 95 | 649.5043 | 1296.9941 | 1296.5867 | 0.4074 | 51 | 1.60E-05 | R.KMoDETDASSAVK.V |
| 4136 | 85 | 95 | 576.8164 | 1151.6183 | 1152.4969 | −0.8786 | 32 | 0.0011 | K.MDETDASSAVK.V |
| 4140 | 85 | 95 | 576.885 | 1151.7555 | 1152.4969 | −0.7414 | 20 | 0.013 | K.MDETDASSAVK.V |
| 4159 | 85 | 95 | 577.0442 | 1152.0738 | 1152.4969 | −0.423 | 22 | 0.008 | K.MDETDASSAVK.V |
| 4161 | 85 | 95 | 577.0544 | 1152.0943 | 1152.4969 | −0.4025 | 24 | 0.014 | K.MDETDASSAVK.V |
| 4173 | 85 | 95 | 577.1104 | 1152.2063 | 1152.4969 | −0.2906 | 50 | 2.10E-05 | K.MDETDASSAVK.V |
| 4196 | 85 | 95 | 577.2958 | 1152.577 | 1152.4969 | 0.0802 | 20 | 0.053 | K.MDETDASSAVK.V |
| 4204 | 85 | 95 | 577.8925 | 1153.7704 | 1152.4969 | 1.2735 | 26 | 0.0035 | K.MDETDASSAVK.V |
| 6390 | 103 | 114 | 671.2714 | 1340.5282 | 1340.7704 | −0.2422 | 54 | 1.80E-05 | K.TSDLIVLGLPWK.T |
| 4096 | 152 | 160 | 572.927 | 1143.8394 | 1143.5448 | 0.2946 | 39 | 0.00021 | R.FTEYETQVK.V |
| 4097 | 152 | 160 | 572.9576 | 1143.9006 | 1143.5448 | 0.3558 | 28 | 0.0032 | R.FTEYETQVK.V |
| 4100 | 152 | 160 | 573.1072 | 1144.1998 | 1143.5448 | 0.655 | 31 | 0.0023 | R.FTEYETQVK.V |
| 5209 | 252 | 263 | 626.5698 | 1251.1251 | 1250.6619 | 0.4632 | 34 | 0.00061 | K.GISVHISNAEPK.H |
| 10703 | 276 | 293 | 864.0738 | 1726.1331 | 1725.7608 | 0.3723 | 108 | 2.10E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 10689 | 276 | 293 | 576.141 | 1725.4011 | 1726.7448 | −1.3437 | 45 | 0.00089 | R.FGGNPGGFG*NQGGFGNSR.G |
| 10714 | 276 | 293 | 864.5613 | 1727.1081 | 1727.7288 | −0.6207 | 103 | 1.20E-09 | R.FGGN*PGGFGNQGGFGN*SR.G |
| fraction5 | chymotrypsin | ||||||||
| 3922 | 114 | 123 | 418.5765 | 1252.7075 | 1253.6139 | −0.9064 | 38 | 0.0024 | W.KTTEQDLKEY.F |
| 5597 | 300 | 313 | 489.2062 | 1464.5969 | 1466.4479 | −1.851 | 24 | 0.061 | L.GN*N*QGS*N*MGGGMNF.G |
| 1088 | 330 | 336 | 413.0149 | 824.0152 | 823.3534 | 0.6618 | 22 | 0.018 | A.LQSSWGMo.M |
| 8438 | 386 | 405 | 658.5479 | 1972.6219 | 1974.5894 | −1.9675 | 32 | 0.014 | W.GSASNAGSGSpGFNGGFGSpSpM.D |
| trypsin | |||||||||
| 15330 | 56 | 79 | 875.8154 | 2624.4245 | 2623.3799 | 1.0446 | 40 | 0.00088 | R.LVEGILHAPDAGWGNLVYVVNYPK.D |
| 15331 | 56 | 79 | 875.8346 | 2624.4819 | 2623.3799 | 1.1021 | 34 | 0.0077 | R.LVEGILHAPDAGWGNLVYVVNYPK.D |
| 5230 | 84 | 95 | 641.4845 | 1280.9544 | 1280.5918 | 0.3626 | 56 | 2.70E-05 | R.KMDETDASSAVK.V |
| 5232 | 84 | 95 | 641.5112 | 1281.0078 | 1280.5918 | 0.416 | 51 | 4.20E-05 | R.KMDETDASSAVK.V |
| 3945 | 85 | 95 | 576.9318 | 1151.8491 | 1152.4969 | −0.6478 | 44 | 0.00018 | K.MDETDASSAVK.V |
| 3952 | 85 | 95 | 577.0027 | 1151.9908 | 1152.4969 | −0.506 | 26 | 0.0054 | K.MDETDASSAVK.V |
| 3958 | 85 | 95 | 577.0422 | 1152.0699 | 1152.4969 | −0.4269 | 50 | 1.90E-05 | K.MDETDASSAVK.V |
| 3980 | 85 | 95 | 577.1116 | 1152.2086 | 1152.4969 | −0.2883 | 31 | 0.004 | K.MDETDASSAVK.V |
| 3983 | 85 | 95 | 577.1158 | 1152.217 | 1152.4969 | −0.2798 | 18 | 0.021 | K.MDETDASSAVK.V |
| 3990 | 85 | 95 | 577.1333 | 1152.252 | 1152.4969 | −0.2448 | 23 | 0.057 | K.MDETDASSAVK.V |
| 4006 | 85 | 95 | 577.1882 | 1152.3619 | 1152.4969 | −0.1349 | 25 | 0.0043 | K.MDETDASSAVK.V |
| 4007 | 85 | 95 | 577.1957 | 1152.3768 | 1152.4969 | −0.12 | 30 | 0.0024 | K.MDETDASSAVK.V |
| 4013 | 85 | 95 | 577.2638 | 1152.513 | 1152.4969 | 0.0162 | 44 | 7.80E-05 | K.MDETDASSAVK.V |
| 4018 | 85 | 95 | 577.4311 | 1152.8476 | 1152.4969 | 0.3508 | 35 | 0.0031 | K.MDETDASSAVK.V |
| 4021 | 85 | 95 | 577.4545 | 1152.8944 | 1152.4969 | 0.3975 | 52 | 2.40E-05 | K.MDETDASSAVK.V |
| 4030 | 85 | 95 | 578.0698 | 1154.125 | 1152.4969 | 1.6281 | 33 | 0.0017 | K.MDETDASSAVK.V |
| 4093 | 85 | 95 | 585.0585 | 1168.1024 | 1168.4918 | −0.3894 | 27 | 0.0031 | K.MoDETDASSAVK.V |
| 4097 | 85 | 95 | 585.7054 | 1169.3962 | 1168.4918 | 0.9044 | 24 | 0.0053 | K.MoDETDASSAVK.V |
| 6018 | 103 | 114 | 671.1033 | 1340.192 | 1340.7704 | −0.5784 | 37 | 0.00067 | K.TSDLIVLGLPWK.T |
| 3868 | 152 | 160 | 573.0732 | 1144.1318 | 1143.5448 | 0.587 | 30 | 0.0028 | R.FTEYETQVK.V |
| 2331 | 182 | 189 | 486.4414 | 970.8682 | 971.4672 | −0.599 | 25 | 0.031 | K.QSQDEPLR.S |
| 2334 | 182 | 189 | 486.5248 | 971.035 | 971.4672 | −0.4322 | 25 | 0.017 | K.QSQDEPLR.S |
| 2345 | 182 | 189 | 487.3027 | 972.5909 | 971.4672 | 1.1237 | 26 | 0.013 | K.QSQDEPLR.S |
| 10280 | 276 | 293 | 864.1884 | 1726.3621 | 1725.7608 | 0.6013 | 106 | 3.40E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 10284 | 276 | 293 | 576.4902 | 1726.4489 | 1726.7448 | −0.296 | 29 | 0.028 | R.FGGNPGGFGNQ*GGFGNSR.G |
