Summary
A complex relationship between the microbiota and the host emerges early at birth and continues throughout life. The microbiota includes the prokaryotes, viruses and eukaryotes living among us, all of which interact to different extents with various organs and tissues in the body, including the immune system. Although the microbiota is most dense in the lower intestine, its influence on host immunity extends beyond the gastrointestinal tract. These interactions with the immune system operate through the actions of various microbial structures and metabolites, with outcomes ranging from beneficial to deleterious for the host. These differential outcomes are dictated by host factors, environment, and the type of microbes or products present in a specific ecosystem. It is also becoming clear that the microbes are in turn affected and respond to the host immune system. Disruption of this complex dialogue between host and microbiota can lead to immune pathologies such as inflammatory bowel diseases, diabetes and obesity. This review will discuss recent advances regarding the ways in which the host immune system and microbiota interact and communicate with one another.
Keywords: host–microbe interactions, immunology, microbiota
Abbreviations
- AIEC
adherent invasive Escherichia coli
- AhR
aryl hydrocarbon receptor
- DC
dendritic cell
- DSS
dextran sulphate sodium
- GF
germ‐free
- GPRs
G protein‐coupled receptors
- IBD
inflammatory bowel diseases
- IECs
intestinal epithelial cells
- IELs
intraepithelial lymphocytes
- IL‐17
interleukin‐17
- ILCs
innate lymphoid cells
- ILC3
group 3 innate lymphoid cell
- iNKT
invariant natural killer T cell
- MHCII
major histocompatibility complex class II
- MyD88
myeloid differentiation primary response protein 88
- NLRs
nod‐like receptors
- NLRP3, NLR family
pyrin domain containing 3
- OMVs
outer membrane vesicles
- PRRs
pattern recognition receptors
- PSA
polysaccharide A from Bacteroides fragilis
- SCFAs
short chain fatty acids
- SFB
segmented filamentous bacteria
- Th17
T helper 17 lymphocyte
- TIGIT
T cell Immunoreceptor with Ig and ITIM domains
- TLRs
Toll‐like receptors
- TRAF6
tumour necrosis factor receptor associated factor 6
- Treg
T regulatory lymphocyte
- TRIF
Toll–interleukin receptor domain‐containing adaptor protein inducing interferon‐β
Introduction
The microbiota refers to the population of microbes (prokaryotes, viruses and eukaryotes) living among us, outnumbering host cells by a factor of 10.1, 2 The host immune system encompasses both the innate and adaptive immune systems, which work together to determine the class of microbial threat and direct the type and degree of immune response to the exposure.3 The immune system and microbiota develop and mature together, beginning at birth, or even potentially in the womb.4 This early coexistence is probably essential in shaping the immune system response to avoid unwanted immune reactions to intestinal microbial components. An inappropriate response to indigenous bacteria could have deleterious consequences for the host as seen with inflammatory bowel diseases (IBD).
The importance of the microbiota in shaping host immunity is best appreciated in germ‐free (GF) models. Germ‐free housing conditions maintain a microorganism‐free environment and are a powerful system within which to dissect various aspects of host–microbe interactions. Germ‐free mice display an ‘underdeveloped’ innate and adaptive immune system: reduced expression of antimicrobial peptides, reduced IgA production, fewer T‐cell types and increased susceptibility to microbial infections.5 The deficits of GF mice highlight the key role of microbes in bringing the immune system into a ‘combat ready’ mode. Studies comparing monozygotic and dizygotic twins suggest that non‐heritable influences from the environment, including the microbiota, determine much of the immune variation seen in humans.6 Alterations in the microbiota, referred to as dysbiosis, have been implicated as risk factors for IBD, cancers, multiple sclerosis, asthma and type I diabetes; reinforcing the impact of the microbiota on host health. Diet also has profound effects on microbiota composition and metabolite production, both of which influence host immunity but this element will not be discussed here (for reviews, see refs 5,7). This review will focus on recent advances in understanding how microbes and microbial components interact with host immunity and how these interactions influence host health.
Broad influence of microbiota on host immunity
Microbiota: intestinal effects
The establishment of a mature microbiota is a dynamic process during the first 2 years of life5 and coincides with the development of the immune system. Throughout the early developmental period innate immune components play key roles in protecting the infant from pathogens and shaping microbiota assembly. IgA is found in breast milk and can prevent immune activation in infants by binding microbial antigens. Similarly, secretory IgA produced along the intestinal tract continues to be important for maintaining mucosal homeostasis through adulthood.8 The development of the mature microbiota is regulated by host immune system components, which can also be influenced by members of the microbiota. Recent work with gnotobiotic mice suggests that proteobacteria, the dominant phylum in newborns, triggers a proteobacteria‐specific IgA response in mice that plays a key role in controlling proteobacteria levels in the adult microbiota.9 Faecal IgA levels (low versus high) are partly controlled by members of the microbiota; a phenotype that is vertically transmissible and independent of host genetic factors.10 16S rRNA sequencing revealed that Sutterella species are partly responsible for variable IgA levels, most probably by degrading IgA secretory component10 (Fig. 1). Noteably, expansion of proteobacteria/Enterobacteriaceae abundance is observed in patients with IBD and in pre‐clinical models.11 Whether this bloom of microorganisms is related to microbe‐mediated faecal IgA levels is unknown.
Figure 1.
