SUMMARY
N6-methyladenosine (m6A) is the most abundant internal modification of messenger RNA. While the presence of m6A on transcripts can impact alternative splicing, a nuclear reader of this mark that mediates the processing of nuclear transcripts has not been identified. We find that the RNA-binding HNRNPA2B1 protein binds m6A-bearing RNAs in vivo and in vitro and its biochemical footprint matches the m6A consensus motif. HNRNPA2B1 directly binds a set of nuclear transcripts and modulates their alternative splicing in a similar manner as the m6A ‘writer’ METTL3. Moreover, HNRNPA2B1 binds to m6A marks in a subset of primary-miRNA transcripts, interacts with the microRNA Microprocessor complex protein DGCR8, and promotes primary miRNA processing—phenocopying the effect of METTL3 depletion on the processing of these precursor transcripts. We propose HNRNPA2B1 to be a nuclear reader of the m6A mark and to mediate, in part, this mark’s effects on primary microRNA processing and alternative splicing.
INTRODUCTION
Nucleotide modifications can expand the information content of nucleic acids. Methylation of cytosines on promoter DNA, for example, can alter the transcriptional output of genes. RNA can also be modified by a myriad of marks and mRNA modifications including among others N6-methyladenosine, 5-methylcytosine, 2n O-methylation of ribose and pseudouridylation (Jaffrey, 2014). The functional roles of the majority of these chemical modifications are unknown. The most common internal modification of eukaryotic messenger RNA is methylation of the N6 nitrogen on adenosine referred to as N6-methyladenosine (m6A). Each transcript contains 1–3 m6A marks on average, which are located in specific positions in introns, exons, and untranslated regions. Methylation of specific positions is non-stoichiometric with only a fraction of transcripts containing this mark at specific sites. While this important modification was discovered over four decades ago, the transcriptome-wide mapping of this mark using next-generation sequencing analysis has recently revived interest in the biological role(s) of m6A (Dominissini et al., 2012; Goodarzi et al., 2012; Meyer et al., 2012; Wang et al., 2014a).
Methyltransferase like 3 (METTL3) was the first methyltransferase implicated in placing m6A on RNA (Dominissini et al., 2012; Meyer et al., 2012; Wei and Moss, 1974). METTL14, which exists in complex with METTL3, has also been shown to exhibit m6A methyltransferase activity (Liu et al., 2014). Fat mass and obesity-associated protein (FTO) was recently identified as an enzyme that can remove this mark (Jia et al., 2011). Classical experiments suggested a role for m6A in the nuclear processing of viral and non-viral transcripts (Carroll et al., 1990; Katz et al., 2015; Liu et al., 2015; Stoltzfus and Dane, 1982). More recent molecular studies have demonstrated m6A to exhibit both a nuclear role in splicing (Dominissini et al., 2012) and a cytoplasmic role in the regulation of RNA stability (Batista et al., 2014; Wang et al., 2014a). We have recently found that m6A plays an additional role in the nucleus in controlling microRNA biogenesis. M6A was found to be enriched in primary microRNA transcripts. Reducing m6A levels by depleting the m6A writer METTL3 reduced the levels of the majority of expressed microRNAs (Alarcon et al., 2015). This effect occurred in the nucleus since METTL3/m6A depletion reduced the association of the DGCR8-containing microRNA processing complex with primary microRNAs and caused unprocessed primary microRNA precursors to accumulate in the nucleus. These findings are consistent with a model wherein the m6A RNA mark is recognized and bound by a nuclear reader protein, which recruits the DGCR8 Microprocessor complex to pri-miRNA containing transcripts resulting in their processing. Recently the YTH domain family 2 (YTHDF2) protein was identified as a ‘reader’ of m6A and found to regulate the stability of m6A-bearing transcripts in the cytoplasm (Wang et al., 2014a). The proposed role of m6A in the control of alternative splicing and our recent findings regarding its impact on primary microRNA processing (Alarcon et al, 2015), suggest the existence of nuclear reader(s) of m6A that bind this mark and mediate nuclear processing events that are mechanistically distinct from RNA stability regulatory events that could occur in the cytoplasm.
We herein identify HNRNPA2B1 as a nuclear reader of m6A. HNRNPA2B1 binds to RGm6AC containing sites on nuclear RNAs in vivo and in vitro. HNRNPA2B1 regulates the alternative splicing of exons in a set of transcripts in a similar manner as METTL3, the m6A ‘writer’. The global impact of HNRNPA2B1 depletion on alternative splicing is highly correlated with the global effect of METTL3 depletion on alternative splicing. HNRNPA2B1 depletion also impairs the nuclear processing of a subset of microRNAs whose maturation is dependent on METTL3 activity. Moreover, HNRNPA2B1 interacts with the DGCR8 protein, a component of the pri-miRNA Microprocessor complex and facilitates the processing of pri-miRNAs. Our findings implicate HNRNPA2B1 as a nuclear reader and effector of the m6A mark.
RESULTS
HNRNPA2B1 binds to m6A-bearing sites in the transcriptome
The presence of the m6A mark has been historically associated with the processing of nuclear transcripts. Our identification of m6A as a regulatory mark that promotes the processing of pri-microRNA transcripts a process that occurs in the nucleus suggested the existence of one or more nuclear readers of this mark that mediate its downstream effector functions. Several approaches that combine UV cross-linking of RNA-binding proteins with immunoprecipitation and deep-sequencing of bound RNAs have proven powerful at mapping the sites of interaction between RNA-binding proteins and their target RNAs (Hafner et al., 2010; Konig et al., 2010; Ule et al., 2005). Our laboratories have in the past few years conducted a number of such high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP; Zhang and Darnell, 2011) studies to map the genome-wide binding of a variety of RNA-binding proteins. Our analysis of the direct binding sites of one such RNA-binding protein, HNRNPA2B1 (Goodarzi et al., 2012), suggested that it might act as a reader of the m6A mark. HNRNPA2B1 is a member of the hnRNP family of RNA-binding proteins that are known to associate with pre-mRNAs in the nucleus. The m6A mark is known to occur within the RGAC motif, with R representing a purine (Schibler et al., 1977). To test the possibility that HNRNPA2B1 also recognizes this motif, we assessed the abundance of this sequence among the HNRNPA2B1 HITS-CLIP peaks relative to randomly generated sequence counterparts with similar dinucleotide frequencies (Giannopoulou and Elemento, 2011). Indeed, using the FIRE analysis pipeline (Elemento et al., 2007), we observed a highly significant enrichment of the RGAC element in HNRNPA2B1 binding sites (Figure 1A).