| 10291 | 276 | 293 | 864.5842 | 1727.1539 | 1726.7448 | 0.4091 | 90 | 2.20E-08 | R.FGGNPGGFGNQGGFGN*SR.G |
| fravtion6 | chymotrypsin | ||||||||
| 3105 | 277 | 289 | 583.4806 | 1164.9467 | 1164.4949 | 0.4519 | 25 | 0.0045 | F.GGNPGGFGNQGGF.G |
| 8141 | 298 | 315 | 594.347 | 1780.0191 | 1778.6389 | 1.3802 | 32 | 0.015 | A.GLGNNQGSpN*MoGGGMNFGA.F |
| 7563 | 300 | 316 | 565.4505 | 1693.3297 | 1693.5985 | −0.2688 | 29 | 0.031 | L.GN*N*QGSNMoGGGMoN*FGAF.S |
| 5625 | 317 | 330 | 486.4602 | 1456.3588 | 1455.6138 | 0.7449 | 18 | 0.041 | F.SpIN*PAMoMAAAQAAL.Q |
| trypsin | |||||||||
| 4254 | 103 | 114 | 671.3554 | 1340.6963 | 1340.7704 | −0.0741 | 36 | 0.00083 | K.TSDLIVLGLPWK.T |
| 7335 | 276 | 293 | 864.0093 | 1726.004 | 1725.7608 | 0.2432 | 65 | 2.40E-06 | R.FGGNPGGFGNQGGFGNSR.G |
| 7340 | 276 | 293 | 864.1556 | 1726.2966 | 1726.7448 | −0.4482 | 87 | 4.90E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
| fraction7 | chymotrypsin | ||||||||
| 4017 | 277 | 289 | 583.4226 | 1164.8307 | 1164.4949 | 0.3358 | 32 | 0.001 | F.GGNPGGFGNQGGF.G |
| 8195 | 375 | 391 | 579.1056 | 1734.2949 | 1733.5372 | 0.7577 | 20 | 0.095 | Y.SpGSN*SGAAIGWGSpASpNA.G |
| 6844 | 389 | 404 | 516.8932 | 1547.6579 | 1548.492 | −0.834 | 23 | 0.065 | A.SpNAGSGSGFNGGFGSSp.M |
| 9620 | 389 | 409 | 673.531 | 2017.5712 | 2017.7361 | −0.1649 | 20 | 0.078 | A.SN*AGSGSGFNGGFGSSpMDSKS.S |
| trypsin | |||||||||
| 7417 | 103 | 114 | 671.6024 | 1341.1903 | 1340.7704 | 0.4199 | 39 | 0.00022 | K.TSDLIVLGLPWK.T |
| 10947 | 276 | 293 | 864.018 | 1726.0215 | 1725.7608 | 0.2606 | 108 | 3.50E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| fraction8 | chymotrypsin | ||||||||
| not detected | |||||||||
| trypsin | |||||||||
| 4254 | 103 | 114 | 671.3554 | 1340.6963 | 1340.7704 | −0.0741 | 36 | 0.00083 | K.TSDLIVLGLPWK.T |
| 7335 | 276 | 293 | 864.0093 | 1726.004 | 1725.7608 | 0.2432 | 65 | 2.40E-06 | R.FGGNPGGFGNQGGFGNSR.G |
| 7340 | 276 | 293 | 864.1556 | 1726.2966 | 1726.7448 | −0.4482 | 87 | 4.90E-08 | R.FGGNPGGFGN*QGGFGNSR.G |
p indicates phosphorylation site; o indicates oxidation site; * indicates deamidation site.
Table 2. List of identifid peptides derived from accumulated TDP-43 in ALS patient case 2.
| Query | Start | End | Observed | Mr(expt) | Mr(calc) | Delta | Score | Expect | Peptide |
|---|---|---|---|---|---|---|---|---|---|
| fraction1 | chymotrypsin | ||||||||
| 4855 | 215 | 226 | 683.3186 | 1364.6226 | 1363.6846 | 0.938 | 17 | 0.066 | Y.GDVMDVFIPKPF.R |
| 20958 | 290 | 313 | 738.3618 | 2212.0636 | 2210.9182 | 1.1455 | 28 | 0.0071 | F.GNSRGGGAGLGNNQ*GSNMGGGMNF.G |
| 21027 | 300 | 321 | 742.2366 | 2223.6879 | 2222.8398 | 0.8481 | 27 | 0.048 | L.GNNQGSpN*MGGGMN*FGAFSINPA.M |
| 21271 | 316 | 336 | 771.2789 | 2310.815 | 2311.92 | −1.1051 | 29 | 0.064 | A.FSIN*PAMoMoAAAQAALQSSpWGMo.M |
| 17965 | 322 | 336 | 556.6434 | 1666.9085 | 1666.6078 | 0.3007 | 26 | 0.056 | A.MMoAAAQ*AALQ*SSpWGMo.M |
| 10760 | 389 | 398 | 489.2573 | 976.5 | 978.2968 | −1.7968 | 23 | 0.057 | A.SpN*AGSGSGFN*.G |
| 10762 | 389 | 398 | 489.3521 | 976.6897 | 978.2968 | −1.6071 | 22 | 0.064 | A.SpN*AGSGSGFN*.G |
| trypsin | |||||||||
| 24329 | 228 | 251 | 898.5277 | 2692.5613 | 2694.2652 | −1.7039 | 38 | 0.0016 | R.AFAFVTFADDQIAQS þ LCGEDLIIK.G |
| 12874 | 252 | 263 | 627.0836 | 1252.1527 | 1250.6619 | 1.4908 | 46 | 7.90E-05 | K.GISVHISNAEPK.H |
| 19990 | 276 | 293 | 863.7753 | 1725.5361 | 1725.7608 | −0.2247 | 103 | 1.50E-09 | R.FGGNPGGFGNQGGFGNSR.G |
| 20016 | 276 | 293 | 864.1752 | 1726.3359 | 1725.7608 | 0.5751 | 121 | 5.20E-12 | R.FGGNPGGFGNQGGFGNSR.G |
| 20030 | 276 | 293 | 864.5692 | 1727.1239 | 1725.7608 | 1.3631 | 100 | 1.30E-09 | R.FGGNPGGFGNQGGFGNSR.G |
| fraction2 | chymotrypsin | ||||||||
| 14560 | 76 | 85 | 725.5255 | 1449.0365 | 1448.7194 | 0.3171 | 23 | 0.0065 | V.NYPKgDNKaRKM.D |
| 18417 | 290 | 311 | 684.6705 | 2050.9898 | 2049.7041 | 1.2857 | 22 | 0.096 | F.GN*SpRGGGAGLGN*N*Q*GSN*MoGGGM.N |
| 15143 | 382 | 397 | 502.4551 | 1504.3433 | 1504.5984 | −0.2551 | 18 | 0.065 | A.AIGWGSASNAGSpGSGF.N |
| 17655 | 389 | 405 | 615.114 | 1842.3202 | 1840.4491 | 1.8711 | 30 | 0.024 | A.SpN*AGSpGSpGFNGGFGSSpM.D |
| 12347 | 398 | 408 | 421.6983 | 1262.0732 | 1261.3723 | 0.7008 | 23 | 0.078 | F.NGGFGSSpMoDSpK.S |
| 14828 | 398 | 411 | 738.5961 | 1475.1776 | 1476.463 | −1.2854 | 17 | 0.045 | F.NGGFGS.pSMDSKSpSG.W |
