Abstract
Repair of tissue wounds is a fundamental process to re-establish tissue integrity and regular function. Importantly, infection is a major factor that hinders wound healing. Multicellular organisms have evolved an arsenal of host-defence molecules, including antimicrobial peptides (AMPs), aimed at controlling microbial proliferation and at modulating the host's immune response to a variety of biological or physical insults. In this brief review we provide the evidence for a role of AMPs as endogenous mediators of wound healing and their promising therapeutic potential for treatment of non-life threatening skin and other epithelial injuries.
Keywords: antibiotic-resistance, antimicrobial peptide, frog skin, innate immunity, skin infections, wound healing
Introduction
Wound healing in animals is an essential process for the repair and restoration of function of tissue after injury (1). If we look at normal skin, both the epidermis and dermis form a protective shield from external physical and chemical environmental challenges (2). However, following surgery, accidental injury, burns, microbial infection, skin diseases or metabolic dysfunction, this barrier can be broken giving rise to a wound (3).
Classically cutaneous wound healing begins with inflammation, the natural response to any injury or insult, followed by a period of tissue regeneration including epithelialization, angiogenesis and laying down of collagen and culminating in a remodeling process to restore the tissue (4–6). Importantly, failure of the mechanisms underlying the recovery of damaged tissues can lead to the formation of non-healing chronic wounds (7, 8). In this context, diabetes, venous or arterial disease and microbial infections are the most common factors favouring the establishment of chronic wounds (7). Opportunistic pathogens, such as the Gram-negative bacterium Pseudomonas aeruginosa or the Gram-positive bacterium Staphylococcus aureus are able to colonize skin wounds forming biofilms, which are characterized by an aggregation of bacterial cells immobilized in an adhesive matrix of extracellular polymeric substances (9, 10). This makes the eradication of bacteria difficult, mainly due to the weak penetration of antibiotics or host clearance mechanisms i.e. antibodies and phagocytes through the microbial biofilm (11–14). Furthermore, toxins released from bacteria contribute to recruitment of immune cells resulting in an excessive, damaging inflammatory response (15, 16). Wound infections have become an increasing cause of death in severely ill hospitalized patients (especially in long-term care facilities) and their treatment has put a significant burden on the health care system and on the economy as a whole (17–20).
To ensure epithelial integrity, multicellular organisms have evolved an arsenal of host-defence molecules, including gene-encoded antimicrobial peptides (AMPs), which are aimed at controlling microbial proliferation and at signaling host cells to change their behaviour in response to injury (21–23). Here we evaluate the evidence for a role for AMPs as endogenous mediators of wound healing and their therapeutic potential.
AMPs and Innate Immunity
AMPs are known to exist in all living kingdoms as evolutionarily conserved products of the innate immune system (24). Pioneering work in the early 1980s resulted in the discovery of "cecropin" peptides, from the hemolymph of the cecropia moth Hyalophora cecropia (25). This was then followed by the isolation and characterization of "defensins" from granules of mammalian granulocytes (26). A third milestone came in 1987 with Michael Zasloff and the discovery that the skin of the African toad Xenopus laevis contains glands rich in AMPs which are discharged by a holocrine mechanism upon alarm or skin lesion (27). Since then, a large variety of AMPs have been identified across species with over two thousand peptides reported so far (28). The current AMP data base can be found at http://aps.unmc.edu/AP/main.php.
Despite their different amino acid sequence (generally comprising from 10 to 50 amino acids), the vast majority of AMPs share a cationic character, due to the prevalence of basic residues, and an amphipathic structure in membrane-mimicking environments (29). Their mechanism of antimicrobial action is largely due to simple electrostatic interaction with the anionic phospholipids of the microbial cell membrane (30) leading to disintegration/perturbation of the latter, and resulting in cell death (31, 32). For this reason, they are less likely to induce resistance in microbes (32) than conventional antibiotics, which are typically directed toward single, mostly enzymatic targets (33, 34). To become resistant to AMPs, microbes would need to "redesign" their membrane lipid composition; but this could not be realized without causing harm to themselves (35). Whilst first recognised for their antimicrobial activity, AMPs have since been found to exert a plethora of activities ranging from neutralization of the pro-inflammatory agent lipopolysaccharide (LPS), to immune cell chemoattraction and cell proliferation (36–40). This wide array of activities has been described for a multiplicity of AMPs leading to widespread hope for their potential development as novel anti-infective molecules with additional beneficial properties, or in some cases for use as stand alone modulators in the absence of overt infection (41–45).
Human Skin AMPs in Health and Disease
In mammals, including humans, keratinocytes within the skin granular layer synthesize and store AMPs along with lipids, within secretory granules called lamellar bodies (46–52). The lamellar bodies are released into the intercellular spaces of the more superficial layers of the epidermis, creating a physical barrier that can constrain microbial growth as well as water loss. The principal AMPs present in healthy human skin are RNase 5, RNase 7, active on both Gram-negative and Gram-positive bacteria (53), and the iron/zinc binding S100 proteins (e.g. psoriasin, calprotectin) (53–55). However, in inflammatory/infectious skin diseases other AMP families with a wide spectrum of antimicrobial activity are additionally expressed by keratinocytes, such as the β-defensins and the cathelicidin hCAP18, which is then converted to the active AMP, LL-37 (2). In particular, the levels of both S100A7 (psoriasin), which is preferentially active against Escherichia coli (47), and the anti-Candida iron/zinc sequestering heterodimeric S100A8/S100A9 complex (calprotectin), become markedly elevated in the epidermis of psoriatic patients (47, 56). On the other hand, in disease conditions such as atopic dermatitis, induction of hCAP18 mRNA transcription in wounds is suppressed, which is hypothesized to underlie an increased rate of microbial superinfections (57, 58).
Pilosebaceous follicles and eccrine glands also represent a further source of skin AMPs, such as histone 4 (mainly active against Gram-positive bacteria) and the antibacterial/antifungal dermcidin, respectively (59, 60). Beside keratinocytes, sebocytes and sweat glands, infiltrating immune cells e.g. neutrophils and natural killer cells also contribute to the pool of AMPs in the skin, mainly by the production of α-defensins and LL-37 (61).
Examples of human AMPs produced by different types of cells in skin or other epithelial tissues with a protective function from environmental insults (see section on “Human AMPs play a crucial role in wound healing”) under normal conditions or upon infection/inflammation are presented in Table 1. It is worth noting that some AMPs are constitutively expressed and detected in both healthy or injured epithelium, whereas some others are negligible in healthy epithelium but produced in abundance in infected or chronically inflamed tissue.
Table 1.
