Summary
Argonaute proteins play a central role in mediating post-transcriptional gene regulation by microRNAs (miRNAs). Argonautes use the nucleotide sequences in miRNAs as guides for identifying target messenger RNAs for repression. Here we used single-molecule FRET to directly visualize how human Argonaute-2 (Ago2) searches for and identifies target sites in RNAs complementary to its miRNA guide. Our results suggest that Ago2 initially scans for target sites with complementarity to nucleotides 2–4 of the miRNA. This initial transient interaction propagates into a stable association when target complementarity extends to nucleotides 2–8. This stepwise recognition process is coupled to lateral diffusion of Ago2 along the target RNA, which promotes target search by enhancing the retention of Ago2 on the RNA. The combined results reveal the mechanisms that Argonaute likely uses to efficiently identify miRNA target sites within the vast and dynamic agglomeration of RNA molecules in the living cell.
Introduction
Post-transcriptional regulation by the microRNA (miRNA) pathway plays an important role in many eukaryotic processes, including brain development (Giraldez et al., 2005), recovery from cardiac stress (van Rooij et al., 2007), and progression of many forms of cancer (Hammond, 2006). In humans, the RNase III enzymes Drosha and Dicer sequentially process long primary miRNA transcripts into ~22 nt miRNA duplexes (Kim et al., 2009). Each duplex is loaded into an Argonaute (Ago) protein. One strand (the mature miRNA, or the guide strand) is selectively retained by Ago to form the functional core of the RNA-induced silencing complex (RISC) (Meister, 2013). Ago uses the sequence information encoded within the loaded miRNA to locate target messenger RNAs (mRNAs) through Watson-Crick base-pairing (Bartel, 2009; Lee et al., 1993). The target-bound Ago elicits a silencing effect through recruitment of GW182/TNRC6 co-factors which, in turn, trigger translational repression or mRNA deadenylation and decay (Meister, 2013). Additionally, human Ago2 can directly cleave target RNAs with extensive guide complementarity using an endonucleolytic activity termed “slicing”.
Biochemical and bioinformatic studies indicate that Ago proteins divide miRNAs into five functionally distinct domains: the 5′ anchor (nt 1), the seed region (nt 2–8), the central region (nt 9–12), the 3′ supplementary region (nt. 13–16), and the 3′ tail (nt 17–22) (Ameres et al., 2007; Bartel, 2009; Khorshid et al., 2013; Wee et al., 2012). The seed appears to be the most important for target recognition, both in terms of Ago2-target affinity as well conservation of vertebrate miRNA target sites (Bartel, 2009; Brennecke et al., 2005; Haley and Zamore, 2004; Krek et al., 2005; Lai, 2002; Lewis et al., 2005; Lim et al., 2005). Structural studies of eukaryotic Ago proteins explain these observations, in part, by revealing that in the absence of target RNAs guide nt 2–6 are splayed out in a helical conformation that is stabilized by protein-mediated contacts to the phosphodiester backbone (Faehnle et al., 2013; Nakanishi et al., 2013; Nakanishi et al., 2012; Schirle and MacRae, 2012). Studies of an archeal MID/PIWI protein (which is structurally related to eukaryotic Ago proteins) showed that pre-ordering of the seed region facilitates target recognition by offsetting the entropic penalty of forming a guide-target duplex (Parker et al., 2009). However, in all eukaryotic Ago crystal structures reported to date, the seed is not presented in a perfect A-form conformation—base stacking between guide nt 6 and 7 is disrupted by a kink in the RNA backbone that appears to be stabilized by a conserved alpha helix (helix-7 in human Ago2). Helix-7 also sterically blocks access to guide nt 6–8, suggesting that target pairing to guide nt 2–5 may promote conformational changes that subsequently facilitate interactions with the full seed (Schirle et al., 2014). However, this stepwise mechanism for target recognition has never been directly tested, and it is not known if distinct segments of the seed contribute differentially to target association and dissociation.
Another essential but poorly understood facet of Ago function, and RNA-protein interactions in general, is the mechanism by which the protein efficiently searches for target sites within the complex milieu of cellular RNAs. Theoretical studies of protein–nucleic acid interactions suggest that target search processes can be orders of magnitude more efficient when three-dimensional diffusion is accompanied by one-dimensional diffusion along long nucleic acid substrates (Berg et al., 1981; Halford and Marko, 2004; Riggs et al., 1970). One-dimensional diffusion has been observed with transcription factors (Hammar et al., 2012; Leith et al., 2012), DNA repair (Blainey et al., 2009; Gorman et al., 2012), and recombination proteins (Graneli et al., 2006; Ragunathan et al., 2012). Because miRNA target sites are typically found in the 3′ untranslated regions of mRNAs, which are often several kilobases in length, it is reasonable to hypothesize that Ago may also use lateral diffusion to identify target sites. However, direct observation of one-dimensional diffusion by Ago has not been reported.
Single-molecule fluorescence has been widely used to study protein–RNA interactions in vitro (Hoskins et al., 2011; Karunatilaka et al., 2010; Kim et al., 2014; Koh et al., 2013; Lee et al., 2012; Shen et al., 2012; Yeom et al., 2011). To gain mechanistic insight into the process of target recognition by human Ago2, we developed a single-molecule FRET (Förster Resonance Energy Transfer) assay that allows real-time visualization of the interactions between Ago2-miRNA and targets. We show that target recognition is initiated by transient pairing to a small segment of the miRNA seed (nt 2–4) and becomes kinetically stable when base pairing is extended to the full length of the seed sequence (nt 2–8). We further demonstrate that the Ago2-miRNA complex can rapidly diffuse between neighboring seed-matched sites on a target RNA. In cases of extensive guide-target complementarity, lateral diffusion is facilitated by the Ago2 PAZ domain. Lateral diffusion also allows neighboring miRNA target sites to cooperatively retain Ago2 on the target RNA.
