Highlight
miRNAs are a new target for genetically improving plant tolerance to abiotic stresses. These miRNAs respond to environmental stresses in a miRNA-, stress-, tissue-, and genotype-dependent manner.
Key words: Abiotic stress, climate change, drought, gene network, microRNA, salinity.
Abstract
MicroRNAs (miRNAs) are an extensive class of endogenous, small RNA molecules that sit at the heart of regulating gene expression in multiple developmental and signalling pathways. Recent studies have shown that abiotic stresses induce aberrant expression of many miRNAs, thus suggesting that miRNAs may be a new target for genetically improving plant tolerance to certain stresses. These studies have also shown that miRNAs respond to environmental stresses in a miRNA-, stress-, tissue-, and genotype-dependent manner. During abiotic stress, miRNAs function by regulating target genes within the miRNA–target gene network and by controlling signalling pathways and root development. Generally speaking, stress-induced miRNAs lead to down-regulation of negative regulators of stress tolerance whereas stress-inhibited miRNAs allow the accumulation and function of positive regulators. Currently, the majority of miRNA-based studies have focused on the identification of miRNAs that are responsive to different stress conditions and analysing their expression profile changes during these treatments. This has predominately been accomplished using deep sequencing technologies and other expression analyses, such as quantitative real-time PCR. In the future, more function and expression studies will be necessary in order to elucidate the common miRNA-mediated regulatory mechanisms that underlie tolerance to different abiotic stresses. The use of artificial miRNAs, as well as overexpression and knockout/down of both miRNAs and their targets, will be the best techniques for determining the specific roles of individual miRNAs in response to environmental stresses.
Introduction
Human population growth and global industrialization are two factors that generate and promote climate change. Both of these factors are exponentially increasing and, therefore, new arable lands for cultivating dedicated food, cash, and biofuel crops will be necessary in order to sustain future generations. However, climate change has the ability to alter atmospheric conditions and modify environmental soils, which can make plant growth and development more difficult. It is well known that environmental abiotic stresses, such as drought and salinity, significantly affect plant survival, growth, and development, and thus decrease plant quality, yield, and biomass production (Wang et al., 2003; Mittler, 2006). The effects of abiotic stresses may also reflect at different suborganismal levels, including at the biochemical, physiological, cellular, molecular, and even finally at the organismal level. Many studies have demonstrated that abiotic stresses inhibit seed germination, seedling development, root development, chlorophyll biosynthesis, and photosynthesis, and that induced oxidative stresses, such as the production of reactive oxygen species (ROS), further damage plant growth and development (Fernandez, 2014; Mathur et al., 2014; Suzuki et al., 2014). Abiotic stresses have also been shown to alter gene expression profiles significantly during different developmental stages; these gene expression programmes changes ultimately regulate plant developmental and timing plasticity (Fernandez, 2014; Mathur et al., 2014; Suzuki et al., 2014). Several important genes, including those encoding transcription factors, have been implicated in response to abiotic stresses, and, when these genes were overexpressed in model plant species, such as Arabidopsis, as well as other agriculturally important crops, they were shown to improve plant tolerance to individual stresses significantly (Ganesan et al., 2012; Diaz-Vivancos et al., 2013; Liu et al., 2013; Gong et al., 2014; Lee et al., 2014; X. Li et al., 2014; Rong et al., 2014; Tamirisa et al., 2014). Over the past decade, the emergence of new technologies, such as microarrays and next-generation sequencing, has led to the discovery of hundreds of protein-coding genes that are associated with a wide range of environmental abiotic stresses (Baxter et al., 2014; Golldack et al., 2014). However, there are many questions that remain unanswered, such as how are these protein-coding genes regulated? What kind of gene networks do plants utilize in order to respond to different abiotic stresses? A recently discovered small regulatory RNA molecule, termed microRNA (miRNA), may be the answer to these questions.
miRNAs are an extensive class of small endogenous RNA molecules that range between 20 and 24 nucleotides in length. It is thought that miRNAs are widely distributed throughout the plant kingdom and are highly evolutionarily conserved from mosses to higher flowering monocots and dicots, such as Arabidopsis, rice, and cotton (Axtell and Bartel, 2005; Zhang et al., 2006). However, the application of new techniques has led scientists to observe not only that an individual plant species contains these conserved miRNAs, such as miR156, but also that they contain a high number of species-specific miRNAs. This suggests that conserved miRNAs may regulate common traits in plants, such as plant morphology and phase change, and that species-specific miRNAs may control unique and variable processes in individual plant species, such as fibre initiation and development in cotton (Xie et al., 2015a ). Both conserved and species-specific miRNAs may be involved in, and play an important role in, plant response to abiotic stress.