Examples of AMPs produced by different types of epithelial cells in healthy tissues or upon tissue infection/inflammation.
| Type of epithelial cell |
Constitutive expression |
Induced production in infected tissues |
Ref. | SOURCE |
|---|---|---|---|---|
| Keratinocytes/Hair follicle | hBD-1 | (73, 135) | 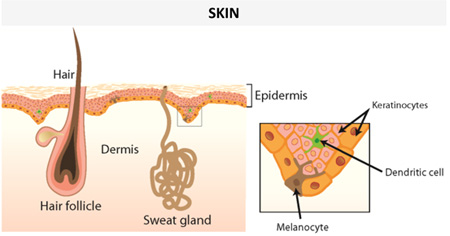 |
|
| hBD-2,3 | (53, 73, 135, 136) | |||
| RNase 7 | RNAse 7 | (46, 73) | ||
| Psoriasin | Psoriasin | (47, 136, 137) | ||
| Calprotectin | Calprotectin | (54, 56) | ||
| LL-37 | (73, 138) | |||
| Sebocytes | Histone 4 | (59) | ||
| Psoriasin | (47) | |||
| hBD2 | (139, 140) | |||
| LL-37 | LL-37 | (141) | ||
| Eccrine sweat glands | Dermcidin | (60, 142) | ||
| LL-37 | LL-37 | (143) | ||
| Corneal epithelial cells | hBD-1 | hBD-2 | (144, 145) | 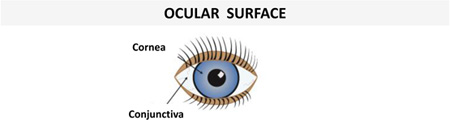 |
| hBD-3 | (144–146) | |||
| LL-37 (low level) | LL-37 | (145, 147) | ||
| Conjunctival epithelial cells | hBD-1, hBD-3 | hBD-2 | (146, 148) | |
| LL-37 (low level) | LL-37 | (147) | ||
| Ciliated airway epithelium | hBD-1 | (149, 150) | 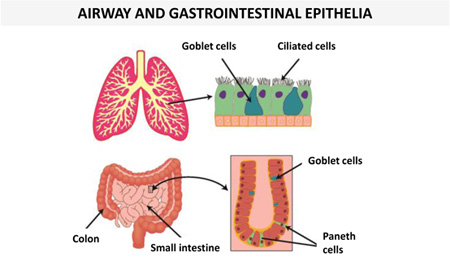 |
|
| hBD-2,3,4 | (150–152) | |||
| LL-37 | LL-37 | (150, 153, 154) | ||
| Paneth epithelial cells (small intestine) | α-defensin 5,6 | (155, 156) | ||
| Colonic epithelial cells | hBD-1 | (157) | ||
| hBD-2,3,4 (low level) | hBD-2,3,4 | (157) | ||
It is important to recall that epithelial surfaces, especially the skin, are continually exposed to both pathogenic and harmless commensal microbes whose composition depends on the anatomical location and the moisture content (62). Indeed, moist sites are generally populated by Staphylococcus and Corynebacterium species (63, 64); sebaceous sites harbor Propionibacterium species, due to their capacity to survive in anaerobic environments, while dry areas of the skin have the most diversity in bacterial inhabitants (65). In addition to the bacteria, members of the skin microbiome also include fungi such as Malassezia species, especially on the scalp (66), and parasitic arthropods (Demodex mites (67)) especially in pilosebaceous units.
Keratinocytes, in particular, are able to sense and discriminate among microbes and to activate a defence response based on the production of AMPs and cytokines (52, 68, 69). It has been suggested that bacteria release factors that diffuse through the stratum corneum of the skin to finally induce the expression of AMPs by keratinocytes of the deeper epidermal layers (70). The correlation between mammalian AMP expression and composition of the commensal microbiota is however actually highly debated (71). Unbalanced skin microflora may alter AMP expression in the skin (62); and a dysregulation of AMPs might be an important factor in the pathogenesis of different skin disorders such as psoriasis, rosacea, atopic dermatitis (72–75), as well as in the host susceptibility to bacterial colonization and wound repair (76, 77).
Studies on the etiology of psoriasis have uncovered another role for AMPs in the control of microbial growth in the setting of epidermal injury (78). Psoriatic skin lesions rarely become infected despite the damaged physical barrier, in large part due to the presence of high concentrations of the AMPs and antimicrobial proteins expressed in the epidermis (47, 79). The stimuli for heightened AMP expression are the cytokines, IL-17 and IL-22, secreted by TH17 and γδ lymphocytes present in the skin, which strongly induce the activity of AMP genes in the basal keratinocytes (78). Recent work has highlighted the role of injury in initiating the process. Cellular damage releases DNA and LL-37, which combine to form a structured complex (80), subsequently recognized by TLR9 receptors in plasmacytoid dendritic cells (pDCs) residing in the dermis (81). Once activated these pDCs secrete γ interferon, which, in turn, promotes the subsequent differentiation of TH17 cells and γδ T cells (82). Under normal conditions, this circuit is eventually turned off, leading to heightened AMP expression in the injured skin. In psoriasis, for reasons still unclear, stimulation continues relentlessly.
