Abstract
Short RNAs mediate gene silencing, a process associated with virus resistance, developmental control and heterochromatin formation in eukaryotes1–5. RNA silencing is initiated through Dicer-mediated processing of double-stranded RNA into small interfering RNA (siRNA)6,7. The siRNA guide strand associates with the Argonaute protein in silencing effector complexes, recognizes complementary sequences and targets them for silencing8–11. The PAZ domain is an RNA-binding module found in Argonaute and some Dicer proteins and its structure has been determined in the free state12–14. Here, we report the 2.6Å crystal structure of the PAZ domain from human Argonaute eIF2c1 bound to both ends of a 9-mer siRNA-like duplex. In a sequence-independent manner, PAZ anchors the 2-nucleotide 3′ overhang of the siRNA-like duplex within a highly conserved binding pocket, and secures the duplex by binding the 7-nucleotide phosphodiester backbone of the overhang-containing strand and capping the 5′-terminal residue of the complementary strand. On the basis of the structure and on binding assays, we propose that PAZ might serve as an siRNA-end-binding module for siRNA transfer in the RNA silencing pathway, and as an anchoring site for the 3′ end of guide RNA within silencing effector complexes.
siRNA is a 19–23-base-paired (bp) duplex with 2-nucleotide (nt) 3′ overhangs at both ends, containing 5′-phosphates and 3′-hydroxyls7,15. siRNA is not merely a consequence of Dicer processing, as both the length and ends are important for mediating RNA interference (RNAi)7,10,15, whereby target messenger RNA is sequence-specifically degraded in an siRNA-programmed effector complex termed the RNA-induced silencing complex (RISC)8,9,16. This suggests that specific structural features of siRNA constitute recognition targets for the protein components within the RNAi machinery. The PAZ domain can bind to single-stranded (ss) RNAs and siRNA duplexes12–14, and requires the 2-nt 3′ overhang for efficient complex formation13,14. We have determined a co-crystal structure of PAZ domain and a 9-mer RNA (5′-CGUGACUCU-3′) in order to illustrate the details of PAZ–RNA interaction and gain functional insights into the recognition process.
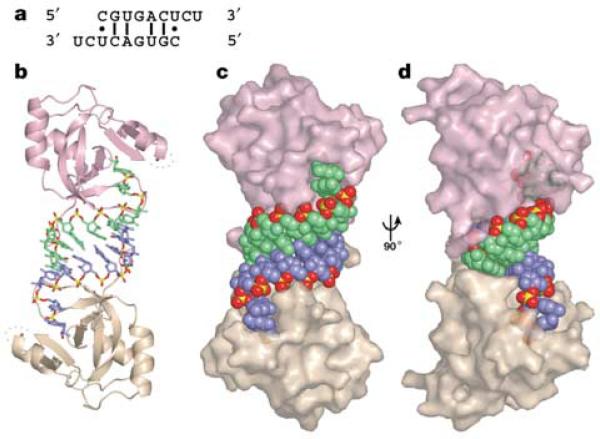
The structure of the PAZ–RNA complex reveals an unanticipated arrangement in which the 9-mer RNA, initially designed as a ssRNA ligand, forms a self-complementary siRNA-like A-form duplex, which is bound by the PAZ domain at each end (Fig. 1). The 3′-shifted pairing of each strand results in 2-nt, single-stranded 3′ overhangs at the duplex ends, which are characteristic of siRNA architecture. However, the short 7-bp duplex is unstable, due to the presence of three non-canonical pairs (Fig. 1a), raising the concern that the observed duplex formation could result from crystal-packing interactions. This is not the case, as the PAZ domain and the 9-mer RNA form a stable 2:2 complex in solution, as judged by gel filtration (Supplementary Fig. S1). Thus, PAZ binding apparently stabilizes siRNA-like duplex formation. Moreover, the absence of contacts between the two PAZ domains in the complex indicates independent binding of PAZ domains to the siRNA-like duplex, and suggests that such an arrangement should also hold for complex formation to the ends of a typical 21-nt siRNA.
Figure 1.