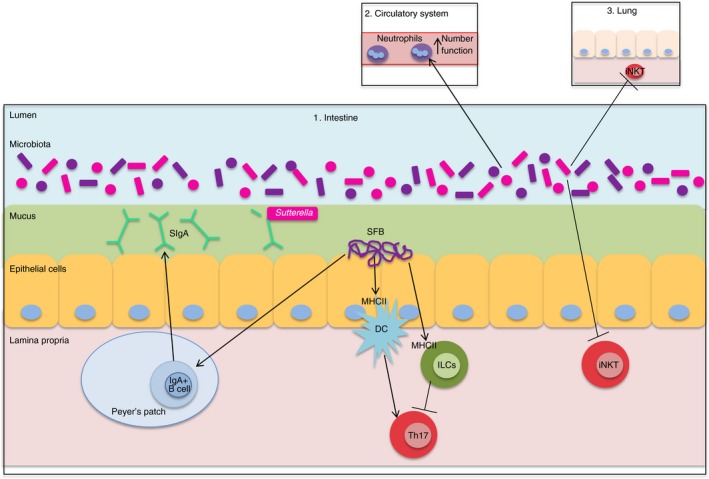
The microbiota affects local and systemic immunity. The intestine (1) contains the greatest number and diversity of microbiota members. Proteobacteria, specifically Sutterella, alter faecal IgA levels, likely through degradation of SIgA. SFB also alter IgA levels by promoting the expansion of germinal centres and inducing IgA‐secreting cells in Peyer's patches, isolated lymphoid follicles, and tertiary lymphoid tissue. MHCII‐dependent SFB antigen presentation on intestinal DCs induces Th17 cell differentiation, while MHCII‐dependent SFB antigen presentation by ILCs constrains Th17 cell differentiation. The intestinal microbiota also influences systemic immunity, including the number and function of circulating neutrophils (2) as well as constraining iNKT levels in the lung (3) and colon (1). DC, dendritic cell; ILCs, innate lymphoid cells; iNKT, invariant natural killer T cell; SFB, segmented filamentous bacteria; SIgA, secretory immunoglobulin A; Th17, T helper 17 lymphocyte; Treg, T regulatory lymphocyte.
The microbiota continues to affect immune function well after development. Studies of Paneth cells using organoids generated from mice reveal that degranulation (release of antimicrobial products) is controlled by immune‐cell‐derived interferon‐γ, which may be induced in vivo during viral or bacterial challenge.12 Thymic and induced T regulatory (Treg) lymphocytes prevent autoimmunity and maintain tolerance to the microbiota, and a recent study suggests that most colonic Treg cells are thymic Treg cells that recognize bacterial antigens, including antigens from Clostridiales, Bacteroides and Lactobacillus. Importantly, antibiotic‐induced alterations in the microbiota, which decrease Clostridiales members among others, reduce intestinal Treg cells and alter colonic thymic Treg T‐cell receptor repertoire, suggesting that microbial composition influences the dynamic response of Treg cells.13
One of the most studied immunomodulatory microbes are segmented filamentous bacteria (SFB), which colonize the terminal ileum in mice, induce IgA production and increase effector T cells, particularly T helper type 17 (Th17) cells.1 Work from several groups suggests that SFB induction of Th17 cells occurs in the intestinal lamina propria rather than in Peyer's patches or the mesenteric lymph nodes.14, 15, 16 Further studies revealed that MHC class II (MHCII) ‐dependent antigen presentation by intestinal dendritic cells (DCs) is essential for SFB‐induced Th17 cells.15, 16 Additionally, Goto et al. provide evidence that MHCII presentation by innate lymphoid cells (ILCs) may constrain Th17 cell differentiation.15 The SFB also stimulate expansion of germinal centres and induce IgA‐secreting cells in Peyer's patches, isolated lymphoid follicles and tertiary lymphoid tissue.14 Recently, Schnupf et al. cultured SFB in vitro and provided evidence that SFB attachment in vivo is required to elicit ileal epithelial responses.17 Future studies examining the specific structural component(s) of SFB responsible for stimulating IgA production and inducing Th17 cells could be aided by the in vitro culture system. Although SFB have not yet been isolated from the human gastrointestinal tract, human cell lines support SFB growth17 and SFB‐specific 16S rRNA has been detected within human stool samples.18 Gram‐stained human ileal–caecal biopsies from a small set of patients with IBD and non‐inflamed patients suggest that SFB is present in patients with ulcerative colitis but absent in those with Crohn's disease.19 Hence, future studies may reveal a role for SFB or SFB‐related bacteria in human immune development and IBD.
In addition to interactions with the immune system, microbes interact with other microorganisms. Although many symbionts have beneficial immune properties, some bacteria facilitate host infection by viruses. For example, human norovirus probably binds to histo‐blood group antigen‐expressing bacteria such as Enterobacter cloacae, which promotes attachment and infection of B cells.20 Conversely, treating mice with the bacterial product, flagellin, prevents rotavirus infection by modulating host innate immune signalling.21 Hence, microbiota composition can promote or inhibit viral infection depending on the type of virus.
Microbiota effects on extra‐intestinal immunity
The effects of the microbiota on host immunity extend beyond the intestine. Germ‐free zebrafish have fewer and less active neutrophils compared with zebrafish colonized with a normal microbiota, as well as impaired neutrophil recruitment in a tail‐fin injury model; a phenomenon linked to microbial induction of serum amyloid A.22 Neonatal mice that are GF or born from antibiotic‐treated dams have fewer circulating and bone marrow neutrophils and are more susceptible to Escherichia coli K1 and Klebsiella pneumoniae sepsis, probably through microbiota induction of granulocytosis.23 Hence, the microbiota contributes to neutrophil development, homeostasis and function in both mice and zebrafish.22, 23 The GF mice have increased invariant natural killer T (iNKT) cells in the lung and colon due to enhanced CXCL16 expression, making them more susceptible to an ovalbumin‐driven model of allergic asthma.24 These studies also suggest that early exposure to microbes is important, as the iNKT cell levels returned to low levels when GF mice were exposed to specific pathogen‐free conditions upon birth but not as adults.24 Consequently, the intestinal microbiota has both local and systemic effects on innate and adaptive immunity (Fig. 1).
Although the majority of the microbiota resides within the intestine, the microbial communities located in extra‐intestinal regions also influence local host immunity.25 Studies comparing GF mice to conventionally raised mice suggest that the skin microbiota regulates expression of complement component C5a receptor, which regulates innate immune defence genes, thereby impacting microbiota diversity and composition.26 Certain skin microbiota community members, particularly Staphylococcus epidermidis interactions with CD103+ DCs, can induce CD8+ T‐cell migration to the epidermis, which enhances barrier function and limits epicutaneous Candida albicans infection through induction of interleukin‐17 (IL‐17).27 In a skin wound healing model, wound closure rate was restored in GF mice conventionalized with microbiota, a phenomenon associated with increased neutrophil accumulation and lower macrophage infiltration into the injured region.28 Microbial dysbiosis has also been implicated in extraintestinal diseases, such as increased Staphylococcus aureus, which is associated with inflammatory skin conditions.29 Hence, microbes occupying extra‐intestinal niches also influence host immunity, although the specific organisms and mechanisms responsible for these responses (Table 1) can differ between regions.