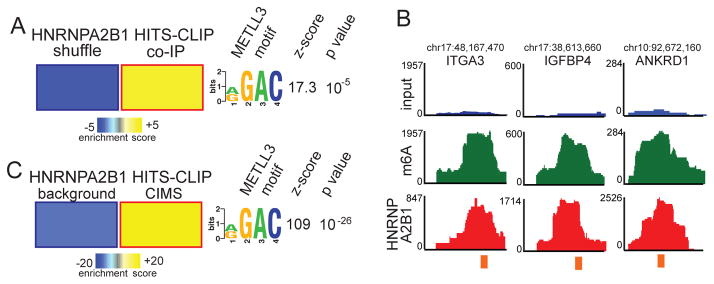
Figure 1. HNRNPA2B1 recognizes m6A methylated sequences.
(A) FIRE, in non-discovery mode, was used to assess enrichment of the RGAC motif among the HNRNPA2B1 HITS-CLIP peaks, obtained from MDA-MB-231 cells, relative to randomly generated sequences with similar dinucleotide frequencies. The figure shows a significant enrichment of the m6A motif (RGAC) in HNRNPA2B1 binding sites, where yellow indicates over-representation and blue represents under-representation. The magnitude of the representation is as per the heat-map scale shown at the bottom. The associated z-score and p-value are also provided.
(B) RNA sequencing read density at exemplary loci where HNRNPA2B1 and m6A peaks intersect. Shown are the input nuclear RNA (blue), m6A-seq (green), and HNRNPA2B1 HITS-CLIP (red) reads. The orange bar denotes the sequence match to the RGAC motif.
(C) FIRE analysis of the enrichment of the RGAC motif in the HNRNPA2B1 footprints. Sequences within 5nt of HNRNPA2B1 cross-linking induced deletions generated by HITS-CLIP protocol were significantly enriched for the RGAC motif when compared to background deletions. The motif, p-value and z-score are also shown.
We next overlapped the m6A peaks from an RNA-seq analysis of m6A-immunoprecipitated nuclear RNA (m6A-seq; Alarcon et al., 2015) with HNRNPA2B1 binding peaks obtained from HITS-CLIP of endogenous nuclear HNRNPA2B1 performed for this study in MDA-MB-231 breast cancer cells. We noted a significant number of cases wherein the HNRNPA2B1 peaks overlapped with m6A peaks (Figure 1B, S1A; p-value< 1E-3), consistent with an enrichment of m6A at HNRNPA2B1 binding sites. Of the 1912 human genes bound by HNRNPA2B1 and containing m6A peaks, 426 contained RGAC instances overlapping both an HNRNPA2B1 HITS-CLIP peak and an m6A-seq peak (FDR<10%). Conducting this analysis on HITS-CLIP data we had previously generated for MBNL1 and TARBP2 revealed zero such cases of overlap for these RNA-binding proteins.
The enrichment of the RGAC motif in HNRNPA2B1 binding sites may be due to general proximity of the RGAC motif to the HNRNPA2B1 binding sites rather than due to direct interactions with the RGAC motif. The RNA-binding protein hnRNPC, for example, has been shown to bind near rather than directly to the RGAC motif. Thus, to study the specificity of overlap between the HNRNPA2B1 footprint and the m6A consensus motif at nucleotide resolution, we took advantage of cross-linking induced deletions that are a hallmark of HITS-CLIP experiments and which provide a nucleotide-level map of physical points of interaction (Zhang and Darnell, 2011). For this, we limited our search to the cross-linking induced deletions that mark the sites of direct interactions between HNRNPA2B1 and its target RNAs (CIMS; Zhang and Darnell, 2011). We also defined a set of control sequences consisting of deletion-containing reads that represent the background occurrence of deletion events during HITS-CLIP sample preparation (i.e. these reads were not part of a peak or cluster; Figure S1). We then used FIRE to re-evaluate the abundance of the RGAC motif in HNRNPA2B1 footprints (i.e. crosslinking-induced deletion sites along with 5nt flanking sequences on either side). We observed a highly significant enrichment of RGAC motifs at these cross-linking induced deletions (p-value <10E-26, z-score=109; Figure 1C). A similar analysis applied to two other RNA-binding proteins for which comparable HITS-CLIP data was available revealed HNRNPA2B1 to be unique among this group in exhibiting a significant enrichment for proximal binding to the RGAC motif (Figure 1C and S1C). Importantly, 6.8% of the nuclear HNRNPA2B1 cross-linking induced deletions overlapped with an m6A peak; whereas, only 0.34% of the background deletions overlapped with m6A sites (20-fold enrichment, Figure S1D). These findings strongly support a role for HNRNPA2B1 in directly binding to a subset of m6A consensus sequences within the transcriptome.