Overview of the PAZ–siRNA-like duplex structure a, The self-complementary siRNA-like duplex formed in the crystal. b, The entire complex of two PAZ domains bound to each end of an siRNA-like duplex. Protein and RNA are presented in ribbon and stick representations, respectively. c, The same view as b, but protein and RNA are presented in semi-transparent surface- and space-filling representations, respectively. d, 90° rotation of c. Note that PAZ domains predominantly contact the 3′-overhang-containing strands and dock with the 5′ ends of the complementary strands. The 2-nt 3′ overhangs are anchored within the pockets, with their base edges facing outwards towards solvent. The PAZ domains are coloured pink and beige and the RNA strands are blue and green, except for the phosphate groups of the RNA strands, in which phosphate is yellow and oxygen is red.
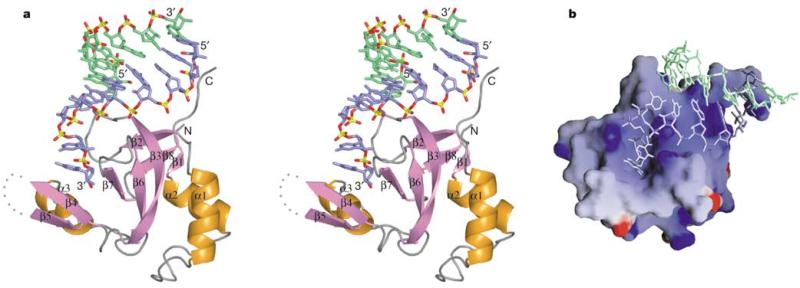
The PAZ domain in the complex adopts a heart-shaped globular topology (Fig. 2a), with a twisted β-barrel consisting of six β-strands (β1–β3, β6–β8), capped by two amino-terminal α-helices (α1, α2) on one side and connected to αβ module (β4–β5–α3) on the other side. The two strands of the siRNA-like duplex interact with a specific PAZ domain in a highly asymmetric manner. The strand bound with its 3′ end contacts the PAZ domain along its full 9-nt length, whereas the complementary strand makes contacts only with the 5′-terminal residue. The first 7 nucleotides constituting the RNA duplex structure, proceeding in the 5′ to 3′ direction, interact with the protein's carboxy-terminal tail and the positively charged surface formed by the strands β2, β3 and the β6–β7 loop (Fig. 2b). The bound RNA adopts a stacked 5′ to 3′ helical trajectory, except for a sharp turn (clockwise rotation of ~110° along the helical axis) in the phosphodiester backbone between the duplex and 2-nt 3′-overhang segments, thereby inserting the 2-nt ends into a central protein pocket formed between the barrel and αβ module. The same central RNA-binding pocket has been proposed previously from an evaluation of NMR chemical shift perturbations, mutagenesis and sequence conservation analysis12–14. The free12–14 and RNA-bound PAZ structures superimpose well around the RNA-binding pocket (Supplementary Fig. S2), indicating that the pocket is largely preformed.
Figure 2.
The PAZ–siRNA-like duplex subcomplex. a, Stereo view showing one PAZ domain interacting with an siRNA-like end. Relative to this PAZ domain, the RNA strand bound by the 3′ end is coloured blue, whereas the strand bound by the 5′-end is coloured green. The protein secondary structure elements are labelled. b, Electrostatic surface of PAZ, with blue and red colours corresponding to positively and negatively charged patches, respectively.
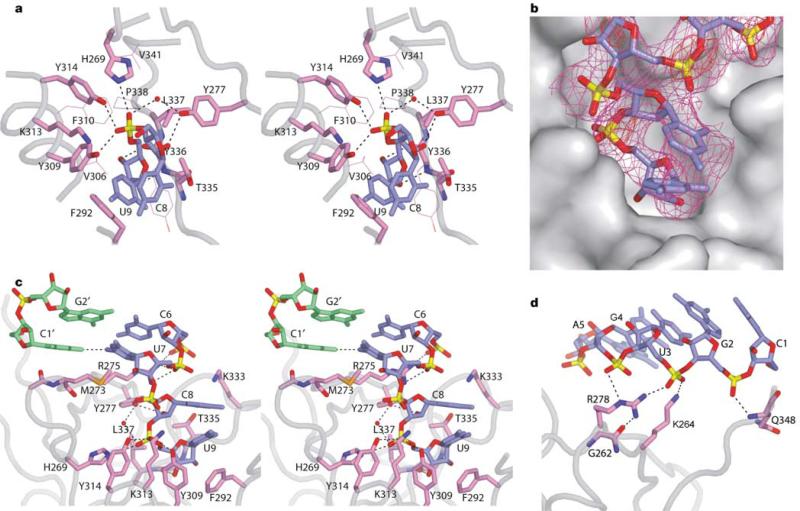
The elongated pocket, which is lined by a large number of aromatic and hydrophobic residues, is generated by helix α3 and strand β4 along one face, and strand β7 and loop β2–β3 along the opposite face (Fig. 2a). The two nucleotides at the 3′ end retain a stacked A-form conformation (Fig. 3a), with the 3′-terminal residue buried deep within the pocket and targeted by numerous protein interactions (Fig. 3b). The non-bridging oxygens of the phosphodiester backbone linking the last two residues make four hydrogen bonds with Y309, Y314 and H269 and a water molecule stabilized by Y277. The sugar ring of the terminal residue is further anchored in place by van der Waals packing against L337 and T335, whereas its 2′- and 3′-hydroxyl groups form hydrogen bonds to the backbone amide and carbonyl groups of Y336, respectively. In addition, portions of the sugar ring and base of the terminal nucleotide are stacked over aromatic residue F292.
