Abstract
Mutations in the hematopoietic transcription factor GATA-1 alter the proliferation/differentiation of hemopoietic progenitors. Mutations in exon 2 interfere with the synthesis of the full-length isoform of GATA-1 and lead to the production of a shortened isoform, GATA-1s. These mutations have been found in patients with Diamond–Blackfan anemia (DBA), a congenital erythroid aplasia typically caused by mutations in genes encoding ribosomal proteins. We sequenced GATA-1 in 23 patients that were negative for mutations in the most frequently mutated DBA genes. One patient showed a c.2T > C mutation in the initiation codon leading to the loss of the full-length GATA-1 isoform.
Keywords: anemia, erythropoiesis, Diamond–Blackfan, Gata-1, ribosomal protein
INTRODUCTION
In mammals, hematopoietic homeostasis is supported by the balance of the expression of multiple genes specific to each process. The GATA-1 gene is located on the X-chromosome (Xp11.23) and encodes a transcription factor that regulates the development of erythrocytes, megakaryocytic cells, eosinophils, mast cells, and dendritic cells [1]. GATA-1 protein has three functional domains (Fig. 1A): an N-terminal transactivation domain (TD), essential for transcriptional activation activity, a N-terminal zinc finger (NF), and a C-terminal zinc finger (CF) responsible for the binding to DNA [1,2]. Accumulating evidence supports the notion that defects in GATA-1 are linked to hematopoietic disorders. Exon 4 mutations have been identified in families with dyserythropoietic anemia, thrombocytopenia, thalassemia, and erythropoietic porphyria [3]. Furthermore, acquired somatic mutations in exon 2, that cause the loss of the N-terminal TD of GATA-1, have been found in individuals with Down Syndrome (DS) who developed either transient myeloproliferative disorder (TMD) or acute megakaryoblastic leukemia (AMKL) [4]. These mutations prevent the synthesis of full-length GATA-1 (GATA-1 FL), but allow for the synthesis of a short isoform of GATA-1 (GATA-1s), that lacks the N-terminal TD [4]. Related germline mutations have also been described. Hollanda et al. [5] reported a c.220G > C mutation in seven-male members of a family all with anemia and trilineage dysplasia. Sankaran et al. [6] identified a mutation in two siblings affected by DBA and another mutation in a third male from a cohort of 62 additional DBA patients (Table I). Both mutations occur at the donor splice site of exon 2 in the GATA-1 gene resulting in exon skipping. Thus only GATA-1s is produced.
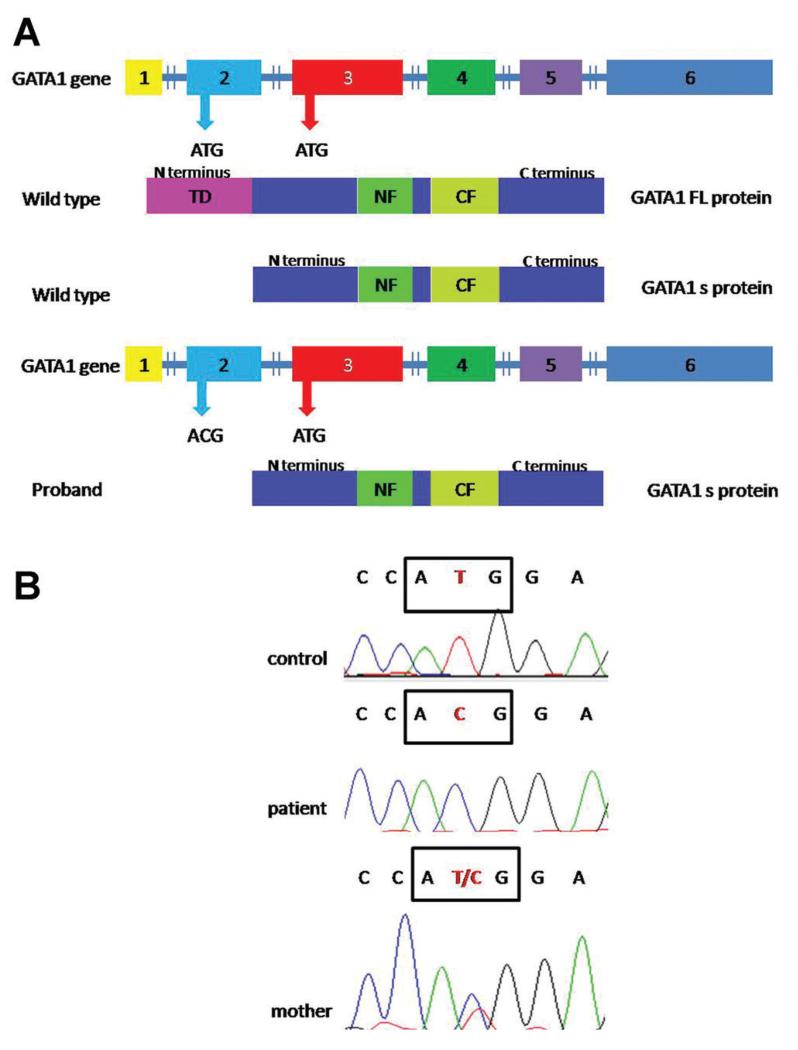
Fig. 1.