Human AMPs play a crucial role in wound healing
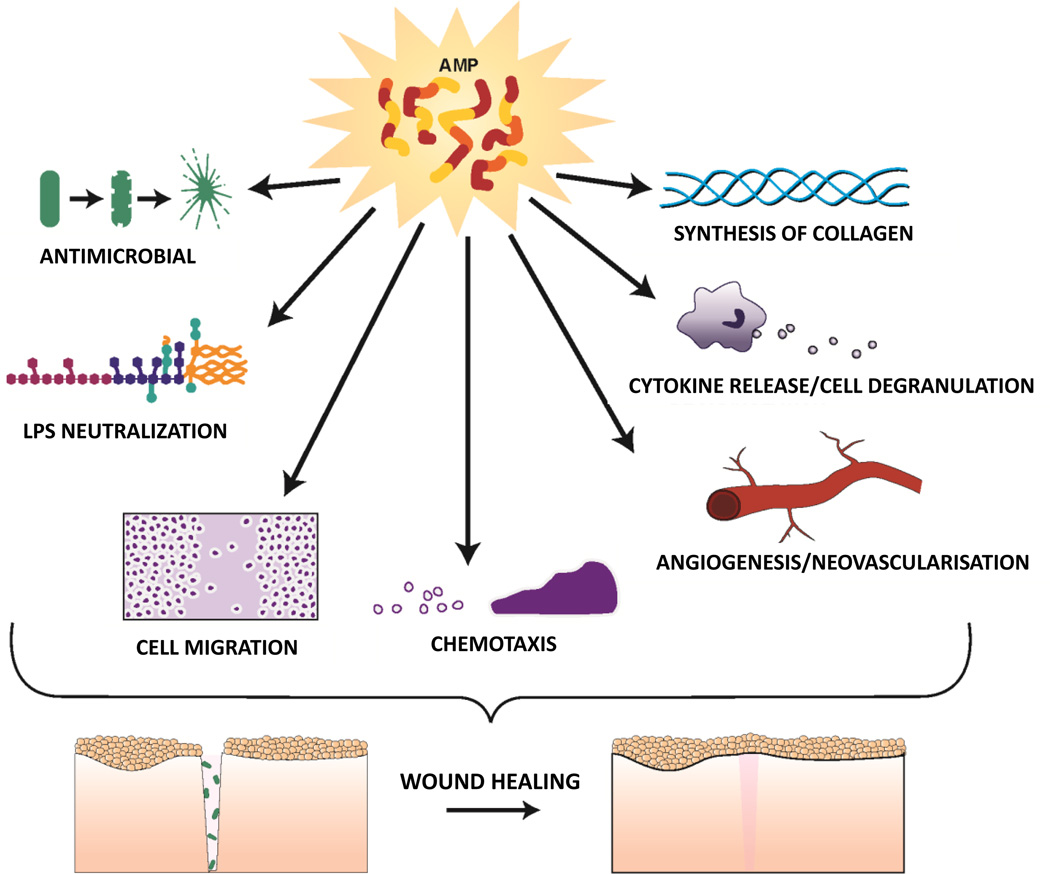
A plethora of studies (76) support the hypothesis that human AMPs promote wound healing in skin, by multiple means including modulation of cytokine production, cell migration, proliferation and in some cases angiogenesis (Figure 1). For example, expression of human β-defensin (hBD)-2 is induced in human skin wounds by epidermal growth factor receptor (EGFR) activation (83) and increases keratinocyte cytokine production and migration (84). Another defensin, hBD-3, is also highly expressed by keratinocytes at wound sites, promotes cytokine secretion, cell migration and proliferation via phosphorylation of EGFR and STAT proteins (84–86), and significantly accelerates wound closure when topically applied in a porcine model of infected skin wounds (87). Importantly, hBD-3 has been shown to exhibit in vitro and in vivo anti-inflammatory activity by inhibiting TLR signaling pathways in immune cells with a resulting transcriptional repression of pro-inflammatory genes (88, 89). This may contribute to resolution of inflammation. In addition, hBD-3 has been shown to act as a ligand for the macrophage receptor CCR2, attracting macrophages to sites of epithelial injury (90). Several actions of LL-37 appear to be mediated via trans activation of EGFR including keratinocyte migration (91). Notably, skin LL-37 expression is also increased after wounding and in an ex vivo human skin wound healing model, antibodies against LL-37 inhibited re-epithelialization (92). Some of LL-37’s beneficial effects appear to involve modulation of angiogenesis as this AMP has been shown to stimulate proliferation and neovascularisation, for example via activation of formyl peptide receptor-like 1 (FPR2/ALX) (93). Also, its porcine analog PR-39 increases the synthesis of the extracellular matrix proteoglycans, important for wound repair (94). Beneficial effects of AMPs on wound healing are not limited to skin but extend to other related tissues such as the stratified non keratinized corneal epithelium, which also derives from the surface ectoderm (95), and where eyelid glands are developmentally similar to skin appendages i.e. hair follicle/sebaceous glands (96). For example, hBD-3 and LL-37 have been shown to stimulate corneal epithelial cell activities necessary to promote healing at the ocular surface (97). Furthermore, they can promote wound healing of other epithelial tissues with a barrier function to the external environment, such as gut and lung (98). Indeed, hBD-2 has been shown to stimulate intestinal wound healing in an in vitro model (99); while hBD-3 promoted intestinal healing in a neonatal rat model (100) and LL-37 stimulated migration and proliferation of airway epithelial cells (101).
Figure 1.
Schematic representation of biological functions of AMPs, beneficial for wound healing
AMP involvement in wound healing is not unique to mammalian tissue
Despite differences in morphology and composition of skin layers among different groups of animals, healing of epidermal wounds is a quite conserved process along the evolutionary scale (102). In Drosophila, epidermal cells under the cuticle of larvae respond to infected wounds by producing the AMP cecropin A (103). Note that in a recent published issue of Experimental Dermatology, the fruit fly Drosophila has been proposed as an advantageous model system for identifying clinically relevant wound-healing genes and understanding the molecular mechanism of how they work (104). In other invertebrates, like the nematode Caenorhabditis elegans, induction of the epidermal AMPs caenacins, upon infection and cuticle damage involves expression of transforming growth factor-β (TGF-β) (105). However, it is not known if wounding elicits neuroimmune or systemic responses as in other organisms (102). With regards to non-mammalian vertebrates, a vast literature supports the belief that amphibian skin contains factors associated with high efficiency wound healing mechanisms (see section on "Therapeutic opportunities of AMPs for wound healing and skin care") up to regeneration of an entire limb or tail in juvenile animals (106–109). In particular, wound healing proceeds very quickly (in less than 10 h) in urodele amphibians (salamanders) compared to the re-epithelialization process in mammalian wounds which normally takes 2–3 days (110).
Therapeutic opportunities of AMPs for wound healing and skin care
Successful development of AMPs for a variety of therapeutic purposes has long been a major goal of those interested in the field. Initially focus was on the development for their antimicrobial properties, particularly the low risk of selecting for resistant strains. More recently attention has been drawn to their potential as immune-modulators, in cancer treatment and in promoting wound healing (111). There are several active pre-clinical and clinical trials of AMP based pharmaceuticals, although most current investigations are focused on anti-infective indications (39, 42). Some examples of AMPs or their derivatives, which hold potential as promoters of wound healing based on published in vitro and animal model data or trials with human beings are discussed below and additional examples are summarized in Table 2.
Table 2.