| trypsin | |||||||||
| 9063 | 76 | 82 | 439.4402 | 876.8658 | 877.4294 | −0.5635 | 21 | 0.065 | V.NYPKDNK.R |
| 12273 | 152 | 160 | 572.6329 | 1143.2513 | 1143.5448 | −0.2935 | 24 | 0.0066 | R.FTEYETQVK.V |
| 13442 | 252 | 263 | 626.5903 | 1251.166 | 1250.6619 | 0.5041 | 60 | 2.20E-06 | K.GISVHISNAEPK.H |
| 18848 | 276 | 293 | 864.1686 | 1726.3226 | 1726.7448 | −0.4222 | 100 | 2.80E-09 | R.FGGNPGGFGNQGGFGN*SR.G |
| 18879 | 276 | 293 | 865.5833 | 1729.1519 | 1727.7288 | 1.4231 | 92 | 2.30E-09 | R.FGGNPGGFGN*QGGFGN*SR.G |
| 14839 | 280 | 293 | 676.5123 | 1351.0101 | 1350.6065 | 0.4036 | 75 | 9.00E-08 | N.PGGFGNQGGFGNSR.G |
| 14842 | 280 | 293 | 676.5812 | 1351.1478 | 1350.6065 | 0.5413 | 46 | 0.00033 | N.PGGFGNQGGFGNSR.G |
| 14844 | 280 | 293 | 676.6381 | 1351.2617 | 1350.6065 | 0.6552 | 64 | 2.80E-06 | N.PGGFGNQGGFGNSR.G |
| 14837 | 280 | 293 | 676.4991 | 1350.9837 | 1351.5905 | −0.6068 | 33 | 0.0068 | N.PGGFGN*QGGFGNSR.G |
| fraction3 | chymotrypsin | ||||||||
| 19885 | 212 | 226 | 880.3099 | 1758.6053 | 1757.8335 | 0.7718 | 22 | 0.0093 | F.SQYGDVMoDVFIPKPF.R |
| 15096 | 215 | 226 | 683.6068 | 1365.1989 | 1363.6846 | 1.5143 | 34 | 0.00061 | Y.GDVMDVFIPKPF.R |
| 4235 | 230 | 234 | 584.3881 | 583.3808 | 583.3006 | 0.0802 | 26 | 0.0086 | F.AFVTF.A |
| 4246 | 230 | 234 | 584.5333 | 583.5261 | 583.3006 | 0.2254 | 26 | 0.0084 | F.AFVTF.A |
| 18976 | 290 | 307 | 831.7477 | 1661.4809 | 1662.7128 | −1.2319 | 40 | 0.00047 | F.GNSRGGGAGLGNNQGSNMo.G |
| 21767 | 290 | 313 | 748.6056 | 2242.7949 | 2242.908 | −0.1131 | 36 | 0.0087 | F.GNSRGGGAGLGNNQGSNMoGGGMoNF.G |
| 20036 | 298 | 315 | 594.1538 | 1779.4396 | 1780.607 | −1.1674 | 26 | 0.065 | A.GLGNN*Q*GSpN*MoGGGMNFGA.F |
| 19286 | 300 | 316 | 564.2299 | 1689.6679 | 1691.6304 | −1.9625 | 23 | 0.057 | L.GNNQGSNMoGGGMoN*FGAF.S |
| 19330 | 300 | 316 | 565.4157 | 1693.2251 | 1691.6304 | 1.5947 | 22 | 0.032 | L.GNNQGSNMoGGGMoN*FGAF.S |
| 21702 | 300 | 321 | 741.259 | 2220.7553 | 2220.8718 | −0.1165 | 20 | 0.068 | L.GNNQGSpNMGGGMNFGAFSINPA.M |
| 21046 | 341 | 359 | 654.4437 | 1960.3094 | 1960.7929 | −0.4836 | 58 | 3.10E-05 | L.ASQQNQSGPSGNNQNQ*GNM.Q |
| 20839 | 386 | 405 | 633.3611 | 1897.0616 | 1895.6071 | 1.4546 | 27 | 0.062 | W.GSASpN*AGSpGSGFNGGFGSSM.D |
| 21630 | 389 | 411 | 727.5361 | 2179.5866 | 2177.7845 | 1.8021 | 32 | 0.027 | A.SNAGSpGSGFN*GGFGSSMoDSKSSG.W |
| trypsin | |||||||||
| 21995 | 209 | 227 | 774.8737 | 2321.5992 | 2321.1191 | 0.4801 | 39 | 0.0013 | R.EFFSQYGDVMDVFIPKPFR |
| 22343 | 228 | 251 | 898.5321 | 2692.5745 | 2694.2652 | −1.6907 | 45 | 0.0003 | R.AFAFVTFADDQIAQSpLCGEDLIIK.G |
| 13384 | 252 | 263 | 625.472 | 1248.9294 | 1250.6619 | −1.7325 | 27 | 0.0032 | K.GISVHISNAEPK.H |
| 13416 | 252 | 263 | 626.5458 | 1251.077 | 1250.6619 | 0.4151 | 28 | 0.0078 | K.GISVHISNAEPK.H |
| 13418 | 252 | 263 | 626.6263 | 1251.238 | 1250.6619 | 0.5761 | 48 | 6.10E-05 | K.GISVHISNAEPK.H |
| 19580 | 276 | 293 | 864.1287 | 1726.2429 | 1725.7608 | 0.4821 | 102 | 5.50E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 19582 | 276 | 293 | 864.1488 | 1726.283 | 1725.7608 | 0.5222 | 122 | 6.00E-12 | R.FGGNPGGFGNQGGFGNSR.G |
| 19600 | 276 | 293 | 865.1992 | 1728.3839 | 1728.7128 | −0.329 | 28 | 0.036 | R.FGGN*PGGFGNQ*GGFGN*SR.G |
| 19637 | 276 | 293 | 866.3511 | 1730.6877 | 1728.7128 | 1.9749 | 75 | 9.20E-08 | R.FGGN*PGGFGN*QGGFGN*SR.G |
| fraction4 | chymotrypsin | ||||||||
| 15701 | 215 | 226 | 683.5077 | 1365.0008 | 1363.6846 | 1.3162 | 34 | 0.0018 | Y.GDVMDVFIPKPF.R |
| 15928 | 317 | 330 | 689.0359 | 1376.0573 | 1375.6475 | 0.4098 | 27 | 0.0096 | F.SIN*PAMMoAAAQAAL.Q |
| 21325 | 324 | 339 | 581.5296 | 1741.567 | 1739.6242 | 1.9428 | 19 | 0.085 | M.AAAQAALQ*SSpWGMoMoGMo.L |
| trypsin | |||||||||
| 14177 | 252 | 263 | 627.0461 | 1252.0777 | 1251.6459 | 0.4318 | 35 | 0.0051 | K.GISVHISN*AEPK.H |
| 23138 | 273 | 293 | 676.4238 | 2026.2497 | 2025.9154 | 0.3342 | 47 | 0.00016 | R.SGRFGGNPGGFGNQGGFGNSR.G |
| 20999 | 276 | 293 | 863.697 | 1725.3794 | 1725.7608 | −0.3815 | 105 | 8.20E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| 21025 | 276 | 293 | 864.4716 | 1726.9286 | 1725.7608 | 1.1678 | 104 | 2.90E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| fraction5 | chymotrypsin | ||||||||
| 7104 | 109 | 113 | 585.5835 | 584.5762 | 584.3322 | 0.244 | 26 | 0.0077 | V.LGLPW.K |
| 11793 | 131 | 139 | 537.3479 | 1072.6812 | 1072.6314 | 0.0498 | 31 | 0.011 | V.LMVQVKKDL.K |