Results
Single-molecule FRET assay for association of Ago2-miRNA with target RNAs
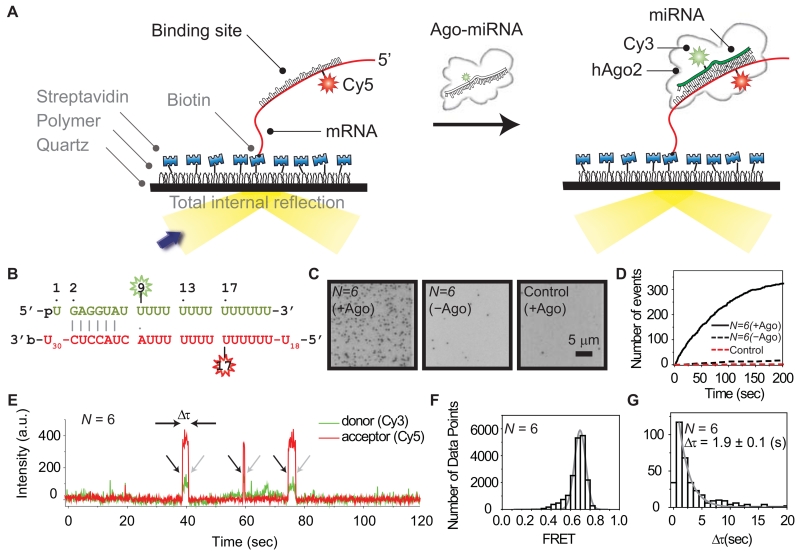
Target recognition by Ago likely involves transient intermediates that are difficult to observe using traditional biochemical assays. For direct observation of this dynamic process, we used single-molecule FRET, which provides high spatio-temporal resolution. We immobilized biotinylated target RNAs on a passivated quartz surface in a microfluidic chamber and introduced human Ago2-miRNA complexes to the same area (Figure 1A, see also Experimental Procedures). Using total-internal-reflection microscopy, we recorded single-molecule fluorescence signals from a donor fluorophore (labeled on nt 9 of the miRNA) and an acceptor (labeled on the target RNA, opposite nt 17 of the miRNA) (Figure 1B). This system leads to high FRET efficiency (E) when the miRNA is paired to the complementary site within the target RNA strand. Bulk experiments showed that dye labeling at nt 9 of the miRNA did not noticeably affect the slicing activity of Ago2, indicating that target binding and cleavage are not significantly perturbed by the modification (Figure S1A). The target RNAs contained a poly-uridine segment (30 nucleotides, U30) downstream of a target site with N continuous base pairs (bp) of complementarity to the miRNA (hsa-let-7a), and (U)39 – N nucleotides upstream. The total length of all target RNAs was 69 nt (30 + N + 39 – N).
Figure 1. Single-Molecule Observation of Ago2-miRNA Target Recognition.
(A) Illustrated schematic representation of our single-molecule FRET assay.
(B) Sequences of miRNA and a target RNA with N (the length of the matched region between miRNA and target RNA) equal to 6 nt. The donor fluorophore (Cy3) is positioned on the 9th nt of miRNA (counting from the 5′ end of miRNA) and the acceptor (Cy5) on target RNA opposite nt 17 of miRNA. Vertical lines denote contiguous base pairs between the guide and target. A dot “.” represents an inconsecutive pair.
(C) CCD images (donor channel) show Ago2-miRNA binding to target RNA. Individual spots represent single-molecule complexes. (Left) Result representative of experiments with N = 6. (Middle) The same as left but without Ago2 included. (Right) U63 used as a negative control while Ago2 was included. Scale bar 5 µm.
(D) The number of accumulated binding events plotted as a function of time for the three cases in [C].
(E) A fluorescence time trace obtained (with a resolution 100 msec) shows three events of docking and dissociation of Ago2-miRNA at a single spot in the microfluidic chamber. Black arrows indicate binding of Ago2-miRNA to a target mRNA. Gray arrows indicate dissociation.
(F) A FRET histogram obtained from 125 single-molecule traces fitted with a Gaussian function.
(G) The dwell time of binding (Δτ) was fitted with a single exponential decay. The first column of the data was not included in the analysis to avoid potential artefacts arising from the limit of the time resolution. Error is the standard deviation (std) of 3 independent experiments that were carried out on different days. The bin size is 1 sec. See also Figure S1.
We first used a target RNA with a complementarity length of N = 6 (matched to nt 2–7 of the miRNA) to observe the interaction of Ago2-miRNA with a minimal seed-matched target (Figure 1B-G). Docking of a single Ago2-miRNA complex to an immobilized target RNA was observed by the sudden appearance of fluorescence signals (Figure 1E, arrows in black). Docking events were marked by a single peak at E ~ 0.75 in a FRET histogram (Figure 1F) indicating a specific interaction between miRNA and seed-matched site within the target RNA. Each docking event was followed by the sudden disappearance of the fluorescence signals (Figure 1E, arrows in gray), indicating dissociation of the Ago2-miRNA complex from the site. The dwell time of this interaction (Δτ, the inverse of the dissociation rate) was 1.9 ± 0.1 sec (Figure 1G), which is two orders of magnitude shorter than photobleaching under the same measurement conditions (~300 sec; Figure S1B). When an immobilized RNA lacking the target sequence was tested, no significant binding events were observed (Figure 1C-D). Likewise, no significant events were observed when the measurement was carried out in the absence of Ago2 (Figure 1C-D), indicating that observed pairing events are due to Ago2-mediated interactions between miRNA molecules and their targets .
Initial steps of microRNA target recognition
To determine the minimal target recognition motif, we tested miRNAs with varied degrees of complementarity to an immobilized target (Figure 2A-B). First, we measured the dwell time of Ago2-miRNA complexes with N lengths equal to 3, 4, 5, 6, 7, 8, 15, 19 (Figure 2C-D). Constructs with N = 3, 4 and 5 displayed observable but short-lived dwell times, averaging about 1 second. Increasing N to 6 increased the dwell time by about a factor of two. A target site that has an adenosine nucleotide opposite miRNA nt 1 increased the dwell time further by 2.5 fold. (Schirle et al, submitted). In contrast, constructs with N ≥ 7 displayed dwell times more than two orders of magnitude longer. This trend was maintained whether or not magnesium ions (1.5 mM) were included in the reaction buffer, revealing that magnesium does not play a significant role in seed pairing (Figure S2A). The same observation was made when a crowding agent (PEG, polyethylene glycol, 8000 Da) was included (Figure S2C). The sharp transition between pairing to nt 2–7 and 2–8 suggests that nt 2–8 is the minimal motif required for long-lived interaction between a miRNA and a target in our system. Notably, the dwell time for N ≥ 7 could not be accurately measured because the majority of Ago2-miRNA remained stably bound to the target RNA beyond our observation time window (approximately 300 sec), which is limited by photobleaching effects (Figure S1B).
Figure 2. Kinetics of MicroRNA Target Recognition.
(A) Sequences of miRNA and a target RNA with various values of N. Vertical lines represent consecutive base paring between the guide and the target RNAs. Colons “:” represent potential GU wobble pairs. Dots “.” represent inconsecutive pairs. “p” represents 5′ phosphate in the guide miRNAs. The biotin at 3′ end of target RNAs is denoted by “b”. The sequence of the miRNA with N = 19 is hsa-let-7a. The sequences of other miRNAs were derived from hsa-let-7a with variations at 5′ and 3′ ends. The sequences of the miRNA with N = 3 and 4 contain A instead of genomic sequence U at nt 6. The sequence of the miRNA with N = 7 contains A instead of genomic sequence U at nt 9 and has a dye conjugated at nt 10.
(B) Representative time traces for the interaction between target RNAs and Ago2-miRNA complexes with different values of N. A time resolution of 100 msec was used for the negative control and N = 3 (nt 5-7), 3 (nt 2-4), 4, 5, and 6. A time resolution of 300 msec was used for N = 7, 8, 15, and 19.
(C) Dwell time histograms for different values of N. The dwell time (Δτ) of N = 3 (nt 5-7), 3 (nt 2-4), 4, 5 and 6 is 0.2 ± 0.1, 0.7 ± 0.2, 0.8 ± 0.1, 1.2 ± 0.3, and 1.9 ± 0.1 sec, respectively. Error is the std of 3 independent experiments carried out on different days. The bin size for N = 3 (nt 5-7) is 0.5 sec. The bin size for N = 3 (nt 2-4), 4, 5 and 6 is 1 sec. The dwell time of N = 7, 8, 15, and 19 was not determined (N. D.) due to photobleaching. The dwell time of the negative control is not available (N.A.) due to a lack of binding events.