Table 1.
Examples of specific bacteria that modulate the host immune system
| Bacteria | Immunomodulatory effect | Mechanism | References |
|---|---|---|---|
| AIEC | Induce inflammatory cytokines in vitro | Activation of NLRP3 in macrophages, inducing IL‐1β production | (55) |
| Bacteroides fragilis | Influence Treg cell development and homeostasis Influence iNKT cell homeostasis | Bacterial PSA/OMVs containing PSA interactions with TLR2 on pDCs | (32–34, 36, 42, 56) |
| Bacterial sphingolipids modulate iNKT development and activation | (37,38) | ||
| Bacteroides thetaiotaomicron | Promote intracellular calcium signaling, nutritional benefits in IECs Promote colitis in CD4‐dnTgfb2;IL10rb −/− mice | Bacterial OMVs containing inositol phosphatase | (40) |
| Bacterial OMVs containing sulphatase, degrade mucin glycans | (35) | ||
| Clostridium scindens | Inhibit Clostridium difficile infection | Bacterial secondary bile acid synthesis | (64) |
| ETBF | Induce Th17 cells, enhance tumorigenesis in mouse models of CRC | Bacterial toxin‐dependent OMVs induce host IECs to secrete sphingolipids | (56) |
| Lactobacillus plantarum | Alter distribution of pro‐ and anti‐inflammatory T‐cell and DC populations | Bacterial teichoic acid d‐alanylation signalling through TLR2 | (45) |
| Proteus mirabilis | Induce intestinal inflammation in DSS model | Dependent on bacterial haemolysin; activation of NLRP3 inflammasome, inducing IL‐1β production | (30,44) |
| SFB | Induce IgA and Th17 | Stimulation of germinal centres and induction of IgA‐secreting cells; MHCII presentation by DCs and ILCs | (14–16) |
| Staphylococcus epidermidis | Induce CD8+ T cells to the epidermis, enhance barrier function | Interactions with CD103+ DCs | (27) |
| Sutterella species | Alter faecal IgA levels | Degradation of the secretory component of IgA | (10) |
AIEC, adherent invasive Escherichia coli; DCs, dendritic cells; ETBF, enterotoxigenic Bacteroides fragilis; IECs, intestinal epithelial cells; IgA, immunoglobulin A; ILCs, innate lymphoid cells; iNKT, invariant natural killer T cell; OMVs, outer membrane vesicles; pDCs, plasmacytoid dendritic cells; PSA, polysaccharide A; SFB, segmented filamentous bacteria; Th17, T helper 17.
Bacterial components that effect innate and adaptive immunity
Structures detected by pattern recognition receptors
The innate immune system detects microbial components or products through several different families of pattern recognition receptors (PRRs), found on numerous cell types including macrophages, DCs and epithelial cells.3 Toll‐like receptors (TLRs) are a class of transmembrane PRRs located on either the cell surface or in endosomes.31 One of the most characterized bacterial immunomodulators is Bacteroides fragilis polysaccharide A (PSA), which is recognized by TLR2 and capable of influencing T‐cell development and homeostasis.32 Recent studies reveal that PSA activates TLR2 on plasmacytoid DCs rather than conventional DCs, leading to the induction of IL‐10 secretion by CD4+ T cells and mucosal protection during a 2,4,6‐trinitrobenzene sulphonic acid model of colitis.33 PSA–TLR2 activation of pDCs and Treg cell induction can also mediate protection in extra‐intestinal inflammatory diseases such as experimental autoimmune encephalomyelitis, a multiple sclerosis animal model.33, 34 Additional work characterizing PSA‐induced Treg cell activation via MHCII‐mediated antigen presentation suggests that the interaction depends on the zwitterionic (carries positive and negative charges) properties of PSA and induces a specific clonal expansion of Treg cells.42 Lactobacillus plantarum teichoic acid d‐alanylation (component of Gram‐positive bacterial envelope) also signals through TLR2 and promotes a pro‐inflammatory cytokine response in DCs, which modulates effector and regulatory T‐cell populations45 (Fig. 2).
Figure 2.
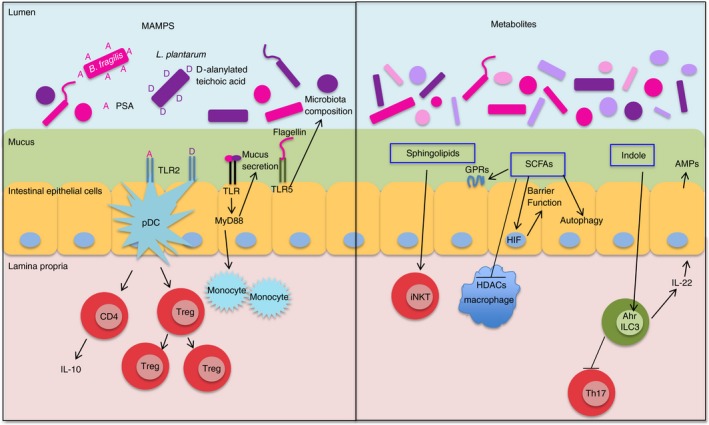
Bacterial components that affect innate and adaptive immunity in the intestine. Bacterial MAMPS signal through host PRRs. PSA (A) from Bacteroides fragilis interacts with TLR2 on plasmacytoid pDCs to induce IL‐10 production from CD4+ T cells and Treg clonal expansion. Lactobacillus plantarum d‐alanylated teichoic acid (D) also signals through TLR2 on DCs to modulate effector and regulatory T‐cell populations. Flagellin activation of TLR5 on epithelial cells alters microbiota composition. Luminal bacteria promote mucus secretion and movement of monocytes closer to epithelial stem cells through a MyD88‐dependent signalling pathway. Sphingolipid metabolites from B. fragilis promote iNKT activation in adults. SCFA metabolites from bacteria impact immunity through multiple mechanisms: activation of GPRs, inhibition of HDACs, and regulation of autophagy. Butyrate exerts anti‐inflammatory effects on macrophages through HDAC inhibition and promotes barrier function in IECs through stabilization of HIF. Lactobacilli produce an AhR ligand, indole‐3‐aldehyde, which induces IL‐22, promoting AMP expression and mucosal homeostasis. AhR signalling on ILC3s also inhibits Th17 cell expansion. AhR, aryl hydrocarbon receptor; AMPs, antimicrobial peptides; GPRs, G protein‐coupled receptors; HIF, hypoxia‐inducible factor; I, indole‐3‐aldehyde, an AhR ligand; IECs, intestinal epithelial cells, ILC3, group 3 innate lymphoid cell; MAMPS, microbe‐associated molecular patterns; MyD88, myeloid differentiation primary response protein 88; PRRs, pattern recognition receptors; pDCs, plasmacytoid dendritic cells; PSA, polysaccharide A from Bacteroides fragilis; SCFAs, short‐chain fatty acids; TLRs, Toll‐like receptors.