| 7071 | 230 | 234 | 583.6779 | 582.6706 | 583.3006 | −0.63 | 26 | 0.0074 | F.AFVTF.A |
| 7086 | 230 | 234 | 584.4906 | 583.4833 | 583.3006 | 0.1827 | 26 | 0.0072 | F.AFVTF.A |
| 12944 | 277 | 289 | 583.3265 | 1164.6384 | 1164.4949 | 0.1435 | 27 | 0.0028 | F.GGNPGGFGNQGGF.G |
| 15760 | 298 | 311 | 696.9571 | 1391.8996 | 1391.437 | 0.4626 | 25 | 0.055 | A.GLGNN*Q*GSpN*MGGGM.N |
| 20561 | 298 | 316 | 649.7761 | 1946.3065 | 1944.6543 | 1.6522 | 25 | 0.051 | A.GLGN*N*Q*GSpN*MoGGGMoNFGAF.S |
| 22061 | 314 | 337 | 863.6423 | 2587.905 | 2586.03 | 1.875 | 33 | 0.0056 | F.GAFSpINPAMoMoAAAQ*AALQSSWGMoMo.G |
| 12943 | 375 | 385 | 583.3155 | 1164.6164 | 1165.3842 | −0.7678 | 19 | 0.078 | Y.SpGSNSpGAAIGW.G |
| 22006 | 375 | 400 | 845.8151 | 2534.4234 | 2532.8023 | 1.6211 | 27 | 0.061 | Y.SGSNSpGAAIGWGSpASpNAGSpGSGFNGG.F |
| 11997 | 389 | 400 | 546.7824 | 1091.5503 | 1092.3397 | −0.7894 | 22 | 0.088 | A.SN*AGSpGSGFN*GG.F |
| trypsin | |||||||||
| 12956 | 152 | 160 | 572.6786 | 1143.3427 | 1143.5448 | −0.2021 | 36 | 0.0034 | R.FTEYETQVK.V |
| 21351 | 209 | 227 | 775.3962 | 2323.1669 | 2322.1031 | 1.0638 | 17 | 0.069 | R.EFFSQ*YGDVMDVFIPKPFR.A |
| 14102 | 252 | 263 | 626.5144 | 1251.0142 | 1250.6619 | 0.3524 | 39 | 0.00022 | K.GISVHISNAEPK.H |
| 18594 | 276 | 293 | 863.6197 | 1725.2249 | 1725.7608 | −0.5359 | 107 | 1.10E-10 | R.FGGNPGGFGNQGGFGNSR.G |
| fraction6 | chymotrypsin | ||||||||
| 23141 | 290 | 321 | 1054.8215 | 3161.4428 | 3162.1846 | −0.7418 | 22 | 8.20E-02 | F.GNSpRGGGAGLGN*N*Q*GSNMoGGGMoNFGAFSpINPA.M |
| 22697 | 297 | 322 | 897.8206 | 2690.4399 | 2689.0162 | 1.4237 | 23 | 5.50E-02 | G.AGLGNNQGSpNMGGGMNFGAFSpINPAMo.M |
| 19240 | 300 | 318 | 659.1209 | 1974.3407 | 1972.6969 | 1.6439 | 28 | 2.10E-02 | L.GNNQ*GSpN*MoGGGMoNFGAFSI.N |
| 22742 | 316 | 340 | 905.2916 | 2712.8529 | 2712.1167 | 0.7362 | 29 | 7.80E-02 | A.FSINPAMMAAAQ*AALQ*SpSWGMoMGML.A |
| 13426 | 317 | 330 | 481.3466 | 1441.0179 | 1439.6189 | 1.3989 | 21 | 4.30E-02 | F.SpINPAMMAAAQ*AAL.Q |
| 10747 | 330 | 340 | 425.4213 | 1273.2422 | 1272.5189 | 0.7233 | 26 | 3.30E-02 | A.LQ*SSWGMMoGMoL.A |
| 16711 | 388 | 404 | 567.5427 | 1699.6063 | 1700.4794 | −0.8731 | 18 | 9.80E-02 | S.ASpN*AGSGSGFNGGFGSpSp.M |
| trypsin | |||||||||
| 8983 | 182 | 189 | 486.6083 | 971.2021 | 971.4672 | −0.2651 | 36 | 2.70E-03 | K.QSQDEPLR.S |
| 12216 | 252 | 263 | 626.5559 | 1251.0973 | 1250.6619 | 0.4354 | 57 | 4.20E-06 | K.GISVHISNAEPK.H |
| 17819 | 276 | 293 | 863.6684 | 1725.3222 | 1725.7608 | −0.4386 | 123 | 7.00E-12 | R.FGGNPGGFGNQGGFGNSR.G |
| fraction7 | chymotrypsin | ||||||||
| 14499 | 56 | 68 | 689.5842 | 1377.1539 | 1376.7088 | 0.4451 | 56 | 9.80E-06 | R.LVEGILHAPDAGW.G |
| 18452 | 56 | 71 | 831.8374 | 1661.6602 | 1660.8573 | 0.8029 | 40 | 0.0006 | R.LVEGILHAPDAGWGNL.V |
| 3963 | 230 | 234 | 584.48 | 583.4727 | 583.3006 | 0.1721 | 26 | 0.0072 | F.AFVTF.A |
| 15174 | 368 | 381 | 474.0898 | 1419.2475 | 1418.4024 | 0.8451 | 33 | 0.0091 | F.GSpGNN*SpYSGSNSGA.A |
| trypsin | |||||||||
| 21694 | 56 | 79 | 876.0681 | 2625.1825 | 2624.3639 | 0.8186 | 61 | 7.10E-06 | R.LVEGILHAPDAGWGN*LVYVVNYPK.D |
| 10852 | 84 | 95 | 641.5242 | 1281.0338 | 1280.5918 | 0.442 | 70 | 1.10E-06 | R.KMDETDASSAVK.V |
| 11727 | 103 | 114 | 671.4896 | 1340.9647 | 1340.7704 | 0.1943 | 38 | 0.00025 | K.TSDLIVLGLPWK.T |
| 17298 | 122 | 136 | 889.2975 | 1776.5804 | 1775.8804 | 0.7 | 69 | 1.90E-06 | K.EYFSTFGEVLMVQVK.K |
| 8866 | 152 | 160 | 573.5646 | 1145.1147 | 1143.5448 | 1.5699 | 26 | 0.0039 | R.FTEYETQVK.V |
| 10398 | 252 | 263 | 626.6704 | 1251.3263 | 1250.6619 | 0.6644 | 37 | 0.0007 | K.GISVHISNAEPK.H |
p indicates phosphorylation site; o indicates oxidation site; * indicates deamidation site; a indicates acetylation site; g indicates ubiqutination site.
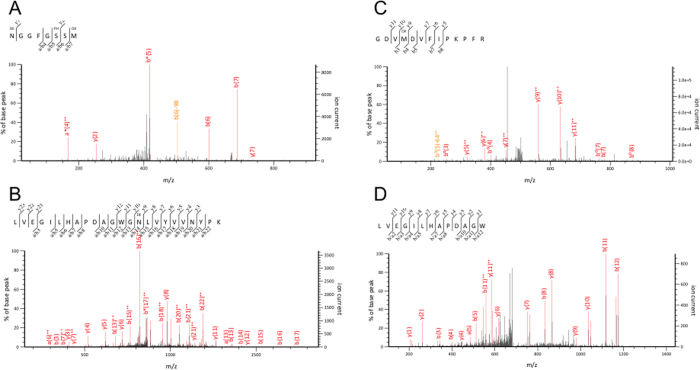
Figure 2. MS/MS identification of phosphorylated peptides and cleavage site peptides.
Representative peptides were shown. (A) Chymotriptic peptide, 398-NGGFGSSM-405 in case 1. The 6th Ser residue was phosphorylated. (B)Triptic peptide, 56-LVEGILHAPDAGWGNLVYVVNYPK-79 in case 1. (C) Tryptic peptide, 215- GDVMDVFIPKPFR-227 in case 1. Trypsin can not cleave N-terminal Tyr214-Gly215 site. Therefore this site is intrinsically cleaved site. (D) Chymotryptic peptide, 56-LVEGILHAPDAGW-68 in case 2. Chymotrypsin can not cleave N-terminal Arg55-Leu56 site. Therefore this site is intrinsically cleaved site.