Examples of naturally occurring peptides or their derivatives with in vitro/in vivo wound healing promoting activities
| Designation | Origin | Features | Ref |
|---|---|---|---|
| AH90 | Odorrana grahami skin | Promotion of wound healing in mice by stimulating TGF-β secretion | (114) |
| CW49 | Odorrana grahami skin | Diabetic wound healing in mice by promoting angiogenesis and preventing excessive inflammatory response | (118) |
| Tylotoin | Skin of salamanders | Enhancement of migration and proliferation of keratinocytes, fibroblasts and vascular endothelial cells in vitro and stimulation of wound closure in mouse models of full-thickness skin wounding | (110) |
| Temporins A and B | Rana temporaria skin | Promotion of wound repair in mice by presumably raising angiogenesis and influencing keratinocytes proliferation/migration | (117, 158) |
| Esculentin-1a(1–21)NH2 | Derivative of the frog skin AMP esculentin-1a | In vitro stimulation of keratinocytes migration through activation of EGFR | (122) |
| Tiger 17 | Frog-based designer | Stimulation of wound healing in mice by recruiting macrophages and by promoting migration/proliferation of keratinocytes and granulation tissue formation | (159) |
| Pexiganan | Derivative of magainin | Promotion of wound healing and reduction of microbial burden in human infected skin ulcers (III Phase Clinical Trial) | (128) |
| IDR-1018 | Derivative of bovine AMP Bactenecin | Promotion of wound healing in S. aureus infected and non-infected porcine ulcers | (160) |
| Nisin | Lantibiotic | Stimulation of non-infected skin ulcers and reduction of bacterial burden in a mouse S. aureus skin infection model | (161) |
| A3APO | Proline-arginine rich peptide dimer | Stimulation of wound closure and reduction of bacterial load in Klebsiellapneumoniae/Acinetobacter baumannii/Proteus mirabilis mouse skin burn | (162, 163) |
| HB-107 | Derivative of the insect AMP cecropin B | Induction of leukocytes infiltration and cytokine secretion in vitro | (164) |
| LL-37 | Human cathelicidin | Induction of neovascularisation, migration and proliferation of epithelial cells; promotion of wound healing in clinical trials | (93, 129) |
| hBD-3 | Human β-defensin | Promotion of cytokine release; intestinal epithelial cells migration and healing in a rat model | (100) |
| PR-39 | Porcine cathelicidin | Increased synthesis of proteoglycan | (94) |
| Lucifensins | Blowfly larvae Lucilia sericata and L.cuprina | Induction of wound healing during maggot debridement therapy, which is routinely used at hospitals worldwide | (165) |
| CaTx-II | Snake toxin | Stimulation of collagen synthesis and neovascularization | (166) |
The application of amphibian skin for treating wounds has been in use since ancient times (112). Beside tyrotropin (113), short peptides (containing from 12 to 24 amino acids) from the skin of frogs have been recently described to promote wound healing in mice by stimulating TGF-β secretion or by promoting angiogenesis and influencing keratinocytes proliferation or migration (110, 112, 114–118). Lately, a short variant of the frog skin AMP esculentin-1a, namely esculentin-1a(1–21)NH2 has been found to be active against bacterial and fungal species (119). It consists of the first 20 amino acids of the AMP esculentin-1a produced by the skin of the green edible frog Pelophylax lessonae/ridibundus, previously known as Rana esculenta complex (120), plus a glycinamide at its C-terminus (119). It rapidly kills both planktonic and biofilms forms of P. aeruginosa strains, via a pronounced membrane-perturbing activity as a plausible mode of action (121). Furthermore, this peptide preserves antimicrobial activity at physiological conditions, unlike most AMPs of mammalian origin, and stimulates in vitro migration of human keratinocytes (through a stereospecific mechanism involving activation of EGFR and STAT3 protein), at a wider peptide concentration range and with a higher cell migration rate than LL-37 (122), thus offering good opportunities for cutaneous tissue restoration uses (see below), namely for the management of P. aeruginosa-induced human skin ulcers. An overview of the role of different AMPs in the timing events of wound healing process are indicated in Table 3.
Table 3.
Role of different AMPs during wound healing events
| Wound healing phases | Peptides | Ref |
|---|---|---|
| Inflammatory phase: | ||
| -Induction of cytokines/ growth factors release | hBD-3 | (100) |
| AH90 | (114) | |
| HB-107 | (164) | |
| Tiger 17 | (159) | |
| Tylotoin | (110) | |
| -Stimulation of neutrophils and macrophage recruitment | Tiger 17 | (159) |
| HB-107 | (164) | |
| -Antimicrobial activity | A3-APO | (163) |
| Nisin | (161) | |
| SR-0379 | (167) | |
| Pexiganan | (126) | |
| Proliferative Phase | ||
| -Stimulation of endothelial cells and fibroblasts proliferation/migration | LL-37 | (168, 169) |
| SR-0379 | (167) | |
| Tylotoin | (110) | |
| -Stimulation of keratinocytes, migration/proliferation | LL-37 | (91) |
| Tylotoin, | (110) | |
| Temporins A and B | (117, 158) | |
| Esculentin -1a(1–21)NH2 | (122) | |
| IDR-1018 | (160) | |
| Granulation Tissue formation | ||
| -Biosynthesis of extracellular matrix | CaTx-II | (166) |
| PR-39 | (94) | |
| -Collagen production | SR-0379 | (167) |
| Tiger 17 | (159) | |
| CaTx-II | (166) | |
| -Neovascularization | LL-37 | (169) |
| CaTx-II | (166) | |
| -Angiogenesis | LL-37 | (93) |
| SR-0379 | (167) | |
| CW49 | (118) | |
| Temporin A | (158) | |
| CaTx-II | (166) | |
| Tissue Remodeling Phase | ||
| -Extracellular matrix remodeling and myofibroblasts differentiation | SR-0379 | (167) |
| Tiger 17 | (159) | |
| Tylotoin | (110) |
The first AMP evaluated for the treatment of human wounds, specifically infected wounds, was pexiganan. It is a synthetic 22 amino acids analogue of the naturally occurring AMP, magainin 2 produced by Xenopus laevis (27, 123, 124) and is currently under study for the treatment of infected diabetic foot ulcers. Pexiganan was initially investigated in the late 1990s as a locally applied alternative to a systemically administered broad spectrum antibiotic for the treatment of infected diabetic ulcers, which remain a major medical problem with some 70,000 lower limb amputations being performed annually in the USA (125). Although the etiology of these wounds is not fully understood, patients are advised to take antibiotics if any indication of infection is evident (126). These are complex polymicrobial infections and no specific microorganism is implicated in the destructive outcome of the initial minor superficial skin wound on the foot (127). The antibiotic spectrum of pexiganan is broad enough to cover the range of microbes known to be present on the diabetic ulcer (128). Furthermore, as a topical agent high local concentrations in the superficial soft tissues could be achieved easily, surpassing the concentrations that could be reached systemically.
Two extensive pivotal Phase III clinical trials involving about 1,000 subjects (128) showed that over two to four weeks of treatment, wounds treated with pexiganan closed at the same rate as those treated with oral ofloxacin. In contrast to ofloxacin, pexiganan did not induce the appearance of resistant bacteria on the treated wounds. Furthermore, those receiving topical treatment reported fewer side effects than those taking the oral therapeutic (128). However, despite this positive outcome, the FDA Advisory panel voted against approval of pexiganan. Their primary concern was the uncertainty over the “placebo” outcome. In response to the Advisory Panel, the FDA requested that a trial be repeated using a placebo (vehicle control) arm. Currently Dipexium Pharmaceuticals is in the midst of conducting such placebo controlled phase III trials the outcome of which will be known in 2016.
