ABSTRACT
PML is the organizer of cellular structures termed nuclear domain 10 (ND10) or PML-nuclear bodies (PML-NBs) that act as key mediators of intrinsic immunity against human cytomegalovirus (HCMV) and other viruses. The antiviral function of ND10 is antagonized by viral regulatory proteins such as the immediate early protein IE1 of HCMV. IE1 interacts with PML through its globular core domain (IE1CORE) and induces ND10 disruption in order to initiate lytic HCMV infection. Here, we investigate the consequences of a point mutation (L174P) in IE1CORE, which was shown to abrogate the interaction with PML, for lytic HCMV infection. We found that a recombinant HCMV encoding IE1-L174P displays a severe growth defect similar to that of an IE1 deletion virus. Bioinformatic modeling based on the crystal structure of IE1CORE suggested that insertion of proline into the highly alpha-helical domain severely affects its structural integrity. Consistently, L174P mutation abrogates the functionality of IE1CORE and results in degradation of the IE1 protein during infection. In addition, our data provide evidence that IE1CORE as expressed by a recombinant HCMV encoding IE1 1-382 not only is required to antagonize PML-mediated intrinsic immunity but also affects a recently described function of PML in innate immune signaling. We demonstrate a coregulatory role of PML in type I and type II interferon-induced gene expression and provide evidence that upregulation of interferon-induced genes is inhibited by IE1CORE. In conclusion, our data suggest that targeting PML by viral regulatory proteins represents a strategy to antagonize both intrinsic and innate immune mechanisms.
IMPORTANCE PML nuclear bodies (PML-NBs), which represent nuclear multiprotein complexes consisting of PML and additional proteins, represent important cellular structures that mediate intrinsic resistance against many viruses, including human cytomegalovirus (HCMV). During HCMV infection, the major immediate early protein IE1 binds to PML via a central globular domain (IE1CORE), and we have shown previously that this is sufficient to antagonize intrinsic immunity. Here, we demonstrate that modification of PML by IE1CORE not only abrogates intrinsic defense mechanisms but also attenuates the interferon response during infection. Our data show that PML plays a novel coregulatory role in type I as well as type II interferon-induced gene expression, which is antagonized by IE1CORE. Importantly, our finding supports the view that targeting of PML-NBs by viral regulatory proteins has evolved as a strategy to inhibit both intrinsic and innate immune defense mechanisms.
INTRODUCTION
Human cytomegalovirus (HCMV), a member of the β-subgroup of herpesviruses, is a widespread human pathogen of high clinical relevance that can cause life-threatening diseases in newborns and people with compromised immune system such as AIDS, transplantation, or malignancy patients. The lytic replication cycle of HCMV is characterized by three sequential phases of viral gene expression, termed the immediate early (IE), early (E), and late (L) phases (1, 2). Expression of the major immediate early genes IE1 (IE1p72) and IE2 (IE2p86) is crucial for initiation of lytic infection, as their gene products stimulate transcription of early genes that replicate viral genomic DNA, which in turn is required for entry into the late phase (3, 4). The IE1 protein, a 72-kDa protein consisting of a globular core domain (IE1CORE) flanked by intrinsically disordered regions, is the most abundant viral protein being expressed at immediate early times (5–7). Characterization of an IE1 deletion virus, derived from the HCMV laboratory strain Towne, revealed that IE1 is essential for virus growth after infection with low viral loads. After high-multiplicity infection, however, the IE1-deleted virus replicates as efficiently as wild-type HCMV, suggesting that virion components can compensate for the lack of IE1 under certain conditions (3, 8).
Studies over the last years identified IE1 as an antagonist of intrinsic and innate immune defenses that target HCMV immediately upon infection (reviewed in reference 9). An important component of the intrinsic immunity is the cellular structure nuclear domain 10 (ND10), also known as PML-nuclear bodies, that senses herpesviral genomes entering the nucleus in order to induce epigenetic silencing of the viral DNA (10–15). ND10 are dynamic intranuclear foci formed by the key component PML, a member of the tripartite motif (TRIM) protein family, and numerous other transiently or permanently localized proteins (16). Covalent and noncovalent interactions of PML with the small ubiquitin-like modifier (SUMO) protein are essential for the assembly and maintenance of these protein complexes (17–20). Besides PML, the ND10 proteins Sp100, hDaxx, and ATRX function as cellular restriction factors and act cooperatively in order to restrict HCMV infection (21–28). An interplay between intrinsic and innate immune mechanisms has been reported, since several cellular restriction factors, including PML and Sp100, are upregulated after interferon (IFN) treatment, which enhances their antiviral activity (29–31). Type I (α and β) IFNs represent an important branch of the innate immune system and are produced as an immediate response to HCMV infection (32). After their secretion, IFNs bind to cognate cell surface receptors, thereby triggering an intracellular signaling cascade that eventually leads to phosphorylation and activation of signal transducer and activator of transcription (STAT) 1 and STAT2. The activated STATs, together with the IFN regulatory factor 9 (IRF9), translocate to the nucleus and activate expression of numerous interferon-stimulated genes (ISGs) via binding to interferon stimulatory response elements (ISRE) in their promoter or enhancer regions. ISG products exert various effector functions contributing to inhibition of virus replication and spread (33).
IE1 promotes lytic infection by interfering with PML-mediated intrinsic immunity and IFN-mediated innate immunity via distinct protein domains. Through its IE1CORE domain, which adopts an elongated, alpha-helical fold as revealed by the recent solution of the crystal structure of primate IE1 proteins, IE1 interacts with PML (6, 34). Subsequently, IE1 induces a depletion of the SUMOylated species of PML, and also of Sp100, resulting in disruption of the ND10 foci within the first 4 h of infection (6, 35–37). Mutations, e.g., the L174P mutation, or deletions within IE1CORE were shown to abolish the interaction with PML and, consequently, the effects on PML SUMOylation and ND10 integrity in transfection assays (34, 37–40). These mutations, however, do not affect the ability of IE1 to interact with STAT proteins through binding sites located in the disordered C-terminal region (5, 41). It has been shown that IE1 interacts with both STAT1 and STAT2 and inhibits DNA binding of these proteins, thereby affecting the level of ISG transcripts. As a result, IE1-deleted HCMV displays increased sensitivity to IFN-α treatment compared to wild-type HCMV (41, 42). In contrast to its influence on type I IFN signaling, expression of IE1 triggers a transcriptional response characterized by an upregulation of immune stimulatory and proinflammatory genes that are normally induced by IFN-γ, the only representative of type II IFNs (43).
Interestingly, PML has recently been implicated in the regulation of IFN-γ-induced gene expression. While one study demonstrated an effect of all nuclear PML isoforms on IFN-γ-induced STAT1 phosphorylation and DNA binding (44), a subsequent study reported that a specific PML isoform, PML II, recruits the major histocompatibility complex class II (MHC-II) transactivator (CIITA) to ND10, resulting in stabilization of CIITA and enhancement of IFN-γ-induced MHC-II expression (45). Furthermore, a contribution of PML isoform IV to IFN-β production was suggested (46). However, the exact role of PML in IFN signaling awaits further clarification.
Here, we characterize a recombinant HCMV expressing the PML-binding-deficient IE1 protein IE1-L174P. We show that L174P mutation in IE1CORE results in a severe defect of HCMV to initiate lytic infection, comparable to the defect of an IE1 deletion virus. Bioinformatic and experimental analyses revealed that L174P mutation affects the structural integrity of IE1CORE and reduces the half-life of the otherwise very stable IE1 protein. Furthermore, our data provide evidence that IE1CORE not only is required to antagonize PML-mediated intrinsic immunity but also affects a stimulatory function of PML in IFN signaling. We conclude that targeting PML represents a viral strategy to perturb both intrinsic and innate immune mechanisms.
MATERIALS AND METHODS
Oligonucleotides and plasmids.
The oligonucleotide primers used for this study were purchased from Biomers GmbH (Ulm, Germany) or Biomol GmbH (Hamburg, Germany) and are listed in Table 1. The eukaryotic expression plasmid pHM3260 encoding IE1-L174P was generated by site-directed mutagenesis of plasmid pHM494, which encodes wild-type IE1 (47), using primers 5′IE1mut-L174P and 3′IE1mut-L174P. Expression plasmids encoding IE1 and IE1-L174P fused to a C-terminal CLIP-tag were obtained by PCR amplification of the respective IE1 sequence using pHM494 or pHM3260 as the template along with primer pair 5′IE1_NheI and 3′IE1_EcoRI. Subsequently, PCR products were inserted into MCS1 of pCLIPf (New England BioLabs, Frankfurt, Germany) via NheI and EcoRI. The prokaryotic expression plasmid encoding glutathione S-transferase (GST)-tagged IE1 was described elsewhere (6). The prokaryotic expression plasmid encoding GST-tagged IE1-L174P was constructed by PCR using pHM3260 as the template and primers 5′IE1-GEX-Bam and 3′IE1_Xho1, followed by ligation into pGEX-6P-1 (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) via BamHI and XhoI. The lentiviral pLVX-shRNA1-based vectors encoding control short hairpin RNA (shRNA) and PML shRNA were described previously (48). Lentiviral vectors that are based on pLVX-Tight-Puro and encode IE1 or IE1 1-382, both fused to a FLAG-tag, were generated by PCR amplification with primer pairs 5′Flag_IE1_BamHI and 3′IE1_EcoRI_stop or 5′Flag_IE1_BamHI and 3′IE1aa382_EcoRI along with pHM494 as the template, followed by insertion into pLVX-Tight-Puro via BamHI and EcoRI.
TABLE 1.