Figure 3.
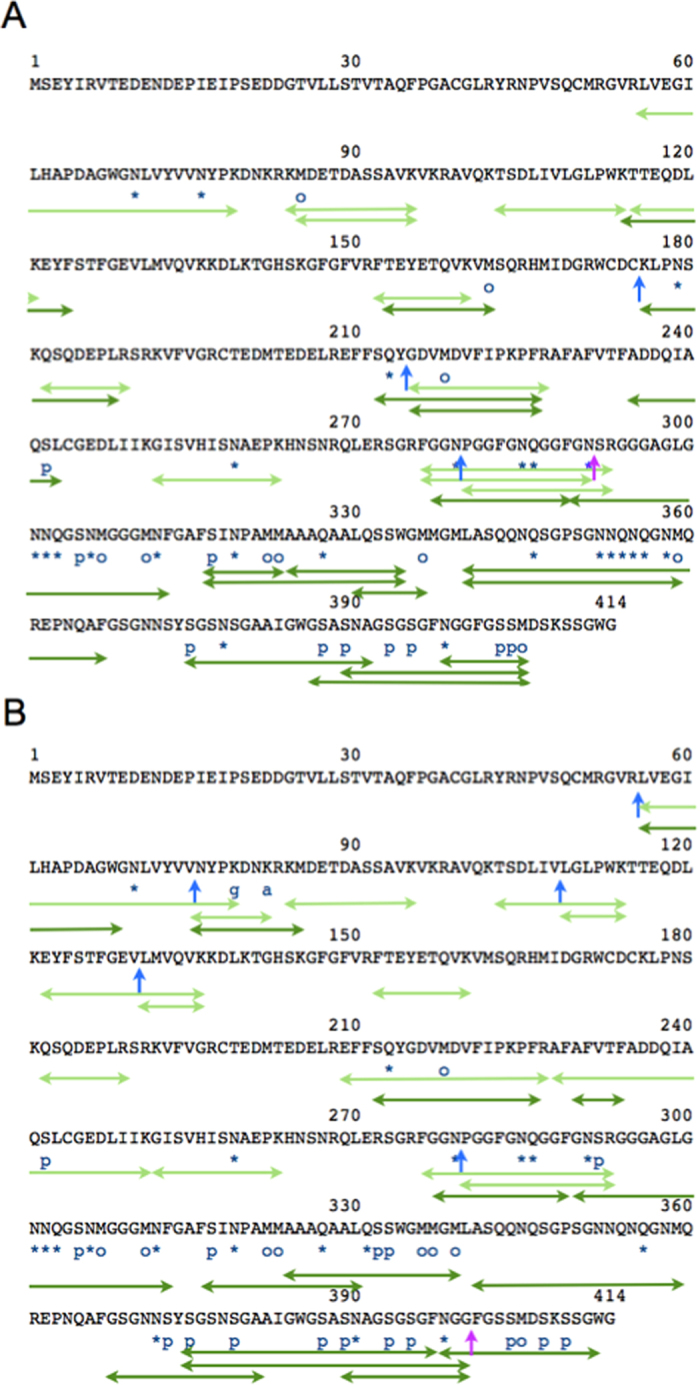
Identification of modification sites in TDP-43 from ALS case 1 (A) and case 2 (B) by LC-MS/MS analysis. Identified peptides from trypsin digestion (light green) and from chymotrypsin digestion (dark green) are shown. p indicates phosphorylation site. o indicates oxidation site. * indicates deamidation site. Blue and pink arrows indicate N-terminal and C-terminal cleavage sites, respectively.
Analysis of Case 1
We identified at least four intrinsically cleaved TDP-43 peptides in the pathological TDP-43, i.e., three N-terminal peptides (blue arrows, Fig. 3A) and one C-terminal peptide (pink arrow). However, no caspase-cleaved peptides were detected in this study (Table 3). Ten serine residues were phosphorylated, as shown in Fig. 3A and Table 4. These phosphorylation sites were the same sites as previously reported27,38. Furthermore, deaminations of Asn and Gln residues and oxidation of Met residues were identified. Almost all modification sites were found in the Gly-rich C-terminal half.
Table 3. N-terminal sequences of abnormally accumulated TDP-43 fragments in human brain.
| Disease | Molecular weight | Sequence | Reference |
|---|---|---|---|
| ALS | 45kDa | 56-LVEGILHA-- | this study |
| 43kDa | 56-LVEGILHA-- | ||
| 30–35kDa | 109-LGLPWK | ||
| 131-LMVQVKK | |||
| 23–25 kDa | 176-KLPNSKQ-- | ||
| 215-GDVMDVF-- | |||
| 280-PGGFGNQ-- | |||
| 15–20 kDa | 76-NYPKDNK-- | ||
| 215-GDVMDVF-- | |||
| FTLD-U | 23–25 kDa | 219-DVFIPKPF-- | Nonaka et al.47 |
| 247-DLIIKGI-- | |||
| FTLD-U | 22 kDa | 208-REFFSQY-- | Igaz et al.44 |
Table 4. List of modifications in cases 1 and 2.
| Residue | Amino acid | Modification | Case1 | Case2 |
|---|---|---|---|---|
| 70 | Asparagine | deamidation | O | O |
| 76 | Asparagine | deamidation | O | |
| 79 | Lysine | ubiquitination | O | |
| 82 | Lysine | acetylation | O | |
| 85 | Methionine | oxidation | O | |
| 162 | Methionine | oxidation | O | |
| 179 | Asparagine | deamidation | O | |
| 213 | Glutamine | deamidation | O | O |
| 218 | Methionine | oxidation | O | O |
| 242 | Serine | phosphorylation | O | O |
| 259 | Asparagine | deamidation | O | O |
| 279 | Asparagine | deamidation | O | O |
| 285 | Asparagine | deamidation | O | O |
| 286 | Glutamine | deamidation | O | O |
| 291 | Asparagine | deamidation | O | O |
| 292 | Serine | phosphorylation | O | |
| 301 | Asparagine | deamidation | O | O |
| 302 | Asparagine | deamidation | O | O |
| 303 | Glutamine | deamidation | O | O |
| 305 | Serine | phosphorylation | O | O |
| 306 | Asparagine | deamidation | O | O |
| 307 | Methionine | oxidation | O | O |
| 311 | Methionine | oxidation | O | O |
| 312 | Asparagine | deamidation | O | O |
| 317 | Serine | phosphorylation | O | O |
| 319 | Asparagine | deamidation | O | O |
| 322 | Methionine | oxidation | O | O |
| 323 | Methionine | oxidation | O | O |
| 327 | Glutamine | deamidation | O | O |
| 331 | Glutamine | deamidation | O | |
| 332 | Serine | phosphorylation | O | |
| 333 | Serine | phosphorylation | O | |
| 336 | Methionine | oxidation | O | O |
| 337 | Methionine | oxidation | O | |
| 339 | Methionine | oxidation | O | |
| 346 | Glutamine | deamidation | O | |
| 352 | Asparagine | deamidation | O | |
| 353 | Asparagine | deamidation | O | |
| 354 | Glutamine | deamidation | O | |
| 355 | Asparagine | deamidation | O | |
| 356 | Glutamine | deamidation | O | O |
| 358 | Asparagine | deamidation | O | |
| 359 | Methionine | oxidation | O | |
| 366 | Glutamine | deamidation | O | |
| 372 | Asparagine | deamidation | O | |
| 373 | Serine | phosphorylation | O | |
| 375 | Serine | phosphorylation | O | O |
| 378 | Asparagine | deamidation | O | |
| 379 | Serine | phosphorylation | O | |
| 387 | Serine | phosphorylation | O | O |
| 389 | Serine | phosphorylation | O | O |
| 390 | Asparagine | deamidation | O | |
| 393 | Serine | phosphorylation | O | O |
| 395 | Serine | phosphorylation | O | O |
| 398 | Asparagine | deamidation | O | O |
| 403 | Serine | phosphorylation | O | |
| 404 | Serine | phosphorylation | O | O |
| 405 | Methionine | oxidation | O | O |
| 407 | Serine | phosphorylation | O | |
| 409 | Serine | phosphorylation | O |
Analysis of Case 2
We identified six intrinsically cleaved TDP-43 peptides in this case, i.e., five N-terminal peptides (blue arrows, Fig. 3B) and one C-terminal peptide (pink arrow). As shown in Fig. 3B and Table 4, 15 serine residues were phosphorylated and 21 Asn/Gln residues were deamidated. Moreover, we found that the 79Lys residue was ubiquitinated and 82Lys was acetylated. However, 145Lys and 192Lys were not detected39. Phosphorylation sites were the same sites as previously reported27,38. Again, almost all of these modification sites were localized in the Gly-rich C-terminal half.
