Abstract
The vast majority of eukaryotic proteins are N-terminally modified by one or more processing enzymes. Enzymes acting on the very first amino acid of a polypeptide include different peptidases, transferases, and ligases. Methionine aminopeptidases excise the initiator methionine leaving the nascent polypeptide with a newly exposed amino acid that may be further modified. N-terminal acetyl-, methyl-, myristoyl-, and palmitoyltransferases may attach an acetyl, methyl, myristoyl, or palmitoyl group, respectively, to the α-amino group of the target protein N-terminus. With the action of ubiquitin ligases, one or several ubiquitin molecules are transferred, and hence, constitute the N-terminal modification. Modifications at protein N-termini represent an important contribution to proteomic diversity and complexity, and are essential for protein regulation and cellular signaling. Consequently, dysregulation of the N-terminal modifying enzymes is implicated in human diseases. We here review the different protein N-terminal modifications occurring co- or post-translationally with emphasis on the responsible enzymes and their substrate specificities.
Keywords: α-amino group, Acetylation, Cell biology, N-terminal, Protein modification, Substrate specificity
1 Introduction
From the moment a eukaryotic nascent polypeptide emerges from the ribosome, a machinery of different enzymes is in place to modify its N-terminal (Nt) amino acid residue. These modifications (Fig. 1) have evolved to substantially increase the cellular protein repertoire. Despite the abundance of Nt-modifications, the specific functions of N-terminally modifying enzymes remain incompletely understood. Initiator methionine excision by methionine aminopeptidases (MetAPs) is very common and essential, but not comprehended in terms of its functional implications [1]. Another highly abundant co-translational modification is Nt-acetylation catalyzed by N-terminal acetyltransferases (NATs) [2]. The NATs also carry out Nt-propionylation, a much rarer and less understood modification [3]. Protein fatty acylation of the N-terminus normally involves Nt-myristoylation catalyzed by N-terminal myristoyltransferases (NMTs) [4]. Nt-palmitoylation is rarer and is carried out by distinct enzymes, the N-terminal palmitoylacyltransferases (PATs) [5]. The abovementioned modifications mostly occur co-translationally, but the N-terminus can also be post-translationally modified. Nt-methylation is a common type of post-translational modification catalyzed by N-terminal methyltransferases (NTMTs) [6,7]. Additionally, Nt-ubiquitylation has emerged as a new scarce member of the Nt-modification family [8]. Together, the various Nt-modifications have profound functional effects. The responsible enzymes act in more or less sequence specific manners in order to establish specific functions to particular substrate proteins. We will here review the major co- and post-translational protein Nt-modifications including their biological impact. We will further address the current knowledge of the responsible enzymes and their substrate sequence requirements.
Figure 1.
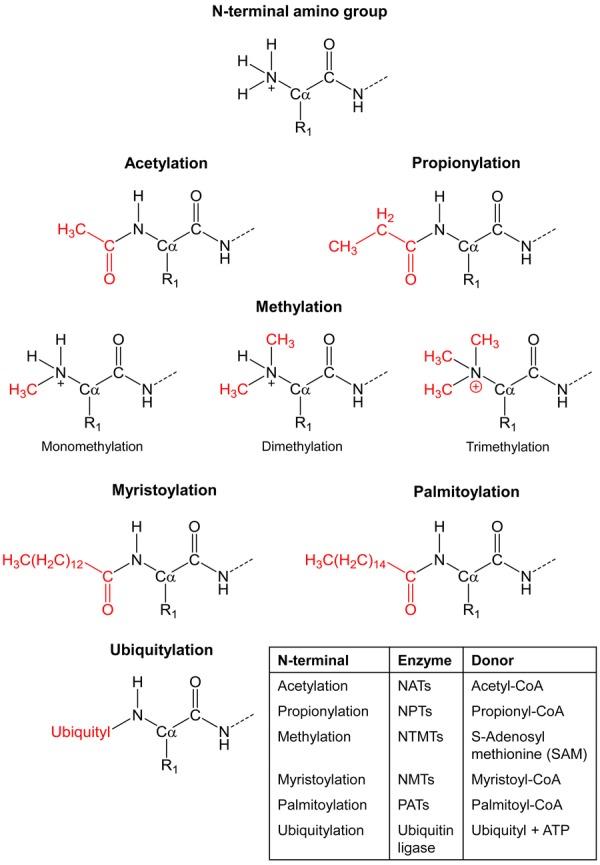
Structural formulae of major N-terminal protein modifications. The N-terminal α-amino group is (usually) positively charged at neutral pH. The chemical character of protein N-termini can be modified by, for example: acetylation, propionylation, methylation, myristoylation, palmitoylation or ubiquitylation (attachment shown in red). The responsible enzymes and their respective donor molecules are listed. NATs, N-terminal acetyltransferases; NPTs, N-terminal propionyltransferases; NTMTs, N-terminal methyltransferases; NMTs, N-terminal myristoyltransferases; PATs, Palmitoylacyltransferases; CoA, Coenzyme A; ⊕ denotes permanent positive charge.
2 N-terminal methionine cleavage
When a nascent polypeptide emerges from the ribosomal exit tunnel it is introduced to a set of different proteins. Amongst them is MetAPs methionine aminopeptidases (MetAPs) [9], destined to co-translationally remove the initiator methionine (iMet) in the case of a favourable second amino acid. Complete cleavage of the iMet is achievable when Ala, Cys, Gly, Pro, Ser, Thr or Val sequesters the second position (Table 1). These amino acids all have an uncharged side chain with a radius of gyration of <1.29 Å as defined by Levitt [10]. In contrast, there is a full iMet-retention when the second residue belongs to any of the other naturally occurring amino acids. These features make up the criteria for iMet removal on a nascent polypeptide [1,10–14]. However, in some cases the amino acid residue in the third position can influence the activity of MetAPs. For example, the iMet of the β-chain of Hemoglobin Long Island [15,16] and Hemoglobin Marseille [17] is retained following His to Pro substitution at position three, possibly due to steric hindrance. Some actins constitute an exception to the general iMet excision pathway. For instance, mammalian cytoplasmic γ-actin (Met-Glu-Glu-) has a large amino acid residue in the second position and the iMet is not processed by the co-translationally acting MetAPs. Instead, this actin is post-translationally processed by an unidentified aminopeptidase [18].
Table 1.
Protein N-terminal modifications specified by the responsible enzymes and their substrate specificity
| Nt-modification | Enzyme | Proteinb) | Alternative name | UniProt | Substrate specificityc) |
|---|---|---|---|---|---|
| iMet excision | MetAP | MetAP1 MetAP2 | MAP1A, Peptidase M 1MAP2, MNPEP, P67EIF2 | P53582 P50579 | Met-Ala-, Met-Cys-, Met-Gly-, Met-Pro-, Met-Ser-, Met-Thr-, Met-Val- |
| ND | Met-Asp-, Met-Glu- | ||||
| Acetylation Propionylationa) | NatA | Naa10cat | ARD1, ARD1A, TE2 | P41227 | Ala-, Cys-, Gly-, Ser-, Thr-, Val-, Aspd)-, Glud)- |
| Naa15aux | NAT1, GA19, NARG1, NATH, TBN | Q9BXJ9 | |||
| NatB | Naa20cat | NAT3, NAT5 | P61599 | Met-Asn-, Met-Asp-, Met-Gln-, Met-Glu- | |
| Naa25aux | MDM20, NAP1 | Q14CX7 | |||
| NatC | Naa30cat | MAK3, NAT12 | Q1473 | Met-Ile-, Met-Leu-, Met-Phe-, Met-Trp- | |
| Naa35aux | MAK10, EGAP, | Q5VZE5 | |||
| Naa38aux | MAK31, LSMD1, PFAAP2 | Q9BRA0 | |||
| NatD | Naa40cat | NAT4, NAT11 | Q86UY6 | Ser-Gly-Gly-, Ser-Gly-Arg- | |
| NatE | Naa50cat | MAK3, NAT13, SAN | Q9GZZ1 | Met-Ala-, Met-Leu-, Met-Lys-, Met-Phe-, Met-Ser-, Met-Thr-, Met-Tyr-, Met-Val- | |
| NatF | Naa60cat | NAT15, HAT4 | Q9H7X0 | Met-Ala-, Met-Gln-, Met-Gly-, Met-Ile-, Met-Leu-, Met-Lys-, Met-Met-, Met-Ser-, Met-Thr-, Met-Tyr-, Met-Val- | |
| Methylation | NTMT | NTMT1 | METTL11A, NRMT1, NRMT1A | Q9BV86 | Ala/Pro/Ser-Pro-Lys- |
| Tae1 (S.c) | NTM1 | P38340 | |||
| NTMT2 | METTL11B, NRMT2, NTM1B | Q5VVY1 | |||
| Myristoylation | NMT | NMT1 | NMT | P30419 | Gly- |
| NMT2 | O60551 | ||||
| Palmitoylation | PAT | Hhat | MART2, SKI1, Skn | Q5VTY9 | Cys- |
| Rasp (D.m) | cmn, sit, ski | Q9VZU2 | |||
| ND | Gly- | ||||
| Ubiquitylation | Ubiquitin | Ube2w | UBC16, UBC-16, | Q96B02 | Unstructured N-terminal backbone |
| ligase | HUWE1 | ARF-BP1, HectH9, LASU1, Mule, UREB1, URE-B1 | Q7Z6Z7 | ND |
N-terminal propionylation is catalyzed by the same enzymes as N-terminal acetylation (NATs), and the substrate specificity is presumably shared.