Bacteria flagellin is recognized by TLR5 expressed on various cells including intestinal epithelial cells (IECs) and DCs. The IEC‐derived TLR5 signalling appears to influence microbiota composition and host response because TLR5ΔIEC mice have an altered microbiota compared with co‐housed sibling wild‐type controls, develop low‐grade inflammation and metabolic syndrome, and have delayed clearance of adherent invasive Escherichia coli (AIEC).46 How TLR5 activation controls composition of the microbiota is unclear but could involve immune cell recruitment to clear pathogens in close proximity to the epithelium, stimulation of epithelial antimicrobial peptide production, or induction of flagellin‐specific IgA.46 The microbiota also impacts vaccine immunity through TLR5 signalling in B cells and macrophages, which is critical for mounting an antibody response to trivalent inactivated influenza vaccine and the inactivated polio vaccine.47
Most of the downstream signalling from TLRs occurs through either myeloid differentiation primary response protein 88 (MyD88) or Toll–interleukin receptor domain‐containing adaptor protein inducing interferon‐β (TRIF) adaptor proteins, resulting in activation of nuclear factor‐κB or interferon regulatory factors, respectively.48 Luminal bacteria promote mucus secretion and movement of monocytes closer to epithelial stem cells through an epithelial MyD88‐signalling pathway. Increased proximity of monocytes to epithelial stem cells results in increased crypt cell proliferation and intestinal stem cell numbers,49 which could be beneficial during intestinal injury response. Studies comparing GF mice to mice colonized with three strains of bacteria (E. coli K‐12, Staphylococcus xylosus and Enterococcus faecalis) reveal that GF mice have delayed microbial clearance, reduced inflammatory responses to intravenous E. coli K12 infection and a decreased myeloid cell pool size.50 Heat‐stable microbial antigens in the serum are able to restore bone marrow myeloid cell numbers through MyD88/Toll‐interleukin 1 receptor domain‐containing adaptor molecule (TICAM)‐dependent TLR signalling.50 MyD88‐dependent TLR signalling also plays a role in microbiota‐mediated tolerance to a non‐invasive strain of Salmonella enterica serovar Typhimurium by preventing CX3CR1hi mononuclear phagocyte‐mediated transport of luminal bacteria to the mesenteric lymph nodes.51 Furthermore, tumour necrosis factor receptor associated factor 6 (TRAF6), a component of TLR signal transduction, has MyD88‐independent effects on immune and microbiota homeostasis. Mice with TRAF6‐deficient DCs develop Th2‐driven small intestine inflammation and have decreased Treg cells, both of which are microbiota‐dependent.52
Nod‐like receptors (NLRs) are a class of cytosolic PRRs that act as intracellular sensors.3 Nod2 recognizes bacterial peptidoglycan through muramyl dipeptide. Studies using Nod2 −/− mice reveal reduced intraepithelial lymphocytes (IELS) and administering muramyl dipeptide to antibiotic‐treated mice restored IEL numbers through up‐regulation of IL‐15, suggesting that Nod2‐mediated recognition of the microbiota affects IEL homeostasis.53 Nod2 has also been implicated in preventing goblet cell dysfunction and restricting expansion of Bacteroides vulgatus in the small intestine, which prevents piroxicam‐induced intestinal inflammation in mice.54 The NLRs also have extra‐intestinal effects, as NLR ligands have been implicated in innate immunity in the lung, which is important for Klebsiella pneumoniae clearance.43
Inflammation or injury can cause members of the microbiota to become pathogenic and stimulate the immune system to induce inflammation. In the context of a dextran sulphate sodium (DSS) mouse model, Proteus mirabilis can induce IL‐1β production via NLR family, pyrin domain containing 3 (NLRP3) inflammasome activation in recruited inflammatory monocytes, promoting intestinal inflammation.44 After comparing different strains, the authors determined that Proteus mirabilis HpmA haemolysin induces K+ efflux, which is required for NLRP3‐induced inflammasome activation.30, 44 In vitro studies show that AIEC isolated from patients with IBD are also able to induce IL‐1β through NLRP3 activation in macrophages.55
Outer membrane vesicles
Outer membrane vesicles (OMVs) are produced by Gram‐negative bacteria and contain various bacterial components, many of which activate PRRs.39 The OMVs can promote immune homeostasis or enhance bacterial pathogenesis; effects that probably depend on the type of bacteria, OMV content and host environment.39 For example, Bacteroides thetaiotaomicron OMVs containing homologues of mammalian inositol phosphatase interact with IECs in vitro to promote intracellular calcium signalling.40 This signalling confers nutritional benefits and potentially anti‐carcinogenic properties, as dietary inositol hexaphosphate administration reduces tumorigenesis in carcinogen (1,2‐dimethylhydrazine or azoxymethane) ‐induced cancers in rats and mice.40 On the other hand, spontaneous colitis‐prone CD4‐dnTgfb2;IL10rb −/− mice exposed to B. thetaiotaomicron develop inflammation due to OMVs containing sulphatase activity, which degrades mucin glycans and allows B. thetaiotaomicron to interact with host macrophages.35 Bacterodies fragilis PSA is also released in OMVs, which can be detected by TLR2.36, 56 Some bacteria produce OMVs that have adverse effects on host immunity. For example, enterotoxigenic B. fragilis secrete B. fragilis toxin‐dependent particles that can induce host IECs to secrete sphingolipids (specifically, sphingosine‐1‐phosphate) in exosome‐like particles, which induce Th17 cells and enhance tumorigenesis in multiple colon cancer mouse models.56