Figure 3.
The details of PAZ–siRNA-like duplex interaction. a, The 2-nt 3′-overhang segment positioned within the conserved pocket. The residues directly contacting RNA are shown in a stick representation, whereas other resides contributing to the hydrophobic pocket are shown as thin lines. b, The RNA in stick representation positioned in the PAZ pocket in a surface representation. Also shown for RNA is the experimental density-modified electron density, contoured at 1.0σ (magenta) and 4.5σ (red). The contour of highest level is strongly indicative of the phosphate groups. c, Interaction of PAZ with the duplex terminus and 3′-overhang segments, viewed from the left side of a. Note the change in the trajectory of RNA phosphodiester backbone between duplex and overhang segments. d, Interactions involving the RNA duplex segment. Potential hydrogen-bonding interactions are shown by dashed lines. a and c are shown in stereo view. Phosphates are coloured in yellow, oxygens in red, nitrogens in blue, protein side chains in violet, the 3′-end-bound RNA strand in blue and the 5′-end-bound RNA strand in green.
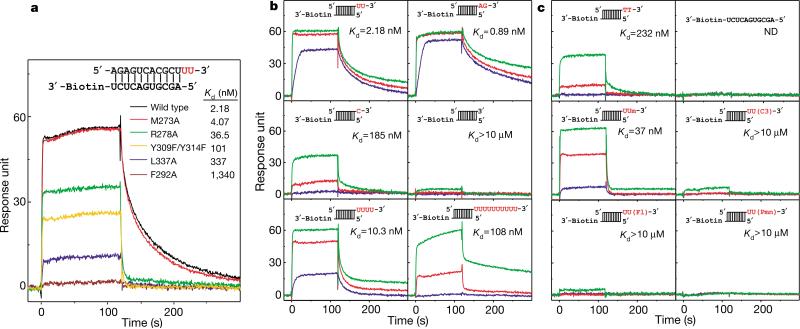
Among residues recognizing the 3′-terminal nucleotide, F292, Y309 and L337 are invariant, and H269, Y277, K313 and Y314 are conserved among PAZ homologues (Supplementary Fig. S3). These residues seem to be critical for efficient binding, as single mutations F292A and L337A cause a 615-fold and 155-fold reduction in binding affinity, and double mutation Y309F/Y314F causes a 46-fold reduction (Fig. 4a). Other conserved residues surrounding the pocket, such as V306, F310, P338 and V341, although not directly contacting the RNA, probably maintain the hydrophobic environment of the pocket. The strong binding of the 3′-terminal nucleotide, as reflected in numerous intermolecular contacts, explains the previous observation that single nucleotide uridine 5′ monophosphate (UMP) can also bind in the same pocket12. By contrast, the penultimate nucleotide from the 3′ end makes only minimal contacts with PAZ—that is, a hydrogen bond between its 2′-hydroxyl group and Y277, and an electrostatic interaction with K313—and is largely exposed to solvent.
Figure 4.
RNA-binding assays monitored using surface plasmon resonance. a, Representative binding curves for wild-type and mutant PAZ domains (100 nM) and a UU 3′-overhang-containing RNA duplex. b, c, Representative binding curves for wild-type PAZ domain at concentrations of 10 nM (blue), 100 nM (red) and 1000 nM (green), and various RNA ligands. RNA ligands contain a common 11-bp duplex, shown explicitly in a and schematically in b and c, which is biotin-labelled at the blunt end and contains either different 3′ extensions (b) or 3′-end modifications (c) at the other end (highlighted in red). Um, 2′-o-methyl; C3, 3′ -o-propanediol; Fl, 3′ -o-fluorescein; Pmn, 3′ -puromycin. The association occurs during the first 120 s, and is followed by disassociation during the next 180 s. The deduced apparent K d values are also listed.