Oligonucleotides used for generation of plasmids, generation of recombinant viruses, and quantitative real-time PCR
| Name of primer | Sequence or source |
|---|---|
| 5′IE1mut-L174P | 5′-GGCTTGTATTAAGGAGCCGCATGATGTGAGCAAGG-3′ |
| 3′IE1mut-L174P | 5′-CCTTGCTCACATCATGCGGCTCCTTAATACAAGCC-3′ |
| 5′IE1_NheI | 5′-CATAGCTAGCATGGAGTCCTCTGCCAAGAG-3′ |
| 3′IE1_EcoRI | 5′-CATAGAATTCCTGGTCAGCCTTGCTTCTAG-3′ |
| 5′IE1-GEX-Bam | 5′-TCACGGATCCATGGAGTCCTCTGCCAAGAG-3′ |
| 3′IE1_Xho1 | 5′-CATACTCGAGTTACTGGTCAGCCTTGCTTC-3′ |
| 5′Flag_IE1_BamHI | 5′-CATAGGATCCATGGACTACAAAGACGATGACGATAAAGAGTCCTCTGCCAAGAGAAA-3′ |
| 3′IE1_EcoRI_stop | 5′-CATAGAATTCTTACTGGTCAGCCTTGCTTC-3′ |
| 3′IE1aa382_EcoRI | 5′-CATAGAATTCTTACTCTTCCTCATCTGACTCCT-3′ |
| 5′BAC_short | 5′-TAGGGATAACAGGGTAATCGATTT-3′ |
| 3′BAC-L174P | 5′-CACCCCCCAACTTGTTAGCGGCGCCCTTGCTCACATCATGCGGCTCCTTAATACAAGCCATCCCAACCAATTAACCAATTCTGATTAG-3′ |
| 5′BAC-L174P | 5′-TGAGGATAAGCGGGAGATGTGGATGGCTTGTATTAAGGAGCCGCATGATGTGAGCAAGGGCGCTAGGGATAACAGGGTAATCGATTT-3′ |
| 3′BAC-L174P_short | 5′-CACCCCCCAACTTGTTAGCG-3′ |
| 5′Intron3/pKD13 | 5′-AAAGATGTCCTGGCAGAACTCGGTAAGTCTGTTGACATGTATGTGATGTAGTGTAGGCTGGAGCTGCTTC-3′ |
| 3′Exon4/pkd13 | 5′-TAGTTTACTGGTCAGCCTTGCTTCTAGTCACCATAGGGTGGGTGCTCTTGATTCCGGGGATCCGTCGACC-3′ |
| 5′gB_forw | 5′-CTGCGTGATATGAACGTGAAGG-3′ |
| 3′gB_rev | 5′-ACTGCACGTACGAGCTGTTGG-3′ |
| CMV gB FAM/TAMRA | 5′-CGCCAGGACGCTGCTACTCACGA-3′ |
| 5′Alb | 5′-GTGAACAGGCGACCATGCT-3′ |
| 3′Alb | 5′-GCATGGAAGGTGAATGTTTCAG-3′ |
| Alb FAM/TAMRA | 5′-TCAGCTCTGGAAGTCGATGAAACATACGTTC-3′ |
| CMV 5′ (IE1) | 5′-AAGCGGCCTCTGATAACCAAG-3′ |
| CMV 3′ (IE1) | 5′-GAGCAGACTCTCAGAGGATCGG-3′ |
| MX1-fw | 5′-CAGCACCTGATGGCCTATCA-3′ |
| MX1-rev | 5′-ACGTCTGGAGCATGAAGAACTG-3′ |
| 5′hISG54 | 5′-ATGTGCAACCTACTGGCCTAT-3′ |
| 3′hISG54 | 5′-TGAGAGTCGGCCCATGTGATA-3′ |
| huGAPDH_TM_for | 5′-GAAGGTGAAGGTCGGAGT |
| huGAPDH_tm_rev | 5′-GAAGATGGTGATGGGATTTC |
| CCL8-fw + CCL8-rev | VHPS-1625 (Biomol GmbH, Hamburg, Germany) |
| IDO1-fw + IDO1-rev | VHPS-4584 (Biomol GmbH, Hamburg, Germany) |
| IFIT3/4-fw + IFIT3/4-rev | VHPS-4466 (Biomol GmbH, Hamburg, Germany) |
| HLA-DRA-fw HLA-DRA-rev | VHPS-4159 (Biomol GmbH, Hamburg, Germany) |
| OASL-fw + OASL-rev | VHPS-6424 (Biomol GmbH, Hamburg, Germany) |
| PML-fw + PML-rev | VHPS-7019 (Biomol GmbH, Hamburg, Germany) |
| TNFSF10-fw + TNFSF10-rev | VHPS-9439 (Biomol GmbH, Hamburg, Germany) |
| UIMC-fw + UIMC-rev | VHPS-7651 (Biomol GmbH, Hamburg, Germany) |
Cell culture, infections, and transfections.
Primary human foreskin fibroblasts (HFFs) were prepared from human foreskin tissue and were maintained in Eagle's minimal essential medium (Gibco/BRL, Eggenstein, Germany) supplemented with 5% fetal calf serum. HFFs with a small interfering RNA-mediated knockdown of PML (siPML) and control HFFs (siC) were cultured in Eagle's minimal essential medium (Gibco/BRL, Eggenstein, Germany) supplemented with 7% fetal calf serum and 5 μg/ml puromycin. HFFs with inducible expression of IE1 or IE1 1-382 were cultured in Eagle's minimal essential medium (Gibco/BRL, Eggenstein, Germany) supplemented with 10% tetracycline-free fetal bovine serum (Clontech, Palo Alto, CA, USA), 5 μg/ml puromycin, and 500 μg/ml Geneticin. Infection experiments were performed with the HCMV laboratory strain AD169 and recombinant viruses AD169/IE1 1-382 (6), AD169/IE1-L174P, and AD169ΔIE1. For this purpose, HFF cells were seeded into six-well dishes at a density of 3 × 105 cells/well. One day later, cells were incubated with 1 ml viral supernatant and, after 1.5 h, provided with fresh medium before they were used for subsequent analyses.
HEK293T cells were cultivated in Dulbecco's minimal essential medium (Gibco/BRL, Eggenstein, Germany) containing 10% fetal calf serum. Transfection of HEK293T cells was performed by applying the standard calcium phosphate precipitation method. For this, HEK293T cells were seeded into six-well dishes (6 × 105 cells/well) 1 day before transfection. For each transfection reaction, 1 to 2 μg of plasmid DNA was used. At about 16 h after transfection, the cells were washed two times with phosphate-buffered saline (PBS) and provided with fresh medium before they were used for further analyses.
Generation of stable HFF cells by lentivirus transduction.
In order to establish HFFs stably expressing the siRNAs siC and siPML, the Lenti-X shRNA system was utilized (Clontech, Palo Alto, CA, USA). For generation of replication-deficient lentiviruses, HEK293T cells were seeded in 10 cm dishes (5 × 106 cells/dish) and cotransfected with pLVX-shRNA1 vectors containing control shRNA or PML shRNA and the Lenti-X HTX packaging mix (Clontech, Palo Alto, CA, USA) using the Lipofectamine 2000 reagent (Invitrogen, Karlsruhe, Germany). Viral supernatants were harvested 48 h after transfection, cleared by centrifugation, filtered, and stored at −80°C. Primary HFFs were incubated for 24 h with lentivirus supernatants in the presence of 7.5 μg/ml Polybrene (Sigma-Aldrich, Deisenhofen, Germany), and stably transduced cell populations were selected by adding 5 μg/ml puromycin to the cell culture medium.
For generation of HFFs with inducible expression of IE1 or IE1 1-382, the Lenti-X Tet-on Advanced Inducible Expression System was utilized (Clontech, Palo Alto, CA, USA). For generation of replication-deficient lentiviruses expressing the tetracycline-controlled transactivator rtTA-Advanced, HEK293T cells were cotransfected with the pLVX-Tet-on Advanced vector and the Lenti-X HTX packaging mix (Clontech, Palo Alto, CA, USA). For generation of lentiviruses expressing IE1 or IE1 1-382 under the control of the inducible pTight promoter, HEK293T cells were cotransfected with pLVX-Tight-Puro vectors containing IE1 or IE1 1-382 and the Lenti-X HTX packaging mix (Clontech, Palo Alto, CA, USA). Viral supernatants were harvested 48 h after transfection, cleared by centrifugation, filtered, and stored at −80°C. Afterwards, primary HFFs were successively transduced with lentiviruses derived from the pLVX-Tet-On Advanced vector and lentiviruses derived from pLVX-Tight-Puro-IE1/IE1 1-382 vectors, followed by selection with 500 μg/ml Geneticin and 5 μg/ml puromycin, respectively.
Generation and reconstitution of recombinant cytomegaloviruses.
The wild-type HCMV bacterial artificial chromosome (BAC) HB15, which is based on the laboratory strain AD169, was manipulated by recombination-based genetic engineering in order to obtain AD169/IE1-L174P and AD169ΔIE1. AD169/IE1-L174P was generated by mutating the leucine residue at position 174 (CTG) in exon 4 of IE1 to proline (CCG). For this purpose, we utilized the two-step red-mediated recombination technique described by Tischer et al. (49), which uses the kanamycin gene as a first selection marker. The linear recombination fragment was generated by PCR using primers 5′BAC_short and 3′BAC-L174P and pEPkanS as the template DNA (kindly provided by K. Osterrieder, Berlin, Germany). The PCR product was treated with DpnI, gel purified, and subjected to a second round of PCR amplification using primers 5′BAC-L174P and 3′BAC-L174P_short. For homologous recombination, the PCR fragment was transformed into Escherichia coli strain GS1783 (a gift of M. Mach, Erlangen) already harboring HB15, and two-step bacteriophage λ red-mediated recombination was performed as described elsewhere (49). To identify positive transformants, the bacteria were plated on agar plates containing kanamycin (first recombination) or chloramphenicol and 1% arabinose (second recombination) and incubated at 32°C for 2 days. BAC DNA was purified from bacterial colonies growing on these plates, and successful recombination was confirmed by PCR, restriction enzyme digestion, and direct sequencing.
AD169ΔhIE1 was generated by deleting exon 4 of IE1 as described previously (6). Briefly, a linear recombination fragment, comprising a kanamycin resistance marker and 5′ and 3′ genomic sequences, was generated by PCR amplification using pKD13 as the template and primers 5′Intron3/pKD13 and 3′Exon4/pkd13. This fragment was used for electroporation of competent Escherichia coli strain DH10B harboring HB15, and homologous recombination was performed as described previously (50). The integrity of the resulting recombinant BAC was confirmed by PCR, restriction enzyme digestion, and direct sequencing.
HFFs stably expressing IE1, which were generated as described elsewhere (6), were utilized for reconstitution of AD169/IE1-L174P and AD169ΔIE1. The cells were seeded in six-well dishes (3 × 105 cells/well) and cotransfected with purified BAC DNA (1 μg), pp71 expression plasmid pCB6-pp71 (0.5 μg) and a vector encoding the Cre recombinase (0.5 μg) using FuGENE6 transfection reagent (Promega, Mannheim, Germany). Transfected HFFs were propagated until viral plaque formation was observed, and the supernatants from these cultures were used for further virus propagation in IE1-expressing HFFs.
Titration of virus stocks.