(D) A bar plot shows the dependence of dwell time on N. The dashed red line indicates the observation time limit (300 sec), which is constrained by photobleaching. Error bars are the std of 3 independent experiments that were carried out on different days.
(E) Binding rate plotted for different values of N. In all the cases, 1 nM Ago2-miRNA was introduced to the chamber. The error bars are the std of 3 independent experiments from different days. See also Figure S2 and Table S1.
We next determined the rate of target binding by Ago2-miRNA complexes with varied N lengths. In contrast to the dissociation kinetics, binding rates were insensitive to values of N. Sub-seed constructs (with N = 3, 4 and 5) showed similar binding rates (kon(obs), “obs” is for observed) as the full seed constructs (N = 6 and 7) (Figure 2E). Again, these results were insensitive to the presence of magnesium ions (Figure S2B). These observations reveal that pairing to miRNA nt 2–4 is sufficient to initiate specific interactions between the Ago2-miRNA complex and a target site.
We next asked if Ago2 primarily uses miRNA nt 2–4 to initiate target recognition, or if pairing to any three consecutive seed nucleotides is sufficient. We measured kon(obs) for an Ago2-miRNA complex with target complementarity at nt 5–7 (N = 3(nt 5-7)). The binding rate of N = 3(nt 5-7) was about an order of magnitude less than N = 3(nt 2-4) (Figure 2E), indicating that initial target recognition by miRNA nt 2–4 is far more efficient than nt 5–7. The combined results suggest that target recognition proceeds in a stepwise fashion, initiating with pairing to the seed 5′ end and then extending in the 3′ direction. Indeed, extending guide:target complementarity to N = 8, 15, and 19 did not increase kon(obs) beyond what we observed for the N = 3 construct..
Lateral diffusion of Ago2-miRNA on target RNAs
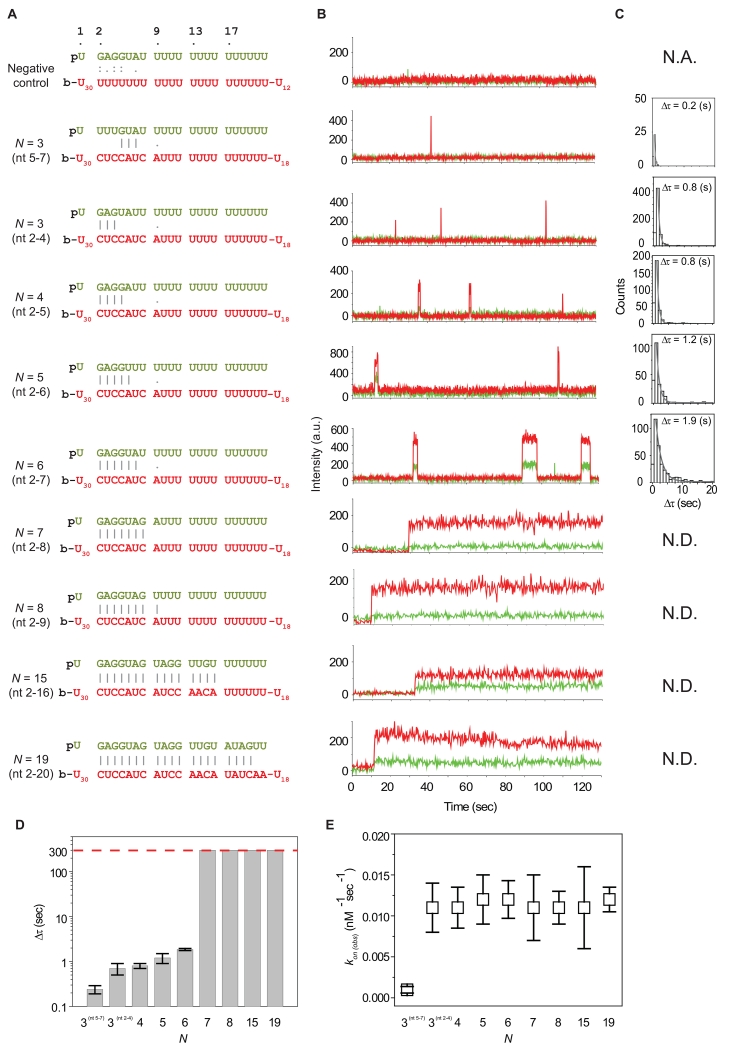
Lateral diffusion of a protein complex on a nucleic acid substrate can enhance the efficiency of a target search by up to two orders of magnitude (Hammar et al., 2012; Ragunathan et al., 2012). In principle, if Ago2-miRNA uses such a mechanism, transient FRET states other than E ~ 0.75 should appear in our assays. However, the FRET signal in our time trajectories is characterized by a single value, with no observable intermediate states (Figure 2). This indicates that either binding events arise from random collisions between the miRNA seed and target sites, or that the Ago2-miRNA complex laterally diffuses on target RNAs faster than the time resolution of our experimental setup (100 msec), and thus escapes our detection. Consistent with the latter possibility, if we assume a lateral diffusion rate of ~3×105 – ~2×107 nt2/sec for a protein on a nucleic acid (Blainey et al., 2009), diffusion of Ago2-miRNA along the entire 69 nt target RNA occurs within 0.1 - 10 msec, which is well beyond our time resolution. To assay for lateral diffusion while working within the temporal resolution of our experimental setup, we designed a target RNA construct with two binding sites (1 and 2), each with the same guide:target complementarity (N1 and N2) (Figure 3A). The two binding sites were separated from each other by (21 – N1) nt (Figure 3B). In this system, only site 1 contains the acceptor fluorophore so that when the Ago2-miRNA complex binds to site 1, the system exhibits a high FRET state (E ~0.75), while binding to site 2 exhibits a lower FRET state. A similar approach was used previously to demonstrate lateral diffusion in RecA-mediated DNA target search processes (Ragunathan et al., 2012).
Figure 3. Lateral Diffusion of Ago2-miRNA.
(A) Illustrated is the schematic representation of the single-molecule FRET assay used to observe lateral diffusion of Ago2-miRNA between tandem binding sites before dissociation occurs. N1 and N2 are the lengths of matched sequence between the miRNA and sites 1 and 2, respectively.
(B) Sequences of N1 = N2 = 6 target sites paired to guide miRNA. The donor (Cy3) is positioned at the 9th nt of miRNA. The acceptor (Cy5) is positioned in site 1, opposite nt 17 of the paired miRNA. When miRNA is bound to site 1, the two dyes are separate by 7 nt, leading to high FRET (E ~ 0.75). When it is bound to site 2, they are separate by 13 nt, leading to low FRET (E ~ 0.5).
(C) Fluorescence signals in a time trace obtained with a time resolution 100 msec reports on docking and dissociation of Ago2-miRNA. Intensity of FRET signal indicates position of Ago2-miRNA complex with respect to sites 1 and 2 (site 1, high FRET; site 2, low FRET).