METTL3 and HNRNPA2B1 regulate common alternative splicing events
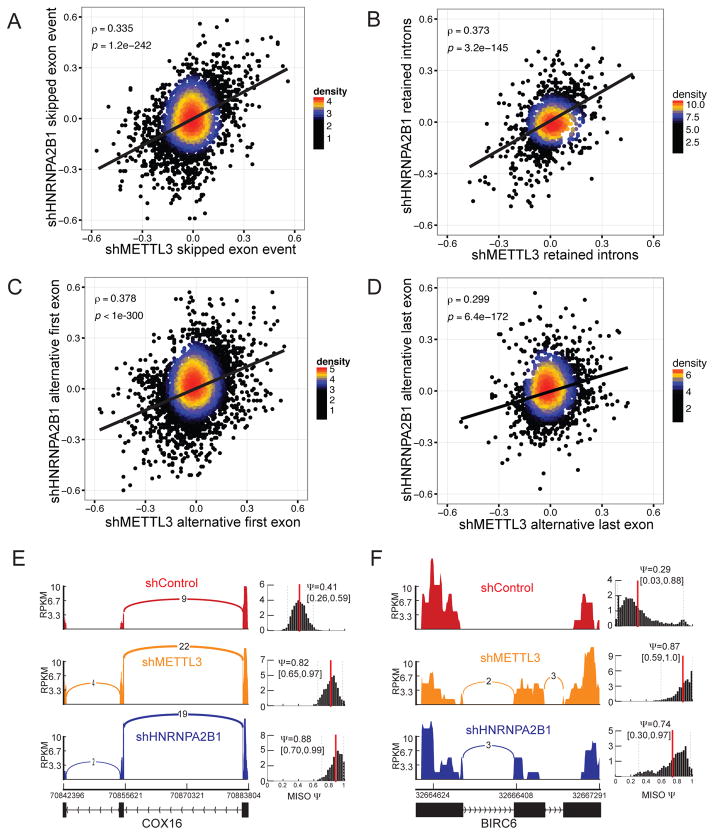
The m6A mark has been associated with (Carroll et al., 1990; Katz et al., 2015; Stoltzfus and Dane, 1982) and causally implicated in pre-mRNA processing (Dominissini et al., 2012). Depletion of METTL3 has also been shown to impact alternative splicing (Dominissini et al., 2012). We hypothesized that HNRNPA2B1, by acting downstream of m6A placement by METTL3, may impact alternative splicing in a similar manner as METTL3. If this were the case, depleting HNRNPA2B1 should result in similar alterations in alternative splicing patterns as depletion of METTL3. To test this, we conducted high-throughput RNA-sequencing (RNA-seq) on nuclear RNA from cells depleted of METTL3 and those depleted of HNRNPA2B1. We then employed MISO, a probabilistic framework (Katz et al., 2010), to quantify the relative expression of alternatively spliced exons in METTL3 and HNRNPA2B1 knockdown cells as well as controls. The differences in MISO-calculated Ψ (psi, percent-spliced in) values between each of the knockdown samples and the control samples were used as a measure of modulations in alternative splicing events upon depletion of each of these factors. Consistent with a role for HNRNPA2B1 in mediating alternative splicing effects downstream of METTL3/m6A, we observed that the global impact on alternative splicing was highly correlated in cells depleted for each of these genes. This effect was observed in skipped exons (ρ= 0.335; p<10−200; Figure 2A), retained introns (ρ= 0.373; p<10−140; Figure 2B), alternative first exons (ρ= 0.378; p<10−300; Figure 2C) and alternative last exons (ρ= 0.299; p<10−170; Figure 2D). The effects on alternative splicing in the context of HNRNPA2B1 and METTL3 depletion were seen in two independent cell lines (Figure 2E–F and Figure S2.). These findings reveal that HNRNPA2B1 depletion has similar genome-wide consequences on alternative splicing as METTL3 depletion. These observations along with our findings that HNRNPA2B1 binds m6A sites support a model whereby HNRNPA2B1 would mediate, in part, the alternative splicing effects of METTL3 and the m6A mark.
Figure 2. Depletion of HNRNPA2B1 and METTL3 similarly affect RNA splicing.
(A–D) Correlation between differential percent spliced in (ψ) in annotated alternative splicing events following HNRNPA2B1 and METLL3 depletion. Annotated skipped exons (A), retained introns (B), alternative first exons (C), and alternative last exons (D) were quantified in HNRNPA2B1 and METTL3 knockdown MDA-MB-231 cells respectively (relative to control cells). Spearman correlation was then used to assess the similarity in splicing modulations following depletion of METTL3 (m6A writer) and HNRNPA2B1 (m6A reader).
(E) Exemplary sashimi plots (Katz et al., 2015) showing concerted alternative splicing changes that occurred in MDA-MB-231 cells depleted of METTL3 or HNRNPA2B1.
(F) Exemplary of sashimi plots as in E but using an independent cell line, HeLa. Shown are the normalized coverage at each exon, along with the estimated ψ value (percent spliced in). For example, in this case, 29% of transcripts were estimated to contain the skipped exon in the control sample, while this estimate was increased to 87% and 74% for METTL3 and HNRNPA2B1 depleted cells, respectively.
HNRNPA2B1 promotes processing of METTL3-dependent microRNAs and interacts with the Microprocessor machinery
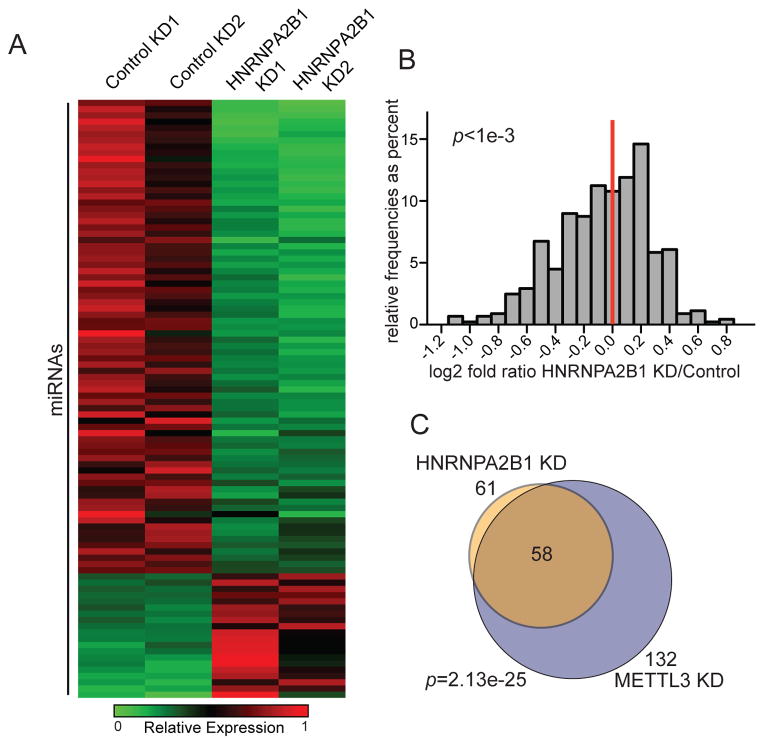
We next asked whether HNRNPA2B1 governs the processing of pri-miRNAs that are dependent on METTL3/m6A for their processing. For this, we performed global microRNA profiling in HEK293 cells depleted of HNRNPA2B1. HNRNPA2B1 knockdown caused a reduction in the levels of a large number of microRNAs (Figure 3A and Figure S3). This reduction could also be visualized as a left-shift in the population-level expression of miRNAs (Figure 3B; p<1e-3). The reduction in mature microRNA expression mirrored what was previously observed upon METTL3 depletion (Alarcon et al., 2015).