In this review, we will first present miRNAs that have been found to respond to different environmental stresses and discuss their expression in a miRNA-, stress-, tissue-, and genotype-dependent manner. Next, we will focus on miRNA-regulated gene networks during plant response to stress. Finally, we will propose the potential application of miRNAs as a new target for genetically improving plant tolerance to abiotic stress. Current existing problems and future directions in this exciting field will also be discussed.
Abiotic stresses induce the aberrant expression of miRNAs
The role of miRNAs in plant response to abiotic stress was initially suggested after data gathered from miRNA target prediction, miRNA expression profile studies during plant response to abiotic stress, and surveys of NCBI expressed sequence tags (ESTs). In one of the earliest plant miRNA papers, Jones-Rhoades and Bartel (2004) predicted and validated that ATP sulphurylase (APS), the enzyme that catalyses the first step of inorganic sulphate assimilation, was one of the targets of miR395, which is responsive to sulphate levels in plants. Based on this initial result, they further analysed the response of miR395 to cellular sulphate levels. Their results showed that in comparison with plants growing under normal sulphate conditions (2mM SO4 2–), miR395 was induced by >100-fold under low sulphate treatment (0.02mM SO4 2–), suggesting that miR395 is involved in sulphate uptake and metabolism in plants. At the same time, Sunkar and Zhu (2004) constructed small RNA libraries from Arabidopsis seedling samples treated with cold stress (0 °C for 24h), salt stress (300mM NaCl for 5h), drought stress (dehydration for 10h), and hormones [100 μM abscisic acid (ABA) for 3 h], as well as from the untreated controls. After both conserved and novel miRNAs were identified from all samples, the authors employed RNA gel blot analysis to study miRNA expression change after all four treatments. Their results showed that miR393 was strongly induced by all four tested stress conditions (cold, dehydration, NaCl, and ABA treatments). In contrast, miR389a.1 was inhibited by all of the stress treatments. Interestingly, other miRNAs showed different responses to the various stress treatments. For example, miR319 was induced by cold but not by salinity, dehydration, or ABA (Sunkar and Zhu, 2004). In one of our early studies, we found that 25.8% of ESTs contained one or more miRNAs (Zhang et al., 2005). Although no experiments had confirmed at that time that these miRNAs were only found in stress-induced tissues, the large percentage of ESTs found that contained miRNAs served as an indicator that miRNAs may play some role in plant response to environmental stresses (Zhang et al., 2005). Since these initial studies, the role of miRNAs in plant response to environmental stresses has been attracting attention from many scientists.
miRNA microarrays and deep sequencing technologies have opened the door for investigating which miRNAs are responsive to certain stresses and how much their expression levels change. To date, hundreds of miRNAs have been identified in single plant species. Therefore, it is almost impossible to employ RNA blotting or regular PCR to analyse their expression unless analysis of an individual miRNA is desired. miRNA microarrays, however, can analyse thousands of miRNAs at the same time. Alternatively, deep sequencing not only sequences all of the known miRNAs in a sample but it also sequences all of the small RNAs that are present. Thus, deep sequencing is the most efficient approach to study miRNA expression profiles because it can be used to find new or novel miRNAs that are induced by an individual stress while simultaneously surveying expression levels. Consequently, next-generation high-throughput deep sequencing is currently a popular approach for identifying stress-responsive miRNAs in any plant species, particularly in plant species for which few genome sequence data are available.
miRNAs respond to abiotic stress in a genotype-dependent manner
Over the course of evolution, plants have evolved complicated physiological and genetic mechanisms in order to cope with and adapt to the harsh environment. It is because of their derivation from a common ancestor that most plants share core gene networks that control plant response to a wide range of environmental factors. However, due to the fact that plants evolved to grow in various environments, plants have also developed numerous regulatory mechanisms for different growth habitats. Even for the same plant species, separate genotypes may show differential gene expression due to deviations in individual plant growth conditions and because of human selection of cultivated crops compared with their wild relatives. Among underlying genetic mechanisms, miRNAs may be one molecule that aids in response to abiotic stress. Like protein-coding genes, many miRNAs also show varied expression from species to species and also from genotype to genotype under certain stress conditions. This difference can be demonstrated by the direction and level of miRNA expression.