Gronberg et al. recently reported the results of a “first in man” randomized, double-blind, placebo controlled, multicenter, prospective trial to investigate the safety of doses of LL-37 and its efficacy in regards to wound healing in hard-to-heal venous leg ulcers (129). Based on the observations that LL-37 is depleted in chronic wounds, the group hypothesized that supplementation with exogenously supplied LL-37 would be beneficial for healing. Indeed, not only was application of LL-37 safe and well tolerated, it also promoted wound healing. Although this study was, by the authors admission, limited by the small number of patients and short treatment time, it shows the potential for LL-37 as a novel wound healing agent for clinical use.
Furthermore it was recently reported that polylactic-co-glycolic acid (PLGA) nanoparticles loaded with LL-37 significantly accelerate wound healing process due to the sustained release of both LL-37 and lactate, compared to LL-37 and PLGA administration alone (130).
This brief overview reveals that AMPs from multiple sources have therapeutic potential as modulators of wound healing. However, as stated earlier, infection is a major factor that hinders wound healing and it is not clear at this point if the beneficial effects of AMPs applied to skin wounds will primarily be due to antimicrobial activity or if direct effects on cellular activities such as migration and proliferation will have an important contribution. Elucidating the relative contributions remains to be determined in future animal studies and clinical trials.
Conclusions
Evidence in the literature supports the hypothesis that endogenously expressed AMPs are important modulators of wound repair in both mammalian and non-mammalian systems and that AMPs and their derivatives have potential as novel therapeutics. AMP beneficial effects may encompass: (i) direct anti-microbial activity, which prevents infection that would otherwise delay healing; (ii) binding and inactivation of molecules such as LPS which reduces the detrimental pro-inflammatory response as well as (iii) one or more direct effects on cellular behaviours such as enhanced migration and proliferation. This multifactorial mechanism of action is very powerful, and coupled with the reduced tendency for AMPs to select for microbial resistance, makes AMPs particularly attractive candidates, possibly superior to conventional antibiotics, for the local treatment of infected skin wounds (131). Importantly, although development of resistance or cross-resistance to AMPs has been reported in vitro (132–134) no comparable results have been found in vivo thus far, likely because the selection of AMP-resistant strains in reality is a much slower process than in vitro conditions and they act in concert with existing endogenous AMPs. However, long term therapeutic use of AMPs should be carefully evaluated to avoid the risk of compromising our innate immune defense and therefore the ability to control commensal microbiome and microbial infections.
Acknowledgments
Special thanks to Kim Thompson of the University of Houston College of Optometry, Audio Visual Department for drawing the figures. This study was supported by grants from Sapienza University of Rome and NIH grant EY13175. MLM wrote the manuscript, which was reviewed critically by AMcD and MZ
Footnotes
Conflict of interests
The authors declare no conflict of interest
References
- 1.Enyedi B, Niethammer P. Trends Cell Biol. 2015;25:398–407. doi: 10.1016/j.tcb.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallo RL, Hooper LV. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kujath P, Kujath C. Eur J Med Res. 2010;15:544–553. doi: 10.1186/2047-783X-15-12-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau K, Paus R, Tiede S, et al. Exp Dermatol. 2009;18:921–933. doi: 10.1111/j.1600-0625.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 5.Hu MS, Maan ZN, Wu JC, et al. Ann Biomed Eng. 2014;42:1494–1507. doi: 10.1007/s10439-014-1010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramot Y, Mastrofrancesco A, Camera E, et al. Exp Dermatol. 2015;24:245–251. doi: 10.1111/exd.12647. [DOI] [PubMed] [Google Scholar]
- 7.Mustoe T. Am J Surg. 2004;187:65S–70S. doi: 10.1016/S0002-9610(03)00306-4. [DOI] [PubMed] [Google Scholar]
- 8.Demidova-Rice TN, Hamblin MR, Herman IM. Adv Skin Wound Care. 2012;25:304–314. doi: 10.1097/01.ASW.0000416006.55218.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancl KA, Kirsner RS, Ajdic D. Wound Repair Regen. 2013;21:352–362. doi: 10.1111/wrr.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vyas KS, Wong LK. Ann Plast Surg. 2015 doi: 10.1097/SAP.0000000000000440. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 11.Wolcott RD, Rhoads DD, Bennett ME, et al. J Wound Care. 2010;19:45–46. doi: 10.12968/jowc.2010.19.2.46966. [DOI] [PubMed] [Google Scholar]
- 12.Breidenstein EB, de la Fuente-Nunez C, Hancock RE. Trends Microbiol. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Mah TF, Pitts B, Pellock B, et al. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 14.Taylor PK, Yeung AT, Hancock RE. J Biotechnol. 2014;191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Ovington L. Ostomy Wound Manage. 2003;49:8–12. [PubMed] [Google Scholar]
- 16.Rajpaul K. Br J Community Nurs. 2015;20:S6. doi: 10.12968/bjcn.2015.20.Sup3.S6. [DOI] [PubMed] [Google Scholar]
- 17.Wilson MA. Am J Surg. 2003;186:35S–41S. doi: 10.1016/j.amjsurg.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Kish TD, Chang MH, Fung HB. Am J Geriatr Pharmacother. 2010;8:485–513. doi: 10.1016/S1543-5946(10)80002-9. [DOI] [PubMed] [Google Scholar]
- 19.Hannigan GD, Pulos N, Grice EA, et al. Adv Wound Care (New Rochelle) 2015;4:59–74. doi: 10.1089/wound.2014.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell-Jones RS, Wilson MJ, Hill KE, et al. J Antimicrob Chemother. 2005;55:143–149. doi: 10.1093/jac/dkh513. [DOI] [PubMed] [Google Scholar]