Metabolites
Besides structural components, bacteria also generate a wide spectrum of metabolites that have the capacity to engage and trigger numerous host responses (Fig. 2). Multiple intestinal Bacteroides species are able to synthesize sphingolipids, which are structurally similar to host lipid agonists of iNKT cells. Bacteroides fragilis sphingolipids have been shown to modulate cellular homeostasis by both promoting iNKT cell activation37 and inhibiting activation and expansion of iNKT cells during mouse neonatal development, which protects against oxazolone‐induced colitis in adulthood.38
Short‐chain fatty acids (SCFAs) are bacterial metabolites generated as by‐products of dietary fibre fermentation. Butyrate, propionate and acetate are the most common intestinal SCFAs and are normally present in the millimolar range in the gut. The mechanisms by which SCFAs impact immunity include activation of G protein‐coupled receptors (GPRs), inhibition of histone deacetylases, and regulation of autophagy.57 Levels of SCFAs depend on two interdependent factors: dietary fibre and microbiota composition. The SCFAs may modulate protection against chemically induced DSS colitis through GPR43 and GPR109A receptor interactions, which are dependent on the NLRP3 inflammasome in non‐haematopoietic cells.58 Butyrate exerts anti‐inflammatory effects on bone marrow‐derived and colonic macrophages via histone deacetylase inhibition. However, gavaging mice with butyrate does not impact the outcome of DSS colitis, suggesting that butyrate promotes bacterial tolerance rather than tissue repair.59 Additionally, butyrate has been shown to increase barrier function by stimulating epithelial metabolism in the colon to promote oxygen depletion, stabilizing hypoxia‐inducible factor and inducing hypoxia‐inducible factor‐dependent target genes that promote barrier function.60 SCFAs from the microbiota can also have systemic effects. In a mouse model of allergic inflammation in the lung, high levels of propionate are protective, probably through GPR41 signalling, which results in DCs with high phagocytic capability and an impaired capacity to induce Th2 differentiation.41
In addition, microbial metabolites can influence host immune responses through an indirect route. For example, SCFAs augment 5‐hydroxytryptamine (serotonin) production from intestinal enterochromaffin cells through up‐regulation of the rate‐limiting biosynthetic enzyme tryptophan hydroxylase.61 The wide impact of serotonin on host biological response including immunity62 suggests that microbes could shape immune responses through complex mechanisms. SCFAs also directly impact innate immune cells in the brain and central nervous system. Germ‐free mice have defects in microglial (tissue macrophages of the brain) maturation, differentiation and function with a diminished response to lipopolysaccharides and viral challenges, while administering a mixture of propionate, butyrate and acetate to the drinking water restores microglial maturation.63 Indigenous bacteria metabolites may have key roles in inhibiting colonization of specific pathogens. For instance, Clostridium scindens inhibition of Clostridium difficile is associated with secondary bile acid synthesis.64
Tryptophan catabolites from the microbiota expand lactobacilli that produce an aryl hydrocarbon receptor (AhR) ligand, indole‐3‐aldehyde. Activation of AhR results in IL‐22 transcription that promotes antimicrobial peptide expression and mucosal homeostasis, providing colonization resistance against gastrointestinal or vaginal Candida albicans infection and DSS colitis models.65 Group 3 ILCs also rely on AhR signalling to inhibit Th17 cell expansion and regulate SFB levels, which could play a role in IBD because ~ 40% of mice that lack AhR signalling abilities in ILCs develop spontaneous colitis between 12 and 20 weeks of age and have exacerbated inflammation in a CD45RBhi T‐cell transfer model of colitis.66 In summary, bacterial components and metabolites affect both innate and adaptive immunity in the intestine (Fig. 2).
Viral, archaeal and eukaryotic microbiota members that influence immunity
Part of how the microbiota impacts host immunity is by limiting pathogen colonization through niche occupation and resource use. These indirect protective effects may extend to the viral members of the microbiota, of which there are an estimated 109 viruses per gram of faeces. Some of these viruses target mammalian cells but bacteriophages, which exclusively infect bacteria, make up the majority of the virus community.67 Bacteriophages displaying immunoglobulin‐like domains on phage capsid proteins adhere to host intestinal mucus and are able to reduce microbial colonization in the mucosal niche by infecting and lysing bacteria.68 Recently, metagenomic sequencing of the human faecal virome from healthy people and patients with IBD revealed an expansion of Caudovirales bacteriophages associated with IBD.69 Hence, microbiota‐associated bacteriophages may impact the pathogenesis of IBD by targeting microbial members with protective or deleterious function.
When it comes to immune system development and function, viral members of the microbiota may be able to confer some of the same immune benefits as bacteria. Murine norovirus infection of GF or antibiotic‐treated mice restores intestinal morphology and lymphocyte function, and suppresses group 2 ILC (ILC2) expansion; additionally RNA‐sequencing revealed transcriptional changes in the intestine associated with immune development and type I interferon signalling.70 Whether viruses contribute to human immune system development and homeostasis remains to be determined.