miRNA response to abiotic stress in a genotype-dependent manner was evidenced by analysing miRNA expression levels in response to certain stresses among several plant species and cultivars. Several technologies, including deep sequencing, microarrays, quantitative real-time PCR (qRT-PCR) analysis, and even the creation of transgenic plants, showed that miRNA expression profiles varied among plant species. Studies have shown that one miRNA may respond to the same stress differently depending on the plant species, while many miRNAs may respond to the same stress in a similar manner. For example, the expression of miR168 and miR396 was induced in Arabidopsis (Liu et al., 2008) and tobacco (Frazier et al., 2011), but was inhibited in rice (Zhou et al., 2010) by drought treatment. On the other hand, drought treatment down-regulated the expression of miR408 in rice (Zhou et al., 2010), peach (Eldem et al., 2012), and cotton (Xie et al., 2015b ) but up-regulated miR408 expression in Arabidopsis (Liu et al., 2008), Medicago (Trindade et al., 2010), and barley (Kantar et al., 2011). Salinity stress induced the overexpression of miR156 in Arabidopsis (Liu et al., 2008) but inhibited expression of that same miRNA in maize (Ding et al., 2009). Similarly, plants treated with NaCl highly expressed miR396 in Arabidopsis (Liu et al., 2008) and maize (Ding et al., 2009), but not in rice (Zhou et al., 2010). Interestingly, the degree of miRNA response to the same stress varied among plant species although it was either induced or inhibited in all plant species. miRNAs may also respond more in one plant species but less in another. Several studies have identified many species-specific miRNAs that respond to stress treatment. Using deep sequencing, 17 drought-specific miRNAs were identified in switchgrass, of which four were conserved and 13 were switchgrass-specific miRNAs (Xie et al., 2014). In a mechanical stress study, Lu and colleagues (2005) identified 21 miRNA gene families that contained 48 miRNA sequences in Populus trichocarpa; among them, only 11 miRNAs were conserved between P. trichocarpa and Arabidopsis. Interestingly, these conserved miRNAs exhibited species-specific developmental expression patterns, suggesting that even conserved miRNAs may differ in their regulatory roles among plant species. Despite their evolutionary origin (conserved miRNAs or tree-specific miRNAs), the expression of the majority of poplar miRNAs was altered in a manner consistent with tree-specific corrective growth against tension and compression stresses, which are two constant mechanical loads in trees (Lu et al., 2005). Additionally, Hackenberg and colleagues (2015) identified novel species-specific miRNAs (hvu-miRX33, hvu-miRX34, and hvu-miRX35) in barley that are significantly induced by drought treatment.
The genotype-dependent response of miRNAs to abiotic stresses is not only different among plant species but also varies among cultivars (genotypes) of the same species. It is well known that the genotypes of one plant species may differ in their capacity to respond to abiotic stress. miRNA-mediated gene regulation may contribute to this difference. Using deep sequencing technology, Barrera-Figueroa and colleagues (2011) investigated the impact of drought treatment on two cowpea cultivars (drought-tolerant IT93K503-1 and drought-sensitive CB46). Their results showed that 20 miRNAs were differentially expressed among the two genotypes. Of these miRNAs, nine were predominantly or exclusively expressed in one of the two genotypes but not in the other. Simultaneously, they also identified 11 drought-regulated miRNAs in one genotype but not in the other (Barrera-Figueroa et al., 2011). Using miRNA microarray technology, Yin and colleagues (2012) analysed the miRNA expression profiles of two cotton cultivars with varying resistance to salinity (SN-011 with high tolerance to salinity and LM-6 with sensitivity to salinity). Based on their results, 12 miRNAs were expressed in a genotype-specific pattern. Under salinity treatment, four miRNAs (miR156, miR169, miR535, and miR827) showed significantly high expression in LM-6 whereas the expression of three miRNAs (miR167, miR397, and miR399) was significantly inhibited in this cultivar (Yin et al., 2012). By comparing 12 salinity-tolerant and 12 salinity-susceptible genotypes in rice, Mondal and Ganie (2014) identified 12 miR-SSRs (simple sequence repeats) that were polymorphic. Only miR172b-SSR, however, proved different between the tolerant and susceptible genotype and could therefore serve as a biomarker for distinguishing cultivars with different responses to salinity stress (Mondal and Ganie, 2014). Their results also showed that miRNA genes were less diverse in the tolerant cultivars than in the susceptible cultivars, as evidenced by their calculated polymorphic index content (Mondal and Ganie, 2014). All of these studies suggest that miRNAs may play a role during cultivar-specific response to abiotic stress conditions.