Common modifications in cases 1 and 2
All modifications are summarized in Table 4. As described above, almost all modifications were localized in the Gly-rich C-terminal half. Furthermore, common modifications focused on 180–330 residues region of these cases, suggesting that the 180–330 region of TDP-43 had the same structure in both case 1 and case 2.
Discussion
Accumulation of filamentous inclusions composed of abnormally phosphorylated full-length TDP-43 (45 kDa) and its fragments (35 and 17–27 kDa) is a defining feature of TDP-43 proteinopathies26. It has been reported that overexpression of full-length TDP-43 with or without mutations in cultured cells and animals leads to fragmentation of the protein, generating 35-kDa (CTF-35) and 25-kDa (CTF-25) C-terminal fragments via caspase-mediated TDP-43 cleavage30,34,40. Furthermore, activation of calpain, a Ca2+-dependent cysteine protease, by upregulation of Ca2+-permeable AMPA receptors generates C-terminally cleaved TDP-43 fragments (~35 kDa) and causes mislocalization of TDP-43 in motor neurons36. Those reports noted that the ~35 kDa fragments were localized in the cytoplasm and formed insoluble aggregates.
However, no such TDP-43 peptides cleaved by caspase or calpain reported were not detected in the pathological TDP-43 in the brains of patients in this study. On the other hand, uncleaved peptides by caspase or calpain were detected as the major molecules in the digests. Furthermore, several novel intrinsically cleaved TDP-43 peptides were identified in the pathological TDP-43, i.e., three and five N-terminal peptides (Cys164-Lys165, Tyr214-Gly215 and Asn279-Pro280 in case 1, and Arg55-Leu56, Val75-Asn76, Val108-Leu109, Val130-Leu131 and Asn279-Pro280 in case 2) (Figs 3A,B and 4 and Table 3) and two C-terminal peptide (Asn291-Ser292 in case 1 and Gly400-Phe401 in case 2) (Figs 3A,B and 4), strongly suggested that these cleaved fragments under pathological conditions was different from that under TDP-43-overexpressing conditions.
Figure 4. Schematic representation of N-terminal and C-terminal cleavage sites on TDP-43.
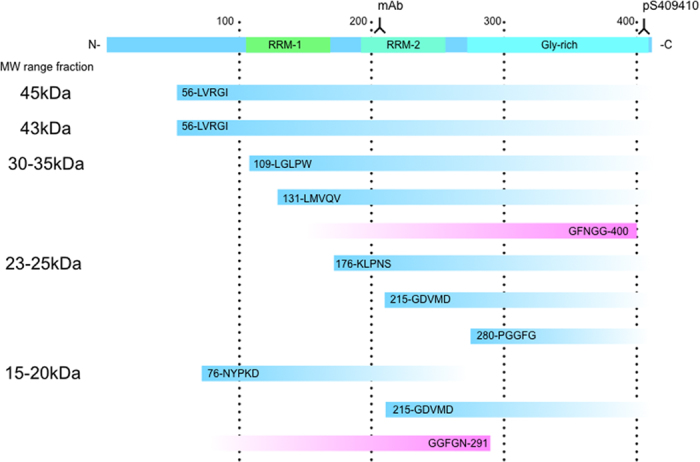
Cleavage sites (amino acid numbering of TDP-43) in gel fractions and antibodies recognition sites were shown.
Cellular expression of TDP-43 is tightly regulated at the transcriptional and post-translational levels. Under pathological conditions, however, formation of TDP-43 aggregates within the cell nucleus or cytoplasm results in reduced free nuclear TDP-43, and therefore the TDP-43 binding-receptor sensor detects a fall in protein levels and responds with increased TDP-43 production; the produced TDP-43 is then sequestered by TDP-43 aggregates2. Hence, TDP-43 production under pathological conditions does not necessarily represent overexpression. It is possible that TDP-43 aggregate formation under pathological conditions is different from that under TDP-43-overexpressing conditions.
It has been recently reported that C9ORF72 repeat expansions in mice cause TDP-43 pathology. In these mice, insoluble phosphorylated TDP-43 was predominantly present in monomeric form and high-molecular-weight or truncated pTDP-43 species were not detected41. Further, recombinant full-length human TDP-43 forms structurally stable, spherical oligomers that are neurotoxic in vitro and in vivo42. Indeed, cellular aggregate formation or accumulation of TDP-43 C-terminal fragments (CTFs) is not primarily responsible for development of the observed disease phenotype in mutant or wild-type TDP-43 mice43,44,45. These findings suggest that full-length TDP-43, not the cleavage fragments, participates at an early stage in TDP-43 pathology. Fragmentation may occur after the accumulation of TDP-43.
We identified some intrinsically cleaved TDP-43 peptides at the N-terminus and C-terminus of Sarkosyl-insoluble TDP-43 in this study. These cleavage sites were neither the caspase cleavage sites nor the calpain cleavage sites of TDP-43 as reported previously46,47. Several cleavage sites deduced from these N-terminal peptides exist in the region of residues 55–280 as shown in Table 3 and Fig. 4. Therefore, full-length TDP-43 may be gradually processed at N-terminal sites after the formation of insoluble aggregates as shown in Fig. 4. Our findings that cleavage site is correlated with the distribution of molecular weight size are consistent with this idea. Further studies using antibodies for these cleavage sites will clarify the gradual processing mechanism.
In our previous studies, we identified phosphorylation sites by immunochemical methods27 and also identified casein kinase-1 phosphorylation sites on recombinant TDP-43 38. Almost all Ser and Thr residues in C-terminal Gly-rich half were able to be phosphorylated in vitro experiment38. In the present work, however, a part of accumulated TDP-43 was phosphorylated, not all. Non-phosphorylated peptides were major products from aggregated TDP-43 digests from these ALS brains. We protein-chemically identified several novel phosphorylation sites by directly analyzing the accumulated TDP-43 from two ALS patients. Phosphorylation sites identified in case 1 and case 2 corresponded to the sites identified in casein kinase-1 phosphorylated TDP-43. Although 9 phosphorylation sites were common to both cases, more phosphorylation sites were identified in case 2, as shown in Fig. 3A,B.
It has known that non-phosphorylated intact TDP-43 forms fibrous aggregate42, suggesting abnormal phosphorylation is not first event in TDP-43 pathology. The level of phosphorylation may change according to the time when TDP-43 aggregates exist in cytoplasm. Phosphorylation-triggered degradation is a common strategy for elimination of regulatory proteins in many important signaling processes. Phosphorylation of TDP-43 may occur in elimination process. The impairment of this process due to TDP-43 aggregation will induce abnormal phosphorylation of TDP-43 aggregate. As the time has passed, the level of phosphorylation seems to become more. Therefore, the individual difference may occur in the level of phosphorylation.