GATA-1 mutation; (A) Schematic representation of GATA-1 protein production, Description of normal and alternative translation initiation sites located at methionine 1 and 84, respectively. The GATA-1 mutation in the first ATG in exon 2 promotes the production of only GATA-1 short isoform while in normal condition both isoforms are produced; (B) Sanger sequencing. The mutation c.2T > C is highlighted in the box.
TABLE I.
Phenotype Associated to the Mutation of Exon 2 of GATA-1
| Reference | Mutation | Patient | Hematology |
|---|---|---|---|
| Sankaran et al. [6] | c.220G>C | Two Brothers | Macrocytic anemia with low reticulocyte counts; mild reduction in neutrophils count |
| Sankaran et al. [6] | c.220delG | 1 | Anemia without other hematologic abnormalities |
| Hollanda et al. [5] | c.220G>C | Seven affected males in a large family | Macrocytic anemia and neutropenia |
| This paper | c.2T>C | 1 | Anemia and progressive three linear hypoplasia |
DBA is a normochromic–macrocytic anemia characterized by impaired proliferation/maturation of the erythroid precursors in the bone marrow. Approximately, 30% of the patients have a heterogeneous mix of congenital malformations [7]. Sixty percent of patients carry mutations in genes encoding ribosomal proteins (RP). Systematic sequencing of all RP genes in patients with DBA has allowed the identification of mutations in 11 RP genes [8–10]. The remaining 40% of patients have mutations in unknown genes.
MATERIALS AND METHODS
Patients
Analysis of the Italian DBA Registry [11] revealed 23 out of 173 patients eligible for GATA-1 screening. These patients were all males and lacked mutations/deletions in the most frequently mutated DBA genes (RPS10, RPS17, RPS19, RPS24, RPS26, RPL5, RPL11, or RPL35A). Clinical characteristics of this cohort are presented in Supplementary Table I. DBA diagnoses were made using the guidelines reported in Vlachos et al. [7] Written informed consent was provided by all subjects participating in this study.
Sanger Sequencing
The six GATA-1 exons were screened for mutations in our cohort using standard PCR-based Sanger sequencing on genomic DNA extracted from peripheral blood. Coding sequences and exon–intron boundaries were PCR amplified using GoTaq® Flexi DNA Polymerase (Promega, Madison, WI). PCR products were sequenced in both directions using a Big Dye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Sequencing data were analyzed using Chromas Lite 2.1.1.
RESULTS
One of the 23 patients studied showed a c.2T > C hemizygous mutation in the initiation codon used to synthetize the GATA-1 FL. This mutation was inherited from his mother (Fig. 1B). As a consequence of this change, GATA-1s is the only isoform expressed. Support for this view comes from a study of DS patients with TMD [4] where mutations in the initiation codon for GATA-1 FL, that cause the loss of the first methionine, including a c.2T > C mutation, were shown to lead to the exclusive synthesis of GATA-1s.
Our is the first study that reports a germline mutation in the first ATG codon of GATA-1 (Fig. 1B).
The proband reported here (Supplementary Table I, patient number 10) was born at term after an uneventful pregnancy. At the age of 9 months he developed a severe hyporegenerative anemia (Hb 5.5 g/dl, MCV 93 fL, reticulocyte count 40,000/μl) and was referred to us based on a suspected diagnosis of DBA. No somatic malformations were evident. His erythrocyte adenosine deaminase activity was slightly increased (1.9 U/g Hb, normal values 1 ± 0.2 U/g Hb). Leukocytes and platelets were in the normal range. The bone marrow aspirate showed a selective deficiency in erythroid precursors without abnormalities of the other hematopoietic lineages. He was treated with a standard dose of prednisone (2 mg/kg/day) for 4 weeks which was gradually reduced. Only a partial response was observed and his hemoglobin level reached 8 g/dl. Treatment was discontinued after 3 months and his hemoglobin level persisted between 8 and 9 g/dl without the need of further treatment including red blood cell transfusion.