The base edges of the 2-nt 3′-overhang residues are facing outwards towards solvent and have no contact with the protein. The absence of specific hydrogen-bonding recognition of base edges as well as the available space along the base edges suggest that the pocket could accommodate 3′-overhang segments containing all nucleotide combinations. This feature is consistent with the potential function of PAZ, which is to recognize siRNAs of random sequence.
The PAZ domain laterally secures the siRNA-like duplex through hydrogen-bond and electrostatic interactions between five phosphodiester groups of the strand bound at the 3′ end, and the basic residues R275, K333, R278, K264 and the C-terminal residues (Fig. 3c, d). The side chain of R278, which is buttressed in place by G262, forms two hydrogen bonds with adjacent phosphodiester groups (Fig. 3d). The importance of this recognition is manifested in the 17-fold loss in binding affinity observed for the R278A mutant (Fig. 4a).
No specific interactions are directed to the 2′-OH groups, which line the minor groove of the RNA helix, consistent with RNAi not generally being affected by 2′-OH group modifications within the siRNA duplex segment17. Nevertheless, the observed contacts between the PAZ domain and adjacent non-bridging phosphate oxygens (Fig. 3d) may provide a mechanism for discriminating the interphosphate separations, which are different between RNA and DNA helices. The PAZ domain also does not contact base edges within either groove of the RNA helix, indicative of non-sequence-specific molecular recognition.
The 5′ end of the complementary strand is capped by loop β2–β3 centred about residue M273 (Fig. 3c). This docking constitutes the only interaction involving the complementary strand, burying 100Å2 of the protein surface, in contrast to 900Å2 of total buried protein surface, upon complex formation. The aliphatic side chain of M273 extends towards the U7 base (Fig. 3c), but seems not to contribute to RNA binding, as no loss of binding was observed for mutant M273A (Fig. 4a). Furthermore, the PAZ domain does not contact the 5′-OH (and presumably 5′-phosphate, were it to be present) of the complementary strand.
To define more accurately the RNA structural determinants for PAZ recognition, we measured apparent dissociation constants (Kd) for RNA ligands with variant 3′ ends, using surface plasmon resonance (Fig. 4b). RNAs containing both UU and AG 3′ overhangs bind PAZ with the highest affinities (Kd values of 0.89–2.18 nM) among all tested constructs (Fig. 4b). The overhang contributes critically to the binding, as removal of one or two 3′-nucleotides reduces the binding affinity by 85- or >5,000-fold, consistent with the crystal structure of the complex, in which the PAZ domain optimally fits the duplex end with 2-nt 3′ overhang in a sequence-independent manner.
In addition, the pocket favours the 2-nt overhang as ribonucleotides, because replacement with deoxyribonucleotides, which eliminates the two hydrogen bonds to the 2′-OH groups of the overhang segment, reduces the binding by 100-fold (Fig. 4c). The pocket is incompatible with bulky 3′-end modifications, such as propanediol, fluorescein and puromycin, which decrease affinity by >104-fold (Fig. 4c), but is fairly tolerant towards accommodation of a smaller 2′-o-methyl group at the terminal nucleotide (Fig. 4c).
The PAZ domain does not offer a strong binding site for the RNA 5′ end, because elongation of the 3′ overhang to 4 nt, which would disrupt the protein interaction with the 5′-terminal residue of the complementary strand, only slightly decreases the binding affinity by 5-fold (Fig. 4b). No binding occurs for ssRNA with a free 5′ end (Fig. 4c), reinforcing this conclusion.
The PAZ domain is able to bind with the 10-nt overhang, in essence an ssRNAwith a free 3′ end, with a Kd of ~100 nM (Fig. 4b). Because of the predominant interaction of PAZ with the 3′-end-bound strand, the ssRNA might conceivably bind the pocket of PAZ by its 3′ end and adopt an extended conformation along the β-barrel surface. The 50-fold reduction in affinity for ssRNA (10-nt overhang), relative to the siRNA end (2-nt overhang), may result from the lack of interaction with the 5′-terminal residue of the complementary strand and the increased flexibility of ssRNA due to the absence of duplex pairing. Together, the structure of the complex and the binding assays establish that the PAZ domain binds to the 3′ end of RNA, requiring an essential 2-nt, single-stranded segment, and has a preference for the remaining RNA in the duplex over the single-stranded form.