miRNAs respond to abiotic stress in a plant tissue-dependent manner
To meet the need of a plant, an initiated plant stem cell differentiates into plant tissues/organs with various functions, such as leaves for photosynthesis and roots for taking up water and nutrients from the soil. Thus, it is not surprising that roots are more sensitive to the majority of stresses, particularly those of the soil environment including drought, high salinity, and exposure to pollutants. Similarly, it is not hard to understand that miRNAs in different tissues respond to stress exposure in different ways. For these reasons, certain stresses in plants tend to induce the expression of miRNAs at higher fold changes in the roots rather than in other plant tissues, such as leaves.
In order to compare root and leaf response to drought stress and to identify genome-wide drought-responsive miRNAs, Eldem and colleagues (2012) constructed four small RNA libraries from both control and treated leaf and root samples and deep sequenced the small RNA populations. The results of this study showed that drought significantly induced the aberrant expression of 262 (104 up-regulated, 158 down-regulated) and 368 miRNAs (221 up-regulated, 147 down-regulated) in leaf and root tissues, respectively (Eldem et al., 2012). Overall, the expression of >100 miRNAs was differentially altered by drought treatment in the roots compared with the leaves (Eldem et al., 2012). Wang and colleagues (2013) also demonstrated that the expression changes of miRNAs were dose and tissue dependent under drought and salinity stress in cotton, in which the tested miRNAs showed altered expression profile patterns in roots compared with the leaves (Wang et al., 2013).
miRNAs respond to abiotic stress in a stress-dependent manner
There are many types of environmental stresses, including drought, salinity, high temperature, low temperature, UV light, high or low light intensity, hypoxia, heavy metal, nanoparticle, and pollutant exposure, and fertilizer deficiency. Many studies have shown that all of these environmental stresses induce the aberrant expression of miRNAs in a dose- and stress-dependent manner. Interestingly, some miRNAs are commonly responsive to all stresses. Accumulating evidence, however, clearly shows that differential expression of certain miRNAs is dependent on the specific stress condition, even in the same plant species. In Arabidopsis, miR169 was inhibited by drought stress (Li et al., 2008) but was found to be induced by salinity treatment (M.Y. Xu et al., 2014). On the other hand, miR398 was induced by UVB light in Arabidopsis but was inhibited by salinity, cold, and oxidative stress (Sunkar et al., 2006; Jia et al., 2009). Further experiments have shown that drought-repressed miR169 expression was through an ABA-dependent pathway (Li et al., 2008). The target of miR166, nuclear factor Y (NF-Y) subunit A 5 (NFYA5), was strongly induced by drought stress at the same time that miR169 was inhibited (Li et al., 2008). Both nfya5 knockout Arabidopsis plants and transgenic plants overexpressing miR169 show the same phenotypes, including increased sensitivity to drought stress and enhanced water loss (Li et al., 2008). In contrast, overexpression of nfya5 in transgenic Arabidopsis enhanced plant resistance to drought stress (Li et al., 2008). Interestingly, miR169 was significantly induced by salinity stress in Arabidopsis, in which the expression of nfya5 was inhibited (M.Y. Xu et al., 2014). A similar phenomenon was also observed in other plant species, including soybean (Ni et al., 2013) and rice (Zhao et al., 2007, 2009). Other studies have shown that miR398 was inhibited by both ABA and salinity treatment (Sunkar et al., 2006; Jia et al., 2009) but was induced by drought treatment (Zhou et al., 2007) in Arabidopsis. Analysis of copper superoxide dismutase (CSD) 1 and 2, both targets of miR398, found that both of these genes were induced by salinity treatment (Sunkar et al., 2006; Jia et al., 2009). This suggests that miRNAs may aid plant tolerance to abiotic stresses in a stress-dependent manner.
miRNAs respond to abiotic stress in a miRNA-dependent manner
While individual miRNAs may respond differently to various stresses in the same plant species, there are many miRNAs whose expression levels can be associated with a particular stress treatment. In Arabidopsis, drought treatment induced the expression of many miRNAs, including miR156, miR319, miR393, miR397, and miR408, although the expression fold change differed from miRNA to miRNA (Liu et al., 2008). Some miRNAs, such as miR169, were down-regulated by drought treatment (Liu et al., 2008). Under salinity stress in Arabidopsis, the expression of miR156, miR159, miR169, miR319, miR393, and miR397 was significantly induced, although with varying fold changes, but the expression of miR398 was significantly inhibited (Liu et al., 2008).