At the age of 4 years he developed a progressive three linear cytopenia. A bone marrow biopsy showed a severe hypocellular marrow. The karyotype showed mosaicism with a 45XY, −7 clone (65%) and a further 50XXY, +3, +8, +21 clone. On this basis, he was diagnosed with Myelodysplastic Syndrome (MDS) after which he underwent Hematopoietic Stem Cell Transplantation (HSCT) from an unrelated donor. Ten years posttransplant, the patient is transfusion independent and in good health.
DISCUSSION
Our study reports a patient with a hyporegenerative macrocytic anemia with a mutation in GATA-1 that interferes with the expression of GATA-1 FL, leaving GATA-1s as the sole isoform expressed. Mutations that abolish the expression of GATA-1 FL have been previously identified in 10 DBA patients from three independent families (Table I) [5,6]. These patients showed various degrees of anemia, with or without an involvement of other hematopoietic lineages (Table I). Our patient displayed a progressive trilinear cytopenia evolving to MDS requiring HSCT. This evolution occurs infrequently in DBA but at an incidence higher than the general population [12]. Progression to MDS is not restricted to GATA-1 genotypes since it has also been observed in patients with mutations in RP genes.
DBA is considered the prototype of ribosomopathies, since the discovery that a mutation in a RP can lead to the impairment of the biogenesis and function of the ribosomes. GATA-1 is the first and so far the only non-ribosomal protein gene that when mutated leads to DBA. In contrast to ribosomal proteins, whose essential function in all cells make it difficult to explain such a selective phenotype, a role for GATA-1 as a causative gene in DBA is much more intuitive. Given its role in the transcription of a number of genes required for erythroid development, loss of its transcription activation domain as occurs in patients with DBA is consistent with the erythroid phenotype.
Unresolved is the relationship between DBA caused by mutations in GATA-1 and RPs. It may be possible that RP haploinsufficiency interferes with the expression of GATA-1 FL. Alternatively, the pathways involving GATA-1 and ribosome function may be independent but result in similar phenotypes. The association of somatic GATA-1 mutations with TMD in patients with DS, raises concerns regarding risks to patients with DBA harboring similar mutations in GATA-1. While the sample size is far too small to calculate relative risks for such progression, the trilineage cytopenia and outgrowth of myelodysplastic clones observed in our patient indicate a need for further studies in this area and the careful surveillance of affected individuals.
Supplementary Material
Acknowledgments
Grant sponsor: Istituto Piemontese per la ricerca sulla Anemia di Diamond–Blackfan (to I.D. and U.R.); Grant sponsor: Diamond–Blackfan Anemia Foundation; Grant sponsor: ENERCA; Grant sponsor: Telethon Grant; Grant number: GGP 10181; Grant sponsor: Cariplo; Grant number: 2011-0554; Grant sponsor: PRIN (to I.D.); Grant sponsor: Banca del Piemonte (to U.R.)
Footnotes
Conflict of interest: Nothing to declare.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- 1.Ferreira R, Ohneda K, Yamamoto M, et al. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calligaris R, Bottardi S, Cogoi S, et al. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc Natl Acad Sci USA. 1995;92:11598–11602. doi: 10.1073/pnas.92.25.11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciovacco WA, Raskind WH, Kacena MA. Human phenotypes associated with GATA-1 mutations. Gene. 2008;427:1–6. doi: 10.1016/j.gene.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanezaki R, Toki T, Terui K, et al. Down syndrome and GATA1 mutations in transient abnormal myeloproliferative disorder: Mutation classes correlate with progression to myeloid leukemia. Blood. 2010;116:4631–4638. doi: 10.1182/blood-2010-05-282426. [DOI] [PubMed] [Google Scholar]
- 5.Hollanda LM, Lima CS, Cunha AF, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38:807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 6.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: Results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan Anemia: Mutation and database update. Hum Mutat. 2010;31:1269–1279. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazda HT, Preti M, Sheen MR, et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in Diamond-Blackfan anemia. Hum Mutat. 2012;33:1037–1044. doi: 10.1002/humu.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landowski M, O’Donohue MF, Buros C, et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132:1265–1274. doi: 10.1007/s00439-013-1326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campagnoli MF, Ramenghi U, Armiraglio M, et al. RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008;29:911–920. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 12.Vlachos A, Farrar JE, Atsidaftos E, et al. Diminutive somatic deletions in the 5q region lead to a phenotype atypical of classical 5q- syndrome. Blood. 2013;122:2487–2490. doi: 10.1182/blood-2013-06-509935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.