The PAZ domain adopts a topology similar to the oligonucleotide/oligosaccharide-binding (OB) fold, despite different connections linking secondary structure elements13,14. The 2-nt overhangs are anchored within a pocket unique to the PAZ domain, with the remainder of the RNA positioned along a surface formed by the β2–β3 strands. This contrasts with OB fold RNA recognition, where the RNA targets the opposite face of the β-barrel18, corresponding to β6–β7 strands of the PAZ domain.
The conservation among PAZ homologues, especially for the RNA-binding residues (Supplementary Fig. S3), predicts that all PAZ family members should similarly recognize siRNA ends, satisfying their exclusive distribution in the RNAi-related proteins Dicer and Argonaute. As an siRNA binder, Dicer–PAZ could hold the product siRNA at one end, after dsRNA cleavage, and transfer the other siRNA end to the PAZ domain of Argonaute in the RISC. Such a transfer would lead to a direct loading of siRNA into RISC, consistent with a previous observation that the activities of Dicer and RISC are coupled through transfer of siRNA7. Because some Dicer proteins, such as Drosophila Dicer-2, do not contain a PAZ domain, alternative pathways are likely to exist, that mediate transfer of siRNA from Dicer to RISC. Drosophila R2D2, which binds to Dicer-2, has been proposed to facilitate loading of siRNA into RISC19.
In view of the predominant interaction of PAZ with one strand of siRNA, and the associated residual binding to ssRNA, the PAZ domain might continue to secure the guide strand following siRNA unwinding, and present it for scanning and pairing with the target sequence in the activated RISC.
PAZ–siRNA recognition seems to provide the molecular basis for previous observations that RNAi efficiency was reduced when the 3′ overhangs of synthetic siRNAs deviated from 2 nt in length7,15, or when they were substituted by deoxyribonucleotide residues20, or chemically modified at the 3′-OH position21. The PAZ domain represents the best candidate to sense these 3′-overhang modifications, which would result in reduced binding as shown experimentally in Fig. 4b, c, and consequently might impair the PAZ-mediated recruitment of siRNA into the RISC. By contrast, many other studies have reported that 3′ modifications of siRNA11,22–24 and commonly used deoxynucleotide-substituted 3′ overhangs15 do not interfere with RNAi, thereby questioning the importance of the PAZ–siRNA interaction in the RNAi pathway. One explanation is that PAZ may not be the only siRNA recognition domain mediating RNAi, with its contribution potentially compensated for by alternative high-affinity binding events during siRNA recruitment. Blunt-end siRNA can induce RNAi in some cases15,25, probably by also by-passing PAZ–siRNA recognition. In addition, the nuclease stability of chemically modified siRNAs may partially compensate for impaired PAZ recognition. Further studies are needed to understand the full diversity of siRNA recognition events in the RNA silencing pathway.
Methods
Protein and RNA preparation
The PAZ domain (residues 225–369) from human eIF2C1 was cloned into pET15b vector with a thrombin-cleavable N-terminal His tag. Escherichia coli strain BL21-Gold(DE3) harbouring the expression plasmid was induced by 1 mM isopropyl-β-d-thiogalactopyranoside and further grown overnight at 25 °C. After sonication-based breakage, the clarified cell lysate was mixed with 0.1% polyethylenimine to precipitate nucleic acid. The PAZ domain was purified by Ni-chelating and heparin Hitrap column, followed by His-tag removal with thrombin, and additional purification using Superdex 75 size-exclusion chromatography. Selenomethionine-labelled protein was cultured by inhibiting the methionine synthesis pathway. Site-directed mutations were made by a two-step PCR-based overlap extension method and confirmed by DNA sequencing. All proteins used in binding assays were purified without the removal of His tag and the use of a final size-exclusion column. RNAs and their 3′ -modified analogues were chemically synthesized, deprotected and purified by denaturing polyacryamide gel electrophoresis, or purchased from Dharmacon.
Crystallization and structure determination
Crystals were grown by the hanging-drop vapour diffusion method at 20 °C. The initial drop consisted of 1 μl of 10 mg ml−1 PAZ and 5′ -CGUGACUCU-3′ 9-mer RNA, 100 mM KCl, 5 mM HEPES-KOH, pH 7.6, 10 mM dithiothreitol (DTT) and 1 μl well solution of 20% PEG 1000, 50 mM HEPES-KOH, pH 7.6. Crystals were briefly cyroprotected in 15% PEG 1000 and 15% (w/v) sucrose (or 20% glycerol) before freezing in liquid nitrogen.