(D) (Top left) A FRET histogram was fit with two Gaussian functions (E ~ 0.5 and 0.75). Dwell time distribution of total binding (Δτ , top right), binding to site 2 (Δτ2, bottom left), and binding to site 1 (Δτ1, bottom right). The distributions were fit with a single exponential decay. The first column of the data was not included in the analysis to avoid artefacts from the limited time resolution. Error is the std of 3 independent experiments that were carried out on different days. The bin size is 1 sec.
(E) A bar graph shows the dependence of the dwell times (Δτ1 and Δτ2) on the length of the matched region between miRNA and each binding site. Error is the std of 3 independent experiments that were carried out on different days. See also Figure S3 and Table S1.
With a tandem target construct of N1 = N2 = 6 nt, we observed two distinct FRET populations (E ~ 0.5 and E ~ 0.75, Figure 3C, 3D top left), with dwell times of 2.5 ± 0.1 sec (total) (Figure 3D, top right), 1.0 ± 0.1 sec (low FRET) and 1.5 ± 0.0 sec (high FRET) (Figure 3D, bottom). The dwell times of the two FRET states were similar to that of the N = 6 nt single site target (Figure 1G), indicating that the FRET signals represent Ago2-miRNA particles occupying either of the two sites, as opposed to photophysical artifacts of dyes. To confirm this idea, we tested a shorter length of Ni (N1 = N2 = 5 nt), corresponding to matches over nt 2–6. As expected, the dwell time of each FRET state (Δτ1 and Δτ2) decreased (Figures 3E, S3A). We conclude that the two observed FRET states result from discrete binding events of the Ago2-miRNA complex at the two seed-matched sites on the target RNA.
The majority of binding events (72.5%, 598 out of 825 events) to the tandem target construct alternated one or more times between the two FRET states. This observation suggests that either: (1) single Ago2-miRNA complexes can rapidly shuttle between adjacent target sites; or (2) dissociation events are often immediately followed by the rapid association of a second Ago2-miRNA complex. Considering the binding rate of Ago2-miRNA to a single site (kon ~0.01 sec−1 at 1 nM Ago2-miRNA), the probability of a second Ago2-miRNA independently binding faster than our time resolution (100 msec) is small (~0.1 %).
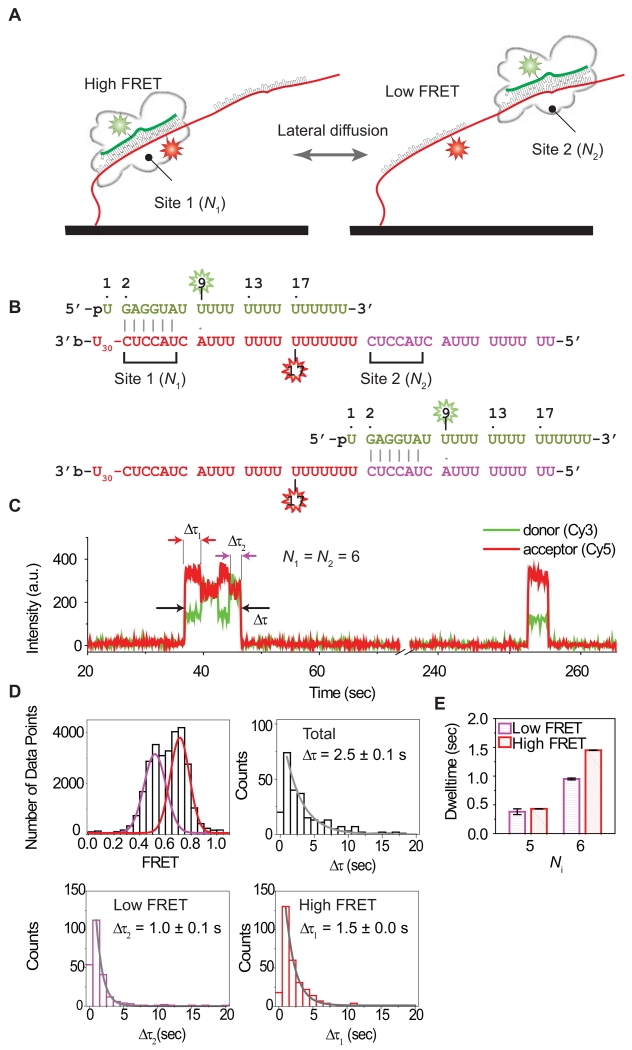
To further determine whether the observed FRET alternation arises from the shuttling of a single Ago2-miRNA complex or the consecutive binding of two different Ago2-miRNA complexes, we designed a three-color FRET assay in which Ago2-miRNA complexes were labeled with one of two possible FRET donors (Cy3 or Cy5) (Figure 4A-B). Our rationale was that, if FRET alternation is due to consecutive binding of different Ago2-miRNA complexes, we should observe binding events that switch from a Cy3 to a Cy5 labeled complex (and vice versa) on single target molecules. The assay used the same tandem target construct as Figure 3A (N1 = N2 = 6 nt) but labeled instead with Cy7, which is an acceptor of both Cy3 and Cy5. We introduced an equal mixture of Cy3 and Cy5 labeled Ago2-miRNA complexes, and used two lasers (532 nm and 637 nm) as excitation sources to image the interaction with the Cy7-labeled target RNA. As expected, the Cy3 (donor) and Cy7 (acceptor) pair exhibited alternations in FRET efficiency (Figure 4C, left), and the same observation was made with Cy5 and Cy7 pair (Figure 4C, right). However, out of 595 shuttling events we recorded, we did not observe any cases in which a Cy3 or Cy5 signal was immediately followed by the other color.
Figure 4. Three-color FRET Assay Detecting Lateral Diffusion.
(A) Illustrated is the schematic representation of the single-molecule three-color FRET assay used to observe lateral diffusion of Ago2-miRNA between tandem binding sites. N1 and N2 are the lengths of matched sequence between the miRNA and sites 1 and 2, respectively. An equal amount of Cy3 (green, donor 1) and Cy5 (red, donor 2) labeled miRNA is added into a chamber which has a Cy7 (blue, acceptor) labeled tandem target construct immobilized.
(B) Sequences of N1 = N2 = 6 target sites paired to guide miRNA. The donor (Cy3 or Cy5) is positioned at the 9th nt of miRNA. The acceptor (Cy7) is positioned in site 1, opposite nt 17 of the paired miRNA.
(C) Fluorescence signals in time traces were obtained with a time resolution 500 msec. (Left) A time trace showing alternations in Cy3-Cy7 FRET efficiency. (Right) A time trace showing alternations in Cy5-Cy7 FRET efficiency. The horizontal lines in FRET efficiency plots indicate two different FRET states.
We carried out an additional experiment to confirm this observation. We first incubated Ago2-miRNA (N = 6) with target RNA and washed free Ago2-miRNA out of the microfluidic chamber. We observed that the FRET alternations persisted even after the flow (Figure S3B). Taken together, we conclude that the observed alternations in FRET states arise from single Ago2-miRNA complexes rapidly diffusing between the neighboring target sites.