Figure 3. Depletion of HNRNPA2B1 impacts miRNA production.
(A) Heat map depicting the miRNAs affected at least by 50% by 2 independent shRNAs targeting HNRNPA2B1 in HEK293 cells. Red represents higher expression and green lower expression levels.
(B) Histogram of the fold change (log2) observed in miRNA expression, as obtained by genome-wide miRNA expression profiling shown in (A). The ratio of the average level for the two independent shRNAs over the average of the two controls is shown. The p-value of the two-sample Kolmogorov-Smirnov test is indicated.
(C) Venn diagram depicting the intersection of miRNAs that were reduced by greater than 50% upon HNRNPA2B1 or METTL3 depletion. 132 miRNAs were downregulated by more than 50% upon METTL3 depletion and 61 miRNAs were downregulated by 50% upon HNRNPA2B1 depletion. The extent of miRNAs detected by microarray in both experiments was 329. The p-value was calculated based on the hypergeometric distribution.
We next investigated the overlap between the set of miRNAs impacted by HNRNPA2B1 depletion and those impacted by METTL3 depletion. Importantly, 95% (58 out of 61) of the miRNAs whose levels decreased by more than 50% when HNRNPA2B1 was depleted were also affected, with similar intensity, upon depletion of METTL3 (p<1e-24, Figure 3C and Table S1). The fraction of miRNAs affected by HNRNPA2B1 depletion was roughly half of the total miRNAs regulated by METTL3 indicating that HNRNPA2B1 regulates a major subset, but not all, of the m6A dependent miRNAs.
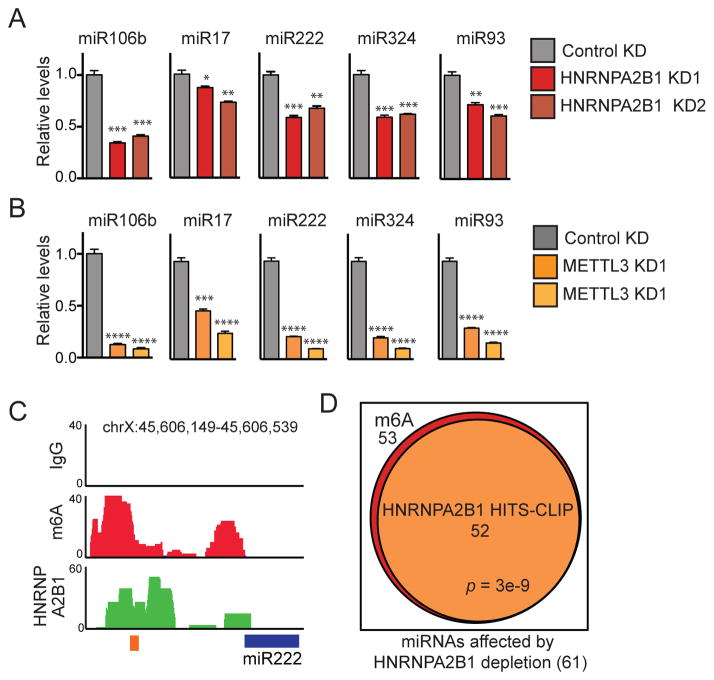
In order to validate these findings using an independent approach, we performed quantitative PCR (qPCR) for specific miRNAs in the setting of HNRNPA2B1 knockdown using additional cell lines. Consistent with the findings described above, HNRNPA2B1 depletion caused a significant reduction in the expression levels of the mature forms of a number of m6A-marked miRNAs in MDA-MB-231 cells (Figure 4A) as well as HEK293 cells (Figure S4). Moreover, the expression levels of these miRNAs were also dependent on METTL3 as their levels were similarly significantly reduced upon METTL3 depletion in both cell lines (Figures 4B and S4).
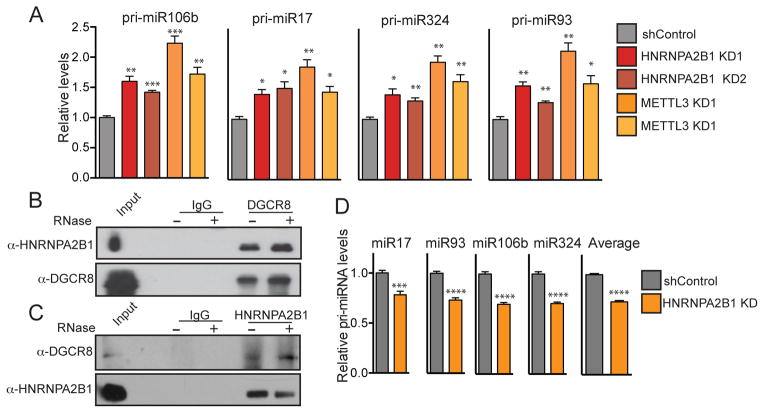
Figure 4. HNRNPA2B1 binds to m6A-methylated pri-miRNA sequences.