All proteins listed are human except where the species is indicated.
The indicated amino acid sequences do not guarantee N-terminal modification.
Naa10 substrates.
ND, not determined; Aux, auxiliary subunit; Cat, catalytic subunit; S.c, Saccharomyces cerevisiae; D.m, Drosophila melanogaster.
The event of iMet removal is conserved throughout evolution [19] and is predicted to occur when the nascent polypeptide has reached a length of 20–40 amino acids [20–22]. It is estimated that more than 50% of all proteins are subjected to iMet removal [23]. Arfin et al. proposed a model for the subfamilies of MetAPs based on the catalytic cobalt-binding domain of the enzymes [24]. The updated version of this model, based on additional sequence features like N-terminal extension, linker region and zinc finger domains, displays two groups of MetAPs, namely MetAP1 and MetAP2. The former is divided into four subgroups, Type 1a-d. Type 1a and 1c are found in prokaryotes, while Type 1b, 1d and Type 2 MetAPs are present in eukaryotes [19,24–32]. Predictions on ribosomal binding of MetAPs suggest that one or several exposed PXXP motifs of the peptidases are involved in protein-protein interactions [25,26,33].
Structural studies of human MetAP1 and MetAP2 revealed a potential difference in the substrate specificity of the respective catalytic sites due to more steric restrictions in MetAP1 [25]. This is supported by the finding that MetAP2, and not MetAP1, is inhibited by the anti-angiogenic agent fumagillin [32]. Growth studies in yeast have shown that the iMet processing provided by MetAP1 and MetAP2 is essential since a double deletion is lethal for yeast while the single deletions are viable [30]. This strongly suggests that both enzymes, at least in part, act on the same substrates in vivo. Significant overlap in substrate specificity was also found for human MetAP1 and MetAP2, although a significant preference of MetAP2 for Met-Val- and Met-Thr- N-termini was observed [34].
3 N-terminal acetylation
One of the most abundant protein modifications occurring in eukaryotes is N-terminal acetylation (Nt-acetylation), where an acetyl moiety is transferred from acetyl-CoA to the α-amino group of a nascent polypeptide. The Nt-acetylome is estimated to include 80–90% of soluble human proteins and 50–70% of yeast proteins [35,36]. N-terminal acetylation of a specific protein can either be complete or partial, and in the latter case the protein exists in both acetylated and non-acetylated forms.
Traditionally, Nt-acetylation has been regarded as a co-translational process taking place on a nascent polypeptide when approximately 25–50 amino acids residues emerge from the ribosomal exit tunnel [37,38]. In addition, there are examples of post-translational Nt-acetylation of proteins [39,40]. The acetylation process neutralizes the positive charge normally associated with the free α-amino group, and thereby efficiently blocks the α-amino group for further ionization and other modifications (Fig. 1).
Nt-acetylation is catalyzed by a set of enzymes, the N-terminal acetyltransferases (NATs). The NATs are members of the GNAT protein superfamily [41], all containing the consensus acetyl-CoA binding sequence (Q/R)XXGXX(G/A) [42]. The enzymatic machinery is conserved from lower to higher eukaryotes suggesting a comparable system for Nt-acetylation [43]. The various NATs display different substrate specificities (Table 1), largely attributed to the identity of the first two amino acids in the polypeptide sequence [44]. However, residues beyond position seven may have an influence [35].
The majority of Nt-acetylation is catalyzed by NatA, NatB and NatC. Here, the NAT-function is dependent upon complex formation between a unique catalytic subunit and one or two auxiliary subunits. The auxiliary subunits have various functions including ribosomal anchoring [45,46]. NatA, the major NAT-complex, is composed of the catalytic subunit Naa10 and the regulatory subunit Naa15 [47–49]. The NatA complex acetylates N-termini starting with Ala, Cys, Gly, Ser, Thr or Val following iMet removal [35,44]. In the absence of Naa15 the substrate specificity profile of Naa10 changes towards acidic N-termini [49]. The NatB complex, which is composed of the catalytic Naa20 and auxiliary Naa25 subunits [50,51], acetylates Met-Asn-, Met-Asp-, Met-Gln- and Met-Glu-starting N-termini [18,44,52]. NatC, on the other hand, is composed of the three subunits Naa30, Naa35 and Naa38. Naa30 is responsible for the acetylation reaction, but all subunits appear to be required for NatC-activity [53]. The activity of NatC is directed towards the iMet when followed by a hydrophobic second residue, that is Ile, Leu, Phe, or Trp [18,44,54,55].
Three additional NATs (NatD, NatE, and NatF) have been identified thus expanding the NAT-family substrate repertoire. The evolutionarily conserved acetyltransferase NatD displays a limited substrate profile by N-terminally acetylating only histones H2A (Ser-Gly-Arg-) and H4 (Ser-Gly-Gly-) [56,57]. However, recent data suggest that NatD may share some Ser-Gly- starting substrates with NatA [58]. NatE was first reported when yeast Naa50 was found to be physically associated with the NatA complex, but no impact on NatA-activity was observed [45]. In vitro studies revealed that human Naa50 is able to perform Nt-acetylation of a Met-Leu- substrate, and the activity was termed NatE [59]. The substrate specificity profile of NatE was later expanded to include Met-starting N-termini: Met-Ala-, Met-Lys-, Met-Met-, Met-Phe-, Met-Ser-, Met-Thr-, Met-Tyr-, and Met-Val- [49]. The evolutionary shift in the degree of Nt-acetylation between yeast and human could partly be explained by the presence of NatF (Naa60) in human (and other multicellular eukaryotes). NatF acetylates Met-Lys- N-termini which are rarely acetylated in yeast, in addition to Met-Ala-, Met-Gln-, Met-Gly-, Met-Ile-, Met-Leu-, Met-Met-, Met-Ser-, Met-Thr-, Met-Tyr- and Met-Val-. NatC, NatE and NatF thus have partially overlapping substrate specificities [36]. Interestingly, Aksnes and colleagues recently reported that NatF is localized to the Golgi, where it specifically Nt-acetylates transmembrane proteins, most likely in a post-translational mode [182]. This study also uncovered that Nt-acetylation is highly abundant among human transmembrane proteins.
Worth mentioning, the Nt-acetylation signatures are not absolute. The specificity can be affected by downstream residues and other determinants, such as secondary structures [18]. This could also explain the existence of both complete and partial Nt-acetylation. Goetze et al. have shown that Nt-acetylation is prevented when the nascent protein has a Pro in the first or second position. Pro as a definite determinant preventing Nt-acetylation is referred to as the (X)PX rule [60]. No N-terminal deacetylase has been identified and thus Nt-acetylation is considered irreversible.
The crystal structures of several NATs have been solved [61–65], including S. pombe NatA [63] and human Naa50/NatE [62]. Structural variability between the two catalytic subunits of NatA and NatE contributes to the substrate specificity. Binding of the auxiliary subunit may introduce conformational changes in the active site of the catalytic subunit and promote sequence-specific Nt-acetylation as seen in NatA [63]. Recent evidence suggests that Nt-acetylation takes place through an ordered ternary-complex (Bi-Bi) mechanism [62–64,66]. Here, binding of acetyl-CoA induces rearrangements in the NAT-enzyme, which subsequently increases the affinity for the peptide substrate allowing the reaction to occur.
4 N-terminal propionylation
The N-terminal protein modification family was recently expanded to include a new member, namely N-terminal propionylation (Nt-propionylation) (Fig. 1). Propionylated N-termini were first discovered in human cells [67,68] and later shown to occur in yeast, demonstrating that this modification is evolutionarily conserved [3].
Studies by Foyn and colleagues show that NATs may also function as N-terminal propionyltransferases (NPTs), both in vivo and in vitro. By challenging purified human Naa10, Naa50, and NatA with propionyl-CoA they demonstrated that all enzymes could indeed perform Nt-propionylation on substrate peptides in vitro. The NPT-activity of purified Naa10 and Naa50 were far less efficient than their acetylation activities using acetyl-CoA. Intriguingly, the NatA complex performed Nt-acetylation and Nt-propionylation with similar rates, particularly for peptides substrate starting with Ser. Furthermore, Nt-proteomics revealed that both yeast and human NatA complexes could perform Nt-propionylation in vivo [3].
Since propionylation of protein N-termini was only recently discovered, virtually nothing is known about its functional importance. The cellular level of acetyl-CoA is 2-20-fold higher compared to propionyl-CoA [69,70]. This could explain the low detection rate of Nt-propionylated compared to Nt-acetylated substrates. In addition, it is uncertain whether Nt-propionylation occurs on specific substrates and conveys a signal different from Nt-acetylation. The propionyl group contains an additional methyl moiety as compared to acetyl and this might result in additional bulkiness and hydrophobicity. Hitherto 18 proteins have been identified as being Nt-propionylated, four of which are processed mitochondrial proteins. This indicates a post-translational as well as a co-translational mode for this modification [3,67,68].