A synergistic effect from neighboring target sites
The observation that Ago2 shuttles between adjacent target sites suggests that neighboring sites could cooperate kinetically to retain the Ago2-miRNA complex on target RNAs. If a target site is isolated, the time that Ago2-miRNA is bound to RNA is determined solely by how quickly Ago2-miRNA dissociates from RNA (on → off). However, with neighboring sites, dissociation is in kinetic competition with lateral diffusion. For example, if Ago2 is bound to one of two neighboring sites (e.g. site 1), the dissociation pathway is diversified to numerous possibilities (site 1 → off; 1 → 2 →off; 1 → 2 → 1 → off; etc.), which would be expected to increase the total dwell time on the target RNA. Indeed, the average total dwell time on the RNA with two N = 6 sites (2.5 ± 0.1 sec, Figure 3D) was longer than dwell time on the equivalent RNA with a single site (1.9 ± 0.1 sec, Figure 1G), revealing a synergistic effect. But the observed difference in dwell times is rather small, indicating that lateral diffusion only moderately helps to retain Ago2 on the target RNA, presumably because the complex has a relatively high dissociation rate in our dilute in vitro condition.
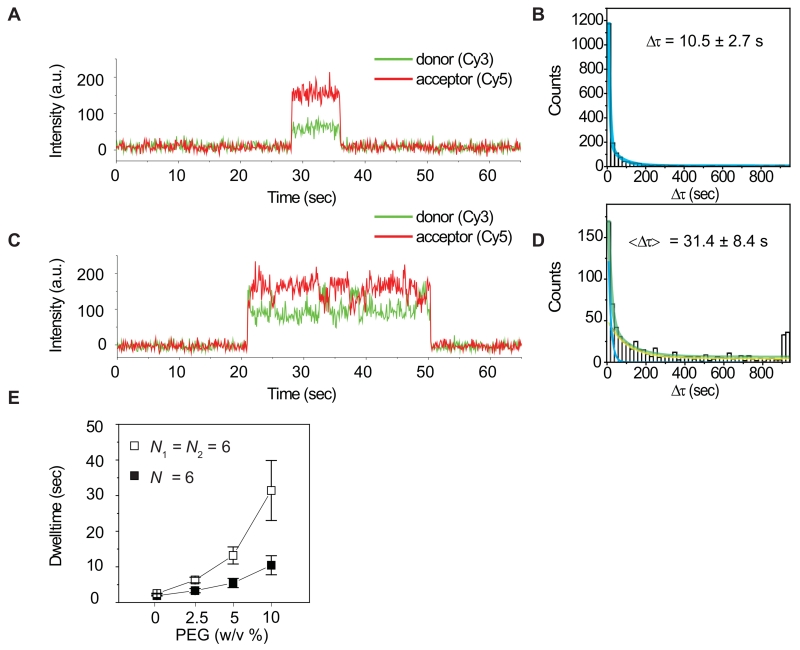
To better mimic the crowded environment of the cell, we measured dwell times of the Ago2-miRNA complex on target RNAs in the presence of various concentrations of a crowding agent (PEG 8000) (Figure 5). We observed that the increasing PEG concentration led to increased dwell times, indicating that crowding effectively repressed dissociation (Figure 5E). Notably, crowding affected dwell times on the RNA with two target sites substantially more than the RNA with a single site. In the presence of 10% PEG the total average time on the two target sites was more than 3 fold greater than that on the single target site. Additionally, we observed two populations of binding events on the RNA with two target sites (Δτ1 = 13.9 sec, 61%; and Δτ2 = 76.0 sec, 39%), with the second population (Δτ2) exhibiting a 7-fold longer dwell time than that of a single site target RNA (Δτ, 10.5 sec) (Figure 5D). Some binding events even lasted longer than 900 seconds. We conclude that neighboring binding sites synergistically increase the dwell time of Ago2 on target RNAs, and suggest that this effect may be substantially enhanced in the crowed molecular environment of the living cell.
Figure 5. A synergistic effect from two neighboring target sites.
(A) A fluorescence time trace obtained using a single target construct (N = 6) in the presence of 10% PEG with a time resolution 300 msec.
(B) A dwell time histogram for the condition used in [A]. The dwell time (Δτ) was 10.5 ± 2.7 sec. Error is the std of 3 independent experiments that were carried out on different days. The bin size is 20 sec.
(C) A fluorescence time trace obtained using a tandem target construct (N1 = N2 = 6) in the presence of 10% PEG with a time resolution 300 msec.
(D) A dwell time histogram for the condition used in [C] was fitted using a double exponential curve (green). The double exponential fit resulted in two characteristic times Δτ1 (61%), 13.9 ± 1.7 s, a blue curve; Δτ2 (39%) = 76.0 ± 22.7 s, a yellow curve). The average, <Δτ> = 31.4 ± 8.4 sec, was estimated from Δτ1 × 61% + Δτ2 × 39%. A peak at Δτ > 900 (sec) is due to a limited observation time and was not included during the fitting. Error is the std of 3 independent experiments that were carried out on different days. The bin size is 20 sec.
(E) The total dwell time plotted for different concentrations (weight to volume) of PEG. Error is the std of 3 independent experiments. See also Figure S5.
Interactions with the miRNA 3′ end facilitate lateral diffusion
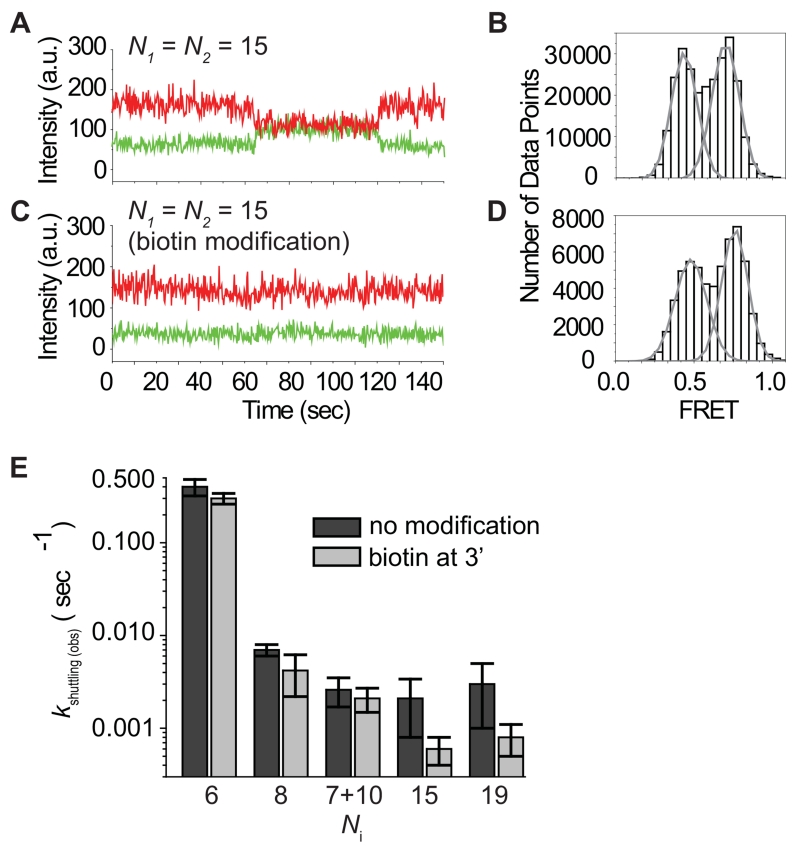
The dwell time between shuttling events increased with increasing values of Ni, indicating that guide-target base pairing is disrupted prior to movement (Figure 3E). To explore this phenomenon further, we tested additional tandem target constructs in which base pairing extended beyond the miRNA seed region. We first tested a tandem target construct that matches over the entire miRNA excluding the central region (nt 9-12) (Figure S4B-C, named “7+10”), which base-pairing structure is frequently observed in vivo (Helwak et al., 2013). Although the total number of the miRNA-target potential base pairs is greater than the minimal number of base pairs required for stable RNA duplex hybridization—7 bp or longer duplexes unzipper very slowly (Cisse et al., 2012), we still observed a dynamic transition between the two FRET states with kshuttling(obs) = 0.0026 ± 0.0009 (sec−1). We made a similar observation with N1 = N2 = 15 and 19 nt (Figure 6A-B, S4D-E). Comparing with the rate of target cleavage (Figure S1C), this suggests that human Ago2 might sample its targets multiple times before cleaving.