Archaeal members of the microbiota can also activate host immune cells. Specifically Methanosphaera and Methanorevibacter have differential capacities to induce pro‐inflammatory cytokine release from human DCs. Activation requires phagocytosis of the archaea, but whether induction involves PRRs that recognize components of the archaeal cell envelope is still unknown.71
Studies examining the immunomodulatory effects of the fungal microbiota have mostly focused on one of the most abundant members, Candida albicans, which can cause severe infections in immunocompromised people.2 The host uses TLRs as well as C‐type lectins, a class of PRRs, to recognize fungal cell wall components such as mannans, β‐glucans and chitin.2, 72 The cell wall components of Candida albicans are immunomodulators, with cell wall glycosylation playing a key role in inducing pro‐inflammatory cytokine expression, proliferation and apoptosis in epithelial cells.72 Chitin induces secretion of the anti‐inflammatory cytokine IL‐10, which is dependent on NOD2, TLR9, and mannose receptors. Anti‐inflammatory cytokines induced by chitin may play a role in resolving immune homeostasis after pathogen clearance and eosinophilia, which is a feature of asthma.73
Helminths are parasitic worms that modulate host immunity by inducing a strong Th2 immune response, Treg cells and regulatory cytokines such as IL‐10 and transforming growth factor‐β.74 Epidemiological evidence and experimental studies suggest that the helminth‐induced immune response may be therapeutic for treating allergies and autoimmune diseases.75 Recent research focused on identifying the helminth immunomodulatory products has revealed that administering excretory/secretory products from Trichinella spiralis adult worms protects mice from DSS‐induced colitis through up‐regulation of Treg cells and reduction of pro‐inflammatory cytokines.74 The Acanthocheilonema viteae product (AvCystatin) modulates MAPK, p38 and ERK pathways in macrophages to induce IL‐10 secretion. Administration of AvCystatin‐treated macrophages to mice with OVA‐induced airway inflammation or DSS‐induced colitis ameliorates disease by suppressing inflammation.76 However, in the context of viral infection, the helminth immune response may be detrimental. Trichinella spiralis‐infected mice induce alternative activation of macrophages, which up‐regulates genes that impair the T‐cell response to murine norovirus.77
Bacterial adaptations to host immune mechanisms
Previous sections have examined how microbes direct immune development and function. However, the immune system also impacts microbes, which includes influencing microbiota composition as well as virulence capacities. Mechanisms by which specific microbes have adapted to coexist with the host immune system have begun to emerge (Table 2). For example, B. thetaiotaomicron uses lipid A dephosphorylation to resist host antimicrobial peptides that target the lipopolysaccharide portion of the bacterial outer membrane.78
Table 2.
Examples of bacterial adaptations to host immune mechanisms
| Bacteria | Host Immune component | Bacterial response | References |
|---|---|---|---|
| Bacteroides thetaiotaomicron | Antimicrobial peptides | Resistance through outer membrane lipid A dephosphorylation | (78) |
| Indigenous bacteria | Fucosylation of IECs triggered by SFB or enteric infection | Metabolic capacity to cleave fucose, use fucose | (79–82) |
| Salmonella enterica serovar Typhimurium | Induction of lipocalin‐2 and calprotectin | Additional siderophores and zinc transporters to overcome host metal ion sequestration | (83) |
| Fusobacterium nucleatum | NK cytotoxicity and T cell effector functions | Binds host inhibitory receptor (TIGIT) on NK and T cells | (85) |
IECs, intestinal epithelial cells; NK, natural killer; SFB, segmented filamentous bacteria; TIGIT, an inhibitory receptor on NK and T cells.
Part of the immune response to infection can promote indigenous microbiota colonization through release of nutrients. TLR agonists induce DC IL‐23 production in a MyD88‐dependent manner, which stimulates ILCs to produce IL‐22, resulting in rapid fucosylation of small intestine epithelial cells.79 Work by Goto et al. demonstrated that bacteria such as SFB, stimulate IL‐22 production by ILC3s, which mediates epithelial fucosylation in the ileum and protects the host from Salmonella typhimurium infection.80 Resident bacteria, such as Bacteroides acidifaciens, have the capacity to cleave fucose, which can subsequently be used by other members of the microbiota; a process that may promote tolerance to intestinal pathogens.79 Host IL‐22RA1 signalling promotes intestinal fucosylation in colonic organoids and the mouse caecum, restoring anaerobic bacterial diversity in the colon to protect against opportunistic pathogens such as Enterococcus faecalis and Citrobacter rodentium.81 Hence, fucosylation seems to be a host response to specific members of the microbiota or infectious challenge, which bolsters the microbiota and protects the host from multiple enteric pathogens. Reinforcing the importance of IL‐22 production in maintaining colonization resistance against enteric pathogens, ID2 (a transcriptional regulator of ILCs) promotes colonization resistance against Citrobacter rodentium by mediating IL‐22 production by ILC3s through an AhR and IL‐23 receptor pathway.82
Some pathogenic bacteria have evolved virulence factors that allow them to better cope with host immune defence mechanisms compared with indigenous bacteria. Interleukin‐22 is part of the immune response to infection and leads to induction of lipocalin‐2 and calprotectin, which sequester iron, zinc and manganese ions. Salmonella enterica serovar Typhimurium overcomes host iron sequestration with the siderophore salmochelin and zinc sequestration through a zinc transporter, giving Salmonella a colonization advantage over resident Enterobacteriaceae that lack additional siderophores.83 Fusobacterium nucleatum, which has previously been linked to colon cancer,84 is able to bind to an inhibitory receptor (TIGIT) on NK and T cells, leading to inhibition of NK cell‐mediated cytotoxicity/T‐cell activities.85 Additionally, the authors demonstrate F. nucleatum OMVs bind TIGIT suggesting a pro‐carcinogenic role for F. nucleatum OMVs by inhibiting host immune function.85 This bilateral communication between microbes and the host clearly highlights the intricate and complex nature of microbe–host interactions.