5 N-terminal myristoylation and palmitoylation
N-terminal myristoylation (Nt-myristoylation) refers to the irreversible transfer of myristoyl (14-carbon saturated fatty acid) from myristoyl-CoA to the N-terminal Gly of the target protein (Fig. 1). N-terminal myristoyltransferase (NMT) catalyzes the chemical reaction [4], following an ordered Bi-Bi reaction mechanism [71–74]. Initially, this protein modification was found to exist as a co-translational event, supported by the isolation of myristoyl-labelled nascent polypeptide chains associated with tRNA and the identification of NMT in the ribosomal subcellular fraction [75–78]. However, Nt-myristoylation has been found to occur as a post-translational event as well, mainly in apoptotic cells [79–81].
Prior to Nt-myristoylation, the iMet needs to be removed by MetAP, thus exposing the consensus sequence recognized by NMT. The consensus sequence has been revealed in the context of several in vivo and in vitro studies [4,82]. Gly in the first position is an absolute requirement. In the second position a charged residue is favored, whereas aromatic residues and Pro are prohibited. There are no special requirements for the third position. Ala, Asn, Cys, Gly or Ser is allowed in the fourth position while in position five Cys, Ser, or Thr is preferred and Pro is prohibited.
Two isozymes of NMT, NMT1 and NMT2, have been isolated and characterized [76,83,84]. The evolutionary conservation of both structure and substrate specificity (Table 1) [82,84–90]. Human NMT1 and NMT2 possess approximately 77% amino acid sequence identity. The majority of divergence between the two NMTs is seen in the N-terminal domain, which is important for ribosomal binding [84,91]. Both unique and overlapping substrate specificities are observed between NMT1 and NMT2 within a given species [84,90,92–94].
It is estimated that about 0.5% of cellular proteins are Nt-myristoylated [95–99]. Different Nt-myristoylation prediction tools exist, e.g. MYR Predictor [100] and Myristoylator [101]. Caution should be exercised because false-positive and false-negative predictions might occur [102], thus demonstrating the necessity of both in vivo and in vitro studies for complete data [4]. Thinon et al. recently established a global profile of the Nt-myristoylome in both normal and apoptotic cells [103].
Unlike the immense amount of data on Nt-myristoylation, far less is known about Nt-palmitoylation. Normally, the palmitoyl group (16-carbon saturated fatty acid) is attached to an internal Cys residue [104], but a few instances of palmitoylated N-termini have been uncovered. A study by Klauss and Krause shows co-translational Nt-palmitoylation of GαS, the α-subunit of the heterotrimeric G protein responsible for activation of adenylyl cyclase. Here, a palmitoyl group from palmitoyl-CoA is attached to the α-amino group of the N-terminal Gly residue (Fig. 1) [105,106].
Interestingly, the secreted vertebrate signaling proteins Hedgehog (Hh) and Sonic Hedgehog (Shh) have been found Nt-palmitoylated at Cys, following cleavage of the N-terminal signal sequence [5,107]. Hhat, an N-terminal palmitoylacyltransferase (PAT) is suggested to palmitoylate Shh [108–111], and this modification constitutes an important regulatory feature for the strength of Shh signaling [112]. Hhat belongs to the family of multipass transmembrane proteins termed MBOAT (membrane-bound O-acyltransferase) [113] and acylates Shh during its passage through the secretory pathway [108]. Rasp, another member of the MBOAT family, is responsible for Nt-palmitoylation of the Hh and Spitz proteins in D. melanogaster [109,114,115].
6 N-terminal methylation
N-terminal methylation (Nt-methylation) is a process by which a methyl group is transferred from S-adenosyl methionine (SAM) to the exposed N-terminal α-amino group following iMet cleavage. The existence of Nt-methylation has been known for decades [6,116–118], however its structural and functional importance has only recently emerged [119–124].
The chemical consequence of Nt-methylation depends on the degree of residue methylation. Monomethylation will probably have a minor effect on the basicity of the α-amino group, by increasing the pKa slightly and cause some steric hindrance that may reduce its reactivity. In contrast, trimethylation (or dimethylation in the case of Pro) will have a profound effect by generating a permanent positive charge on the N-terminal amino group (Fig. 1). Consequently, the nucleophilicity normally associated with the α-amino nitrogen is abolished [6]. Nt-methylation is assumed irreversible as no N-terminal demethylase has been identified [7].
Recently, two independent studies finally identified the first N-terminal methyltransferases (NTMTs). The orthologues yeast Tae1 and human METTL11A were reported to respectively catalyze the stoichiometric Nt-methylation of the ribosomal proteins Rpl12a/b and Rps25a/b [125] and RCC1, RB and SETα [126]. To reflect their unique role in Nt-methylation the enzymes were renamed NTMT1. NTMT1, which is a member of the seven-beta-strand class I methyltransferase family, is conserved across eukaryotes and have one close human homologue. A study by Petkowski et al. confirmed that NTMT2 displays N-terminal methyltransferase activity as well [127].
The substrate consensus sequence for the eukaryotic NTMTs was initially thought to be X-Pro-Lys-, where X can be Ala, Pro or Ser (Table 1) [6]. Further in vitro studies have shown that NTMT1 is somewhat promiscuous concerning the identity of the first amino acid. A recombinant NTMT1 is able to methylate RCC1 peptides as long as the first position is not occupied by the acidic residues Asp or Glu, Trp, or the hydrophobic residues Ile or Leu [126]. An expanded peptide library methylation assay furthermore showed that the presence of a Pro in the second position is not an absolute requirement [128]. Efficient Nt-methylation requires Lys in the third position, but can in rare cases be replaced by Arg [123,126,128]. Taken together this implies that NTMT1 may have a broader specificity than previously acknowledged, and this could also be the case for NTMT2. Interestingly, it was recently shown that CENP-A [123] and CENP-B [121] with the Gly-Pro-starting N-termini are Nt-methylated in vivo.
Given both sequence and structural similarity [127], it is not unreasonable to believe that NTMT1 and NTMT2 have similar localization patterns and catalytic activities. In fact, both enzymes are expressed at low levels and localizes predominantly to the nucleus [127]. In contrast, the enzymes display different methylation mechanisms. NTMT1 is a distributive trimethylase, which can mono-, di-, and trimethylate its substrates whereas NTMT2 is primarily an Nt-monomethylase. A synergistic NTMT mechanism has been proposed where NTMT2 primes substrates for subsequent di- and trimethylation by NTMT1. Hence, NTMT2 would confer aid to NTMT1 when the substrate burden is too high [127].
7 N-terminal ubiquitylation
The addition of ubiquityl to a substrate protein proceeds through a three-step process that is achieved by the combined activity of ubiquitin activating (E1), conjugating (E2), and ligating (E3) enzymes. Most commonly, ubiquityl is conjugated to the ε-amino group of an internal Lys residue. N-terminal ubiquitylation (Nt-ubiquitylation) refers to the addition of an ubiquityl moiety to the free α-amino group of the first residue of a protein (Fig. 1). In both cases, ubiquityl may serve as a target for polyubiquitylation, which is a well-known degradation signal recognized by the proteasome [129,130].
An N-terminal residue was initially found by Breitschopf et al. to act as a novel site for ubiquitylation when Lys replacement in the protein MyoD did not significantly affect its susceptibility for either in vitro or in vivo ubiquitylation or degradation [131]. The first direct evidence of Nt-ubiquitylation came when MS analysis revealed that ubiquityl was indeed fused to the N-terminal α-amino group of HPV-58 oncoprotein E7 [132]. Previously, there had been strong indications that a handful of proteins underwent degradation mediated by Nt-ubiquitylation [133–139]. However, in these cases the stability is presumably modulated through interplay between ubiquitylation at the N-terminus and on internal Lys residues. Seeing that HPV-58 E7 is a naturally occurring lysine-less protein its degradation is more likely to be completely dependent upon Nt-ubiquitylation.
The known substrates for Nt-ubiquitylation do not share any homology in their N-terminal region [8]. Thus, it is not unreasonable to consider that Nt-ubiquitylation is facilitated by several enzymes that provide different substrate specificity, subcellular localization and modes of regulation. Hitherto, E2 Ube2w [140,141] and E3 HUWE1 [142] are the only enzymes with a reported ability to ubiquitylate the N-terminus of substrates. HUWE1 was shown to ubiquitylate the N-terminus of lysine-less MyoD, but, interestingly, favors an internal Lys in wild-type MyoD and leaves the N-terminus unubiquitylated [142]. Ube2w, on the other hand, is able to successfully ubiquitylate the N-terminus of a lysine-less version of Ataxin-3 and Tau [140]. When comparing the active site of Ube2w to that of classical E2s there are some distinctive differences. Together, these features make the novel active site of Ube2w more suitable to accommodate a neutral α-amino group rather than a positively charged Lys side chain [140,141]. Interestingly, Vittal et al. recently reported that Ube2w recognizes the peptide backbone of unstructured N-termini, and that the presence of Pro in position two to four has an inhibitory effect on Ube2w-activity [143].