Figure 6. Role of the PAZ Domain in Lateral Diffusion.
(A) A time trace obtained with a time resolution 300 msec shows lateral diffusion that occurs between two binding sites (N1, high FRET; N2, low FRET, each 15 nt).
(B) A FRET histogram was obtained under the condition of [A] and was fit with Gaussian functions (E ~ 0.5 and 0.75).
(C) A time trace was obtained with a time resolution 300 msec using a miRNA construct that was modified at its 3′ end with biotin. (N1, high FRET; N2, low FRET, each 15 nt).
(D) A FRET histogram was obtained under the condition of [C] and was fitted with Gaussian functions (E ~ 0.5 and 0.75).
(E) The kinetic rate of shuttling between two binding sites (N1 and N2) was measured with Ni = 6, 8, “7+10”, 15, and 19 using unmodified and 3′ biotinylated miRNA constructs. Error is the std of 3 independent experiments that were carried out on different days. See also Figure S4.
We presume that Ago2 must unzip the extended miRNA:target RNA duplex, a process that requires energy far larger than thermal fluctuation energy, such as ATP hydrolysis, prior to lateral diffusion. Remarkably, in our system this process occurs in the absence of any external energy source. These results indicate that Ago2 perturbs the interaction between miRNA and target such that not all the nucleotides of miRNA can stably pair to the target RNA. This conclusion is reminiscent to bulk studies showing that in fly Ago2-RISC guide RNA nucleotides 3′ of the seed make little contributions to the slicing reaction Km (Wee et al., 2012). Structural studies of bacterial Ago proteins suggest that target association is coupled to conformational changes in both protein and guide, whereby release of the guide 3′-end from the PAZ domain is required for extensive guide-target pairing (Wang et al., 2009; Zander et al., 2014). Target dissociation by Thermus thermophilus Ago is accelerated by anchoring of the guide 3′-end in the PAZ domain (Jung et al., 2013). We speculated that anchoring of the miRNA 3′-end in the PAZ domain of human Ago2 might similarly reduce the binding affinity between miRNA and target RNA and thereby facilitate shuttling between adjacent target sites.
To test this hypothesis, we biotinylated the 3′ end of the miRNA in order to disrupt 3′ end interactions with the PAZ domain (Ma et al., 2005; Yan et al., 2003). To prevent the 3′ biotinylated miRNA (named miRNA3′ biotin) from also becoming immobilized, we saturated any unbound streptavidin with 1.7 mM free biotin. In case of Ni = 15 and 19, the Ago2-miRNA3′ biotin complexes alternated between the two FRET states 3.5 and 3.8 times less frequently than the unmodified complex, respectively (Figure 6C-D, S4F-G). In contrast, shuttling of Ago2-miRNA3′ biotin on targets with complementarity to the seed region (Ni = 6, nt 2–7; Ni = 8, nt 2–9) was essentially unaffected by the modification. The correlation between the fold change of the shuttling frequency and the extent of guide:target base pairing suggests that interactions with the guide 3′ end, presumably via the Ago2 PAZ domain (Jung et al., 2013; Xia et al., 2013; Yan et al., 2003), can destabilize pairing to the 3′ supplementary and tail regions (nt 13–16) when Ago2-miRNA is bound to a fully complementary target RNA.
DISCUSSION
Using single-molecule FRET, we investigated miRNA-target binding by human Ago2. Our results show that Ago2 can efficiently recognize target sites as small as three consecutive nucleotides and forms a long-lived interaction if pairing extends over the full 7 nt seed region (Figure 2). We further demonstrate that the Ago2-miRNA complex often shuttles between neighboring target sites on a single RNA molecule, indicating that Ago2 may rapidly diffuse along single stranded target RNAs when searching for target sites (Figure 3). Shuttling between adjacent sites also increases the dwell time on target RNAs, particularly in crowded molecular environments. Modification of the miRNA 3′ end reduces the frequency of shuttling events at extensively paired target sites (Figure 6), suggesting that interactions with the PAZ domain facilitate unwinding of the guide-target duplex in the 3′ supplemental region.
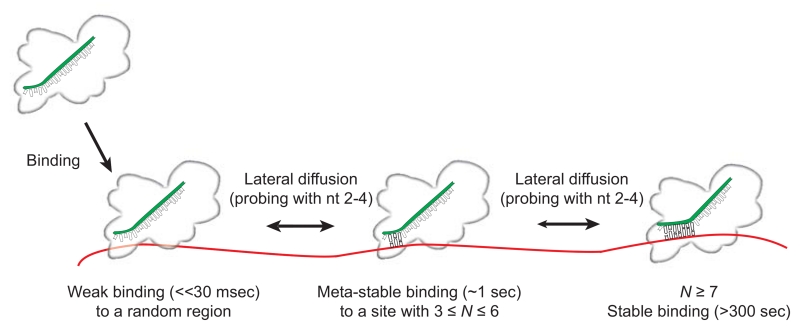
Based on these results, we propose a mechanism for miRNA target identification by Ago2. In this model, Ago2 initially binds to and diffuses along target RNAs while scanning for sequences complementary to a sub-seed recognition motif (guide nt 2–4) (Figure 7). We hypothesize that, by limiting the initial target search to only a few guide nucleotides, Ago2 effectively flattens the target search energy landscape, thereby facilitating lateral diffusion and enabling efficient target scanning. Ago2 can then convert transient interactions with the sub-seed into stable interactions when the base pairing extends the length of the entire seed (nt 2–8). When base pairing propagates further down the miRNA (into the 3′ supplementary region), interactions of the guide 3′ end facilitate unwinding of the guide:target duplex.
Figure 7. A model for target search and recognition by human Ago2-miRNA.