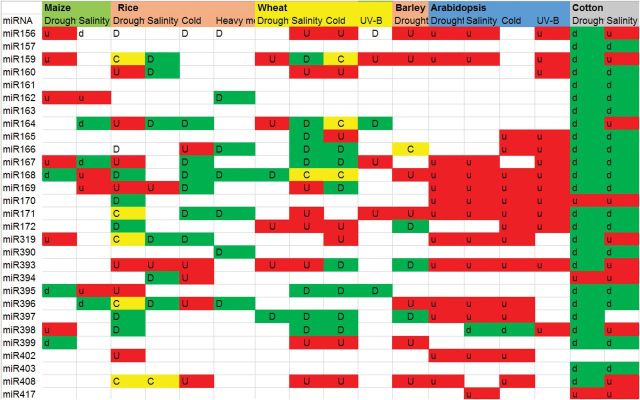
In summary, studies have shown that environmental abiotic stresses (drought, salinity, high and low temperature, and osmotic stress) induce significant differential expression of miRNAs in a variety of plant species (Fig. 1), including Arabidopsis (Liu et al., 2008; Jagadeeswaran et al., 2009; Jia et al., 2009), rice (Zhao et al., 2007), corn (Ding et al., 2009; Kong et al., 2014), poplar (Lu et al., 2005; Jia et al., 2009), and others (Barrera-Figueroa et al., 2011; Budak and Akpinar, 2011; Kulcheski et al., 2011; Li et al., 2011; Qin et al., 2011; Wang et al., 2011). Currently, a number of miRNAs have been reported to be induced by drought and salinity stresses in several different plant species. These miRNAs include miR156, miR159, miR165, miR167, miR168, miR169, miR319, miR393, miR395, miR396, miR398, miR399, and miR402. Almost all of these stress-induced miRNAs are evolutionarily conserved, although evidence supports that miRNAs respond to environmental stresses in a miRNA-, stress-, and genotype-dependent manner. Taken together, this information suggests that miRNAs play a versatile role in plant response to environmental abiotic stresses.
Fig. 1.
A miRNA responds to an environmental abiotic stress in a stress-, species-, and miRNA-dependent manner. Various miRNAs were aberrantly expressed under different abiotic stress treatments in a multiple plant species. Red colour indicates up-regulated (u). Green colour indicates down-regulated (d). Yellow colour indicates that both up-regulation and down-regulation were observed among different tissues or at different developmental stages. The data were based on current literature of Arabidopsis, rice, wheat, barley, switchgrass, and cotton.
miRNA–target gene networks involved in plant response to abiotic stress
miRNAs do not function directly in plant growth and development or in plant response to environmental stress. Instead, miRNAs participate in plant response to abiotic stresses through regulating key components of complex gene networks. After mature miRNAs are generated, they are loaded into the RNA-induced silencing complex, which contains an argonaute protein, and they then bind to the mRNAs of targeted genes (Winter and Diederichs, 2011; Iwakawa and Tomari, 2013). Based on the complementarity between a miRNA and its targeted mRNA, miRNAs regulate gene expression either by targeting mRNAs for cleavage or by inhibiting protein translation. Usually, when a miRNA perfectly or nearly perfectly aligns with a targeted mRNA, the mRNA will be cleaved at the site on the mRNA corresponding to the site between the 10th and 11th nucleotide of the targeting miRNA. Alternatively, if a miRNA imperfectly binds to the targeted mRNA, the mRNA cannot be translated into protein. In plants, almost all miRNAs perfectly or nearly perfectly bind to their targeted mRNAs; thus, the majority of miRNAs cleave their targeted mRNAs. There have been several reports, however, of plant miRNAs inhibiting protein translation (Rhoades et al., 2002; Bartel, 2004; Zhang et al., 2007). The high degree of complementarity between miRNAs and their targets provides a powerful approach for predicting and validating miRNA target genes in plants. In most cases, a simple BLASTn search can be performed against protein-coding genes using a known miRNA and the results validated using RACE (rapid amplification of cDNA ends)-PCR. Currently, degradome sequencing has been developed and widely used for both miRNA target identification and validation in plants. Table 1 lists the confirmed targets for conserved miRNAs associated with different environmental abiotic stresses.
Table 1.