Furthermore, it seems reasonable to speculate that heavily phosphorylated peptides may not be detected because of low recovery from the column and low ionization efficiency in mass analysis. S409/S410-phosphorylated TDP-43 was immunochemically included in almost all fractions as shown in Fig. 1. However, we found small amount of peptide including S409 with phosphorylation in case 2, while we found no peptide S409/S410 with phosphorylation in case1. A peptide including S409/S410, which derived from chymotrypsin digestion, has at least 5 phosphorylation sites. We have already reported that TDP-43 C-terminal region including these phosphorylation sites was heavily phosphorylated26,27,32. Therefore, we might not detect C-terminal region peptide including S409/S410 residues. It is possible that there are some differences in phosphorylation states of accumulated TDP-43 and in the detection efficiency in the mass analysis between the cases.
Indeed, there were individual differences in other modifications, as shown in Fig. 3A,B. However, we found that common modifications in case 1 and case 2 were focused in the 180–330 region (Fig. 3A,B, and Table 4). This suggests that the 180–330 region of TDP-43 had the same structure in both case 1 and case 2.
Interestingly, inclusions within the brain of ALS and FTLD-TDP patients are readily labelled with antibodies that recognize the C-terminus of TDP-43, but not with N-terminal TDP-43 antibodies. In contrast, spinal cord inclusions are labelled with both N- and C-terminal TDP-43 antibodies, suggesting that they are composed of full-length TDP-43 48. This regional heterogeneity in terms of C-terminal fragment formation suggests that these fragments may not be necessary for TDP43-mediated neurodegeneration49. Furthermore, TDP-43 fragments generated during neurodegeneration were not C-terminal fragments, but rather were derived from a central portion of human TDP-43 45. These results indicate that a common structure region, residues 180–330 of TDP-43, is significant for neurodegeneration
It has already reported that a Gly-rich region (287–322), which contains multiple glycine repeats, may contribute significantly to fiber formation as well as aggregation propensity50,51. Recently, it has also been reported that the 274–414 GQN-rich region (C-terminal region) has prion-like properties33,35, and that RRM2 domain of TDP-43 plays a key role in forming proteinaceous aggregates52. The common structure region, residues 180–330 of TDP-43, includes RRM2 and a part of the GQN-rich region. Therefore, this common structure region may be the core region of TDP-43 aggregates. The identification of N-terminal and C-terminal peptides in this study suggests that most of the cleavage sites exist in the N-terminal and C-terminal non-core regions. Proteolytic enzymes may readily access these regions and cleavage may occur gradually from distal regions. Our data provide important insight into the mechanism of TDP-43 accumulation, though further studies will be needed to clarify the molecular pathology of TDP-43 in detail.
Methods
Fractionation of accumulated TDP-43 in ALS brain
Sarkosyl-insoluble abnormal TDP-43 was prepared from brains of two ALS cases with abundant and widespread TDP-43 pathologies. Brain samples (0.5 g) from patients with ALS (case 1, 77-year-old male, disease duration 2.5 years and case 2, 57-year-old female, disease duration 2 years) and from vascular dementia (disease control, 88-year-old female) were homogenized in 10 ml of homogenization buffer (10 mM Tris–HCl, pH 7.5, containing 0.8 M NaCl, 1 mM EGTA, 1 mM dithiothreitol). Sarkosyl was added to the homogenates (final concentration: 2%), which were then incubated for 30 min at 37 °C and centrifuged at 20,000 g for 10 min at 25 °C. The supernatants were centrifuged at 100,000 g for 20 min at 25 °C. The pellets were further washed with sterile saline and centrifuged at 100,000 g for 20 min. The resulting pellets were used as Sarkosyl-insoluble fraction. This study was approved by the research ethics committee of Tokyo Metropolitan Institute of Medical Science (number: 15–5), and carried out in accordance with the approved guidelines. Informed consent about the brain donation was obtained from all subjects.
SDS-polyacrylamide gel electrophoresis (PAGE), immunoblotting and in-gel digestion
The Sarkosyl-insoluble, SDS/urea-soluble fractions were separated on 4 ~ 20% polyacrylamide gradient gels by SDS-PAGE and immunochemically detected as described26. Briefly, for immunoblotting, mAb anti-TDP-43 (60019-2-Ig, 1:3000, ProteinTech Group), polyclonal anti-TDP (10782-1-AP, 1:3000, ProteinTech Group) and pS409/410 (rabbit polyclonal, 1:1000) were used. Bands of TDP-43 and its derivatives were excised and soaked in 50 mM Tris-HCl, pH 8.0, containing 50% acetonitrile for 30 min. The gel was dried in a Speed-Vac (Savant) and incubated in 50 mM Tris-HCl, pH 8.0 containing 125–250 ng of modified trypsin (Roche Diagnostics, Mannheim, Germany) or chymotrypsin (Roche Diagnostics, Mannheim, Germany) at 37 °C for 6–20 hours. The digests were extracted from the gel twice with 100 μl of 0.1% TFA containing 60% acetonitrile. These two extracts were combined, evaporated in a Speed-Vac, and stored at −80 °C until assayed.
Nano-flow liquid chromatography-ion trap mass spectrometry (LC-MS/MS)
The sample was resuspended in 0.1% formic acid containing 2% acetonitrile and introduced into a nano-flow HPLC system, DiNa fitted with an automatic sampler (KYA Technology Corporation, Tokyo, Japan). A packed nano-capillary column NTCC-360/75-3-123 (0.075 mm I.D. ×125 mm L, particle diameter 3 μm, Nikkyo Technos Co., Ltd., Tokyo, Japan) was used at a flow rate of 300 nl / min with a 2–80% linear gradient of acetonitrile for 60 min. Eluted peptides were directly detected with an ion trap mass spectrometer, Velos Pro (Thermo Fisher Scientific Inc., Waltham, USA) at a spray voltage of 1.9 kV and a collision energy of 35%. The mass acquisition method consisted of one full MS survey scan followed by an MS/MS scan of the most abundant precursor ions from the survey scan. Dynamic exclusion for the MS/MS was set to 30 seconds. An MS scan range of 400–2000 m/z was employed in the positive ion mode, followed by data-dependent MS/MS using the CID or HCD operating mode on the top 10 ions in order of abundance. The data were analyzed with Proteome Discoverer (Thermo Fisher Scientific Inc., Waltham, USA), Mascot software (Matrix Science Inc., Boston, USA) and Scaffold software (Proteome Software, Inc., Oregon, USA). Swiss prot and GenBank databases were used.
Additional Information
How to cite this article: Kametani, F. et al. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281; doi: 10.1038/srep23281 (2016).
Supplementary Material
Acknowledgments
This work was supported by MEXT KAKENHI Grant Numbers 23228004, 26117005, 15H02356 (to M.H.) and MHLW Grant ID Number 12946221 (to M.H.)
Footnotes
Author Contributions All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: F.K. and M.H. Acquisition of data: F.K. and M.H. Analysis and interpretation of data: F.K. Drafting of the manuscript: F.K. Critical revision of the article for important intellectual content: M.H. Obtained funding: M.H. Material support: T.O., T.S., H.A., S.M., Y.S. and M.Y. Study supervision: M.H.