(A) qRT-PCR quantification of exemplary miRNAs that were modulated by HNRNPA2B1 depletion in MDA-MB-231 cells. Stable cell lines expressing shControl vector or two independent shRNAs targeting HNRNPA2B1 were generated and total RNA was extracted and quantified. ***, p-value <1E-3; **, p-value <1E-2; *, p-value <5E-2.
(B) Quantification of the expression levels of miRNAs shown in (A) when METTL3 was depleted in MDA-MB-231 cells by 2 independent shRNAs, as measured by qRT-PCR. P-values as in (A). ****, p-value <5E-4; ***, p-value <1E-3.
(C) Genome tracks depicting sequencing read coverage from m6A-seq (red) and HNRNPA2B1 HITS-CLIP (green) within an exemplary pri-miRNA obtained from MDA-MB-231 cells. The upper track represents the reads from the IgG immunoprecipitated control sample. The blue box indicates the position of the pre-miRNA and the orange box indicates the position of the RGAC motif. The chromosomal location of the sequence depicted in the figure is shown at the top of the panel.
(D) Venn diagram showing the overlap between m6A-seq tags and HNRNPA2B1-HITS-CLIP tags within pri-miRNA regions of miRNAs affected by HNRNPA2B1 depletion. The p-value was calculated based on the hypergeometric distribution.
We next asked whether HNRNPA2B1 binds to pri-miRNA sites and whether it regulates the processing of METTL3/m6A dependent microRNAs. To answer these questions, we identified m6A peaks in pri-miRNA transcripts from m6A-seq data and searched for co-occurrence of HNRNPA2B1 binding sites. Consistent with a direct role for HNRNPA2B1 in regulating the processing of these pri-miRNA transcripts, we noted overlaps between m6A peaks and HNRNPA2B1 binding sites detected by HITS-CLIP (Figure 4C). Importantly, of the 61 miRNAs affected by HNRNPA2B1 depletion, 53 contained m6A-seq tags and 52 of those exhibited overlapping HNRNPA2B1 HITS-CLIP tags (p=3e-9, Figure 4D). These findings are consistent with recognition of the m6A mark by HNRNPA2B1 at miRNA loci.
If HNRNPA2B1 mediates its effects in the nucleus prior to the processing of pri-miRNAs to pre-miRNAs, then its depletion should result in the accumulation of pri-miRNA transcripts within the nucleus. Consistent with this, depletion of HNRNPA2B1 resulted in the accumulation of specific pri-miRNAs in the nucleus phenocopying the effect of METTL3 depletion (Figure 5A). HNRNPA2B1 thus positively regulates the nuclear processing of a set of pri-miRNAs. The processing of these pri-miRNAs is dependent on METTL3, the m6A writer, and HNRNPA2B1, a putative reader of this mark. These findings are consistent with a model wherein METTL3 and HNRNPA2B1 proteins comprise a pathway that controls the processing of a set of miRNA precursors.
Figure 5. HNRNPA2B1 regulates miRNA processing and interacts with the Microprocessor.
(A) Pri-miRNA expression levels for the miRNAs shown in (4B–C) upon HNRNPA2B1 and METTL3 depletion as measured by qRT-PCR. All experiments were performed in biological triplicates. ***, p-value <1E-3; **, p-value <1E-2; *, p-value <5E-2.
(B) In vivo interaction between DGCR8 and HNRNPA2B1. HEK293 cells were chemically crosslinked before antibody-mediated immunoprecipitation of endogenous DGCR8. After immunoprecipitation, samples were washed and incubated with RNAse as indicated. Western blots for HNRNPA2B1 and DGCR8 are shown.
(C) Same as B, but the reciprocal immunoprecipitation was performed. In this case endogenous HNRNPA2B1 was immunoprecipitated and its interaction with DGCR8 was detected by Western blot under similar conditions.
(D) Quantification of DGCR8-bound pri-miRNAs. Endogenous DGCR8 was immunoprecititated after UV-crosslinking and pri-miRNAs bound to it were extracted and quantified by qRT-PCR. The bar graph shows the qRT-PCR of a panel of HNRNPA2B1-target miRNAs for which expression was shown to be reduced upon HNRNPA2B1 depletion. ****, p-value <5E-4; ***, p value-<1E-3.
A reader of m6A should serve as a bridge between this mark and effector proteins, or adaptor proteins that would ultimately interact with effectors. The effector involved in pri-miRNA processing is the Microprocessor complex, composed of the proteins DGCR8 and DROSHA. DGCR8 is an RNA-binding protein that recognizes the junction between the stem region and the flanking single-stranded RNA of the pri-miRNA hairpin. Hairpin recognition and binding allow for recruitment of the ribonuclease type III DROSHA, which cleaves the double stranded RNA to release the pre-miRNA molecule (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Han et al., 2006; Landthaler et al., 2004). We had previously proposed the existence of one or more readers that could mediate the interaction between the m6A mark present in pri-miRNA regions and the Microprocessor to facilitate the recognition of the pri-miRNA among the large landscape of secondary structures that populate the transcriptome (Alarcon et al, 2015). Since HNRNPA2B1 recognizes m6A sequences and phenocopies the effects of METTL3, we investigated if it could link the m6A mark and the Microprocessor. Consistent with a direct role for HNRNPA2B1 in the regulation of pri-miRNA processing, immunoprecipitation of endogenous DGCR8 co-precipitated HNRNPA2B1, while reciprocal immunoprecipitation also revealed this interaction (Figure 5B–C). Importantly, this interaction persisted despite ribonuclease treatment, suggesting that the interaction between these two proteins is mediated by protein-protein interactions (Figure 5B–C). If HNRNPA2B1 recruits DGCR8 to a subset of pri-miRNA loci, then HNRNPA2B1 depletion should reduce the interaction of DGCR8 with pri-miRNA substrates. Consistent with this, HNRNPA2B1 depletion reduced the binding of endogenous DGCR8 with HNRNPA2B1-dependent pri-miRNA substrates (Figure 5D). HNRNPA2B1 thus recognizes m6A sequences, interacts with the Microprocessor, and facilitates the processing of a set of METTL3-dependent pri-miRNAs. These collective findings are consistent with its proposed role as a reader of the m6A mark.