The diffraction data sets were collected at the Advanced Photon Source and processed using the HKL package26. The crystals belong to the space group P64, with unit cell parameters of a = b = 99.8Å, c = 34.9 Å, and contain 51% solvent. The asymmetric unit contains one-half of the complex (one PAZ monomer and one 9-mer ssRNA) with the crystallographic dyad coinciding with the dimeric axis in the complex.
The structure was solved by single anomalous diffraction (SAD) implemented in CNS27 for the complex of selenomethionine-labelled PAZ and 9-mer RNA. After density modification by solvent flipping, the map calculated to 3.0Å showed clear electron density for both protein and RNA in the complex (Fig. 3b). The model was built using the O program28. Nevertheless, refinement was primarily undertaken against a better data set associated with a complex of PAZ and 9-mer RNA with the 2-nt, 3′ -ribonucleotide overhang substituted by its deoxy counterpart. The refined PAZ–RNA/DNA complex structure, following conversion of the 2-nt 3′ overhangs from deoxyribo to ribo residues, was used as the starting model for the refinement of the all-RNA structure. Both complex structures are essentially identical except for the conformation of the sugar puckers of the 2-nt 3′ overhangs, which shift from 2′ -endo pucker in deoxyribonucleotides to 3′ -endo pucker in ribonucleotides. The conformation of sugar puckers was not constrained during refinement. The protein residues exhibit good stereochemistry with 85% located in the most favourable region, whereas the remaining fall in additional allowed regions of the Ramachandran plot. Detailed crystallographic statistics can be found in Supplementary Table 1. The symmetry-related guanines in the centre of the RNA duplex are tilted relative to each other, and fit well into the electron density. The final structures contain PAZ residues 225–295 and 302–349, 9 nucleotides and 15/16 water molecules. The figures were prepared with PyMOL (http://pymol.sourceforge.net/) and GRASP29.
Biacore-based measurements of dissociation constants
Binding experiments30 were performed on BIAcore 2000 biosensor (BIAcore) at 25 °C. The RNA constructs were designed to consist of an 11-bp duplex, with a biotin attached at one 3′ end (blunt end) and different length 3′ extensions at the opposite end. Our choice was based on the anticipation that PAZ would not bind to the blunt end and the short length of the duplex would minimize nonspecific complex formation. The 3′ biotinylated 11-mer RNA (5′ -AGCGUGACUCU-biotin-3′) was first immobilized on a streptavidin-coated biosensor chip. The duplex was formed by injecting 500 nM of specific complementary strands in 1 M NaCl, followed by washing away unbound RNAs. The association was monitored over a 2-min period during which wild-type or mutant proteins, at concentrations ranging from 1 nM to 5 μM and in a buffer of 0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% Surfactant P20 and 1 mM DTT, were flown over immobilized RNAs at a rate of 30 μl min−1, and dissociation monitored by flowing the blank buffer over a subsequent 3-min period. The residual bound protein was removed by injecting 20 μl of 2 M NaCl at 20 μl min−1. Experiments at each concentration of proteins were repeated at least twice. The apparent Kd values were derived from the global fit of the association and disassociation curves to a simple 1:1 Langmuir interaction model with a correction for mass transport effects using BIAevaluation 3.0 software.
Supplementary Material
Acknowledgements
We thank K. Saigo for providing us with the eIF2C1 complementary DNA clone. This research was supported by the NIH. We thank Y. Cheng and personnel at the Advanced Photon Source (APS) beamlines 19BM and 14IDB for help in collecting the X-ray diffraction data. Use of the APS beamline was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Denli AM, Hannon GJ. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 2003;28:196–201. doi: 10.1016/S0968-0004(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 4.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 5.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 9.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 10.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 11.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 12.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 13.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 14.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 17.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 20.Hohjoh H. RNA interference (RNAi) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 2002;521:195–199. doi: 10.1016/s0014-5793(02)02860-0. [DOI] [PubMed] [Google Scholar]
- 21.Harborth J, et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 22.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 23.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czauderna F, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Jones T, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;227:174–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 30.Katsamba PS, Park S, Laird-Offringa IA. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.