Nt-ubiquitylation is not to be confused with the N-end rule, which relates the in vivo half-life of a protein to the identity of its N-terminal residue. Specific E3 ubiquitin ligases, called N-recognins, target protein substrates through their destabilizing N-terminal residues for polyubiquitylation and proteasomal degradation [144,145]. In eukaryotes, these N-terminal degradation signals, called N-degrons, comprise a destabilizing N-terminal residue as well as an internal lysine(s) residue within an unstructured (flexible) segment of the protein's polypeptide chain [146]. Primary destabilizing residues are positively charged (basic) and bulky hydrophobic N-terminal residues that are directly recognized by N-recognins [147,148]. A recent study reported that an unacetylated N-terminal Met could also act as a primary destabilizing residue if this Met is followed by a hydrophobic residue [149]. The secondary and tertiary destabilizing residues Asn, Asp, Cys, Gln, and Glu require preliminary enzymatic modifications, including Nt-deamidation and Nt-arginylation, before the recognition by N-recognins of the N-end rule pathway [147,148]. Yet another mechanistically distinct branch of the N-end rule pathway was identified in 2010 [150]. In this branch, termed the Ac/N-end rule pathway (the previously known branch was termed the Arg/N-end rule pathway), destabilizing N-terminal residues (those that can be Nt-acetylated by NATs) are recognized by distinct ubiquitin ligases (Doa10 and Not4) termed Ac/N-recognins [150,151]. The N-end rule pathway continues to be deciphered and as novel features are revealed, the framework of this proteolytic system keeps expanding (recommended in-depth reviews for further reading [145,147,148,152]).
8 Biological functions of N-terminal modifications
While the presence and abundance of Nt-modifications have been thoroughly demonstrated during the last decades, the functional roles of these modifications are now beginning to emerge. Very recent studies demonstrated that iMet cleavage and Nt-acetylation might be important players in the expanded N-end rule pathway, linking the identity of the protein N-terminus to its in vivo stability [149–151,153]. Nt-acetylation may further be important for targeting specific proteins to intracellular membranes like the Golgi-membrane or the inner nuclear membrane [154–156]. In a global survey, N-termini of cytosolic proteins were found to be more prone to be Nt-acetylated as compared to secreted proteins [157]. Interestingly, mutating the N-termini of specific proteins with signal sequences to become Nt-acetylated inhibited their post-translational translocation to the endoplasmic reticulum. This suggested that Nt-acetylation prevents this type of subcellular targeting. In some cases, Nt-acetylation is crucial for proper protein complex formation [158,159]. A very recent investigation revealed that deficiency in NatA-mediated Nt-acetylation most likely causes misfolding of a variety of NatA substrates thus suggesting Nt-acetylation to be a general factor mediating protein folding [160–162]. The (patho)physiological importance of Nt-acetylation only recently emerged, as the Ogden syndrome was found to be caused by a NAT mutation [58,163,164]. Clinical features of the Ogden syndrome include postnatal growth failure, developmental delays and death during infancy [164]. The genetic cause of the Ogden syndrome, a Ser37Pro mutation in NAA10, results in impaired enzymatic activity and NatA complex formation [58,163]. Further characterization of Naa10 Ser37Pro revealed specific downstream Nt-acetylation defects in vivo as well as abnormal cell migration and proliferation capacity of affected fibroblasts [163]. In addition, de novo missense mutations in the NAA10 gene have been identified in two independent cases of global developmental delay [165], and a truncated Naa10 protein is implicated in Lenz microphthalmia syndrome [166]. Finally, several NATs, in particular the NatA subunits Naa10 and Naa15, are dysregulated in various human cancers [167]. In most types of cancers investigated, NAT-overexpression mediates increased survival and proliferation of cancer cells. Because of its implication in human disease and cancer, NatA is a potential drug target and specific NAT-inhibitors are under development [168]. The role of Nt-acetylation during development is reviewed in another article in this issue [169].
The main feature of protein fatty acylation is to provide the target protein with hydrophobicity thus promoting membrane binding. Nt-palmitoylation is quite rare, but seems to be an important protein modification involved in signal transduction [106]. Indeed, the hydrophobic nature of palmitoyl has been found indispensable for the strength of Shh signaling [112]. A reduced pattering activity in mouse limb is observed for Shh in the absence of Nt-palmitoylation, while in D. melanogaster the Hh is found inactive [170,171]. In humans, an uncontrolled activation of Hh signaling pathway is linked to different types of cancer [172]. Compared to Nt-palmitoylation, Nt-myristoylation is more prevalent and the common denominator for many proteins modified in this way is their participation in cellular signaling pathways [173]. This involves subcellular targeting, protein-protein and protein-membrane interactions [4] and possibly protein structural stability [174].
To date, protein specific consequences of Nt-methylation have only been described for a subset of eukaryotic proteins [119–123]. Similar to NatA subunits, aberrant expression of NTMT1 has been reported in numerous cancer types. However, the functional understanding of NTMT1's role in cancer progression and prognosis is limited making it a focus of research [7]. Nt-methylation is inter alia involved in regulating protein function, specifically protein-DNA interactions, and several studies have suggested a role in chromatin conformation and segregation [119,121,123,126] and DNA repair [120]. Nt-methylation of RCC1 (regulator of chromosome condensation 1) was shown to promote association with chromatin. Both methylation-defective RCC1 mutants and NTMT1 knockdown results in decreased affinity for DNA, which causes mislocalization from chromatin and subsequent defects in spindle pole organization and chromosome missegregation [119,126]. RCC1 binds to chromatin through a bimodal attachment mechanism, where the methylated N-terminus associates with negatively charged DNA and the main protein body binds to histones H2A/H2B [119,124].
Due to limited knowledge on the Nt-ubiquitylome, the biological understanding of Nt-ubiquitylation remains limited. Vittal et al. proposed that Nt-ubiquitylation could be linked to protein quality control on stalled ribosomes, where it primes nascent polypeptides for subsequent polyubiquitylation [143]. On the contrary, Ube2w is predicted to harbor a nuclear localization signal and may therefore regulate nuclear proteins [175]. Ube2w has been found to be overexpressed in certain human cancers [176]. In that regard, Mittag and Marzahn have suggested that Nt-ubiquitylation could be involved in maintaining homeostasis of important regulatory proteins [177].
9 Discussion
The majority of eukaryotic protein N-termini is chemically modified by at least one of the many Nt-modifying enzymes. Multiple factors determine whether and how a given protein substrate will be modified including: i) the N-terminal amino acid sequence of the protein relative to the different specificities of the modifying enzymes, ii) the availability and subcellular localization of the different Nt-modifying enzymes under given conditions, and iii) the availability of the donor substrate (e.g., acetyl-CoA, SAM, etc.). Some of the Nt-modifying enzymes have overlapping substrate specificities (Table 2), so what governs the modification faith of individual proteins?
Table 2.
Protein N-terminal modifications sorted by the N-terminal sequence
| AA1 | AA2 | % abundance in pos. 2b) | Nt-modificationc) | N-terminus | Enzyme | Substrate |
|---|---|---|---|---|---|---|
| Met | Ala | 23.1 | Ac | Met-Ala- | NatE/F | |
| iMet exc. | Met-Ala- | MetAP1/2 | ||||
| Ac | Ala- | NatA | ||||
| Methyl | Ala-Pro-Lys- | NTMT1/2 | DDB2, SET | |||
| Arga) | 4.5 | Ac | Met-Arg- | NatE/F | ||
| Asn | 3.2 | Ac | Met-Asn- | NatB | ||
| Asp | 5.7 | Ac | Met-Asp- | NatB | ||
| iMet exc. | Met-Asp- | ND | Cyto. β-actin | |||
| Ac | Asp- | Naa10 | Cyto. β-actin | |||
| Cys | 0.9 | iMet exc. | Met-Cys- | MetAP1/2 | ||
| Ac | Cys- | NatA | ||||
| Palm | Cys- | Hhat | Shh | |||
| Gln | 2.3 | Ac | Met-Gln- | NatB/F | ||
| Glu | 9.6 | Ac | Met-Glu- | NatB | ||
| iMet exc. | Met-Glu- | ND | Cyto. γ-actin | |||
| Ac | Glu- | Naa10 | Cyto. γ-actin | |||
| Gly | 7.9 | Ac | Met-Gly | NatF | ||
| iMet exc. | Met-Gly- | MetAP1/2 | ||||
| Ac | Gly- | NatA | ||||
| Methyl | Gly-Pro-Lys- | NTMT1/2 | CENP-A/B | |||
| Myr | Gly- | NMT1/2 | ||||
| Palm | Gly- | PAT | GαS | |||
| Hisa) | 1.0 | Ac | Met-His- | NatE/F | ||
| Ile | 1.5 | Ac | Met-Ile- | NatC/F | ||
| Leu | 5.3 | Ac | Met-Leu- | NatC/E/F | ||
| Lys | 4.3 | Ac | Met-Lys- | NatE/F | ||
| Met | 1.6 | Ac | Met-Met- | NatE/F | ||
| Phe | 1.8 | Ac | Met-Phe- | NatC | ||
| Pro | 5.4 | iMet exc. | Met-Pro- | MetAP1/2 | ||
| Methyl | Pro-Pro-Lys- | NTMT1/2 | RB | |||
| Ser | 11.4 | Ac | Met-Ser- | NatE/F | ||
| iMet exc. | Met-Ser | MetAP1/2 | ||||
| Ac | Ser- | NatA | ||||
| Ac | Ser-Gly-Arg- | NatD | H2A | |||
| Ac | Ser-Gly-Gly- | NatD | H4 | |||
| Methyl | Ser-Pro-Lys- | NTMT1/2 | RCC1 | |||
| Thr | 4.4 | Ac | Met-Thr- | NatE/F | ||
| iMet exc. | Met-Thr- | MetAP1/2 | ||||
| Ac | Thr- | NatA | ||||
| Trp | 1.3 | Ac | Met-Trp- | NatC | ||
| Tyr | 0.8 | Ac | Met-Tyr- | NatE/F | ||
| Val | 4.0 | Ac | Met-Val- | NatE/F | ||
| iMet exc. | Met-Val- | MetAP1/2 | ||||
| Ac | Val- | NatA |
Arg and His presumable follow the same modification pattern as Lys.