HNRNPA2B1 binds methylated RNA
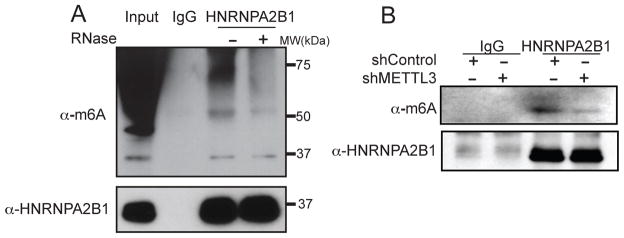
We next sought biochemical evidence for the in vivo interaction of HNRNPA2B1 with methylated RNA. MDA-MB-231 cells were UV-crosslinked, their nuclear fractions isolated, and endogenous HNRNPA2B1 was immunoprecipitated with a specific antibody. Upon HNRNPA2B1 immunoprecipitation and SDS-PAGE, immunoblotting with the m6A antibody revealed HNRNPA2B1 to interact with methylated RNA (Figure 6A). Importantly, ribonuclease treatment diminished the m6A signal of high molecular weight corresponding to long RNAs associated with HNRNPA2B1 but did not affect the m6A signal at the molecular weight of HNRNPA2B1 (37 kilodaltons) suggesting that HNRNPA2B1 directly associates with m6A methylated RNA and protects its RNA target sites from ribonuclease degradation (Figure 6A). The direct association between HNRNPA2B1 and m6A appears to be specific, since similar direct protection of m6A-containing RNA by another hnRNP RNA-binding protein was not observed (Figure S6A). If the methylated RNA that is bound by HNRNPA2B1 contains m6A, then depleting cells of the m6A writer METTL3 should reduce the HNRNPA2B1-bound m6A signal detected by western blot. Consistent with HNRNPA2B1 binding to cellular m6A, the m6A signal bound by endogenous HNRNPA2B1 recovered from METTL3 depleted cells was reduced relative to control cells (Figure 6B). Interestingly, Dominissini and colleagues used mass-spectrometry to identify candidate proteins that bound m6A modified RNA but not unmethylated RNA. Although not functionally studied, HNRNPA2B1 was amongst the set of proteins identified that preferentially bound m6A-modified RNA (Dominissini et al., 2012). Taken together, our findings reveal that endogenous HNRNPA2B1 directly interacts with m6A-modified RNA in vivo.
Figure 6. HNRNPA2B1 binds m6A methylated RNA.
(A) Immunoprecipitation of endogenous HNRNPA2B1 from MDA-MB-231 cells. Cells were UV-crosslinked before immunoprecipitation and samples were treated with RNase-A or left untreated as indicated after the immunoprecipitation. Western blotting was performed using the indicated antibodies.
(B) Immunoprecipitation of endogenous HNRNPA2B1 and associated RNA from control cells or cells depleted of METTL3. Cells were UV-crosslinked prior to immunoprecipitation and Western blotting was done using the antibodies depicted in the figure.
We next studied the interaction between HNRNPA2B1 and methylated RNA in vitro. Purified full length HNRNPA2B1 exhibited some preference for binding methylated RNA probes relative to unmethylated ones as shown by gel shift assays (Figures S6B-C). The probes used for these assays correspond to sequences obtained from endogenous pri-miRNAs regions that contained m6A-seq tags as well as endogenous HNRNPA2B1 binding sites (HITS-CLIP tags) (Figure S6D–E). Importantly, this preference was not observed using a different hnRNP protein (Figure S6F). We then generated a series of constructs containing GST-tagged individual domains. In isolation and under the conditions used in this study, only RRM1 exhibited a modest preference for m6A-containing RNA (Figure S6G), however we cannot exclude the possibility that in vivo and in the context of full-length protein, additional protein domains could mediate or facilitate recognition and binding of the m6A mark. These results suggest that HNRNPA2B1 can preferentially associate with m6A-containing RNA in vitro and that the RRM1 motif could, in part, mediate this preference.
DISCUSSION
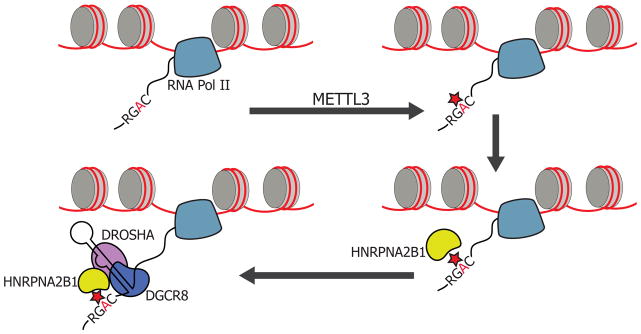
Our findings reveal that the HNRNPA2B1 RNA-binding protein binds the m6A consensus motif and also directly binds the m6A mark in vivo and in vitro. Loss-of-function experiments with HNRNPA2B1 and METTL3 resulted in similar effects on global alternative splicing patterns. HNRNPA2B1 was found to exhibit preferential in vitro binding to m6A-modified RNA substrates used in this study over unmodified substrates. Additionally, HNRNPA2B1 interacts with the Microprocessor protein DGCR8, enhances binding of DGCR8 to pri-miRNA transcripts, and positively regulates pri-miRNA processing in a similar manner as METTL3, the m6A writer. Moreover, we have identified a set of miRNAs whose processing is dependent on both METTL3 and HNRNPA2B1. These observations as a whole are consistent with a model wherein HNRNPA2B1 acts as a reader of the m6A mark in the nucleus and mediates, in part, the effects of m6A/METTL3 on microRNA processing (Figure 7).