Titers of wild-type AD169 were determined by IE1 fluorescence. For this purpose, HFFs were infected with various dilutions of virus stocks. After 24 h of incubation, cells were fixed and stained with monoclonal antibody p63-27, directed against IE1 (51). Subsequently, the number of IE1-positive cells was determined by quantifying IE1 expression in >200 cells using ImageJ software. Results were used to calculate viral titers, expressed as IE protein-forming units (IEU). To ensure infection with equal genome copy numbers of wild-type and recombinant viruses, AD169/IE1-L174P and AD169ΔIE1 were titrated via determination of genome equivalents in virus stocks and in infected cells by quantitative real-time PCR. Samples of virus stocks were subjected to treatment with proteinase K, and TaqMan-PCR was performed as described below in order to determine the amount of the gB gene corresponding to HCMV genome equivalents. For determination of genome copies having entered the cells, HFFs were infected with various dilutions of AD169 and recombinant viruses. Sixteen hours later, viral and cellular DNA was extracted from infected cells using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) and subjected to quantitative real-time PCR (TaqMan-PCR) using HCMV gB-specific primers and primers specific for the cellular albumin gene as internal control. Results were used to calculate the number of intracellular HCMV genomes resulting from infection with 1 ml of input virus.
Multistep growth curve analysis.
HFF cells were seeded into six-well dishes at a density of 3 × 105 cells/well and infected the following day with wild-type AD169 at a multiplicity of infection (MOI) of 0.1 and equivalent genome copy numbers of AD169/IE1-L174P and AD169ΔIE1. Triplicate samples of infected cell supernatants were harvested at 2, 4, 6, 8, 10, and 13 days after inoculation and lysed by proteinase K treatment. Afterwards, quantitative real-time PCR (TaqMan-PCR) of the HCMV gB gene was conducted as described below to analyze the amount of viral genome copies in the supernatants.
Quantitative TaqMan real-time PCR.
For quantitative real-time PCR (TaqMan-PCR), an Applied Biosystems 7500 real-time PCR system was utilized (Applied Biosystems, Foster City, CA, USA) together with the corresponding software SDS (sequence detection system) (52). To quantify viral DNA, a sequence region within the HCMV gB gene region was amplified using primers 5′gB_forw and 3′gB_rev along with the fluorescence labeled probe CMV gB FAM/TAMRA, also directed against the gB gene locus. When intracellular genomes where examined, quantification of cellular albumin genes was conducted in parallel using primers 5′Alb and 3′Alb together with a fluorescence-labeled probe, Alb FAM/TAMRA (6-carboxyfluorescein/6-carboxytetramethylrhodamine). For determination of reference CT values (cycle threshold), serial dilutions of the respective standards (107 to 101 DNA molecules of gB or albumin) were analyzed by PCRs in parallel. The 20-μl reaction mixture contained 5 μl sample or standard DNA solution together with 10 μl 2× TaqMan PCR Mastermix (Applied Biosystems, Foster City, CA, USA), 1.5 μl of each primer (5 μM stock solution), 0.4 μl of probe (10 μM stock solution), and 1.6 μl of H2O. The thermal cycling conditions consisted of two initial steps of 2 min at 50°C and 10 min at 95°C followed by 40 amplification cycles (15 s at 95°C, 1 min at 60°C). The viral genome copy numbers and albumin copy numbers were subsequently calculated using the sample-specific CT value when set into relation to the standard serial dilutions.
RNA isolation and quantitative SYBR green reverse transcription-PCR (qRT-PCR).
Total RNAs from 3 × 105 to 6 × 105 infected or transduced HFFs were isolated in triplicate using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany). To exclude any DNA contaminations, 10 units of RNase-free DNase I (Roche, Mannheim, Germany) together with 20 units of RNaseOUT RNase Inhibitor (Invitrogen, Karlsruhe, Germany) were added to each sample and incubated at 37°C for 45 min, followed by DNase I inactivation at 75°C for 10 min. RNAs were further purified by phenol extraction and ethanol precipitation, and the RNA pellets were resuspended in RNase-free H2O. Afterwards, first-strand cDNAs were synthesized from 1 μg of total RNA using oligo(dT)18 and random hexamer primers provided by the Maxima First Strand cDNA synthesis kit (Thermo Scientific, Waltham, MA, USA). First-strand cDNAs were diluted 4-fold with DNase-free water, and 2 μl was used as the template for quantitative real-time PCR mixtures containing 12.5 μl Maxima SYBR green/Rox qPCR master mix (Thermo Scientific, Waltham, MA, USA), 10.35 μl nuclease-free H2O, and 0.15 μl of the following primer pairs: CMV 5′ (IE1) and CMV 3′ (IE1), PML-fw and PML-rev, MX1-fw and MX1-rev, TNFSF10-fw and TNFSF10-rev, OASL-fw and OASL-rev, IFIT3/4-fw and IFIT3/4-rev, CCL8-fw and CCL8-rev, IDO1-fw and IDO1-rev, 5′hISG54 and 3′hISG54, HLA-DRA-fw and HLA-DRA-rev, and UIMC-fw and UIMC-rev. For normalization of the qPCR values, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was amplified as a housekeeping gene using primers huGAPDH_TM_for and huGAPDH_tm_rev. SYBR green PCR was conducted in 96-well plates compatible with the ABI Prism 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) together with the corresponding software SDS (sequence detection system). The initial denaturation step (95°C, 10 min) was subsequently followed by 40 cycles of denaturation (95°C, 15 s), primer annealing (60°C, 35 s), and strand elongation, followed by a dissociation stage to ascertain the specificity of the reactions. Data were analyzed using the Applied Biosystems SDS software, and relative quantification of mRNA levels was performed.
CLIP labeling.
For labeling of CLIP-tagged IE1 proteins, transfected HEK293T cells were incubated with 3 μM red fluorescent substrate CLIP-Cell TMR STAR in cell culture medium (Dulbecco's minimal essential medium supplemented with 10% fetal calf serum) for 30 min at 37°C. Afterwards, the cells were washed three times with cell culture medium and incubated for another 30 min at 37°C before they were washed again. The cells were pooled and reseeded into 12-well plates to ensure equal expression and labeling efficiency in every sample. Cell lysates were prepared immediately (0 h) or at later times (2 h to 30 h) by resuspending cell pellets in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiling at 95°C for 5 min, and sonication. Labeled proteins were separated on SDS-containing 8% polyacrylamide gels and visualized by in-gel fluorescence scanning using an INTAS Advanced Fluorescence Imager (INTAS, Göttingen, Germany).
Western blotting, indirect immunofluorescence, and antibodies.
For Western blot analysis, total lysates from transfected or infected cells were prepared in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated on SDS-containing 8 to 12% polyacrylamide gels, and transferred to nitrocellulose membranes. Chemoluminescence detection was performed according to the manufacturer's instructions (ECL Detection kit; Amersham Pharmacia Biotech Europe, Freiburg, Germany).
Indirect immunofluorescence analysis of infected HFF cells was performed by fixation of infected cells with 4% paraformaldehyde and fluorescence staining as described elsewhere (6). Samples were analyzed using a Leica TCS SP5 confocal microscope with 488-nm, 543-nm, and 633-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported, processed with Adobe Photoshop CS5, and assembled using CorelDraw X5.
The following monoclonal antibodies were used for immunofluorescence and Western blot analyses: α-IE1 p63-27 (51), α-UL44 BS510 (kindly provided by B. Plachter, Mainz, Germany), α-MCP 28-4 (53), α-FLAG 1804 (Sigma-Aldrich, Deisenhofen, Germany), and α-β-actin AC-15 (Sigma-Aldrich, Deisenhofen, Germany). The following polyclonal antibodies were used for immunofluorescence and Western blot analyses: α-IE2 pHM178 (produced by immunizing rabbits with prokaryotically expressed protein); α-PML #4 (a kind gift from P. Hemmerich, Jena, Germany), used for immunofluorescence analysis; α-PML A301-167A (Bethyl Laboratories, Montgomery, TX, USA) in combination with α-PML A301-168A (Bethyl Laboratories), applied for Western blot analysis; α-Sp100 GH3 (kindly provided by H. Will, Hamburg, Germany); and α-STAT2 H190, obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Alexa Fluor 488-/555-conjugated secondary antibodies for indirect immunofluorescence experiments were purchased from Life technologies (Karlsruhe, Germany), and horseradish peroxidase-conjugated anti-mouse/anti-rabbit secondary antibodies for Western blot analysis were obtained from Dianova (Hamburg, Germany).
Recombinant protein production and purification.
Recombinant IE1 and IE1-L174P were expressed in E. coli and purified as described elsewhere (6). Briefly, LB medium was inoculated with E. coli colonies freshly transformed with pGEX-6P-1-based expression plasmids encoding GST-tagged IE1 or IE1-L174P. Cell cultures were grown at either 30°C or 37°C, and expression of recombinant proteins was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 to 6 h. Cell pellets were resuspended in phosphate buffer and lysed by sonication. Protein purification was achieved by GST affinity chromatography, proteolytic cleavage with PreScission protease, a second GST affinity chromatography, and size exclusion chromatography using a Superdex 200 prepgrade column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Bioinformatic analyses.
Modeling of the L174P mutation was performed with SwissModel, and the lowest-energy conformer of P174 was selected (54). WhatCheck was used for structure analysis and VMD for visualization (55, 56).
RESULTS
Insertion of L174P mutation into IE1 of HCMV results in a severe growth defect comparable to a deletion of IE1.