References
- Buratti E. et al. TDP-43 Binds Heterogeneous Nuclear Ribonucleoprotein A/B through Its C-terminal Tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulatior exon 9 splicing. J. Biol. Chem. 280, 37572–37584, 10.1074/jbc.M505557200 (2005). [DOI] [PubMed] [Google Scholar]
- Baralle M., Buratti E. & Baralle F. E. The role of TDP-43 in the pathogenesis of ALS and FTLD. Biochem Soc Trans 41, 1536–1540, 10.1042/bst20130186 (2013). [DOI] [PubMed] [Google Scholar]
- Ayala Y. M., Misteli T. & Baralle F. E. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA 105, 3785–3789, 10.1073/pnas.0800546105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala Y. M., Pagani F. & Baralle F. E. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett 580, 1339–1344, 10.1016/j.febslet.2006.01.052 (2006). [DOI] [PubMed] [Google Scholar]
- Corrado L. et al. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Human Mutation 30, 688–694 (2009). [DOI] [PubMed] [Google Scholar]
- Buratti E. & Baralle F. E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 13, 867–878 (2008). [DOI] [PubMed] [Google Scholar]
- Buratti E. et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20, 1774–1784, 10.1093/emboj/20.7.1774 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J. K., Wang I. F., Hung L., Tarn W.-Y. & Shen C. K. J. TDP-43 overexpression enhances exon-7 inclusion during SMN Pre-mRNA splicing. J. Biol. Chem. 283, 28852–28859, 10.1074/jbc.M805376200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S. H., Wu F., Harrich D., Garcia-Martinez L. F. & Gaynor R. B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69, 3584–3596 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y., Wang I. F., Bose J. & Shen C. K. J. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83, 130–139 (2004). [DOI] [PubMed] [Google Scholar]
- Wang I. F., Reddy N. M. & Shen C. K. Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA 99, 13583–13588, 10.1073/pnas.212483099 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton C. F. et al. TDP-43 Is a Developmentally Regulated Protein Essential for Early Embryonic Development. Journal of Biological Chemistry 285, 6826–6834, 10.1074/jbc.M109.061846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-S. et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. genesis 48, 56–62, 10.1002/dvg.20584 (2010). [DOI] [PubMed] [Google Scholar]
- Neumann M. et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 314, 130–133, 10.1126/science.1134108 (2006). [DOI] [PubMed] [Google Scholar]
- Arai T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and Biophysical Research Communications 351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- Yokota O. et al. Phosphorylated TDP-43 pathology and hippocampal sclerosis in progressive supranuclear palsy. Acta Neuropathol 120, 55–66, 10.1007/s00401-010-0702-1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T. et al. Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol 117, 125–136, 10.1007/s00401-008-0480-1 (2009). [DOI] [PubMed] [Google Scholar]
- Hasegawa M. et al. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain 130, 1386–1394, 10.1093/brain/awm065 (2007). [DOI] [PubMed] [Google Scholar]
- Tan C. F. et al. Selective occurrence of TDP-43-immunoreactive inclusions in the lower motor neurons in Machado-Joseph disease. Acta Neuropathol 118, 553–560, 10.1007/s00401-009-0552-x (2009). [DOI] [PubMed] [Google Scholar]
- Toyoshima Y. et al. Spinocerebellar ataxia type 2 (SCA2) is associated with TDP-43 pathology. Acta Neuropathol 122, 375–378, 10.1007/s00401-011-0862-7 (2011). [DOI] [PubMed] [Google Scholar]
- Kabashi E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40, 572–574 (2008). [DOI] [PubMed] [Google Scholar]
- Sreedharan J. et al. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science 319, 1668–1672, 10.1126/science.1154584 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I. R., Rademakers R. & Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9, 995–1007, 10.1016/S1474-4422(10)70195-2 (2010). [DOI] [PubMed] [Google Scholar]
- Tamaoka A. et al. TDP-43 M337V mutation in familial amyotrophic lateral sclerosis in Japan. Intern Med 49, 331–334 (2010). [DOI] [PubMed] [Google Scholar]
- Pesiridis G. S., Lee V. M. & Trojanowski J. Q. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet 18, R156–162, 10.1093/hmg/ddp303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H. et al. Molecular analysis and biochemical classification of TDP-43 proteinopathy. Brain 135, 3380–3391, 10.1093/brain/aws230 (2012). [DOI] [PubMed] [Google Scholar]
- Hasegawa M. et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Annals of Neurology 64, 60–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Kwong L. K., Sampathu D. M., Trojanowski J. Q. & Lee V. M. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol 64, 1388–1394, 10.1001/archneur.64.10.1388 (2007). [DOI] [PubMed] [Google Scholar]
- Neumann M. et al. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol 66, 152–157, 10.1097/nen.0b013e31803020b9 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA 106, 7607–7612, 10.1073/pnas.0900688106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M. et al. Molecular analysis and biochemical classification of TDP-43 proteinopathy. Dementia and Geriatric Cognitive Disorders 33, 103–104 (2012). [Google Scholar]
- Inukai Y. et al. Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Letters 582, 2899–2904 (2008). [DOI] [PubMed] [Google Scholar]
- Nonaka T. et al. Prion-like Properties of Pathological TDP-43 Aggregates from Diseased Brains. Cell Reports 4, 124–134, 10.1016/j.celrep.2013.06.007 (2013). [DOI] [PubMed] [Google Scholar]
- Li Q., Yokoshi M., Okada H. & Kawahara Y. The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat Commun 6, 10.1038/ncomms7183 (2015). [DOI] [PubMed] [Google Scholar]
- Wang I. F. et al. The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat Commun 3, 766, 10.1038/ncomms1766 (2012). [DOI] [PubMed] [Google Scholar]
- Yamashita T. et al. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat Commun 3, 1307, 10.1038/ncomms2303 (2012). [DOI] [PubMed] [Google Scholar]
- Aggad D., Vérièpe J., Tauffenberger A. & Parker J. A. TDP-43 toxicity proceeds via calcium dysregulation and necrosis in aging Caenorhabditis elegans motor neurons. J Neurosci 34, 12093–12103, 10.1523/JNEUROSCI.2495-13.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametani F. et al. Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochemical and Biophysical Research Communications 382, 405–409, 10.1016/j.bbrc.2009.03.038 (2009). [DOI] [PubMed] [Google Scholar]
- Cohen T. J. et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun 6, 10.1038/ncomms6845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Lee K. & Matsuoka M. TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. J Biol Chem 286, 13171–13183, 10.1074/jbc.M110.197483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J. et al. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154, 10.1126/science.aaa9344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y. S. et al. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat Commun 5, 4824, 10.1038/ncomms5824 (2014). [DOI] [PubMed] [Google Scholar]
- Janssens J. et al. Overexpression of ALS-associated p.M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol Neurobiol 48, 22–35, 10.1007/s12035-013-8427-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz L. M. et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 121, 726–738, 10.1172/JCI44867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alton S. et al. Divergent Phenotypes in Mutant TDP-43 Transgenic Mice Highlight Potential Confounds in TDP-43 Transgenic Modeling. PLoS ONE 9, e86513, 10.1371/journal.pone.0086513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz L. M. et al. Expression Of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 284, 8516-8524, M809462200, 10.1074/jbc.M809462200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T., Kametani F., Arai T., Akiyama H. & Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353–3364, 10.1093/hmg/ddp275 (2009). [DOI] [PubMed] [Google Scholar]
- Igaz L. M. et al. Enrichment of C-Terminal Fragments in TAR DNA-Binding Protein-43 Cytoplasmic Inclusions in Brain but not in Spinal Cord of Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Am J Pathol 173, 182–194, 10.2353/ajpath.2008.080003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. B., Lee V. M. Y. & Trojanowski J. Q. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 13, 38–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol 18, 822–830, 10.1038/nsmb.2053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. K. et al. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J Am Chem Soc 132, 1186–1187, 10.1021/ja9066207 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Y.-T. et al. The Truncated C-terminal RNA Recognition Motif of TDP-43 Protein Plays a Key Role in Forming Proteinaceous Aggregates. Journal of Biological Chemistry 288, 9049–9057, 10.1074/jbc.M112.438564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.