Figure 7. HNRNPA2B1 is a mediator of the m6A mark.
Schematic representation of the nuclear role of HNRNPA2B1 in miRNA processing. HNRNPA2B1 is shown as a reader of the m6A methylation mark. Pri-miRNA processing is depicted in the model. The red star represents m6A mark on RNA; and the yellow shape represents the HNRNPA2B1 RNA-binding protein. A similar model could be use to depict alternative splicing, in which HNRNPA2B1 binding to m6A methylated pre-mRNAs would facilitate the engagement of splicing factors.
YTHDF2 was recently identified as a cytoplasmic ‘reader’ protein of the m6A mark. YTHDF2 was found to regulate the degradation of specific mRNAs by localizing them to processing bodies. The historical association of the m6A mark with pre-mRNA processing (Carroll et al., 1990; Finkel and Groner, 1983; Katz et al., 2015; Stoltzfus and Dane, 1982) and the more recent molecular studies linking m6A or components of the m6A-methylation machinery with splicing (Dominissini et al., 2012; Granadino et al., 1990; Horiuchi et al., 2013; Little et al., 2000; Liu et al., 2014; Ortega et al., 2003; Ping et al., 2014; Schwartz et al., 2014; Wang et al., 2014b; Zhao et al., 2014) suggested the existence of a nuclear factor that could recognize this modification. Moreover, our recent work implicating m6A/METTL3 in the processing of pri-miRNAs an event occurring in the nucleus also supported the existence of a nuclear ‘reader’ and effector of this mark (Alarcon et al., 2015). The role of HNRNPA2B1 in m6A-dependent nuclear pri-miRNA processing and alternative splicing events, its direct binding to m6A and the RGAC motif in vivo, as well as its in vitro preference for specific m6A-modified substrates relative to unmodified substrates support a role for this RNA-binding protein as a nuclear ‘reader’ and effector of this mark.
A natural question that arises is why there would exist distinct readers of m6A that act within the nucleus or the cytoplasm? The fates of RNA molecules within the nucleus and cytoplasm are fundamentally distinct. Within the nucleus, nascent transcripts are internally processed to yield mature messages lacking introns. Similarly, primary microRNA transcripts are processed within the nucleus to yield shorter pre-miRNA forms the precursors for mature miRNAs. Within the cytoplasm, mRNAs can be localized to distinct compartments, translated, stabilized, or destabilized. The divergent processes in which transcripts participate in within the nucleus or cytoplasm are mediated by distinct protein complexes, such as the microRNA and splicing machineries in the nucleus or the deadenylation complex in the cytoplasm. The existence of distinct nuclear and cytoplasmic readers that bind m6A but have additional protein-protein interaction domains would allow for each reader to interact with specific downstream effector proteins to mediate requisite events in the nucleus and cytoplasm. Consistent with this, YTHDF2 and HNRNPA2B1 contain distinct protein domains in addition to their RNA-binding domains P/Q/N-rich domain for YTHDF2 and RGG and M9 domains for HNRNPA2B1. These domains likely mediate interactions with specific cytoplasmic and nuclear effector proteins, respectively, or could form binding sites for adaptor proteins that recruit these machineries. In the case of HNRNPA2B1, its interaction with the Microprocessor represents an example of such selectivity. YTHDF2, on the other hand, has been shown to co-localize with components of the processing bodies in the cytoplasm (Wang et al., 2014a).
A long-standing question regarding m6A has been the molecular basis for the localization of m6A marks to specific infrequent (roughly one modification per 1000 bases) sites in transcripts (Rottman et al., 1994). The basis for this specificity is unknown, given that the m6A consensus motif is short and therefore inherently non-specific. It has been proposed that local structural elements might contain the requisite contextual information that enables m6A marks to be deposited (Rottman et al., 1994). Indeed, the methylation site in arguably the best-studied m6A containing transcript (prolactin) falls in a predicted stem-loop/bulge structure (Rottman et al., 1994). Moreover, similar stem-loop/bulge structures were predicted for multiple m6A consensus sites found in RSV RNA (Csepany et al., 1990; Kane and Beemon, 1985). With respect to ‘writing’ of this mark, METTL3 has been shown to methylate single-stranded RNA substrates in vitro to a similar degree as structured substrates (Liu et al., 2014). While others and we have identified many m6A marks to also occur in regions that are not predicted to form structures, these historical findings suggest the presence of at least some m6A marks in structured contexts. Future in vivo RNA structural mapping studies aimed at establishing the fraction of structured versus unstructured sites that contain the m6A mark could provide important insights (Ding et al., 2014; Rouskin et al., 2014).
Another question that arises is whether the structured context within which m6A occurs is important for its ‘reading’ by proteins such as HNRNPA2B1. While this question is beyond the scope of the current work and will require future genome-wide in vivo structural analyses, we speculate that HNRNPA2B1 could potentially bind to m6A at both unstructured/linear sites as well as in structured sites given its multiple RRM domains. At linear sites, the RRM1 domain could potentially bind m6A-containing locations in linear RNA. At structured sites, the RRM1 domain could bind to an m6A in a single-stranded loop of a stem-loop structure while the RRM2 domain could bind to a single-stranded bulge irrespective of m6A-modification. Conversely, the RRM1 domain could bind an m6A in the bulge of a stem-loop while the RRM2 domain could bind to a loop irrespective of its m6A-modification. This speculative model is consistent with previously characterized transcriptome-wide binding of HNRNPA2B1 to AU-rich structural elements, which are less thermodynamically stable structures (Goodarzi et al., 2012). Another possibility is that the presence of the m6A mark in double-stranded RNA could increase its open-conformation probability, thereby facilitating RRM1 binding to the m6A-bearing strand (Roost et al., 2015). Thus, the presence of multiple RRM motifs in HNRNPA2B1 could enable it to bind cooperatively to two distinct sites such as a loop and a stem or a loop and a bulge within a stem-loop. Future structural and molecular studies are needed to shed light on the details and dynamics of the events that follow m6A placement at linear and structured sites.