(A) Illustration of proposed steps in target recognition by Ago2-miRNA. Ago2-miRNA binds the single stranded target RNA and diffuses along the RNA rapidly—sliding over 69 nt (the length of our target construct) in less than 100 msec (our time resolution). Complementary sites of N ≥ 3 cause the complex to pause and remain meta-stably bound (on the order of 1 second). When site complementarity extends the full length of the seed (nt 2–8) or longer, the complex remains bound stably for extended times (>>300 sec). See also Figure S6.
We hypothesize that this mechanism is most effective in scanning unstructured regions of potential target mRNAs because RNA secondary structures and RNA binding proteins likely interfere with the ability of Ago2 to make base pairing contacts to the miRNA seed. Such obstacles may also place limits on the extent to which Ago2 can laterally diffuse along any given mRNA. Indeed, Ameres et al. previously showed that target site accessibility correlates directly with Ago2 cleavage efficiency and suggested that transient interactions with single-stranded regions may facilitate target searches (Ameres et al., 2007). It will be of interest to determine if Ago2 moves primarily by sliding or microscopic hopping as it diffuses along target RNAs, and if it has mechanisms to bypass different types of roadblocks.
Our results also illuminate new levels of detail in existing models of mechanisms underlying RNA silencing. In contrast to annealing rates of naked RNAs, which are highly dependent on duplex length (Cisse et al., 2012), the kinetics of Ago2-miRNA:target pairing are insensitive to the length of the guide:target duplex for all targets pairing to guide nt 2–4 and beyond (Figure 2E). Pairing to nt 5–7 alone, however, was nearly an order of magnitude slower. These results suggest that Ago2 controls RNA pairing such that nucleation of the guide-target duplex occurs at the 5′ end of the seed with the actual “seed” for nucleation being guide nucleotides 2–4.
Baek et al. showed that N = 6 miRNA target sites generally have a marginal impact on the expression levels of target proteins, while N ≥ 7 sites are far more potent (Baek et al., 2008). Our dwell time measurements provide a reasonable mechanistic explanation for this observation: Ago2-miRNA remained associated with the N = 7 target >100 times longer than the N = 6 target (Figure 2D). These results contrast naked 7 bp RNA duplexes, which melt about 10 times more slowly than 6 bp duplexes (Cisse et al., 2012). The dissociation rate of Ago2-miRNA from an N = 7 target (<<0.003 sec−1) was also at least an order of magnitude slower than the melting rate of a naked 7 base pair duplex (0.014 sec−1), indicating that Ago2 stabilizes the N = 7 duplex to increase the dwell time on the target site well beyond what would be expected of a simple RNA-RNA pairing. We therefore suggest that an extended dwell time of Ago2 on the target RNA (N ≥ 7) may be an important feature of effective miRNA-mediated repression.
We speculate that lateral diffusion may also contribute to maintaining Ago occupancy on target RNAs. During lateral diffusion, seed and sub-seed sequences are repeatedly recognized by the Ago2-miRNA complex. We suggest that lateral diffusion accompanied by these brief interactions kinetically competes with dissociation, and thus the Ago2-miRNA complex likely associates longer with RNAs that have clustered seed or sub-seed matched sites than RNAs with isolated sites. These interactions may contribute to cooperative repression that has been observed with neighboring miRNA target sites (Broderick et al., 2011; Grimson et al., 2007; Saetrom et al., 2007). A hint of such synergistic effect was observed from the tandem target construct (Figure 5). Additionally, it was recently reported that kilobase-long circular RNAs (circRNAs) with densely packed miRNA binding sites (e.g. as-CDR1 circRNA containing ~1 seed matched site per 20 nt) function as potent miRNA sponges (Hansen et al., 2013; Memczak et al., 2013). Close inspection of the circRNA sequence reveals that miRNA sub-seed motifs (complementary to nt 2–4) are also over-represented in the regions between seed-matched sites (3-4 times more abundant than expected by chance) (Figure S5). Given the densely packed seed and sub-seed matched sites in the circRNA, we suggest that lateral diffusion of Ago2-miRNA may allow the circRNA to function as a far more effective sponge than the simple sum of its independent binding sites.
To conclude, our study shows that miRNA targeting by human Ago2 is a highly dynamic process that has been optimized for fast and efficient target recognition. We expect that the other three human Ago proteins use the same mechanism because all four Agos are highly homologous in sequence and are essentially identical in the areas surrounding the miRNA seed region (Figure S6A-B). Budding yeast Ago also displays the sub-seed sequence of its guide RNA, (Figure S6C), indicating that the target scanning mechanism may be common among diverse Ago proteins. We also note that general features of miRNA targeting are similar to other nucleic acid target search processes. Homologous recombination by RecA uses lateral diffusion to probe potential target sites for complementarity to a “sub-seed” of six constrained nucleotides (Ragunathan et al., 2012). Additionally, Cas9 identifies potential target sites by recognition of a small three-nucleotide protospacer adjacent motif (Sternberg et al., 2014). These features may thus be recurring themes in the mechanisms by which proteins identify complementary sites to nucleic acid guides. It will be of interest to determine if other small DNA or small RNA-mediated target search systems, including those in which DNA targets DNA (Swarts et al., 2014), DNA targets RNA (Olovnikov et al., 2013), RNA targets DNA (Blosser et al., 2015; Rutkauskas et al., 2015; Sternberg et al., 2014), and RNA targets RNA (Samai et al., 2015) also function through processes involving lateral diffusion, sub-seed scanning and modulation of guide:target pairing.
EXPERIMENTAL PROCEDURES
Single-molecule two-color FRET
Single-molecule fluorescence measurements were performed with a prism-type total internal reflection fluorescence microscope. The details are as published elsewhere (Selvin and Ha, 2007). In brief, Cy3 molecules were excited using a 532 nm diode laser (Compass 215M/50mW, Coherent). Fluorescence signals of Cy3 and Cy5 were collected through a 60× water immersion objective (UplanSApo, Olympus) with an inverted microscope (IX73, Olympus). The 532 laser scattering was blocked out by a 532 nm long pass filter (LPD01-532RU-25, Semrock). The Cy3 and Cy5 signals were separated with a dichroic mirror (635 dcxr, Chroma) and imaged using an EM-CCD camera (iXon Ultra, DU-897U-CS0-#BV, Andor Technology).
Single-molecule three-color FRET
Three-color measurements were performed using an analogous experimental setup as above. Cy3 and Cy5 molecules were excited using a 532 nm diode laser (Sapphire 532-100mW CW CDRH, Coherent) and a 637 nm diode laser (OBIS 637 nm LX 140 mW), respectively. The 532 nm and 637 nm laser scattering was blocked out by a 532 nm and 633 nm notch filters (NF03-532E-25 and NF03-633E-25, Semrock), respectively. The Cy3 signal was separated from the other signals using a dichroic mirror (635 dcxr, Chroma). The remaining Cy5 and Cy7 signals were split using a dichroic mirror (740dcxr, Chroma). The fluorescence signals were imaged using an EM-CCD camera (iXon3, DU-897U-C00-#BV, Andor Technology).