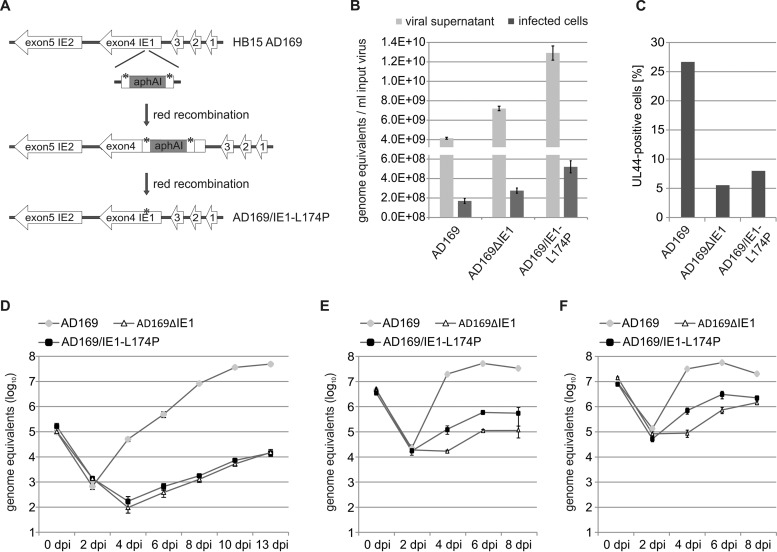
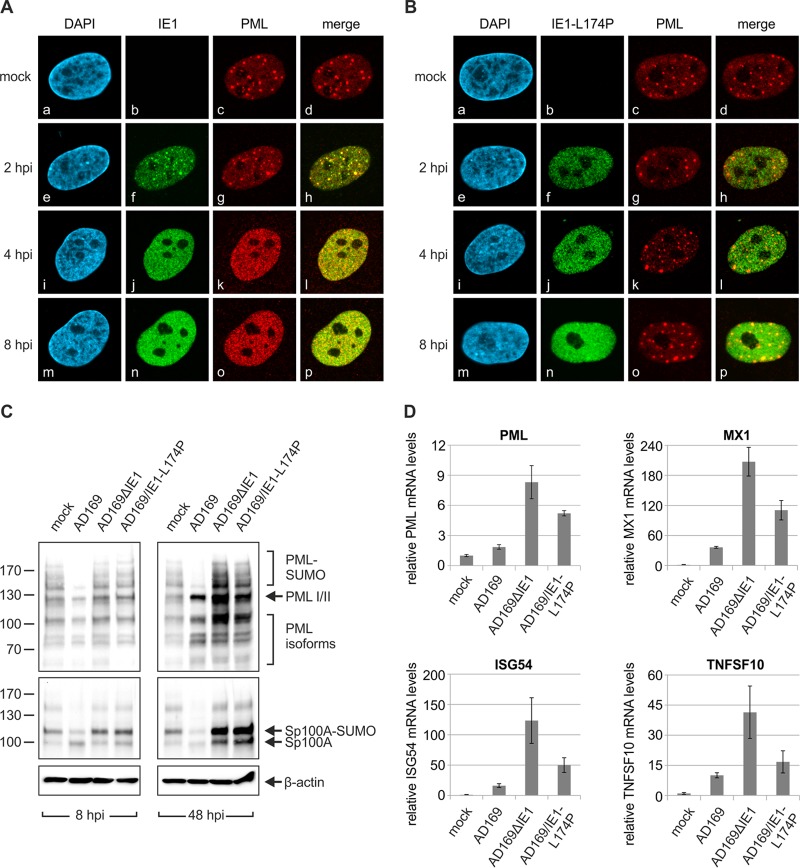
After entry of HCMV genomes into the nucleus, the immediate early protein IE1 disrupts the subnuclear, antiviral structure ND10 in order to initiate lytic HCMV infection. Previous studies have demonstrated that a leucine-to-proline mutation (L174P) within the globular core domain of IE1 (IE1CORE) abolishes the interaction with the major ND10 protein PML and, consequently, abrogates the dispersal of ND10 in transient-transfection assays (38–40). To investigate the impact of this mutation on lytic HCMV infection, we generated a recombinant HCMV expressing IE1-L174P, referred to as AD169/IE1-L174P. As illustrated in Fig. 1A, the L174P mutation was introduced into the IE1 gene of HCMV laboratory strain AD169 using a two-step, markerless BAC recombination technique (49). In addition, a recombinant virus harboring a deletion of IE1 (AD169ΔIE1), which was generated as previously described (6), was included in this study, allowing comparative analysis of replication defects caused by IE1 mutation and deletion. Since it is known that IE1-deleted viruses display a nonlinear relationship between the multiplicity of infection (MOI) and the extent of virus growth (3, 8), titration of the newly generated viruses was performed by real-time PCR quantification of genome equivalents in viral supernatants and in infected cells. Figure 1B shows that considerably more viral genomes were detected in virus stocks of AD169/IE1-L174P and AD169ΔIE1 than in those of wild-type AD169, and a similar ratio of genome copies was determined in infected primary human foreskin fibroblast (HFF) cells. In contrast, lytic replication was initiated in significantly more cells infected with wild-type AD169 than in cells infected with AD169/IE1-L174P or AD169ΔIE1 using the same amount of input virus, illustrating the silencing of viral genomes in the absence of the ND10-antagonistic activity of IE1 (Fig. 1C). Next, we analyzed the growth kinetics of the recombinant viruses by performing multistep growth curve analysis. For this purpose, HFF cells were infected either with wild-type AD169 at MOIs of 0.1, 4, or 10, respectively, as determined by IE1 fluorescence, or with equal genome copy numbers of AD169/IE1-L174P and AD169ΔIE1. Aliquots of cell supernatants were collected at various times after infection and subjected to quantitative real-time PCR to determine the amount of the gB gene corresponding to HCMV genome equivalents. At an MOI of 0.1, AD169/IE1-L174P displayed a strongly reduced growth, which was comparable to the reduction caused by deletion of the IE1 gene (Fig. 1D). Release of mutant progeny virions was not detectable until 6 days postinfection (dpi) and, compared to the release of wild-type virions, was 3,000-fold to 7,000-fold decreased on the following days. At an MOI of 4 (Fig. 1E) or 10 (Fig. 1F), growth of AD169/IE1-L174P was still significantly reduced compared to wild-type virus. Furthermore, AD169/IE1-L174P exhibited a slightly increased replication in comparison to the IE1-deleted virus, suggesting residual growth-promoting functions of IE1-L174P under high-MOI conditions. The observed growth reduction of AD169/IE1-L174P underlines the importance of the PML-antagonistic function of IE1 for lytic replication; however, the severity of the defect observed at low MOI was surprising to us since IE1 was shown to promote replication through both interfering with PML-mediated intrinsic immunity via IE1CORE and inhibiting interferon (IFN) signaling via its disordered C-terminal domain.
FIG 1.
Generation and characterization of a recombinant HCMV expressing IE1-L174P. (A) Schematic representation of the HCMV major immediate early gene region in which the L174P mutation (indicated by asterisks) was introduced. The HCMV BAC HB15, which is based on the laboratory strain AD169, was manipulated using a two-step, red-mediated recombination technique (49), in order to obtain AD169/IE1-L174P. (B) Determination of HCMV genome copies in viral supernatants and infected cells. Viral supernatants were subjected to protease K treatment followed by HCMV gB-specific real-time PCR (light gray bars). Standard deviations of four replicate measurements are indicated. To analyze viral genomes having entered the cells (dark gray bars), HFFs were infected with different dilutions of AD169, AD169/IE1-L174P, and the IE1 deletion virus AD169ΔhIE1. At 16 hpi, viral and cellular DNA was extracted from infected cells and subjected to HCMV gB-specific quantitative real-time PCR. Values of viral genomes were extrapolated to 1 ml of input virus. Standard deviations of results from four experiments and two replicate measurements are indicated. (C) Quantification of cells initiating lytic infection. HFF cells were infected with the same amount of AD169 (MOI, 0.4), AD169/IE1-L174P, and AD169ΔhIE1 genomes. The cells were fixed at 24 hpi, followed by immunofluorescence staining of early protein UL44 and quantification of viral gene expression in >400 cells. (D, E, F) Multistep growth curves of AD169, AD169/IE1-L174P, and AD169ΔhIE1. HFF cells were infected with wild-type AD169 at an MOI of 0.1 (D), 4 (E), or 10 (F) and equivalent genome copy numbers of AD169/IE1-L174P and AD169ΔhIE1. Cell supernatants were harvested at the indicated times after infection and analyzed for genome equivalents by HCMV gB-specific quantitative real-time PCR.
Recombinant AD169 encoding IE1-L174P displays reduced viral protein expression under low- and high-MOI conditions.
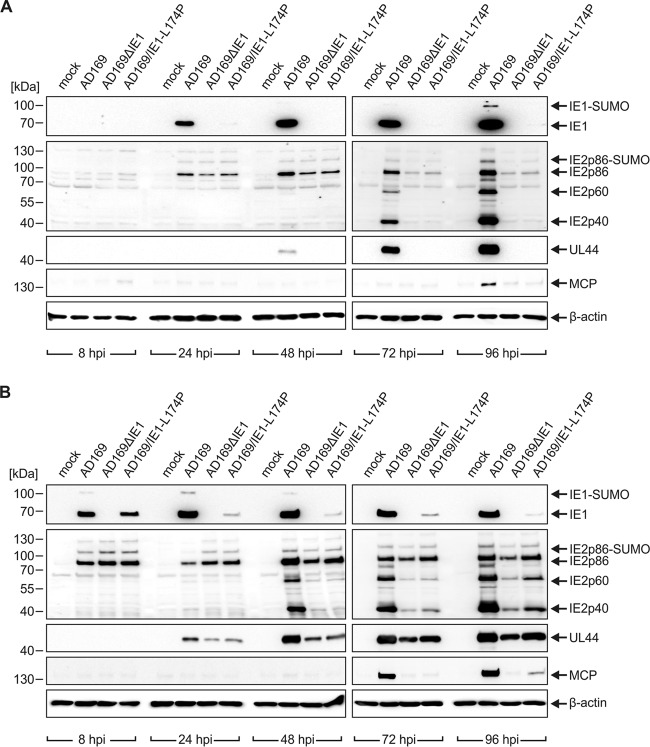
In order to examine the observed growth defect of AD169/IE1-L174P more precisely, viral protein accumulation was monitored during one round of replication. To this end, HFF cells were infected either at low MOI (0.1) (Fig. 2A) or high MOI (2) (Fig. 2B) with wild-type AD169 and equivalent genome copy numbers of AD169/IE1-L174P and AD169ΔIE1. Total cell lysates were prepared at different times after infection and expression patterns of viral immediate early (IE1, IE2 – p86, p60, p40), early (UL44), and late proteins (MCP) were analyzed by Western blotting. Intriguingly, hardly any IE1-L174P was present throughout the replication cycle (Fig. 2A and B, top panels). Only initially after high-multiplicity infection, comparable levels of IE1-L174P and wild-type IE1 were detected (Fig. 2B, top panels, 8 hpi), but the abundance of IE1-L174P declined as infection progressed (Fig. 2B, top panels, 24 hpi to 96 hpi). This loss of IE1-L174P during infection provides a plausible explanation for the similar growth characteristics of AD169/IE1-L174P and the IE1 deletion virus AD169ΔIE1. Similar to IE1, levels of the second major IE protein IE2p86 were comparable initially upon infection (Fig. 2A, second panels from the top, 24 hpi; Fig. 2B, second panels from the top, 8 hpi). However, expression of IE2p86 and the smaller isoforms IE2p60 and IE2p40 was strongly reduced at later times after infection with AD169/IE1-L174P and AD169ΔIE1 compared to wild-type AD169. Furthermore, no expression of early and late proteins was observed until 96 h after low-multiplicity infection with both of the recombinant viruses (Fig. 2A, third and fourth panels from the top). This is consistent with the delayed and reduced virus release determined in growth curve analysis (Fig. 1D). Notably, although IE1 was shown to be dispensable during high-multiplicity infections with mutants derived from the HCMV laboratory strain Towne (3), we detected a substantial attenuation of viral early and late gene expression in cells infected at a high MOI with AD169/IE1-L174P and AD169ΔIE1 (Fig. 2B, third and fourth panels from the top). We conclude that the low levels of IE1-L174P during AD169/IE1-L174P infection, which may be caused by transcriptional dysregulation or reduced protein stability, lead to a defect in viral gene expression—under low- and high-MOI conditions—that is similar to the defect caused by IE1 deletion.