While we have implicated HNRNPA2B1, an RNA-binding protein implicated in several aspects of RNA biology (He and Smith, 2009; Villarroya-Beltri et al., 2013), as a downstream mediator of the effects of the m6A mark/METTL3 in the nucleus, our findings do not exclude the possibility that there exist additional ‘readers’ of this mark within the nucleus that could mediate similar processing effects at other miRNA, exonic, or intronic loci or to mediate other aspects of RNA processing or localization. In fact, we have found that roughly half of the m6A regulated miRNAs studied by us were dependent on HNRNPA2B1. This finding suggests the existence of additional nuclear ‘readers’ of this mark that mediate miRNA processing as well as other nuclear processing events. It would be quite interesting to determine whether other RNA-binding proteins of the hnRNP A/B family, that share similar RRM domains, recognize the m6A modification and mediate, in part, its effects on nuclear processing events.
Our findings have identified HNRNPA2B1 as a nuclear ‘reader’, a downstream effector of the m6A mark and an adaptor for the Microprocessor complex. Future proteomic and biochemical studies are needed to elucidate the molecular networks and mechanisms by which this reader mediates other nuclear processing events downstream of the m6A mark.
EXPERIMENTAL PROCEDURES
Tissue culture
MDA-MB-231, HeLa, and HEK293T cell lines were purchased from ATCC. All cells were cultured in vitro in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 2mM L-Glutamine, 1mM sodium pyruvate, and 2.5 ug/mL fungizone. All experiments were conducted with cells from passage 2 to 5.
Stable cell lines
Generation of lentivirus-mediated knockdowns was performed as described previously (Tavazoie et al., 2008). Commercially available shRNAs purchased from Sigma were co-transfected with lentivirus packaging plasmids in HEK293T cells to generate lentivirus. Two shRNAs targeting HNRNPA2B1 (TRCN0000001058, TRCN0000010582), two shRNAs specific for METTL3 (TRCN0000034715, TRCN0000034717), and a control shRNA (SHC002) were used for experiments. We generated stable fresh knock down cell-lines after 2 or 3 passages in order to avoid compensatory mechanisms of loss of knockdown.
qRT-PCR
Mature and pri-miRNAs were quantified by Taqman microRNA and Pri-miRNA assays. RNU44, U6 snRNA, GAPDH, and GusB were used as endogenous controls. Quantitative miRNAs expression data were obtained using the ABI Prism 7900HT Fast Real-Time PCR System and data was analyzed by Relative Quantification (RQ) Manager software.
Small RNA isolation
mirVana (Applied Biosystems) and Total RNA Purification Kit (Norgen Biotek) were used to extract total RNA from cells according to the manufacturer’s instructions.
RNA sequencing
Samples were lysed using LB1 buffer (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton x-100, and protease inhibitors). RNA was extracted from the nuclear fraction using the Total RNA Purification Kit (Norgen Biotek). RNA was subjected to RiboZero treatment (Epicentre), barcoded using ScriptSeq V2 kit (Epicentre) and sent for sequencing.
microRNA expression profiling (microarray)
The RNA extracted from two independent stable knockdowns of HNRNPA2B1 and two control cells expressing a control hairpin were labeled and hybridized on miRNA microarrays by LC sciences. Of all the probes included in the array, those corresponding to 329 miRNAs revealed a signal above background in at least two of the samples and were also expressed in the microarray previously performed using METTL3 depleted cells.
Supplementary Material
RESEARCH HIGHLIGHTS.
HNRNPA2B1 binds m6A containing sites and RGAC motif in nuclear transcripts
HNRNPA2B1 mediates m6A-dependent primary-microRNA processing events
HNRNPA2B1 modulation causes similar effects on alternative splicing as METTL3 modulation
Acknowledgments
We are grateful to Alexander Nguyen for insightful comments and technical advice. We thank C. Zhao and C. Lai of the Rockefeller Genomics Resource Center for expert assistance with next-generation RNA sequencing. C.A is the recipient of the Robert S. Bennett Postdoctoral Fellowship and supported by the Debra and Leon Black Challenge Grant. H.G. is currently the recipient of a Ruth L. Kirschstein National Research Service Award from the NIH (T32CA009673-36A1). S.F.T. is a Department of Defense Era of Hope Scholar and a Department of Defense Breast Cancer Collaborative Scholars and Innovators Award recipient. S.T. was supported by the National Human Genome Research Institute (R01 HG003219).
Footnotes
Accession Numbers
High-throughput sequencing data were deposited at GEO under the accession number GSE70061.
References
- Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ, Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. The Journal of biological chemistry. 1990;265:20117–20122. [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel D, Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, Salavati R, Cristea IM, Tavazoie S. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485:264–268. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino B, Campuzano S, Sanchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. The EMBO journal. 1990;9:2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. Journal of visualized experiments : JoVE. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cellular and molecular life sciences : CMLS. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. The Journal of biological chemistry. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR. An expanding universe of mRNA modifications. Nature structural & molecular biology. 2014;21:945–946. doi: 10.1038/nsmb.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Silterra J, Schwartz S, Wong B, Thorvaldsdottir H, Robinson JT, Mesirov JP, Airoldi EM, Burge CB. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Human molecular genetics. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, Niksic M, Bachi A, Wilm M, Sanchez L, Hastie N, Valcarcel J. Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. The Journal of biological chemistry. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. Journal of the American Chemical Society. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76:1109–1114. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. Journal of molecular biology. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell reports. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. Journal of virology. 1982;42:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature communications. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N(6)-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Moss B. Methylation of newly synthesized viral messenger RNA by an enzyme in vaccinia virus. Proc Natl Acad Sci U S A. 1974;71:3014–3018. doi: 10.1073/pnas.71.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.