Distribution based on human proteins listed in Swiss-Prot 56.0.
Most likely, N-terminal propionylation follows the same patterns as N-terminal acetylation.
AA, amino acid; ND, Not determined
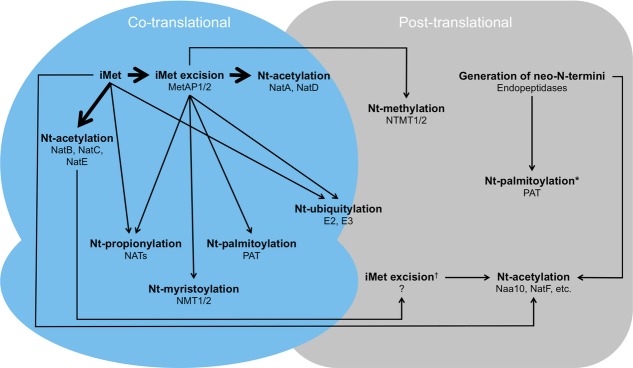
First, it is important to distinguish between co- and post-translational N-terminal modifications (Fig. 3). The enzymes capable of contacting the nascent N-termini, on the ribosome like MetAPs, NATs and NMTs, will be the first to target the pool of substrates. The non-ribosomal enzymes are however only capable of modifying the substrates not modified co-translationally, in addition to the neo-N-termini generated by endopeptidases. For instance, the Ser-Pro- or Gly-Pro- N-termini that are never co-translationally Nt-acetylated by NATs [60] are specifically targeted by NTMT-mediated post-translational Nt-methylation [6,123]. The substrate specificities of the NTMTs have possibly evolved to prefer those N-termini that are available (thus those that have not been modified by co-translationally acting NATs or NMTs).
Figure 3.
Overview of co- and post-translational N-terminal modifications. Co-translational protein modifications (left) taking place on the ribosomes include iMet excision, Nt-acetylation, Nt-propionylation, Nt-myristoylation or Nt-palmitoylation. Post-translational modifications (right) include Nt-methylation, Nt-palmitoylation or Nt-acetylation. Regarding Nt-ubiquitylation, it is uncertain whether it is a co- or post-translational event. Listed in relation to each modification are the responsible enzymes. The arrows are weighed according to the presumed extent of the individual modifications.*Preceding post-translational Nt-palmitoylation of Cys, a signal sequence is removed by endopeptidases thus generating neo-N-termini. †The case of mammalian cytoplasmic β-actin and γ-actin (Met-Asp and Met-Glu, respectively) involves co-translational Nt-acetylation of iMet, presumably by NatB, followed by post-translational Ac-iMet excision by an unknown aminopeptidase and finally Nt-acetylation by Naa10.
This simple picture of mutually exclusive modifications may be true given that all Nt-modifications are irreversible; one modification event will normally block for further modifications, except in the case of iMet excision. However, are these N-terminal modifications truly irreversible? In most cases the Nt-modifications are probably permanent throughout the lifetime of a protein. However, it is plausible to think that at least a small fraction of the Nt-modified proteins are subjected to hitherto unidentified Nt-demodifying enzymes that act to regulate critical substrates. If so, Nt-modifications may serve a dynamic regulatory role for protein function, and different modifications may be interchangeable to serve specific functions. Substrates suggested to be subjected to Nt-demodifying enzymes include Histone H4 [178] and myosin regulatory light chain 9 (MYL9) [7,128].
Among the enzymes acting on the ribosome, there may be competition for the emerging polypeptides. Sterically, the different modifying enzymes are unlikely to bind simultaneously to the ribosome [94]. Thus a specific order of modification events could be envisioned. However, this does not always seem to be the case. For example, competition between co-translational Nt-acetylation and Nt-myristoylation has been observed and found to depend on amino acid sequences beyond the targeted Gly residue [179]. Further, a recent study suggested a kinetic competition between MetAPs and NATs: If the NAT (NatE) acts first, then the iMet will be retained since further MetAP action is precluded by the acetylated iMet, while if the MetAP acts first, then iMet excision will occur and could potentially be followed by downstream Nt-acetylation by NatA [180].
Within a single class of enzymes like the NATs there is an apparent overlap in the in vitro substrate specificity; NatC, NatE and NatF all Nt-acetylate Met-starting N-termini with an aliphatic or hydrophilic amino acid in the second position (Fig. 2). This could perhaps be explained by specialization towards different types of substrates in vivo. NatC co-translationally acetylates hydrophobic N-termini that are normally never subjected to iMet excision (Met-Ile-, Met-Leu-, etc.). NatE (Naa50 bound to Naa15 and Naa10) co-translationally acetylates proteins that are often subjected to iMet excision (Met-Ala-, Met-Val-, etc.), but when targeted by NatE will retain its iMet in its acetylated state [180]. NatE/Naa50 may also have a post-translational role in the cytosol or nucleus towards Met-Leu- starting N-termini and similar substrates [49,59]. NatF, on the other hand, was very recently revealed to specifically Nt-acetylate transmembrane proteins [57]. This unique activity is most likely mediated in a post-translational manner via the membrane anchoring properties of NatF.
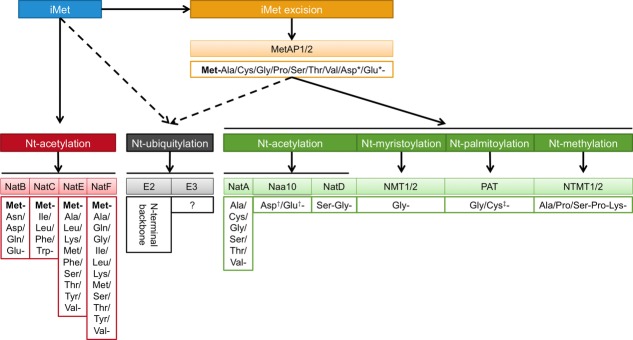
Figure 2.
Substrate specificity of N-terminal modifying enzymes. Proteins are synthesized with an initiator methionine (iMet). The iMet can remain at the N-terminus (blue) or be removed by MetAPs (yellow). A retained iMet can undergo Nt-acetylation (red) by one of four NATs (NatB/NatC/NatE/NatF), depending upon the subsequent amino acid (listed). Following iMet removal, the N-terminal amino acid residue can become Nt-acetylated, Nt-myristoylated, Nt-palmitoylated or Nt-methylated (green). It is not known whether Nt-ubiquitylation (grey) takes place on iMet and/or the exposed amino acid residue following iMet removal. A consensus sequence for Nt-ubiquitylation has not been established. *For Met-Asp and Met-Glu of mammalian cytoplasmic β-actin and γ-actin, respectively, iMet excision is catalyzed by an unidentified aminopeptidase. †Asp and Glu of mammalian cytoplasmic β-actin and γ-actin, respectively, are Nt-acetylated by Naa10. ǂCys is post-translationally Nt-palmitoylated after generation of protein neo-N-termini by endopeptidases.
Significant knowledge on the different N-terminal modifications and their responsible enzymes has been gained in the past decades. N-terminal modifications undoubtedly have a pivotal role in protein regulation and cellular signaling. However, future efforts are required to fully comprehend the functional role and the in vivo impact of these modifications, which includes understanding their dynamics, potential reversibility and interplay. Dysfunction or dysregulation of N-terminal modifying enzymes is implicated in human diseases including cancer, and this further stresses the importance of continuing research efforts in this field.
Acknowledgments
The authors acknowledge support of research grants from the Research Council of Norway (Grants 197136 and 230865 to TA), the Norwegian Cancer Society, the Bergen Research Foundation (BFS), The Western Norway Regional Health Authority, and the Meltzer Research Fund (to SV).
The authors have declared no conflict of interests.