FIG 2.
Impaired viral gene expression of recombinant HCMV encoding IE1-L174P. HFFs were either not infected (mock) or infected at an MOI of 0.1 (A) or an MOI of 2 (B) with AD169 and equivalent genome copy numbers of AD169ΔhIE1 and AD169/IE1-L174P. At the indicated times after virus inoculation (8, 24, 48, 72, and 96 hpi), cell lysates were prepared and the expression levels of viral immediate early (IE1 and IE2 isoforms IE2p86, IE2p60, and IE2p40), early (UL44), and late (MCP) proteins were compared by Western blotting. Cellular β-actin levels served as internal loading control.
IE1-L174P has a reduced half-life compared to wild-type IE1.
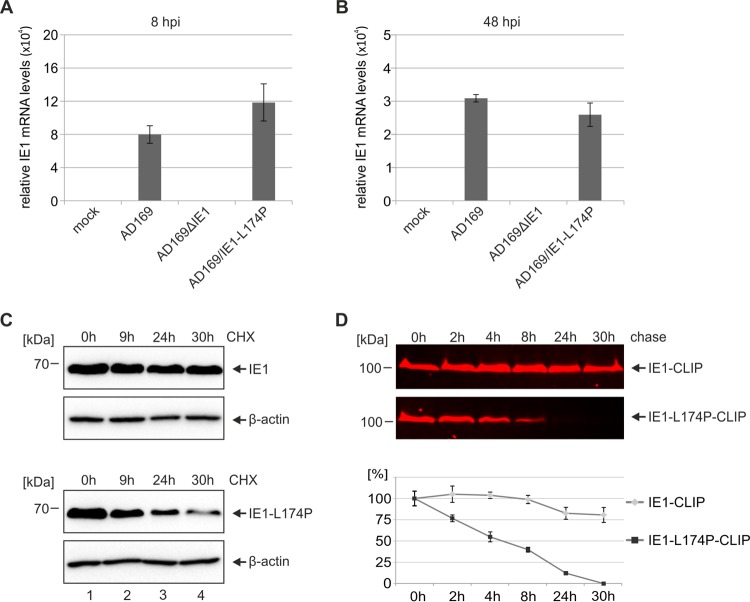
Since IE1 has been suggested to positively regulate its own promoter and, moreover, transactivation activities of IE1 were mapped to IE1CORE containing leucine 174 (40, 57), the question arose whether the low protein levels of IE1-L174P may result from impaired transcription. To test this, we measured IE1 mRNA levels at 8 h (Fig. 3A) and 48 h (Fig. 3B) after high-MOI infection with wild-type AD169 or recombinant viruses. Quantitative RT-PCR analysis revealed slightly enhanced expression of IE1-L174P early after infection (Fig. 3A) and similar mRNA levels of IE1-L174P and wild-type IE1 at 48 hpi (Fig. 3B), indicating that the loss of IE1-L174P during infection is not due to deregulation at the transcriptional level. Therefore, we speculated that a leucine-to-proline mutation within the highly alpha-helical IE1CORE domain may result in misfolding and degradation of the IE1 protein. To investigate the stability of IE1-L174P in living cells, we transfected HEK293T cells with expression constructs for wild-type IE1 or IE1-L174P and analyzed the protein levels at different times after addition of the protein synthesis inhibitor cycloheximide (CHX). While the protein level of wild-type IE1 remained almost unchanged after 30 h of CHX treatment (Fig. 3C, upper panels), a clear decline of IE1-L174P was observed over time (Fig. 3C, lower panels) indicating that mutation of leucine 174 to proline reduces the half-life of the IE1 protein. Since CHX does not fully inhibit de novo protein synthesis and since its toxicity can lead to erroneous results, we next utilized a pulse-chase assay based on CLIP-tag technology (New England BioLabs). For this, wild-type IE1 and IE1-L174P fused to a C-terminal CLIP-tag were transiently expressed in HEK293T cells, followed by labeling with the red fluorescent substrate CLIP-Cell TMR-Star. At sequential time points, the cells were harvested and the amount of labeled IE1-CLIP and IE1-L174P-CLIP was determined by in-gel fluorescence scanning of SDS-PAGE gels. Consistent with the results from CHX experiments, no significant reduction of IE1 was observed within 30 h after the labeling period, demonstrating a high stability of the wild-type protein (Fig. 3D, top panel, upper box). In contrast, labeled IE1-L174P was rapidly degraded (Fig. 3D, top panel, lower box) and displayed an approximate half-life of 5 h, as determined by quantification of the fluorescent signal (Fig. 3D, lower panel [graph]). These findings strongly point toward a negative effect of the leucine-to-proline mutation on the structural integrity and, thereby, on the stability of IE1.
FIG 3.
Reduced half-life of IE1-L174P compared to wild-type IE1. (A, B) Analysis of the transcription levels of IE1 and IE1-L174P during infection. HFF cells were either not infected (mock) or infected with AD169 at an MOI of 2 and equivalent genome copy numbers of AD169ΔhIE1 and AD169/IE1-L174P. At 8 hpi (A) or 48 hpi (B), total mRNA was isolated and relative expression levels of IE1 were analyzed by qRT-PCR. (C) Determination of the half-lives of IE1 and IE1-L174P using cycloheximide (CHX). HEK293T cells were transfected with expression plasmids encoding wild-type IE1 or IE1-L174P. Twenty-four hours after transfection, cells were treated with 25 μg/ml of CHX and harvested at the indicated times in order to analyze IE1 levels by Western blotting. Cellular β-actin levels were detected as internal control. (D) Determination of the half-lives of IE1 and IE1-L174P using the CLIP-tag technology. HEK293T cells were transfected with expression plasmids encoding wild-type IE1 or IE1-L174P in fusion with a C-terminal CLIP-tag. Twenty-four hours after transfection, IE1-CLIP proteins were labeled with 3 μM red fluorescent substrate CLIP-Cell TMR-star. Cell lysates were prepared at the indicated times, followed by separation of IE1-CLIP proteins on SDS-PAGE gels and visualization by in-gel fluorescence scanning (top two panels). Densitometric quantification of fluorescent signals was performed using the AIDA Image Analyzer v.4.22 software, and the mean values with standard deviations from three experiments are shown (bottom panel [graph]).
L174P mutation affects the structural integrity of the IE1 fold.
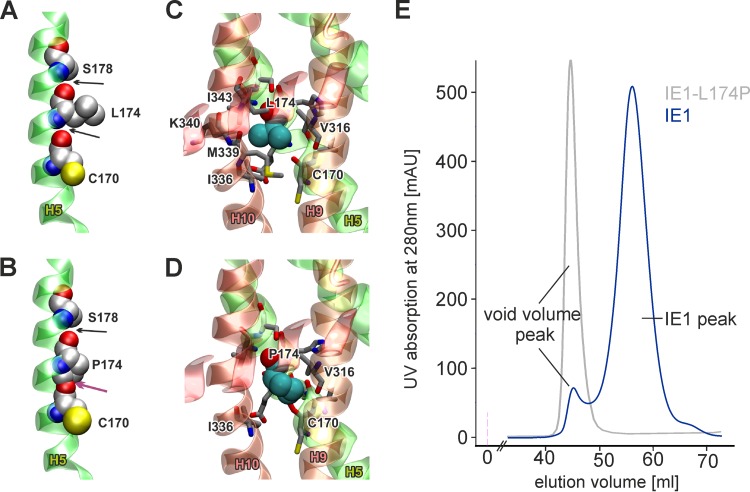
To evaluate possible effects of L174P mutation on the IE1 fold, we performed bioinformatic analysis based on the recently determined crystal structure of IE1CORE. IE1CORE consists of 11 α-helices and adopts an elongated, femur-shaped fold that can be divided into an N-terminal head region and a C-terminal head region interconnected by a stalk (6). The L174P mutation is located within helix 5 (H5) of the C-terminal head region and most likely disrupts the helical structure of H5 by multiple unfavorable structural effects. Due to the missing amide proton, the hydrogen bond interaction with C170 is lost and, additionally, clashes with the oxygen atom of C170 backbone are caused by the carbon atoms of the proline imidazole ring (Fig. 4A and B; Table 2. Moreover, L174 forms an extensive hydrophobic packing with side chains of adjacent residues and with residues of neighboring helices, namely, I336, M339, and I343 of H10 and V316 of H9 (Fig. 4C). Upon mutation to proline, most of these contacts are lost due to missing side chain atoms, thus destabilizing the tight helix packing of H4, H5, H9, and H10 in the C-terminal head region and presumably resulting in misfolding of the entire IE1CORE (Fig. 4D). In order to experimentally confirm the perturbation of the IE1 fold by L174P mutation, we produced recombinant proteins in bacteria and purified them by affinity chromatography and gel filtration. While gel filtration chromatography of wild-type IE1 resulted in one symmetric main peak, IE1-L174P eluted with the void volume, which contains proteins and protein aggregates with a molecular mass of more than 600 kDa (Fig. 4E), indicating an aggregated state of IE1-L174P. The aggregation of IE1-L174P impeded further investigation of the protein fold by additional methods but allowed the conclusion that the leucine-to-proline mutation within H5 indeed affects the structural integrity of the IE1CORE fold, hereby inducing rapid degradation of IE1-L174P in living cells.
FIG 4.
Unfavorable effects of L174P mutation on the structural integrity of IE1. (A) Structure of helix 5 showing the interactions of L174 in wild-type IE1. L174 forms backbone hydrogen bonds with C170 and S178, indicated by black arrows (the side chain oxygen of S178 has been omitted for clarity). (B) In the L174P mutation, no backbone hydrogen bond can be formed between P174 and C170 and clashes with the proline imidazole ring occur (indicated by the magenta arrow). Both effects are expected to destabilize helix H5 significantly. (C) Interactions of L174 (space filled) with helices H9 and H10. Residues that lie within 4 Å of any atom of L174 are shown as sticks. Most of the stabilizing contacts are formed to V316, I336, M339, K340, and I343. (D) Same presentation as described for panel C but for the L174P mutant, indicating that most of the stabilizing interactions to helices H9 and H10 are lost. (E) Gel filtration chromatography of recombinant IE1 and IE1-L174P. The chromatogram shows an overlay of the elution profiles of wild-type IE1 and IE1-L174P in blue and gray, respectively. The void volume peak, which contains proteins larger than 600 kDa, is indicated.
TABLE 2.
Clashes caused by the L174P mutation in IE1CORE
| Atom in P174 | Other atom involved in clash | Size of clash (Å) |
|---|---|---|
| CD | C170-O | 1.19 |
| CG | C170-O | 0.57 |
| CD | C170-C | 0.44 |
| CD | I171-C | 0.38 |
| CD | E173-N | 0.34 |
Effects of IE1 mutants IE1-L174P and IE1 1-382 on the inhibition of interferon signaling during viral infection.