- 21.Lai Y, Gallo RL. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakatsuji T, Gallo RL. J Invest Dermatol. 2012;132:887–895. doi: 10.1038/jid.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon JM. Peptides. 2015;67:29–38. doi: 10.1016/j.peptides.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 25.Steiner H, Hultmark D, Engstrom A, et al. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 26.Ganz T, Selsted ME, Szklarek D, et al. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zasloff M. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, Mishra B, Lau K, et al. Pharmaceuticals (Basel) 2015;8:123–150. doi: 10.3390/ph8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangoni ML, Shai Y. Cell Mol Life Sci. 2011;68:2267–2280. doi: 10.1007/s00018-011-0718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epand RM, Epand RF. Biochim Biophys Acta. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 31.Wimley WC. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hale JD, Hancock RE. Expert Rev Anti Infect Ther. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 33.Fjell CD, Hiss JA, Hancock RE, et al. Nat Rev Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen LT, Haney EF, Vogel HJ. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Shai Y. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld Y, Papo N, Shai Y. J Biol Chem. 2006;281:1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 37.Brandenburg K, Schromm AB, Gutsmann T. Subcell Biochem. 2010;53:53–67. doi: 10.1007/978-90-481-9078-2_3. [DOI] [PubMed] [Google Scholar]
- 38.Choi KY, Chow LN, Mookherjee N. J Innate Immun. 2012;4:361–370. doi: 10.1159/000336630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayana JL, Chen JY. Peptides. 2015 S0196-9781(15)00168-0. [Google Scholar]
- 40.Bhattacharjya S. Curr Top Med Chem. 2016;16:4–15. doi: 10.2174/1568026615666150703121943. [DOI] [PubMed] [Google Scholar]
- 41.Yeung AT, Gellatly SL, Hancock RE. Cell Mol Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox JL. Nat Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 43.Mansour SC, Pena OM, Hancock RE. Trends Immunol. 2014;35:443–450. doi: 10.1016/j.it.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Cruz J, Ortiz C, Guzman F, et al. Curr Med Chem. 2014;21:2299–2321. doi: 10.2174/0929867321666140217110155. [DOI] [PubMed] [Google Scholar]
- 45.Dutta P, Das S. Curr Top Med Chem. 2016;16:99–129. doi: 10.2174/1568026615666150703121819. [DOI] [PubMed] [Google Scholar]
- 46.Harder J, Schroder JM. J Biol Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 47.Glaser R, Harder J, Lange H, et al. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 48.Braff MH, Di Nardo A, Gallo RL. J Invest Dermatol. 2005;124:394–400. doi: 10.1111/j.0022-202X.2004.23443.x. [DOI] [PubMed] [Google Scholar]
- 49.Yamasaki K, Gallo RL. Eur J Dermatol. 2008;18:11–21. doi: 10.1684/ejd.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroder JM. Curr Opin Infect Dis. 2010;23:106–110. doi: 10.1097/QCO.0b013e328335b004. [DOI] [PubMed] [Google Scholar]
- 51.Gallo RL. Exp Dermatol. 2013;22:517. doi: 10.1111/exd.12197. [DOI] [PubMed] [Google Scholar]
- 52.Simanski M, Glaser R, Harder J. Exp Dermatol. 2014;23:230–231. doi: 10.1111/exd.12365. [DOI] [PubMed] [Google Scholar]
- 53.Schroder JM, Harder J. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckert RL, Broome AM, Ruse M, et al. J Invest Dermatol. 2004;123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x. [DOI] [PubMed] [Google Scholar]
- 55.Nakashige TG, Zhang B, Krebs C, et al. Nat Chem Biol. 2015;11:765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abtin A, Eckhart L, Glaser R, et al. J Invest Dermatol. 2010;130:2423–2430. doi: 10.1038/jid.2010.158. [DOI] [PubMed] [Google Scholar]
- 57.Jensen JM, Ahrens K, Meingassner J, et al. Exp Dermatol. 2011;20:783–788. doi: 10.1111/j.1600-0625.2011.01322.x. [DOI] [PubMed] [Google Scholar]
- 58.Mallbris L, Carlen L, Wei T, et al. Exp Dermatol. 2010;19:442–449. doi: 10.1111/j.1600-0625.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- 59.Lee DY, Huang CM, Nakatsuji T, et al. J Invest Dermatol. 2009;129:2489–2496. doi: 10.1038/jid.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rieg S, Garbe C, Sauer B, et al. Br J Dermatol. 2004;151:534–539. doi: 10.1111/j.1365-2133.2004.06081.x. [DOI] [PubMed] [Google Scholar]
- 61.Agerberth B, Charo J, Werr J, et al. Blood. 2000;96:3086–3093. [PubMed] [Google Scholar]
- 62.Grice EA, Segre JA. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grice EA, Kong HH, Conlan S, et al. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Costello EK, Lauber CL, Hamady M, et al. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanford JA, Gallo RL. Semin Immunol. 2013;25:370–377. doi: 10.1016/j.smim.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paulino LC, Tseng CH, Strober BE, et al. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jarmuda S, O'Reilly N, Zaba R, et al. J Med Microbiol. 2012;61:1504–1510. doi: 10.1099/jmm.0.048090-0. [DOI] [PubMed] [Google Scholar]
- 68.Harder J, Schroder JM, Glaser R. Exp Dermatol. 2013;22:1–5. doi: 10.1111/exd.12046. [DOI] [PubMed] [Google Scholar]
- 69.Wanke I, Skabytska Y, Kraft B, et al. Exp Dermatol. 2013;22:153–155. doi: 10.1111/exd.12083. [DOI] [PubMed] [Google Scholar]
- 70.Percoco G, Merle C, Jaouen T, et al. Exp Dermatol. 2013;22:800–806. doi: 10.1111/exd.12259. [DOI] [PubMed] [Google Scholar]
- 71.Gallo RL, Nakatsuji T. J Invest Dermatol. 2011;131:1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ong PY, Ohtake T, Brandt C, et al. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 73.Brogden NK, Mehalick L, Fischer CL, et al. Skin Pharmacol Physiol. 2012;25:167–181. doi: 10.1159/000337927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinz K, Schubert S, Harder J, et al. Exp Dermatol. 2014;23:862–864. doi: 10.1111/exd.12538. [DOI] [PubMed] [Google Scholar]
- 75.Elgarhy LH, Shareef MM, Moustafa SM. Clin Exp Dermatol. 2015;40:361–366. doi: 10.1111/ced.12560. [DOI] [PubMed] [Google Scholar]
- 76.Steinstraesser L, Koehler T, Jacobsen F, et al. Mol Med. 2008;14:528–537. doi: 10.2119/2008-00002.Steinstraesser. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salzer S, Ruzicka T, Schauber J. Exp Dermatol. 2014;23:379–381. doi: 10.1111/exd.12401. [DOI] [PubMed] [Google Scholar]