Data acquisition and analysis
Using a custom-made program written in Visual C++ (Microsoft), a series of CCD images of time resolution 0.1 - 0.5 sec were recorded. The time traces were extracted from the CCD image series using IDL (ITT Visual Information Solution) employing an algorithm that looked for fluorescence spots with a defined gaussian profile and with signals above the average of the background signals. Colocalization between Cy3, Cy5 and Cy7 signals was carried out with a custom-made mapping algorithm written in IDL. The extracted time traces were processed using Matlab (MathWorks) and Origin (Origin Lab). Cy7 signals were multiplied by a gamma factor of 2 to correct for the low sensitivity of an EM-CCD camera in the near-infrared region.
Single-molecule sample preparation
To reduce the nonspecific binding of proteins, the quartz slides (G.Flinkenbeiner) of microfluidic chambers were coated with polyethylene glycol (mPEG-Succinimidyl Valerate, MW 5000, Laysan). The details of the procedure are described in our video protocol (Chandradoss et al., 2014). A microfluidic chamber was incubated with 20 µL Streptavidin (0.1 mg/mL, Sigma) for 30 sec. Unbound Streptavidin was washed with 100 µL of buffer T50 (10 mM Tris-HCl [pH8.0], 50 mM NaCl buffer). The fifty microliters of 50 pM acceptor-labelled mRNA construct were introduced into chamber and incubated for 1 min. Unbound labeled constructs were washed with 100 µL of buffer T50.
The effector complex was formed by incubating 10 nM purified recombinant hAgo2 (see Protein purification in EXTENDED EXPERIMENTAL PROCEDURES) with 1 nM of donor-labeled hsa-let-7a miRNA (see RNA preparation in EXTENDED EXPERIMENTAL PROCEDURES) in a buffer containing 50 mM Tris-HCl [pH 8.0] (Ambion), 50 mM NaCl (Ambion) and 60 mM KCl (Ambion) at 31°C for 20 min. When an extended observation was needed, NaCl was not included to promote longer binding (Figures 4C, S3B). Unless specified otherwise, magnesium ions were not included in the buffer throughout this work to avoid any target cleavage. For experiments involving PEG (8000 Da, Sigma), 1.25× final concentration of PEG in H2O was pre-incubated at 31°C and then mixed with the other components. An imaging buffer for single-molecule FRET was added before the mixture was injected to a microfluidic chamber. The final concentration of the imaging buffer consists of the 0.8 % dextrose (Sigma), 0.5 mg/mL glucose oxidase (Sigma), 85 µg/mL Catalase (Merck), and 1 mM Trolox ((±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, 238813, Sigma). The experiments were performed at the room temperature (23 ± 2°C). For three-color experiments, 0.5 nM of Cy3-labeled hsa-let-7a miRNA and 0.5 nM of Cy5-labelled hsa-let-7a miRNA was incubated with hAgo2. H2O was replaced with deuterated water (D2O) to enhance the photophysics of Cy7 (Klehs et al., 2014).
Bulk biochemical experiments
See Preparation of FLAG-Ago2 Agarose and Bulk slicing reactions in EXTENDED EXPERIMENTAL PROCEDURES
Supplementary Material
ACKNOWLEDGMENTS
We thank Anna Haagsma, Margreet Docter, Pawel Tuliniski and Inge Geuzebroek for their technical support. We thank Anna Haagsma, Mohamed Fareh, Inha Heo, Margreet Docter, and Sung Hyun Kim for their critical reading this manuscript and V. Narry Kim, Jessica Sheu-Gruttadauria, Robert A. Luenberger, and Mahipal Ganji for stimulating conversations. I.J.M. and N.T.S. were supported by NIH grant R01 GM104475. C.J. was funded by the Open Program of the Division for Earth and Life Sciences (822.02.008) of the Netherlands Organization for Scientific Research and European Research Council under the European Union’s Seventh Framework Programme [FP7/2007-2013] / ERC grant agreement n° [309509].
REFERENCES
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nature structural & molecular biology. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blosser TR, Loeff L, Westra ER, Vlot M, Kunne T, Sobota M, Dekker C, Brouns SJ, Joo C. Two Distinct DNA Binding Modes Guide Dual Roles of a CRISPR-Cas Protein Complex. Molecular cell. 2015;58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss SD, Haagsma AC, Lee YK, Hwang JH, Nam JM, Joo C. Surface passivation for single-molecule protein studies. Journal of visualized experiments : JoVE. 2014 doi: 10.3791/50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Kim H, Ha T. A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nature structural & molecular biology. 2012;19:623–627. doi: 10.1038/nsmb.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell reports. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci U S A. 2012;109:E3074–3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graneli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc Natl Acad Sci U S A. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature structural & molecular biology. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SR, Kim E, Hwang W, Shin S, Song JJ, Hohng S. Dynamic anchoring of the 3′-end of the guide strand controls the target dissociation of Argonaute-guide complex. J Am Chem Soc. 2013;135:16865–16871. doi: 10.1021/ja403138d. [DOI] [PubMed] [Google Scholar]
- Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 2010;467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–255. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson SA. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334–338. doi: 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Klehs K, Spahn C, Endesfelder U, Lee SF, Furstenberg A, Heilemann M. Increasing the brightness of cyanine fluorophores for single-molecule and superresolution imaging. Chemphyschem : a European journal of chemical physics and physical chemistry. 2014;15:637–641. doi: 10.1002/cphc.201300874. [DOI] [PubMed] [Google Scholar]
- Koh HR, Kidwell MA, Ragunathan K, Doudna JA, Myong S. ATP-independent diffusion of double-stranded RNA binding proteins. Proc Natl Acad Sci U S A. 2013;110:151–156. doi: 10.1073/pnas.1212917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science. 2012;336:1726–1729. doi: 10.1126/science.1216848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leith JS, Tafvizi A, Huang F, Uspal WE, Doyle PS, Fersht AR, Mirny LA, van Oijen AM. Sequence-dependent sliding kinetics of p53. Proc Natl Acad Sci U S A. 2012;109:16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nature reviews Genetics. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell reports. 2013;3:1893–1900. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Molecular cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Molecular cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragunathan K, Liu C, Ha T. RecA filament sliding on DNA facilitates homology search. eLife. 2012;1:e00067. doi: 10.7554/eLife.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. Journal of molecular biology. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell reports. 2015 doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Saetrom P, Heale BS, Snove O, Jr., Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Gene regulation. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin PR, Ha T. Single-Molecule Techniques: A Laboratory Manual. 1st edn Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- Shen K, Arslan S, Akopian D, Ha T, Shan SO. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature. 2012;492:271–275. doi: 10.1038/nature11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Huynh T, Ren P, Zhou R. Large domain motions in Ago protein controlled by the guide DNA-strand seed region determine the Ago-DNA-mRNA complex recognition process. PLoS One. 2013;8:e54620. doi: 10.1371/journal.pone.0054620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO reports. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander A, Holzmeister P, Klose D, Tinnefeld P, Grohmann D. Single-molecule FRET supports the two-state model of Argonaute action. RNA biology. 2014;11:45–56. doi: 10.4161/rna.27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.