Prior studies have demonstrated that L174P mutation in IE1 prevents targeting, de-SUMOylation, and dispersal of PML in transfection assays. To confirm this finding in HCMV-infected HFF cells, we next analyzed the subcellular localization of IE1 and PML after infection with wild-type AD169 or AD169/IE1-L174P. As illustrated in Fig. 5A, infection of HFFs with wild-type virus resulted in the typical, transient colocalization of IE1 and PML (Fig. 5A, panels e to h) followed by disruption of ND10 integrity within the first hours of infection (Fig. 5A, panels i to p). In contrast, IE1-L174P neither localized to ND10 nor induced dispersal of ND10 foci during infection (Fig. 5B, panels f, j, n). Consistently, Western blot analysis revealed that IE1-L174P fails to induce depletion of the SUMOylated forms of PML (Fig. 5C, upper panels) and of Sp100 (Fig. 5C, middle panels) but produces a similar pattern of the various PML and Sp100 species as detected in AD169ΔIE1-infected cells.
FIG 5.
Less-efficient suppression of IFN signaling by HCMV encoding PML binding-deficient IE1-L174P than that of wild-type HCMV. (A, B) Immunofluorescence analysis of PML dispersal after infection with wild-type or recombinant HCMV. HFF cells were either not infected (mock) or infected with the same genome copy numbers of AD169 (MOI, 2) (A) and AD169/IE1-L174P (B). At the indicated times after infection (2 h, 4 h, and 8 h), the cells were fixed for immunofluorescence staining of IE1 and endogenous PML. Cell nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). (C) Western blot analysis of the SUMOylation state of PML and Sp100 after infection with wild-type or recombinant HCMV. HFF cells were infected with the same genome copy numbers of AD169 (MOI, 2), AD169ΔhIE1, and AD169/IE1-L174P. At 8 hpi (left panels) or 48 hpi (right panels), lysates were prepared and analyzed for expression of endogenous PML (top panels), Sp100 (middle panels), and β-actin as internal loading control (bottom panels). (D) Determination of ISG transcription after infection with wild-type or recombinant HCMV. HFF cells were either not infected (mock) or infected with the same genome copy numbers of AD169 (MOI, 2), AD169ΔhIE1, and AD169/IE1-L174P for 48 h. Relative mRNA levels were determined by qRT-PCR using primers specific for a set of ISGs (PML, MX1, ISG54, and TNFSF10). Standard deviations from three biological replicates are indicated.
Since it has been shown that PML binding by IE1CORE occurs in a manner independent of the interaction with signal and transducer of transcription (STAT) proteins by the disordered C terminus of IE1 (5, 41), we next explored the influence of IE1-L174P on STAT-mediated IFN signaling during infection. To address this issue, mRNA levels of interferon-stimulated genes (ISGs) were determined in HFF cells infected with high multiplicities of wild-type AD169 and equivalent genome copy numbers of AD169/IE1-L174P and AD169ΔIE1 for 48 h. We observed that transcription of PML, which is known to be IFN inducible, was slightly elevated in AD169-infected cells compared to uninfected cells but highly upregulated after infection with IE1-deleted virus AD169ΔIE1 (Fig. 5D, upper left panel). Infection with AD169/IE1-L174P resulted in an intermediate effect (Fig. 5D, upper left panel, and Fig. 5C, 48 hpi, top panel). Induction of other IFN-α/β-stimulated genes was also efficiently suppressed by wild-type AD169 but not by AD169ΔIE1 (Fig. 5D, upper right panel and lower panels), being in accordance with the described role of IE1 as an antagonist of type I IFN-dependent STAT signaling (42). Again, IE1-L174P displayed an intermediate inhibition of ISG upregulation, although the disordered C terminus harboring the STAT interaction motif is not affected by L174P mutation. This intermediate effect may arise from the low levels of IE1-L174P during infection. However, since PML recently emerged as a positive regulator of interferon signaling (44–46), it is also possible that the loss of PML binding caused by L174P mutation accounts for the inefficient suppression of the interferon response.
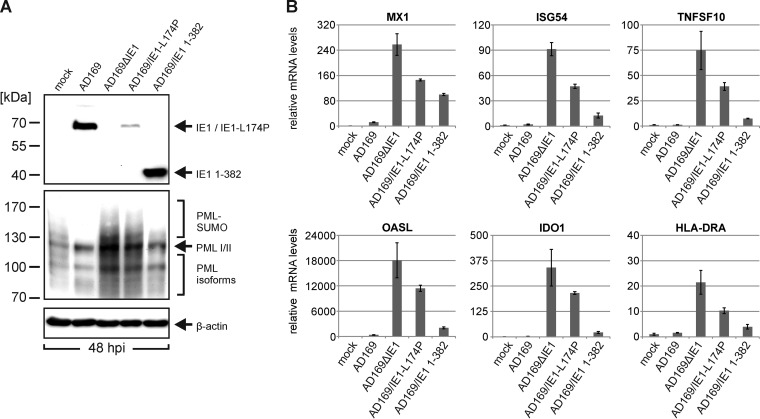
In order to obtain further evidence for a role of IE1-mediated PML modification in attenuation of ISG induction during viral infection we directly compared wild-type AD169 with the recombinant viruses AD169ΔIE1, AD169/IE1-L174P, and AD169/IE1 1-382 (Fig. 6). AD169/IE1 1-382 expresses a C-terminally truncated IE1 protein lacking the STAT-binding region and has previously been demonstrated to efficiently antagonize ND10-mediated intrinsic immunity during viral infection (6). As shown in Fig. 6A, IE1 1-382 is expressed at levels similar to those of wild-type IE1 at 48 h postinfection while the abundance of IE1-L174P was significantly reduced (Fig. 6A, top panel). Furthermore, while AD169/IE1 1-382 induced PML deSUMOylation, this was not the case for AD169/IE1-L174P (Fig. 6A, middle panel). When mRNA levels of ISGs were determined in HFF cells infected with the various viruses, we observed that AD169/IE1 1-382 efficiently suppressed ISG induction (Fig. 6B). For most investigated ISGs, the inhibitory effect exerted by IE1 1-382 was only slightly reduced compared to that of wild-type IE1, suggesting that PML modification by IE1 is of major importance for the inhibition of interferon signaling during viral infection.
FIG 6.
Efficient suppression of IFN signaling by HCMV encoding IE1 1-382, which contains an intact PML binding domain but lacks the STAT2 interaction region. (A) Detection of IE1 variants and endogenous PML protein after infection with wild-type or recombinant viruses. HFF cells were either not infected (mock) or infected with wild-type AD169, AD169ΔhIE1, AD169/IE1-L174P, or AD169/IE1 1-382 for 48 h and were subjected to Western blot analysis using antibodies directed against IE1 (top panel), PML (middle panel), and β-actin (bottom panel), which served as internal loading control. (B) Determination of ISG transcription after infection with wild-type or recombinant HCMV. HFF cells were either not infected (mock) or infected with the same genome copy numbers of AD169 (MOI, 2), AD169ΔhIE1, AD169/IE1-L174P, or AD169/IE1 1-382. At 48 hpi, total RNA was isolated, and relative mRNA levels were determined by qRT-PCR using primers specific for a set of ISGs (MX1, ISG54, TNFSF10, OASL, IDO1, HLA-DRA). Standard deviations from three replicates are indicated.
PML positively regulates type I and type II IFN signaling.
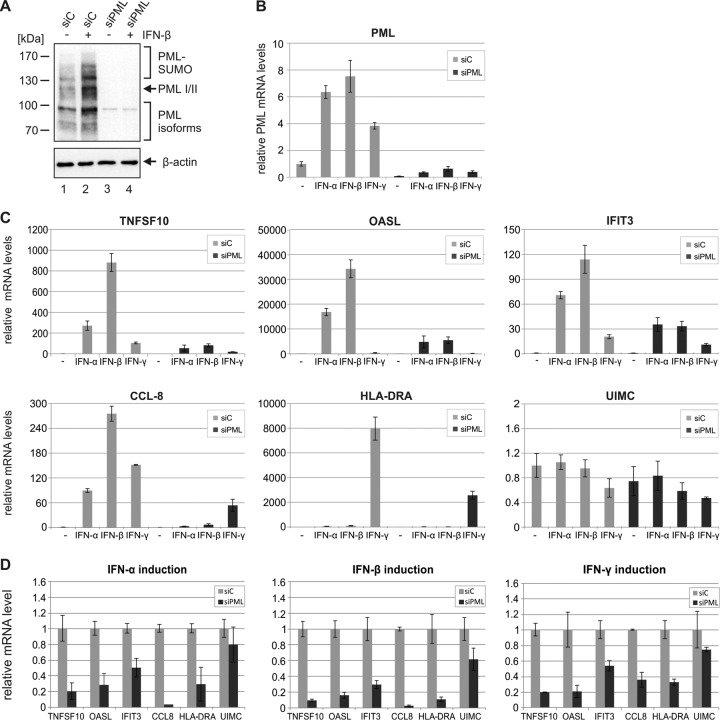
Recent evidence suggested a positive effect of PML on transcription of IFN-γ-stimulated genes and an involvement of one specific PML isoform in IFN-β production (44–46); however, the exact role of PML is far from being understood. To further characterize the influence of PML on interferon signaling, we measured ISG induction in HFFs with an shRNA-mediated depletion of PML (siPML) in comparison to that observed in control cells (siC). As shown in Fig. 7, siPML cells exhibited a downregulation of all PML variants under unstimulated and IFN-stimulated conditions, as assessed by Western blot (Fig. 7A) and qRT-PCR (Fig. 7B) analysis. Intriguingly, stimulation of PML-depleted cells with IFN-α, IFN-β, or IFN-γ resulted in a less efficient upregulation of ISG expression than stimulation of control cells (Fig. 7C). This was true for ISGs that are predominantly induced by type I IFNs, for example TNFSF10, OASL, or IFIT3, and also for ISGs primarily induced by type II IFNs, like HLA-DRA (Fig. 7C). Expression levels of other genes, such as UIMC, did not show major changes, thus excluding a generally reduced transcription in siPML cells. Figure 7D summarizes the effects of IFN-α, IFN-β, and IFN-γ stimulation in siPML cells compared to siC cells and highlights the positive role of PML in both type I and type II IFN-induced gene expression. Taken together, our results indicate that PML not only is encoded by an ISG but also contributes to the interferon response by acting as a coregulator of ISG induction.
FIG 7.