- 78.Morizane S, Gallo RL. J Dermatol. 2012;39:225–230. doi: 10.1111/j.1346-8138.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harder J, Schroder JM. J Leukoc Biol. 2005;77:476–486. doi: 10.1189/jlb.0704409. [DOI] [PubMed] [Google Scholar]
- 80.Schmidt NW, Jin F, Lande R, et al. Nat Mater. 2015;14:696–700. doi: 10.1038/nmat4298. [DOI] [PubMed] [Google Scholar]
- 81.Lande R, Gregorio J, Facchinetti V, et al. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 82.Elloso MM, Gomez-Angelats M, Fourie AM. J Leukoc Biol. 2012;92:1187–1197. doi: 10.1189/jlb.0212101. [DOI] [PubMed] [Google Scholar]
- 83.Roupe KM, Nybo M, Sjobring U, et al. J Invest Dermatol. 2010;130:1167–1177. doi: 10.1038/jid.2009.284. [DOI] [PubMed] [Google Scholar]
- 84.Niyonsaba F, Ushio H, Nakano N, et al. J Invest Dermatol. 2007;127:594–604. doi: 10.1038/sj.jid.5700599. [DOI] [PubMed] [Google Scholar]
- 85.Sorensen OE, Thapa DR, Rosenthal A, et al. J Immunol. 2005;174:4870–4879. doi: 10.4049/jimmunol.174.8.4870. [DOI] [PubMed] [Google Scholar]
- 86.Niyonsaba F, Ushio H, Nagaoka I, et al. J Immunol. 2005;175:1776–1784. doi: 10.4049/jimmunol.175.3.1776. [DOI] [PubMed] [Google Scholar]
- 87.Hirsch T, Spielmann M, Zuhaili B, et al. J Gene Med. 2009;11:220–228. doi: 10.1002/jgm.1287. [DOI] [PubMed] [Google Scholar]
- 88.Semple F, Webb S, Li HN, et al. Eur J Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Semple F, MacPherson H, Webb S, et al. Eur J Immunol. 2011;41:3291–3300. doi: 10.1002/eji.201141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin G, Kawsar HI, Hirsch SA, et al. PLoS One. 2010;5:e10993. doi: 10.1371/journal.pone.0010993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tokumaru S, Sayama K, Shirakata Y, et al. J Immunol. 2005;175:4662–4668. doi: 10.4049/jimmunol.175.7.4662. [DOI] [PubMed] [Google Scholar]
- 92.Heilborn JD, Nilsson MF, Kratz G, et al. J Invest Dermatol. 2003;120:379–389. doi: 10.1046/j.1523-1747.2003.12069.x. [DOI] [PubMed] [Google Scholar]
- 93.Koczulla R, von Degenfeld G, Kupatt C, et al. J Clin Invest. 2003;111:1665–1672. doi: 10.1172/JCI17545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gallo RL, Ono M, Povsic T, et al. Proc Natl Acad Sci USA. 1994;91:11035–11039. doi: 10.1073/pnas.91.23.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shalom-Feuerstain Ruby AD. Exp Rev Dermatol. 2008;3:357–366. [Google Scholar]
- 96.Parfitt GJ, Geyfman M, Xie Y, et al. J Invest Dermatol. 2015;135:1175–1177. doi: 10.1038/jid.2014.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kolar SS, McDermott AM. Cell Mol Life Sci. 2011;68:2201–2213. doi: 10.1007/s00018-011-0713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Di Meglio P, Perera GK, Nestle FO. Immunity. 2011;35:857–869. doi: 10.1016/j.immuni.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 99.Otte JM, Werner I, Brand S, et al. J Cell Biochem. 2008;104:2286–2297. doi: 10.1002/jcb.21787. [DOI] [PubMed] [Google Scholar]
- 100.Sheng Q, Lv Z, Cai W, et al. Pediatr Res. 2014;76:269–279. doi: 10.1038/pr.2014.93. [DOI] [PubMed] [Google Scholar]
- 101.Shaykhiev R, Beisswenger C, Kandler K, et al. Am J Physiol Lung Cell Mol Physiol. 2005;289:L842–L848. doi: 10.1152/ajplung.00286.2004. [DOI] [PubMed] [Google Scholar]
- 102.Chisholm AD. Adv Wound Care (New Rochelle) 2015;4:264–271. doi: 10.1089/wound.2014.0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Onfelt Tingvall T, Roos E, Engstrom Y. EMBO Rep. 2001;2:239–243. doi: 10.1093/embo-reports/kve048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anderson AE, Galko MJ. Exp Dermatol. 2014;23:809–810. doi: 10.1111/exd.12498. [DOI] [PubMed] [Google Scholar]
- 105.Zugasti O, Ewbank JJ. Nat Immunol. 2009;10:249–256. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- 106.Alibardi L. Adv Anat Embryol Cell Biol. 2010;207:1–109. [PubMed] [Google Scholar]
- 107.Raghavan KV, Babu M, Rajaram R, et al. Lipids Health Dis. 2010;9:74. doi: 10.1186/1476-511X-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Campbell LJ, Crews CM. Cell Mol Life Sci. 2008;65:73–79. doi: 10.1007/s00018-007-7433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Satoh A, Graham GM, Bryant SV, et al. Dev Biol. 2008;319:321–335. doi: 10.1016/j.ydbio.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 110.Mu L, Tang J, Liu H, et al. FASEB J. 2014;28:3919–3929. doi: 10.1096/fj.13-248476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Otvos L, Ostorhazi E., Jr Expert Rev Anti Infect Ther. 2015;13:871–881. doi: 10.1586/14787210.2015.1033402. [DOI] [PubMed] [Google Scholar]
- 112.Mashreghi M, Rezazade Bazaz M, Mahdavi Shahri N, et al. J Ethnopharmacol. 2013;145:793–797. doi: 10.1016/j.jep.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 113.Meier NT, Haslam IS, Pattwell DM, et al. PLoS One. 2013;8:e73596. doi: 10.1371/journal.pone.0073596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu H, Mu L, Tang J, et al. Int J Biochem Cell Biol. 2014;49:32–41. doi: 10.1016/j.biocel.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 115.Simonetti O, Cirioni O, Goteri G, et al. Peptides. 2008;29:520–528. doi: 10.1016/j.peptides.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 116.Rezazade Bazaz M, Mashreghi M, Mahdavi Shahri N, et al. Pharm Biol. 2013;51:1600–1606. doi: 10.3109/13880209.2013.804846. [DOI] [PubMed] [Google Scholar]
- 117.Di Grazia A, Luca V, Segev-Zarko LA, et al. Antimicrob Agents Chemother. 2014;58:2520–2527. doi: 10.1128/AAC.02801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu H, Duan Z, Tang J, et al. FEBS J. 2014;281:4633–4643. doi: 10.1111/febs.12968. [DOI] [PubMed] [Google Scholar]
- 119.Islas-Rodriguez AE, Marcellini L, Orioni B, et al. J Pept Sci. 2009;15:607–614. doi: 10.1002/psc.1148. [DOI] [PubMed] [Google Scholar]
- 120.Conlon JM. Peptides. 2008;29:1815–1819. doi: 10.1016/j.peptides.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 121.Luca V, Stringaro A, Colone M, et al. Cell Mol Life Sci. 2013;70:2773–2786. doi: 10.1007/s00018-013-1291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Di Grazia A, Cappiello F, Imanishi A, et al. PLoS One. 2015;10:e0128663. doi: 10.1371/journal.pone.0128663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ge Y, MacDonald D, Henry MM, et al. Diagn Microbiol Infect Dis. 1999;35:45–53. doi: 10.1016/s0732-8893(99)00056-5. [DOI] [PubMed] [Google Scholar]