Glossary
- CoA
coenzyme A
- iMet
initiator methionine
- MetAP
methionine aminopeptidase
- Naa
Nα-acetyltransferase
- NAT
N-terminal acetyltransferase
- NMT
N-terminal myristoyltransferase
- NPT
N-terminal propionyltransferase
- Nt
N-terminal
- NTMT
N-terminal methyltransferase
- PAT
palmitoylacyltransferase
- SAM
S-adenosyl methionine
10 References
- 1.Sherman F, Stewart JW, Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985;3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- 2.Starheim KK, Gevaert K, Arnesen T. Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci. 2012;37:152–161. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Foyn H, Van Damme P, Stove SI, Glomnes N, et al. Protein N-terminal acetyltransferases act as N-terminal propionyltransferases in vitro and in vivo. Mol. Cell. Proteomics. 2013;12:42–54. doi: 10.1074/mcp.M112.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin DD, Beauchamp E, Berthiaume LG. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie. 2011;93:18–31. doi: 10.1016/j.biochi.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Buglino JA, Resh MD. Palmitoylation of Hedgehog proteins. Vitam. Horm. 2012;88:229–252. doi: 10.1016/B978-0-12-394622-5.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stock A, Clarke S, Clarke C, Stock J. N-Terminal Methylation of Proteins – Structure, Function and Specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 7.Tooley JG, Schaner Tooley CE. New roles for old modifications: emerging roles of N-terminal post-translational modifications in development and disease. Protein Sci. 2014;23:1641–1649. doi: 10.1002/pro.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Kerwar SS, Weissbach H, Glenner GG. An aminopeptidase activity associated with brain ribosomes. Arch. Biochem. Biophys. 1971;143:336–337. doi: 10.1016/0003-9861(71)90215-3. [DOI] [PubMed] [Google Scholar]
- 10.Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- 11.Boissel JP, Kasper TJ, Bunn HF. Cotranslational amino-terminal processing of cytosolic proteins. Cell-free expression of site-directed mutants of human hemoglobin . J. Biol. Chem. 1988;263:8443–8449. [PubMed] [Google Scholar]
- 12.Burstein Y, Schechter I. Primary structures of N-terminal extra peptide segments linked to the variable and constant regions of immunoglobulin light chain precursors: implications on the organization and controlled expression of immunoglobulin genes. Biochemistry. 1978;17:2392–2400. doi: 10.1021/bi00605a022. [DOI] [PubMed] [Google Scholar]
- 13.Huang S, Elliott RC, Liu PS, Koduri RK, et al. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry. 1987;26:8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- 14.Tsunasawa S, Stewart JW, Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J. Biol. Chem. 1985;260:5382–5391. [PubMed] [Google Scholar]
- 15.Barwick RC, Jones RT, Head CG, Shih MF, et al. Hb Long Island: a hemoglobin variant with a methionyl extension at the NH2 terminus and a prolyl substitution for the normal histidyl residue 2 of the beta chain. Proc. Natl. Acad. Sci. USA. 1985;82:4602–4605. doi: 10.1073/pnas.82.14.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prchal JT, Cashman DP, Kan YW. Hemoglobin Long Island is caused by a single mutation (adenine to cytosine) resulting in a failure to cleave amino-terminal methionine. Proc. Natl. Acad. Sci. USA. 1986;83:24–27. doi: 10.1073/pnas.83.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blouquit Y, Arous N, Lena D, Delanoe-Garin J, et al. Hb Marseille [alpha 2 beta 2 N methionyl-2 (NA2) His—-Pro]: a new beta chain variant having an extended N-terminus. FEBS Lett. 1984;178:315–318. doi: 10.1016/0014-5793(84)80624-9. [DOI] [PubMed] [Google Scholar]
- 18.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 19.Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 1998;23:263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 20.Jackson R, Hunter T. Role of methionine in the initiation of haemoglobin synthesis. Nature. 1970;227:672–676. doi: 10.1038/227672a0. [DOI] [PubMed] [Google Scholar]
- 21.Palmiter RD, Gagnon, Walsh J, Ovalbumin KA. a secreted protein without a transient hydrophobic leader sequence. Proc. Natl. Acad. Sci. USA. 1978;75:94–98. doi: 10.1073/pnas.75.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arfin SM, Bradshaw RA. Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry. 1988;27:7979–7984. doi: 10.1021/bi00421a001. [DOI] [PubMed] [Google Scholar]
- 23.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell. Mol. Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arfin SM, Kendall RL, Hall L, Weaver LH, et al. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc. Natl. Acad. Sci. USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Addlagatta A, Hu X, Liu JO, Matthews BW. Structural basis for the functional differences between type I and type II human methionine aminopeptidases. Biochemistry. 2005;44:14741–14749. doi: 10.1021/bi051691k. [DOI] [PubMed] [Google Scholar]
- 26.Addlagatta A, Quillin ML, Omotoso O, Liu JO, Matthews BW. Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome. Biochemistry. 2005;44:7166–7174. doi: 10.1021/bi0501176. [DOI] [PubMed] [Google Scholar]
- 27.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giglione C, Serero A, Pierre M, Boisson B, Meinnel T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000;19:5916–5929. doi: 10.1093/emboj/19.21.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leszczyniecka M, Bhatia U, Cueto M, Nirmala NR, et al. MAP1D, a novel methionine aminopeptidase family member is overexpressed in colon cancer. Oncogene. 2006;25:3471–3478. doi: 10.1038/sj.onc.1209383. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Chang YH. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serero A, Giglione C, Sardini A, Martinez-Sanz J, Meinnel T. An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem. 2003;278:52953–52963. doi: 10.1074/jbc.M309770200. [DOI] [PubMed] [Google Scholar]
- 32.Sin N, Meng L, Wang MQ, Wen JJ, et al. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. USA. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giglione C, Fieulaine S, Meinnel T. Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem. Sci. 2009;34:417–426. doi: 10.1016/j.tibs.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnesen T, Van Damme P, Polevoda B, Helsens K, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Damme P, Hole K, Pimenta-Marques A, Helsens K, et al. NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation. PLoS Genet. 2011;7:e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strous GJ, Berns AJ, Bloemendal H. N-terminal acetylation of the nascent chains of alpha-crystallin. Biochem. Biophys. Res. Commun. 1974;58:876–884. doi: 10.1016/s0006-291x(74)80498-5. [DOI] [PubMed] [Google Scholar]
- 38.Strous GJ, van Westreenen H, Bloemendal H. Synthesis of lens protein in vitro. N-terminal acetylation of alpha-crystallin. Eur. J. Biochem./FEBS. 1973;38:79–85. doi: 10.1111/j.1432-1033.1973.tb03036.x. [DOI] [PubMed] [Google Scholar]
- 39.Helbig AO, Gauci S, Raijmakers R, van Breukelen B, et al. Profiling of N-Acetylated Protein Termini Provides In-depth Insights into the N-terminal Nature of the Proteome. Mol. Cell. Proteomics. 2010;9:928–939. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helsens K, Van Damme P, Degroeve S, Martens L, et al. Bioinformatics Analysis of a Saccharomyces cerevisiae N-Terminal Proteome Provides Evidence of Alternative Translation Initiation and Post-Translational N-Terminal Acetylation. J. Proteome Res. 2011;10:3578–3589. doi: 10.1021/pr2002325. [DOI] [PubMed] [Google Scholar]
- 41.Vetting MW, LP SdC, Yu M, Hegde SS, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 43.Polevoda B, Sherman F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 2003;308:1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
- 44.Polevoda B, Norbeck J, Takakura H, Blomberg A, Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautschi M, Just S, Mun A, Ross S, et al. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J. Cell. Biochem. 2008;103:492–508. doi: 10.1002/jcb.21418. [DOI] [PubMed] [Google Scholar]
- 47.Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnesen T, Anderson D, Baldersheim C, Lanotte M, et al. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochemical J. 2005;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Damme P, Evjenth R, Foyn H, Demeyer K, et al. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(alpha)-acetyltransferases and point to hNaa10p as the post-translational actin N(alpha)-acetyltransferase. Mol. Cell. Proteomics. 2011;10:M110 004580. doi: 10.1074/mcp.M110.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J. Biol. Chem. 2003;278:30686–30697. doi: 10.1074/jbc.M304690200. [DOI] [PubMed] [Google Scholar]
- 52.Starheim KK, Arnesen T, Gromyko D, Ryningen A, et al. Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem. J. 2008;415:325–331. doi: 10.1042/BJ20080658. [DOI] [PubMed] [Google Scholar]
- 53.Van Damme P, Lasa M, Polevoda B, Gazquez C, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. USA. 2012;109:12449–12454. doi: 10.1073/pnas.1210303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polevoda B, Sherman F. NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J. Biol. Chem. 2001;276:20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- 55.Starheim KK, Gromyko D, Evjenth R, Ryningen A, et al. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell. Biol. 2009;29:3569–3581. doi: 10.1128/MCB.01909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tercero JC, Wickner RB. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 1992;267:20277–20281. [PubMed] [Google Scholar]