Reduction of IFN-induced ISG expression in PML-depleted cells compared to control cells. (A) Detection of endogenous PML protein in control cells and PML-depleted cells by Western blotting. HFF cells stably expressing control shRNAs (siC) or shRNAs directed against PML (siPML) either were left untreated (−) or were treated with IFN-β (+) for 24 h and were harvested for Western blot analysis using antibodies directed against PML and β-actin, which served as internal loading control. (B) Determination of PML mRNA levels in control cells and PML-depleted cells by qRT-PCR. HFF cells stably expressing control shRNAs (siC) or shRNAs directed against PML (siPML) either were left untreated (−) or were treated with IFN-α, IFN-β, or IFN-γ for 24 h. Subsequently, the cells were harvested for total mRNA isolation, and qRT-PCR analysis was performed using primers specific for PML. Results were normalized to GAPDH and are shown as mean values with standard deviations from three biological replicates. (C, D) Effects of PML depletion on IFN-induced ISG expression. Control HFFs (siC) and PML-depleted HFFs (siPML) were either left untreated (−) or treated with IFN-α, IFN-β, or IFN-γ for 24 h. Subsequently, total mRNAs were prepared and qRT-PCR was performed to determine transcription of type I IFN-responsive ISGs (TNFSF10, OASL, IFIT3, CCL-8), type II IFN-responsive ISGs (HLA-DRA), and control genes (UIMC). Results were normalized to GAPDH, and standard deviations from three biological replicates are indicated. ISG expression is related to unstimulated siC cells (values set to 1) (C) or to IFN-stimulated siC cells (values set to 1) (D).
The PML-binding domain of IE1 suppresses IFN-induced expression of specific ISGs.
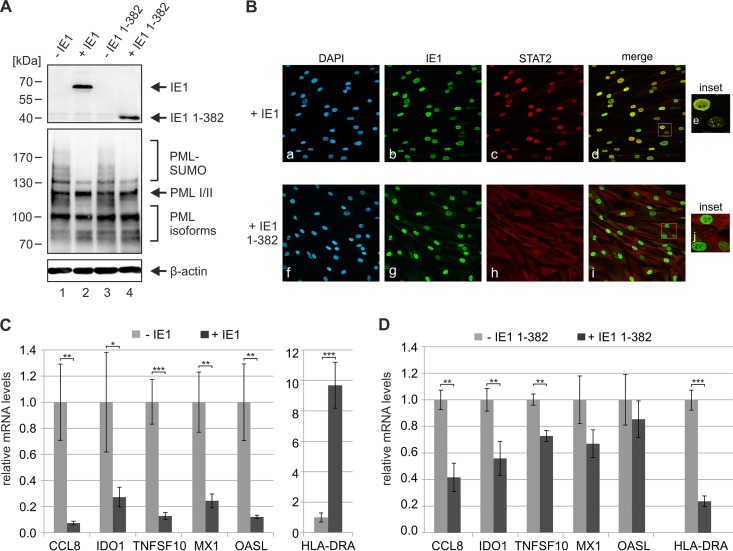
Having shown that PML positively regulates ISG expression, we wanted to further confirm that IE1 influences IFN signaling not only through the interaction with STAT proteins but also through its interaction with PML. To address this issue, we utilized HFF cells with doxycycline-inducible expression of wild-type IE1 or truncated IE1 1-382, which contains the PML-binding domain IE1CORE but lacks the STAT-binding region. Western blot analysis of PML levels in the absence and presence of doxycycline revealed an efficient deSUMOylation of PML by wild-type IE1 and IE1 1-382 under IFN-induced conditions, confirming the expression of active IE1 proteins (Fig. 8A). Furthermore, indirect immunofluorescence experiments were performed in order to investigate the previously described recruitment of STAT2 by IE1 (5, 42). While full-length IE1 exhibited a distinct nuclear colocalization with STAT2 (Fig. 8B, panels a to e), this could not be observed for IE1 1-382, thus confirming that deletion of the C-terminal STAT-binding region abrogates the interaction with STAT2 (Fig. 8B, panels f to j). Next, the newly generated HFFs were stimulated with IFN-β and ISG induction was analyzed in the absence and presence of IE1 proteins. As illustrated in Fig. 8C, wild-type IE1 suppressed induction of type I IFN-responsive genes (Fig. 8C, left panel) but induced expression of HLA-DRA, an ISG usually controlled by IFN-γ (Fig. 8C, right panel). These opposed effects on type I and type II IFN-responsive genes are consistent with results from previous studies and were shown to depend on binding of STAT2 and STAT1, respectively (41–43). Intriguingly, expression of IE1 1-382, which binds PML but is no longer able to interact with STAT proteins, still had an influence on ISG induction. We detected a significant reduction of HLA-DRA mRNA levels (Fig. 8D), while expression of type I IFN-controlled genes was diminished to various degrees (Fig. 8D). To sum up, although the different extents of inhibition suggest complex underlying mechanisms that will require further studies to become fully elucidated, our data provide evidence for an inhibition of ISG induction by the PML interaction domain of IE1. We conclude that IE1 targets PML in order to manipulate intrinsic as well as innate immune responses.
FIG 8.
Inhibition of IFN-induced ISG expression by the PML interaction domain of IE1. (A) Effects of wild-type IE1 and truncated IE1 1-382 on the SUMOylation state of PML under IFN-induced conditions. HFF cells with doxycycline-inducible expression of wild-type IE1 or IE1 1-382 either were left untreated (−IE1, −IE1 1-382) or were treated with doxycycline (+ IE1, + IE1 1-382) for 24 h. Afterwards, the cells were stimulated with IFN-β for 24 h, and cell lysates were analyzed by Western blotting for the expression of IE1 (top panel), endogenous PML variants (middle panel), and β-actin as protein loading control (bottom panel). (B) Effects of wild-type IE1 and truncated IE1 1-382 on the subcellular localization of STAT2 under IFN-induced conditions. HFF cells with doxycycline-inducible expression of wild-type IE1 (upper panels) or IE1 1-382 (lower panels) were treated with doxycycline for 24 h. Afterwards, the cells were stimulated with IFN-β for 24 h and fixed for immunofluorescence staining of endogenous STAT2 and IE1. Cell nuclei were counterstained with DAPI. Insets show magnified cells exhibiting a dot-like intranuclear distribution of IE1. (C, D) Effects of IE1 proteins on IFN-induced ISG expression. HFF cells with doxycycline-inducible expression of wild-type IE1 (C) or IE1 1-382 (D) either were left untreated (light gray bars) or were treated with doxycycline (dark gray bars) for 24 h. Afterwards, the cells were stimulated with IFN-β for 24 h and qRT-PCR was performed to determine transcription of type I IFN-responsive ISGs (CCL-8, IDO1, MX1, TNFSF10, OASL) and a type II IFN-responsive ISG (HLA-DRA). Results were normalized to GAPDH and are shown as mean values with standard deviations from three biological replicates. ISG expression in cells treated with doxycycline is related to the respective untreated cells (values set to 1). Statistical significances were determined using the Student t test and are indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
In common with many DNA viruses, HCMV has evolved multifaceted strategies to overcome intrinsic and innate host defenses. The immediate early protein 1 (IE1) plays a major role in initiation of lytic infection, since it acts as an antagonist of PML-mediated intrinsic immunity as well as IFN-based innate immunity. In the present study, we found that a leucine-to-proline mutation (L174P) in the PML-binding domain IE1CORE affects virus growth almost to the same extent as a deletion of the IE1 gene. Contrary to the general opinion that IE1 is required only under low-MOI conditions, we detected a clear attenuation of early and late gene expression after low- and high-multiplicity infection with IE1-defective AD169 (Fig. 1 and 2). Similar findings were reported by two recent studies on recombinant viruses derived from the laboratory strain Towne and the clinical isolate TB40/E (58, 59), suggesting a greater relevance of IE1 for lytic HCMV infection than inferred from earlier experiments. The profound growth defect of AD169/IE1-L174P can be explained by the low abundance of IE1-L174P during infection. Only at 8 hpi, when transcription of IE1 reaches its peak level (60), were similar protein amounts of IE1 and IE1-L174P detected (Fig. 2B). Due to the high stability of wild-type IE1 (Fig. 3C and D), the protein level remains constant at later times after infection, when the major immediate early promoter (MIEP) is repressed and IE1 transcription is diminished (60). However, abundances of IE1-L174P protein, which has a severely reduced half-life (Fig. 3C and D), rapidly decline during the replication cycle. We found that reduced stability of IE1-L174P results from structural perturbation by helix breaker proline being inserted into IE1CORE (Fig. 4). Recent determination of the crystal structure of IE1CORE by our group revealed an elongated, highly alpha-helical fold that can easily be affected by small deletions and point mutations (6). This is in line with the previous observation that also various deletions within IE1CORE abrogate the functionality of IE1 (34, 37, 40, 61). Until now, the residues mediating the interaction between IE1 and PML have not been identified. However, the fact that IE1CORE shares structural elements with coiled-coil domains of TRIM proteins and targets TRIM family member PML through coiled-coil interactions suggests the involvement of multiple residues along the entire surface of IE1CORE (6, 62–64). Thus, the unusual, elongated structure of IE1CORE may have developed during evolution to accommodate efficient binding of PML.
Our data provide evidence that the association of IE1CORE with PML not only is a prerequisite for antagonizing ND10-mediated immunity but also affects the function of PML in IFN signaling. While initial studies provided controversial results on the role of PML in regulation of MHC-I expression (65–68), more-recent data convincingly demonstrated that PML acts as a positive regulator of IFN-γ-induced MHC-II expression. Attempts to elucidate the mechanistic basis of this activity revealed that PML induces activation and DNA binding of STAT1 immediately after IFN induction, thereby enhancing transcription of genes such as the MHC-II transactivator (CIITA) (44, 45). Furthermore, PML isoform II was shown to directly interact with CIITA, leading to stabilization and prolonged activity of the protein (45). In addition to effects of PML on type II IFN signaling, PML isoform IV was suggested to participate in IFN-β production during vesicular stomatitis virus infection by increasing the stability of interferon regulatory factor 3 (IRF3), indicating that PML is involved in multiple stages of the interferon response. Here, we show that PML not only is required for efficient induction of the MHC-II gene HLA-DRA but also positively regulates the induction of several type I IFN-stimulated genes (Fig. 7). This finding confirms and complements the results of a study by Kim and Ahn on the role of PML in type I interferon response, which was published while this paper was in preparation (59). By using PML knockdown cells, they demonstrated that PML is required for an efficient accumulation of STAT proteins and their phosphorylated forms. Furthermore, they detected an association of PML with STAT1 and STAT2 as well as ISG promoters, suggesting that PML regulates the STAT1/STAT2/IRF9 complex in order to activate ISG transcription. An even more recent report implicated specifically PML isoform II in the activation of ISG expression. They detected an interaction of STAT1 with the unique C terminus of PML II resulting in a recruitment of STAT1 to ISG promoters (69). The impact of other PML isoforms on ISG transcription, however, is still unknown.