Conclusions/Perspective
The impact of the microbiota on myriad components of innate and adaptive immunity has been well established, especially in the intestine. Recent studies have moved on to characterizing how members of the microbiota and the host immune system communicate with one another. It is likely that the dialogue between microbes and host is a dynamic phenomenon, taking place from birth and evolving over time, even though the phylogenic microbial composition is quite stable for the majority of life. Therefore, microbial bioactivities rather than composition may account for most of the host response, including immunity. Clearly, more information is needed on the specific nature of this dialogue and how communication breakdowns result in disrupted host immune homeostasis. It will be important to continue detailing the relationship between microbially derived metabolites and host immune homeostasis. These studies should include transcriptomic and metabolomic approaches to better understand the impact of microbial activities on the host immune response. The continuous characterization of microbial communities at the genomic and proteomic levels, in conjunction with specific culture methods, will contribute to understanding how specific microbes shape the immune response. Ahern et al. offer a possible approach for identifying members of the human microbiota that impact the immune system using gnotobiotic mice.86 The persistent mapping and annotation of microbial genes in conjunction with the establishment of tools to genetically modify these genes will enable molecular dissection of the contributions of bacteria to host immunity. Additionally, it is likely that more interactions occur between members of the microbiota (viruses, prokaryotes and eukaryotes) that influence host immune health than are currently known. Continued research focused on the signalling that occurs between microbiota components and the immune system may lead to the development of new or improved strategies to restore or reset altered communication networks between the host and microbes.
Disclosures
The authors declare that they have no competing interests.
Acknowledgements
CJ acknowledges support from the NIH (RO1DK047700, RO1DK073338 and RO1 AT08623). We would like to thank Dr Christina Ohland for critical reading of the manuscript.
References
- 1. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mukherjee PK, Sendid B, Hoarau G, Colombel J‐F, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2015; 12:77–87. [DOI] [PubMed] [Google Scholar]
- 3. Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Nat Immunol 2015; 16:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jain N, Walker WA. Diet and host–microbial crosstalk in postnatal intestinal immune homeostasis. Nat Rev Gastroenterol Hepatol 2015; 12:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJL, Furman D et al Variation in the human immune system is largely driven by non‐heritable influences. Cell 2015; 160:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tilg H, Moschen AR. Food, immunity, and the microbiome. Gastroenterology 2015; 148:1107–19. [DOI] [PubMed] [Google Scholar]
- 8. Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol 2013; 14:646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC et al Proteobacteria‐specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2013; 5:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moon C, Baldridge MT, Wallace M A, Burnham C‐AD, Virgin HW, Stappenbeck TS. Vertically transmitted faecal IgA levels determine extra‐chromosomal phenotypic variation. Nature 2015; 521:90–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winter SE, Bäumler AJ. Why related bacterial species bloom simultaneously in the gut: principles underlying the “Like will to like” concept. Cell Microbiol 2014; 16:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farin HF, Karthaus WR, Kujala P, Rakhshandehroo M, Schwank G, Vries RGJ et al Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell‐derived IFN‐γ . J Exp Med 2014; 211:1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL et al Thymus‐derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013; 497:258–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lécuyer E, Rakotobe S, Lengliné‐Garnier H, Lebreton C, Picard M, Juste C et al Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 2014; 40:608–20. [DOI] [PubMed] [Google Scholar]
- 15. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG et al Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014; 40:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geem D, Medina‐Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. Specific microbiota‐induced intestinal Th17 differentiation requires MHC Class II but not GALT and mesenteric lymph nodes. J Immunol 2014; 193:431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schnupf P, Gaboriau‐Routhiau V, Gros M, Friedman R, Moya‐Nilges M, Nigro G et al Growth and host interaction of mouse segmented filamentous bacteria in vitro . Nature 2015; 520:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M et al Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 2013; 7:615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caselli M, Tosini D, Gafà R, Gasbarrini A, Lanza G. Segmented filamentous bacteria‐like organisms in histological slides of ileo‐cecal valves in patients with ulcerative colitis. Am J Gastroenterol 2013; 108:860–1. [DOI] [PubMed] [Google Scholar]
- 20. Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR et al Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014; 346:755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL et al Prevention and cure of rotavirus infection via TLR5/NLRC4‐mediated production of IL‐22 and IL‐18. Science 2014; 346:861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanther M, Tomkovich S, Xiaolun S, Grosser MR, Koo J, Flynn EJ et al Commensal microbiota stimulate systemic neutrophil migration through induction of Serum amyloid A. Cell Microbiol 2014; 16:1053–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE et al The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 2014; 20:524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Surana NK, Kasper DL. Deciphering the tête‐à‐tête between the microbiota and the immune system. J Clin Invest 2014; 124:4197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci U S A 2013; 110:15061–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naik S, Bouladoux N, Linehan JL, Han S‐J, Harrison OJ, Wilhelm C et al Commensal–dendritic cell interaction specifies a unique protective skin immune signature. Nature 2015; 520:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Canesso MCC, Vieira AT, Castro TBR, Schirmer BGA, Cisalpino D, Martins FS et al Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J Immunol 2014; 193:5171–80. [DOI] [PubMed] [Google Scholar]
- 29. Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH et al Dysbiosis and Staphyloccus aureus colonization drives inflammation in atopic dermatitis. Immunity 2015; 42:756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muñoz‐Planillo R, Kuffa P, Martínez‐Colón G, Smith B, Rajendiran T, Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 2013; 38:1142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci 2010; 15:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dasgupta S, Erturk‐Hasdemir D, Ochoa‐Reparaz J, Reinecker H‐C, Kasper DL. Plasmacytoid dendritic cells mediate anti‐inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 2014; 15:413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Telesford KM, Ochoa‐Reparaz J, Haque‐Begum S, Christy M, Kasper EJ et al An intestinal commensal symbiosis factor controls neuroinflammation via TLR2‐mediated CD39 signaling. Nat Commun 2014; 5:4432–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hickey CA, Kuhn KA, Martens EC, Stappenbeck TS, Hickey CA, Kuhn KA et al Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase‐dependent manner via outer membrane vesicles short article colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase‐de. Cell Host Microbe 2015; 17:672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012; 12:509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M et al Production of α‐galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol 2013; 11:e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk‐Hasdemir D et al Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014; 156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaparakis‐Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 2015; 15:375–87. [DOI] [PubMed] [Google Scholar]