- 57.Song OK, Wang XR, Waterborg JH, Sternglanz R. An N-alpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J. Biol. Chem. 2003;278:38109–38112. doi: 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- 58.Hole K, Van Damme P, Dalva M, Aksnes H, et al. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PloS One. 2011;6:e24713. doi: 10.1371/journal.pone.0024713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Damme P, Stove SI, Glomnes N, Gevaert K, Arnesen T. A Saccharomyces cerevisiae model reveals in vivo functional impairment of the Ogden syndrome N-terminal acetyltransferase NAA10 Ser37Pro mutant. Mol. Cell. Proteomics. 2014;13:2031–2041. doi: 10.1074/mcp.M113.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evjenth R, Hole K, Karlsen OA, Ziegler M, et al. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 2009;284:31122–31129. doi: 10.1074/jbc.M109.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aksnes H, Van Damme P, Goris M, Starheim KK, et al. An Organellar N-alpha Acetyltransferase, Naa60, Acetylates Cytosolic N-termini of Transmembrane Proteins and Maintains Golgi Integrity. Cell Rep. 2015;10:1362–1374. doi: 10.1016/j.celrep.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 62.Goetze S, Qeli E, Mosimann C, Staes A, et al. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vetting MW, Bareich DC, Yu M, Blanchard JS. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008;17:1781–1790. doi: 10.1110/ps.035899.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liszczak G, Arnesen T, Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 2011;286:37002–37010. doi: 10.1074/jbc.M111.282863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liszczak G, Goldberg JM, Foyn H, Petersson EJ, et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 2013;20:1098–1105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liszczak G, Marmorstein R. Implications for the evolution of eukaryotic amino-terminal acetyltransferase (NAT) enzymes from the structure of an archaeal ortholog. Proc. Natl. Acad. Sci. USA. 2013;110:14652–14657. doi: 10.1073/pnas.1310365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magin RS, Liszczak GP, Marmorstein R. The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD. Structure. 2015;23:332–341. doi: 10.1016/j.str.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evjenth RH, Brenner AK, Thompson PR, Arnesen T, et al. Human protein N-terminal acetyltransferase hNaa50p (hNAT5/hSAN) follows ordered sequential catalytic mechanism: combined kinetic and NMR study. J. Biol. Chem. 2012;287:10081–10088. doi: 10.1074/jbc.M111.326587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dormeyer W, Mohammed S, Breukelen B, Krijgsveld J, Heck AJ. Targeted analysis of protein termini. J. Proteome Res. 2007;6:4634–4645. doi: 10.1021/pr070375k. [DOI] [PubMed] [Google Scholar]
- 70.Zhang X, Ye J, Hojrup P. A proteomics approach to study in vivo protein N(alpha)-modifications. J. Proteomics. 2009;73:240–251. doi: 10.1016/j.jprot.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 71.Hosokawa Y, Shimomura Y, Harris RA, Ozawa T. Determination of short-chain acyl-coenzyme A esters by high-performance liquid chromatography. Anal. Biochem. 1986;153:45–49. doi: 10.1016/0003-2697(86)90058-8. [DOI] [PubMed] [Google Scholar]
- 72.King MT, Reiss PD. Separation and measurement of short-chain coenzyme-A compounds in rat liver by reversed-phase high-performance liquid chromatography. Anal. Biochem. 1985;146:173–179. doi: 10.1016/0003-2697(85)90412-9. [DOI] [PubMed] [Google Scholar]
- 73.Bhatnagar RS, Schall OF, Jackson-Machelski E, Sikorski JA, et al. Titration calorimetric analysis of AcylCoA recognition by myristoylCoA:protein N-myristoyltransferase. Biochemistry. 1997;36:6700–6708. doi: 10.1021/bi970311v. [DOI] [PubMed] [Google Scholar]
- 74.Farazi TA, Waksman G, Gordon JI. Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis. Biochemistry. 2001;40:6335–6343. doi: 10.1021/bi0101401. [DOI] [PubMed] [Google Scholar]
- 75.Rudnick DA, McWherter CA, Adams SP, Ropson IJ, et al. Structural and functional studies of Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase produced in Escherichia coli. Evidence for an acyl-enzyme intermediate. J. Biol. Chem. 1990;265:13370–13378. [PubMed] [Google Scholar]
- 76.Rudnick DA, McWherter CA, Rocque WJ, Lennon PJ, et al. Kinetic and structural evidence for a sequential ordered Bi Bi mechanism of catalysis by Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Biol. Chem. 1991;266:9732–9739. [PubMed] [Google Scholar]
- 77.Deichaite I, Casson LP, Ling HP, Resh MD. In vitro synthesis of pp60v-src: myristylation in a cell-free system. Mol. Cell. Biol. 1988;8:4295–4301. doi: 10.1128/mcb.8.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glover CJ, Hartman KD, Felsted RL. Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 1997;272:28680–28689. doi: 10.1074/jbc.272.45.28680. [DOI] [PubMed] [Google Scholar]
- 79.Olson EN, Spizz G. Fatty acylation of cellular proteins. Temporal and subcellular differences between palmitate and myristate acylation. J. Biol. Chem. 1986;261:2458–2466. [PubMed] [Google Scholar]
- 80.Wilcox C, Hu JS, Olson EN. Acylation of proteins with myristic acid occurs cotranslationally. Science. 1987;238:1275–1278. doi: 10.1126/science.3685978. [DOI] [PubMed] [Google Scholar]
- 81.Martin DD, Vilas GL, Prescher JA, Rajaiah G, et al. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Utsumi T, Sakurai N, Nakano K, Ishisaka R. C-terminal 15 kDa fragment of cytoskeletal actin is posttranslationally N-myristoylated upon caspase-mediated cleavage and targeted to mitochondria. FEBS Lett. 2003;539:37–44. doi: 10.1016/s0014-5793(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 83.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 84.Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J. Biol. Chem. 1987;262:1030–1036. [PubMed] [Google Scholar]
- 85.Duronio RJ, Reed SI, Gordon JI. Mutations of human myristoyl-CoA:protein N-myristoyltransferase cause temperature-sensitive myristic acid auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1992;89:4129–4133. doi: 10.1073/pnas.89.9.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J. Biol. Chem. 1998;273:6595–6598. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 87.Towler DA, Adams SP, Eubanks SR, Towery DS, et al. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. USA. 1987;84:2708–2712. doi: 10.1073/pnas.84.9.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Towler DA, Adams SP, Eubanks SR, Towery DS, et al. Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 1988;263:1784–1790. [PubMed] [Google Scholar]
- 89.Lodge JK, Johnson RL, Weinberg RA, Gordon JI. Comparison of myristoyl-CoA:protein N-myristoyltransferases from three pathogenic fungi: Cryptococcus neoformans, Histoplasma capsulatum, and Candida albicans. J. Biol. Chem. 1994;269:2996–3009. [PubMed] [Google Scholar]
- 90.Ntwasa M, Egerton M, Gay NJ. Sequence and expression of Drosophila myristoyl-CoA: protein N-myristoyl transferase: evidence for proteolytic processing and membrane localisation. J. Cell Sci. 1997;110:149–156. doi: 10.1242/jcs.110.2.149. [DOI] [PubMed] [Google Scholar]
- 91.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 92.Rioux V, Beauchamp E, Pedrono F, Daval S, et al. Identification and characterization of recombinant and native rat myristoyl-CoA: protein N-myristoyltransferases. Mol. Cell. Biochem. 2006;286:161–170. doi: 10.1007/s11010-005-9108-0. [DOI] [PubMed] [Google Scholar]
- 93.Takamune N, Kuroe T, Tanada N, Shoji S, Misumi S. Suppression of human immunodeficiency virus type-1 production by coexpression of catalytic-region-deleted N-myristoyltransferase mutants. Biol. Pharm. Bull. 2010;33:2018–2023. doi: 10.1248/bpb.33.2018. [DOI] [PubMed] [Google Scholar]
- 94.Yang SH, Shrivastav A, Kosinski C, Sharma RK, et al. N-myristoyltransferase 1 is essential in early mouse development. J. Biol. Chem. 2005;280:18990–18995. doi: 10.1074/jbc.M412917200. [DOI] [PubMed] [Google Scholar]
- 95.Ducker CE, Upson JJ, French KJ, Smith CD. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res. 2005;3:463–476. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giglione C, Fieulaine S, Meinnel T. N-terminal protein modifications: Bringing back into play the ribosome. Biochimie. 2014 doi: 10.1016/j.biochi.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 97.Boisson B, Giglione C, Meinnel T. Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 2003;278:43418–43429. doi: 10.1074/jbc.M307321200. [DOI] [PubMed] [Google Scholar]
- 98.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 99.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- 100.Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugii M, Okada R, Matsuno H, Miyano S. Performance improvement in protein N-myristoyl classification by BONSAI with insignificant indexing symbol. Genome Inform. Int. Conf. Genome Inform. 2007;18:277–286. [PubMed] [Google Scholar]
- 102.Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, et al. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 104.Suzuki T, Moriya K, Nagatoshi K, Ota Y, et al. Strategy for comprehensive identification of human N-myristoylated proteins using an insect cell-free protein synthesis system. Proteomics. 2010;10:1780–1793. doi: 10.1002/pmic.200900783. [DOI] [PubMed] [Google Scholar]
- 105.Thinon E, Serwa RA, Broncel M, Brannigan JA, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Linder ME, Middleton P, Hepler JR, Taussig R, et al. Lipid modifications of G proteins: alpha subunits are palmitoylated. Proc. Natl. Acad. Sci. USA. 1993;90:3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kleuss C, Krause E. Galpha(s) is palmitoylated at the N-terminal glycine. EMBO J. 2003;22:826–832. doi: 10.1093/emboj/cdg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 109.Pepinsky RB, Zeng C, Wen D, Rayhorn P, et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 110.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 2008;283:22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chamoun Z, Mann RK, Nellen D, von Kessler DP, et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293:2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 112.Lee JD, Treisman JE. Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr. Biol. 2001;11:1147–1152. doi: 10.1016/s0960-9822(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 113.Micchelli CA, The, Selva I, Mogila E, Perrimon V, Rasp N. a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development. 2002;129:843–851. doi: 10.1242/dev.129.4.843. [DOI] [PubMed] [Google Scholar]
- 114.Taylor FR, Wen D, Garber EA, Carmillo AN, et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry. 2001;40:4359–4371. doi: 10.1021/bi002487u. [DOI] [PubMed] [Google Scholar]