Having identified PML as a positive regulator of IFN signaling, we speculated that IE1 may affect this function through its interaction with PML. We observed that expression of an IE1 variant harboring the PML interaction domain IE1CORE but lacking the STAT interaction region significantly reduces induction of HLA-DRA and also exerts effects on some type I IFN-regulated genes (Fig. 8D), to a lesser extent, however, than wild-type IE1 (Fig. 8C). Interestingly, the inhibitory effect of IE1CORE on HLA-DRA gene expression drastically differs from the stimulation observed in the presence of wild-type IE1 (compare Fig. 8C and D). This may be related to the previously published finding that IE1 is able to elicit a type II interferon-like host cell response which strictly depends on STAT1 and thus requires the presence of the C-terminal STAT binding region (43). Of note, stimulation of HLA-DRA was observed only after isolated expression of IE1 but not after infection of primary human fibroblasts with either wild-type AD169 or AD169/IE1 1-382, suggesting major differences concerning the modulation of the interferon response during infection and the isolated expression of viral proteins (compare Fig. 6B and 8C). Thus, these results complement the experiments of Kim and Ahn (59), which analyzed the consequences of deletions within the PML-binding domain IE1CORE or the STAT interaction region on the capacity of IE1 to inhibit type I IFN signaling. They suggested a requirement of both PML binding and STAT binding for an efficient inhibition of the interferon response by IE1 and proposed sequestration of PML/STAT complexes as a mechanistic basis. Although one would expect that this action on PML results in a general reduction of ISG transcription by IE1CORE, we detected effects only on individual ISGs (Fig. 8D). A more global analysis may be required to further characterize the influence of IE1 on PML-based ISG regulation. Moreover, it will be a future challenge to unravel the underlying mechanisms in detail, since this may involve different PML isoforms acting at various steps of the interferon signaling pathway. In this context, it is worth mentioning that, in addition to binding PML, IE1 was found to interact with further members of the TRIM protein family, namely, TRIM5α and TRIM33 (6, 70). Since activation of innate immune signaling pathways is a common feature of this protein family, it is possible that IE1 influences also other TRIM proteins in order to modulate cellular IFN signaling.
In summary, the data of our and other groups clearly demonstrate a coregulatory function of PML in innate immune signaling. Besides HCMV, several other viruses, including herpes-, adeno-, and papillomaviruses were found to affect PML by using diverse strategies such as degradation, relocalization, or deSUMOylation of the cellular protein (14). Thus, targeting PML by effector proteins may represent a common viral strategy to overcome intrinsic as well as innate immune mechanisms.
ACKNOWLEDGMENTS
We thank P. Hemmerich (Jena), M. Mach (Erlangen), K. Osterrieder (Berlin), B. Plachter (Mainz), and H. Will (Hamburg) for providing valuable reagents for this study.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB796 projects A2, A3, and B4) and the Interdisciplinary Center for Clinical Research Erlangen (IZKF Erlangen; projects A62 and J45).
REFERENCES
- 1.Wathen MW, Stinski MF. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol 41:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonough SH, Spector DH. 1983. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology 125:31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- 3.Greaves RF, Mocarski ES. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol 72:366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krauss S, Kaps J, Czech N, Paulus C, Nevels M. 2009. Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J Virol 83:12854–12870. doi: 10.1128/JVI.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherer M, Klingl S, Sevvana M, Otto V, Schilling EM, Stump JD, Muller R, Reuter N, Sticht H, Muller YA, Stamminger T. 2014. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog 10:e1004512. doi: 10.1371/journal.ppat.1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michelson-Fiske S, Horodniceanu F, Guillon JC. 1977. Immediate early antigens in human cytomegalovirus infected cells. Nature 270:615–617. doi: 10.1038/270615a0. [DOI] [PubMed] [Google Scholar]
- 8.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci U S A 93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherer M, Stamminger T. 2014. The human cytomegalovirus IE1 protein: past and present developments. Future Virol 9:415–430. doi: 10.2217/fvl.14.20. [DOI] [Google Scholar]
- 10.Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res 157:128–133. doi: 10.1016/j.virusres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Everett RD. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 12.Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol 11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 14.Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783:2207–2221. doi: 10.1016/j.bbamcr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Tavalai N, Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–1264. doi: 10.3390/v1031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardi R, Pandolfi PP. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 17.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2753. [PubMed] [Google Scholar]
- 19.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. 2006. The mechanisms of PML-nuclear body formation. Mol Cell 24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de Thé H. 2001. Role of promyelocytic leukemia (PML) sumoylation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med 193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol 82:126–137. doi: 10.1128/JVI.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler M, Tavalai N, Muller R, Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J Gen Virol 92:1532–1538. doi: 10.1099/vir.0.030981-0. [DOI] [PubMed] [Google Scholar]
- 24.Glass M, Everett RD. 2013. Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J Virol 87:2174–2185. doi: 10.1128/JVI.02950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J Biol Chem 281:37652–37660. doi: 10.1074/jbc.M604273200. [DOI] [PubMed] [Google Scholar]
- 26.Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol 80:3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukashchuk V, McFarlane S, Everett RD, Preston CM. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J Virol 82:12543–12554. doi: 10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YE, Lee JH, Kim ET, Shin HJ, Gu SY, Seol HS, Ling PD, Lee CH, Ahn JH. 2011. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J Virol 85:11928–11937. doi: 10.1128/JVI.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guldner HH, Szostecki C, Grotzinger T, Will H. 1992. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol 149:4067–4073. [PubMed] [Google Scholar]
- 30.Stadler M, Chelbi-Alix MK, Koken MH, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin MC, Schindler C. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565–2573. [PubMed] [Google Scholar]
- 31.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871–876. [PubMed] [Google Scholar]
- 32.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn JH, Brignole EJ III, Hayward GS. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol 18:4899–4913. doi: 10.1128/MCB.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 36.Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71:4599–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson GW, Kelly C, Sinclair JH, Rickards C. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol 79(Part 5):1233–1245. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- 38.Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Ahn JH, Cheng M, apRhys CM, Chiou CJ, Zong J, Matunis MJ, Hayward GS. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J Virol 75:10683–10695. doi: 10.1128/JVI.75.22.10683-10695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, Hayward GS, Ahn JH. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol 82:10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulus C, Krauss S, Nevels M. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A 103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knoblach T, Grandel B, Seiler J, Nevels M, Paulus C. 2011. Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-gamma. PLoS Pathog 7:e1002016. doi: 10.1371/journal.ppat.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Bougrini J, Dianoux L, Chelbi-Alix MK. 2011. PML positively regulates interferon gamma signaling. Biochimie 93:389–398. doi: 10.1016/j.biochi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Ulbricht T, Alzrigat M, Horch A, Reuter N, von Steimle MA, Schmitt E, Kramer OH, Stamminger T, Hemmerich P. 2012. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J Cell Biol 199:49–63. doi: 10.1083/jcb.201112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Asmi F, Maroui MA, Dutrieux J, Blondel D, Nisole S, Chelbi-Alix MK. 2014. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog 10:e1003975. doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hofmann H, Floss S, Stamminger T. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol 74:2510–2524. doi: 10.1128/JVI.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagenknecht N, Reuter N, Scherer M, Reichel A, Muller R, Stamminger T. 2015. Contribution of the major ND10 proteins PML, hDaxx and Sp100 to the regulation of human cytomegalovirus latency and lytic replication in the monocytic cell line THP-1. Viruses 7:2884–2907. doi: 10.3390/v7062751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreoni M, Faircloth M, Vugler L, Britt WJ. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods 23:157–167. doi: 10.1016/0166-0934(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 52.Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res 6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 53.Waldo FB, Britt WJ, Tomana M, Julian BA, Mestecky J. 1989. Non-specific mesangial staining with antibodies against cytomegalovirus in immunoglobulin-A nephropathy. Lancet i:129–131. [DOI] [PubMed] [Google Scholar]
- 54.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 1 4:33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- 56.Hooft RW, Vriend G, Sander C, Abola EE. 1996. Errors in protein structures. Nature 381:272. [DOI] [PubMed] [Google Scholar]
- 57.Cherrington JM, Mocarski ES. 1989. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol 63:1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zalckvar E, Paulus C, Tillo D, Asbach-Nitzsche A, Lubling Y, Winterling C, Strieder N, Mucke K, Goodrum F, Segal E, Nevels M. 2013. Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc Natl Acad Sci U S A 110:13126–13131. doi: 10.1073/pnas.1305548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim YE, Ahn JH. 2015. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog 11:e1004785. doi: 10.1371/journal.ppat.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamminger T, Puchtler E, Fleckenstein B. 1991. Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J Virol 65:2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee HR, Huh YH, Kim YE, Lee K, Kim S, Ahn JH. 2007. N-terminal determinants of human cytomegalovirus IE1 protein in nuclear targeting and disrupting PML-associated subnuclear structures. Biochem Biophys Res Commun 356:499–504. doi: 10.1016/j.bbrc.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. 2014. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci U S A 111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y, Cheng M, Chen YG, Gao N, Yu H, Wang L, Li W, Yang M. 2014. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res 24:762–765. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldstone DC, Walker PA, Calder LJ, Coombs PJ, Kirkpatrick J, Ball NJ, Hilditch L, Yap MW, Rosenthal PB, Stoye JP, Taylor IA. 2014. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci U S A 111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng P, Guo Y, Niu Q, Levy DE, Dyck JA, Lu S, Sheiman LA, Liu Y. 1998. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature 396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 66.Bruno S, Ghiotto F, Fais F, Fagioli M, Luzi L, Pelicci PG, Grossi CE, Ciccone E. 2003. The PML gene is not involved in the regulation of MHC class I expression in human cell lines. Blood 101:3514–3519. doi: 10.1182/blood-2002-11-3335. [DOI] [PubMed] [Google Scholar]
- 67.Larghero J, Zassadowski F, Rousselot P, Padua RA, Degos L, Chomienne C. 1999. Alteration of the PML proto-oncogene in leukemic cells does not abrogate expression of MHC class I antigens. Leukemia 13:1295–1296. [DOI] [PubMed] [Google Scholar]
- 68.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Wright J, Meng X, Leppard KN. 2015. Promyelocytic leukemia protein isoform II promotes transcription factor recruitment to activate interferon beta and interferon-responsive gene expression. Mol Cell Biol 35:1660–1672. doi: 10.1128/MCB.01478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez FP, Tang Q. 2014. Identification of cellular proteins that interact with human cytomegalovirus immediate-early protein 1 by protein array assay. Viruses 6:89–105. doi: 10.3390/v6010089. [DOI] [PMC free article] [PubMed] [Google Scholar]