- 40. Stentz R, Osborne S, Horn N, Li AWH, Hautefort I, Bongaerts R et al A bacterial homolog of a eukaryotic inositol phosphate signaling enzyme mediates cross‐kingdom dialog in the mammalian gut. Cell Rep 2014; 6:646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom‐Bru C et al Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20:159–66. [DOI] [PubMed] [Google Scholar]
- 42. Johnson JL, Jones MB, Cobb BA. Polysaccharide a from the capsule of Bacteroides fragilis induces clonal CD4+ T cell expansion. J Biol Chem 2015; 290:5007–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via NLR ligands. Infect Immun 2014; 82:4596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seo S‐U, Kamada N, Muñoz‐Planillo R, Kim Y‐G, Kim D, Koizumi Y et al Distinct commensals induce interleukin‐1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity 2015: 744–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smelt MJ, De Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM et al The Impact of Lactobacillus plantarum WCFS1 teichoic acid d‐alanylation on the generation of effector and regulatory T‐cells in healthy mice. PLoS ONE 2013; 8:e63099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chassaing B, Ley RE, Gewirtz AT. Intestinal epithelial cell toll‐like receptor 5 regulates the intestinal microbiota to prevent low‐grade inflammation and metabolic syndrome in mice. Gastroenterology 2014; 147:1363–77e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M et al TLR5‐mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014; 41:478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll‐like receptor signalling complexes. Nat Rev Immunol 2014; 14:546–58. [DOI] [PubMed] [Google Scholar]
- 49. Skoczek DA, Walczysko P, Horn N, Parris A, Clare S, Williams MR et al Luminal microbes promote monocyte–stem cell interactions across a healthy colonic epithelium. J Immunol 2014; 193:439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Balmer ML, Schurch CM, Saito Y, Geuking MB, Li H, Cuenca M et al Microbiota‐derived compounds drive steady‐state granulopoiesis via MyD88/TICAM Signaling. J Immunol 2014; 193:5273–83. [DOI] [PubMed] [Google Scholar]
- 51. Diehl GE, Longman RS, Zhang J‐X, Breart B, Galan C, Cuesta A et al Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 2013; 494:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J et al Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota‐dependent immune tolerance. Immunity 2013; 38:1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang W, Wang X, Zeng B, Liu L, Tardivel A, Wei H et al Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med 2013; 210:2465–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus . Immunity 2014; 41:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. De la Fuente M, Franchi L, Araya D, Díaz‐Jiménez D, Olivares M, Álvarez‐Lobos M et al Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol 2014; 304:384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deng Z, Mu J, Tseng M, Wattenberg B, Zhuang X, Egilmez NK et al Enterobacteria‐secreted particles induce production of exosome‐like S1P‐containing particles by intestinal epithelium to drive Th17‐mediated tumorigenesis. Nat Commun 2015; 6:6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 2013; 14:676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S et al Metabolite‐sensing receptors GPR43 and GPR109A facilitate dietary fibre‐induced gut homeostasis through regulation of the inflammasome. Nat Commun 2015; 6:6734. [DOI] [PubMed] [Google Scholar]
- 59. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014; 111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ et al Crosstalk between microbiota‐derived short‐chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015; 17:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L et al Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015; 161:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baganz NL, Blakely RD. A dialogue between the immune system and brain, spoken in the language of serotonin. ACS Chem Neurosci 2013; 4:48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E et al Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18:965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A et al Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile . Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zelante T, Iannitti R, Cunha C, DeLuca A, Giovannini G, Pieraccini G et al Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity 2013; 39:372–85. [DOI] [PubMed] [Google Scholar]
- 66. Qiu J, Guo X, Chen ZE, He L, Sonnenberg GF, Artis D et al Group 3 innate lymphoid cells inhibit t‐cell‐mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 2013; 39:386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cadwell K. The virome in host health and disease. Immunity 2015; 42:805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J et al Bacteriophage adhering to mucus provide a non‐host‐derived immunity. Proc Natl Acad Sci U S A 2013; 110:10771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC et al Disease‐specific alterations in the enteric virome in article disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015; 160:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014; 516:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE 2014; 9:e99411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wagener J, Weindl G, de Groot PWJ, de Boer AD, Kaesler S, Thavaraj S et al Glycosylation of Candida albicans cell wall proteins is critical for induction of innate immune responses and apoptosis of epithelial cells. PLoS ONE 2012; 7:e50518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wagener J, Malireddi RKS, Lenardon MD, Köberle M, Vautier S, MacCallum DM et al Fungal chitin dampens inflammation through IL‐10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog 2014; 10:e1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang X, Yang Y, Wang Y, Zhan B, Gu Y, Cheng Y et al Excretory/secretory products from Trichinella spiralis adult worms ameliorate DSS‐induced colitis in mice. PLoS ONE 2014; 9:e96454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Girgis NM, Gundra UM, Loke P. Immune regulation during helminth infections. PLoS Pathog 2013; 9:e1003250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ziegler T, Rausch S, Steinfelder S, Klotz C, Hepworth MR, Kühl AA et al A novel regulatory macrophage induced by a helminth molecule instructs IL‐10 in CD4+ T cells and protects against mucosal inflammation. J Immunol 2015; 194:1555–64. [DOI] [PubMed] [Google Scholar]
- 77. Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR et al Virus–helminth coinfection reveals a microbiota‐independent mechanism of immunomodulation. Science 2014; 345:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS et al Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 2015; 347:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV et al Rapid fucosylation of intestinal epithelium sustains host – commensal symbiosis in sickness. Nature 2014; 514:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goto Y, Domino SE, Kulig P, Becher B. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014; 345:1254009–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pham TAN, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP et al Epithelial IL‐22RA1‐mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 2014; 16:504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Guo X, Liang Y, Zhang Y, Lasorella A, Kee BL, Fu Y‐X. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2‐dependent regulation of the microbiota. Immunity 2015; 42:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W et al The cytokine IL‐22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 2014; 40:262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Allen‐Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett 2014; 162:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M et al Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015; 42:344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity 2014; 40:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