- 115.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 116.Miura GI, Buglino J, Alvarado D, Lemmon MA, et al. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev. Cell. 2006;10:167–176. doi: 10.1016/j.devcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 117.Miura GI, Treisman JE. Lipid modification of secreted signaling proteins. Cell Cycle. 2006;5:1184–1188. doi: 10.4161/cc.5.11.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brosius J, Chen R. The primary structure of protein L16 located at the peptidyltransferase center of Escherichia coli ribosomes. FEBS Lett. 1976;68:105–109. doi: 10.1016/0014-5793(76)80415-2. [DOI] [PubMed] [Google Scholar]
- 119.Chang CN, Schwartz M, Chang FN. Identification and characterization of a new methylated amino acid in ribosomal protein L33 of Escherichia coli. Biochem. Biophys. Res. Commun. 1976;73:233–239. doi: 10.1016/0006-291x(76)90698-7. [DOI] [PubMed] [Google Scholar]
- 120.Wittmann-Liebold B, Pannenbecker R. Primary structure of protein L33 from the large subunit of the Escherichia coli ribosome. FEBS Lett. 1976;68:115–118. doi: 10.1016/0014-5793(76)80417-6. [DOI] [PubMed] [Google Scholar]
- 121.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, et al. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 2007;9:596–U203. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cai Q, Fu L, Wang Z, Gan N, et al. alpha-N-methylation of damaged DNA-binding protein 2 (DDB2) and its function in nucleotide excision repair. J. Biol. Chem. 2014;289:16046–16056. doi: 10.1074/jbc.M114.558510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dai X, Otake K, You C, Cai Q, et al. Identification of novel alpha-n-methylation of CENP-B that regulates its binding to the centromeric DNA. J. Proteome Res. 2013;12:4167–4175. doi: 10.1021/pr400498y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kimura Y, Kurata Y, Ishikawa A, Okayama A, et al. N-Terminal methylation of proteasome subunit Rpt1 in yeast. Proteomics. 2013;13:3167–3174. doi: 10.1002/pmic.201300207. [DOI] [PubMed] [Google Scholar]
- 125.Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA. 2013;110:11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J. Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. Identification of Protein N-Terminal Methyltransferases in Yeast and Humans. Biochemistry. 2010;49:5225–5235. doi: 10.1021/bi100428x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, et al. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466:1125–1128. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, et al. NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. Biochem. J. 2013;456:453–462. doi: 10.1042/BJ20131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, et al. Substrate specificity of mammalian N-terminal alpha-amino methyltransferase NRMT. Biochemistry. 2012;51:5942–5950. doi: 10.1021/bi300278f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ciechanover A, Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta. 2014;1843:86–96. doi: 10.1016/j.bbamcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 132.Varshavsky A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 133.Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ben-Saadon R, Fajerman I, Ziv T, Hellman U, et al. The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 2004;279:41414–41421. doi: 10.1074/jbc.M407201200. [DOI] [PubMed] [Google Scholar]
- 135.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 136.Aviel S, Winberg G, Massucci M, Ciechanover A. Degradation of the epstein-barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 2000;275:23491–23499. doi: 10.1074/jbc.M002052200. [DOI] [PubMed] [Google Scholar]
- 137.Ikeda M, Ikeda A, Longnecker R. Lysine-independent ubiquitination of Epstein-Barr virus LMP2A. Virology. 2002;300:153–159. doi: 10.1006/viro.2002.1562. [DOI] [PubMed] [Google Scholar]
- 138.Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S. N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol, Cell. Biol. 2004;24:6140–6150. doi: 10.1128/MCB.24.14.6140-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 140.Fajerman I, Schwartz AL, Ciechanover A. Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem. Biophys. Res. Commun. 2004;314:505–512. doi: 10.1016/j.bbrc.2003.12.116. [DOI] [PubMed] [Google Scholar]
- 141.Trausch-Azar JS, Lingbeck J, Ciechanover A, Schwartz AL. Ubiquitin-Proteasome-mediated degradation of Id1 is modulated by MyoD. J. Biol. Chem. 2004;279:32614–32619. doi: 10.1074/jbc.M403794200. [DOI] [PubMed] [Google Scholar]
- 142.Scaglione KM, Basrur V, Ashraf NS, Konen JR, et al. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 2013;288:18784–18788. doi: 10.1074/jbc.C113.477596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tatham MH, Plechanovova A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to alpha-amino groups of protein N-termini. Biochem. J. 2013;453:137–145. doi: 10.1042/BJ20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Noy T, Suad O, Taglicht D, Ciechanover A. HUWE1 ubiquitinates MyoD and targets it for proteasomal degradation. Biochem. Biophys. Res. Commun. 2012;418:408–413. doi: 10.1016/j.bbrc.2012.01.045. [DOI] [PubMed] [Google Scholar]
- 145.Vittal V, Shi L, Wenzel DM, Scaglione KM, et al. Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat. Chem. Biol. 2015;11:83–89. doi: 10.1038/nchembio.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 147.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 149.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 2014;24:603–611. doi: 10.1016/j.tcb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 150.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim HK, Kim RR, Oh JH, Cho H, et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156:158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu. Rev. Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Giglione C, Vallon O, Meinnel T. Control of protein life-span by N-terminal methionine excision. EMBO J. 2003;22:13–23. doi: 10.1093/emboj/cdg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Behnia R, Panic B, Whyte JR, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 157.Murthi A, Hopper AK. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: roles for N-acetylation and an integral membrane protein. Genetics. 2005;170:1553–1560. doi: 10.1534/genetics.105.043620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 159.Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J. Biol. Chem. 1994;269:24310–24315. [PubMed] [Google Scholar]
- 162.Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat. Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arnesen T, Starheim KK, Van Damme P, Evjenth R, et al. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol, Cell. Biol. 2010;30:1898–1909. doi: 10.1128/MCB.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dikiy I, Eliezer D. N-terminal acetylation stabilizes N-terminal helicity in lipid- and micelle-bound alpha-synuclein and increases its affinity for physiological membranes. J. Biol. Chem. 2014;289:3652–3665. doi: 10.1074/jbc.M113.512459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Myklebust LM, Van Damme P, Stove SI, Dorfel MJ, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Human Mol. Genet. 2015;24:1956–1976. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rope AF, Wang K, Evjenth R, Xing J, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Human Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Popp B, Stove SI, Endele S, Myklebust LM, et al. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur. J. Human Genet. 2015;23:602–609. doi: 10.1038/ejhg.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Esmailpour T, Riazifar H, Liu L, Donkervoort S, et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet. 2014;51:185–196. doi: 10.1136/jmedgenet-2013-101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–276. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 170.Foyn H, Jones JE, Lewallen D, Narawane R, et al. Design, synthesis, and kinetic characterization of protein N-terminal acetyltransferase inhibitors. ACS Chem. Biol. 2013;8:1121–1127. doi: 10.1021/cb400136s. [DOI] [PubMed] [Google Scholar]
- 171.Silva DR, Martinho RG. Developmental roles of Protein N-terminal acetylation. Proteomics. 2015;15:2402–2409. doi: 10.1002/pmic.201400631. [DOI] [PubMed] [Google Scholar]
- 172.Dawber RJ, Hebbes S, Herpers B, Docquier F, van den Heuvel M. Differential range and activity of various forms of the Hedgehog protein. BMC Dev. Biol. 2005;5:21. doi: 10.1186/1471-213X-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lee JD, Kraus P, Gaiano N, Nery S, et al. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev. Biol. 2001;233:122–136. doi: 10.1006/dbio.2001.0218. [DOI] [PubMed] [Google Scholar]
- 174.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 175.Hayashi N, Titani K. N-myristoylated proteins, key components in intracellular signal transduction systems enabling rapid and flexible cell responses. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:494–508. doi: 10.2183/pjab.86.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 177.Yin G, Ji C, Wu T, Shen Z, et al. Cloning, characterization and subcellular localization of a gene encoding a human Ubiquitin-conjugating enzyme (E2) homologous to the Arabidopsis thaliana UBC-16 gene product. Front. Biosci. 2006;11:1500–1507. doi: 10.2741/1899. [DOI] [PubMed] [Google Scholar]
- 178.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:p11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mittag T, Marzahn MR. Protein disorder: wagging a tail at ubiquitin. Nat. Chem. Biol. 2015;11:7–8. doi: 10.1038/nchembio.1716. [DOI] [PubMed] [Google Scholar]
- 180.Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A, Kirmizis A. N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet. 2013;9:e1003805. doi: 10.1371/journal.pgen.1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Utsumi T, Sato M, Nakano K, Takemura D, et al. Amino acid residue penultimate to the amino-terminal gly residue strongly affects two cotranslational protein modifications, N-myristoylation and N-acetylation. J. Biol. Chem. 2001;276:10505–10513. doi: 10.1074/jbc.M006134200. [DOI] [PubMed] [Google Scholar]
- 182.Van Damme P, Hole K, Gevaert K, Arnesen T. N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics. 2015;15:2436–2446. doi: 10.1002/pmic.201400575. [DOI] [PubMed] [Google Scholar]