- 124.Ge Y, MacDonald DL, Holroyd KJ, et al. Antimicrob Agents Chemother. 1999;43:782–788. doi: 10.1128/aac.43.4.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Foster D, Lauver LS. Ostomy Wound Manage. 2014;60:16–22. [PubMed] [Google Scholar]
- 126.Lipsky BA, Berendt AR, Deery HG, et al. Clin Infect Dis. 2004;39:885–910. doi: 10.1086/424846. [DOI] [PubMed] [Google Scholar]
- 127.Ge Y, MacDonald D, Hait H, et al. Diabet Med. 2002;19:1032–1034. doi: 10.1046/j.1464-5491.2002.00696_1.x. [DOI] [PubMed] [Google Scholar]
- 128.Lipsky BA, Holroyd KJ, Zasloff M. Clin Infect Dis. 2008;47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- 129.Gronberg A, Mahlapuu M, Stahle M, et al. Wound Repair Regen. 2014;22:613–621. doi: 10.1111/wrr.12211. [DOI] [PubMed] [Google Scholar]
- 130.Chereddy KK, Her CH, Comune M, et al. J Control Release. 2014;194:138–147. doi: 10.1016/j.jconrel.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 131.Do N, Weindl G, Grohmann L, et al. Exp Dermatol. 2014;23:326–331. doi: 10.1111/exd.12384. [DOI] [PubMed] [Google Scholar]
- 132.Shireen T, Singh M, Das T, et al. Antimicrob Agents Chemother. 2013;57:5134–5137. doi: 10.1128/AAC.00780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Habets MG, Brockhurst MA. Biol Lett. 2012;8:416–418. doi: 10.1098/rsbl.2011.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Perron GG, Zasloff M, Bell G. Proc Biol Sci. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ali RS, Falconer A, Ikram M, et al. J Invest Dermatol. 2001;117:106–111. doi: 10.1046/j.0022-202x.2001.01401.x. [DOI] [PubMed] [Google Scholar]
- 136.Kesting MR, Stoeckelhuber M, Holzle F, et al. Br J Dermatol. 2010;163:121–127. doi: 10.1111/j.1365-2133.2010.09781.x. [DOI] [PubMed] [Google Scholar]
- 137.Dressel S, Harder J, Cordes J, et al. Exp Dermatol. 2010;19:628–632. doi: 10.1111/j.1600-0625.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- 138.Yamasaki K, Di Nardo A, Bardan A, et al. Nat Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 139.Nakatsuji T, Kao MC, Zhang L, et al. J Invest Dermatol. 2010;130:985–994. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nagy I, Pivarcsi A, Kis K, et al. Microbes Infect. 2006;8:2195–2205. doi: 10.1016/j.micinf.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 141.Lee DY, Yamasaki K, Rudsil J, et al. J Invest Dermatol. 2008;128:1863–1866. doi: 10.1038/sj.jid.5701235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schittek B. J Innate Immun. 2012;4:349–360. doi: 10.1159/000336844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Murakami M, Ohtake T, Dorschner RA, et al. J Invest Dermatol. 2002;119:1090–1095. doi: 10.1046/j.1523-1747.2002.19507.x. [DOI] [PubMed] [Google Scholar]
- 144.Narayanan S, Miller WL, McDermott AM. Invest Ophthalmol Vis Sci. 2003;44:3795–3801. doi: 10.1167/iovs.02-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McDermott AM. Ophthalmic Res. 2009;41:60–75. doi: 10.1159/000187622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Garreis F, Schlorf T, Worlitzsch D, et al. Histochem Cell Biol. 2010;134:59–73. doi: 10.1007/s00418-010-0713-y. [DOI] [PubMed] [Google Scholar]
- 147.McIntosh RS, Cade JE, Al-Abed M, et al. Invest Ophthalmol Vis Sci. 2005;46:1379–1385. doi: 10.1167/iovs.04-0607. [DOI] [PubMed] [Google Scholar]
- 148.Haynes RJ, Tighe PJ, Dua HS. Lancet. 1998;352:451–452. doi: 10.1016/s0140-6736(05)79185-6. [DOI] [PubMed] [Google Scholar]
- 149.Bals R, Hiemstra PS. Eur Respir J. 2004;23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 150.Diamond G, Beckloff N, Ryan LK. J Dent Res. 2008;87:915–927. doi: 10.1177/154405910808701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Singh PK, Jia HP, Wiles K, et al. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ganesan S, Comstock AT, Sajjan US. Tissue Barriers. 2013;1:e24997. doi: 10.4161/tisb.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tecle T, Tripathi S, Hartshorn KL. Innate Immun. 2010;16:151–159. doi: 10.1177/1753425910365734. [DOI] [PubMed] [Google Scholar]
- 154.Bals R, Wang X, Zasloff M, et al. Proc Natl Acad Sci U S A. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Selsted ME, Ouellette AJ. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 156.Bevins CL. Dig Dis. 2013;31:299–304. doi: 10.1159/000354681. [DOI] [PubMed] [Google Scholar]
- 157.Kim JM. Intest Res. 2014;12:20–33. doi: 10.5217/ir.2014.12.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ghiselli R, Giacometti A, Cirioni O, et al. J Vasc Surg. 2002;36:1027–1030. doi: 10.1067/mva.2002.127530. [DOI] [PubMed] [Google Scholar]
- 159.Tang J, Liu H, Gao C, et al. PLoS One. 2014;9:e92082. doi: 10.1371/journal.pone.0092082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Steinstraesser L, Hirsch T, Schulte M, et al. PLoS One. 2012;7:e39373. doi: 10.1371/journal.pone.0039373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Heunis TD, Smith C, Dicks LM. Antimicrob Agents Chemother. 2013;57:3928–3935. doi: 10.1128/AAC.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ostorhazi E, Holub MC, Rozgonyi F, et al. Int J Antimicrob Agents. 2011;37:480–484. doi: 10.1016/j.ijantimicag.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 163.Ostorhazi E, Rozgonyi F, Sztodola A, et al. J Antimicrob Chemother. 2010;65:2416–2422. doi: 10.1093/jac/dkq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee PH, Rudisill JA, Lin KH, et al. Wound Repair Regen. 2004;12:351–358. doi: 10.1111/j.1067-1927.2004.012303.x. [DOI] [PubMed] [Google Scholar]
- 165.Cerovsky V, Bem R. Pharmaceuticals (Basel) 2014;7:251–264. doi: 10.3390/ph7030251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Samy RP, Kandasamy M, Gopalakrishnakone P, et al. PLoS One. 2014;9:e80199. doi: 10.1371/journal.pone.0080199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tomioka H, Nakagami H, Tenma A, et al. PLoS One. 2014;9:e92597. doi: 10.1371/journal.pone.0092597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duplantier A, Jvan Hoek ML. Front Immunol. 2013;4:143. doi: 10.3389/fimmu.2013.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ramos R, Silva JP, Rodrigues AC, et al. Peptides. 2011;32:1469–1476. doi: 10.1016/j.peptides.2011.06.005. [DOI] [PubMed] [Google Scholar]