Common stress-responsive miRNAs and their targetsa
| miRNAs | Targets | References |
|---|---|---|
| 156/157 | SPL | Stief et al. (2014) |
| 159 | MYB/TCP | Achard et al. (2004); Reyes and Chua (2007) |
| 160 | ARF | Guo et al. (2005) |
| 164 | NAC | Rhoades et al. (2002); Fang et al. (2014) |
| 169 | NFY | Ni et al. (2013) |
| 173 | TAS | Li et al. (2014) |
| 319 | TCP | Sunkar and Zhu (2004) |
| 393 | TIR | Jones-Rhoades and Bartel (2004); Sunkar and Zhu (2004) |
| 394 | F-box | Jones-Rhoades and Bartel (2004) |
| 395 | AST | Allen et al. (2005) |
| 395 | APS | Jones-Rhoades and Bartel (2004) |
| 396 | GRF | Sunkar and Zhu (2004) |
| 397 | Laccase | Lu et al. (2013) |
| 398 | CSD | Guan et al. (2013); Naya et al. (2014) |
| 402 | DML3 DEMETER-LIKE protein 3) | Kim et al. (2010b ) |
| 828 | MYB | Luo et al. (2012) |
a There have been multiple studies on the targets of individual miRNAs. Here, we only list one or two references for each.
It is well known that the majority of stress-responsive miRNAs target transcription factors. Transcription factors play an important role during plant response to different environmental stresses. NAM, ATAF, and CUC transcription factors comprise the extensive class of NAC plant-specific transcription factors. To date, >100 NAC transcription factors have been identified in numerous plant species, including Arabidopsis and rice (Nakashima et al., 2012). Recently, several studies have shown that NAC transcription factors play an important role in plant response to various environmental stresses, including drought, salinity, and harsh temperature; correspondingly, overexpression of certain NAC transcription factors significantly enhanced plant tolerance to different abiotic stresses (Mao et al., 2012, 2014; Al Abdallat et al., 2014; Jiang et al., 2014; Pandurangaiah et al., 2014; Q. Xu et al., 2014). More interestingly, the NAC transcription factors are widely targeted by miRNAs. Several studies in Arabidopsis and rice have shown that miR164 cleaves NAC mRNAs that modulate plant developmental processes and responses to abiotic stress (Rhoades et al., 2002; Fang et al., 2014). Fang and colleagues (2014) tested six miR164-targeted NAC genes (OMTN1–OMTN6) in rice and found that four of them negatively regulated drought tolerance. SPL transcription factors are master regulators of plant developmental timing and phase change, and have been shown to be post-transcriptionally regulated by miR156 (Wang et al., 2009; Chen et al., 2010). A recent study in Arabidopsis found that miR156-mediated down-regulation of SPL increased plant response to environmental stresses, including heat stress and heat stress memory (Stief et al., 2014). Almost all of these miRNAs, as well as their targeted transcription factors, are highly evolutionarily conserved in the plant kingdom, and both the miRNAs and their targeted transcription factors are important for plant development. Therefore, plants may utilize regulatory mechanisms, such as miRNAs and transcription factors, in order to respond quickly to development cues and stress conditions, and it may be a common mechanism for all plant species to respond to environmental stresses through miRNA-mediated gene targeting of transcription factors.
miRNAs respond to different abiotic stresses by targeting stress-responsive genes. Besides transcription factors, there are also lots of other genes associated with and/or responsive to different environmental abiotic stresses, and many of these stress-responsive genes are targeted by an individual miRNA. Laccases are multi-copper-containing glycoproteins that play a role in plant response to abiotic stress by regulating cell wall function during stress conditions (Liang et al., 2006). A recent study showed that laccase genes were targeted by miR397 (Lu et al., 2013). Superoxide dismutase (SOD) is an important enzyme that scavenges ROS, which are usually induced under oxidative stress caused by heavy metal contamination and other abiotic stresses. According to the metal ligands that they bind, there are three types of SODs in plants, iron SOD (Fe-SOD), manganese SOD (Mn-SOD), and copper/zinc SOD (CSD). Loss-of-function mutants of csd1, csd2, and ccs (a copper chaperone of CSD1 and CSD2) exhibited enhanced heat-responsive gene expression in Arabidopsis and, overall, the transgenic plants were more heat tolerant (Guan et al., 2013). Additionally, several studies have shown that all three CSD genes are targeted by miR398. Guan and colleagues (2013) generated transgenic Arabidopsis plants that expressed miR398-resistant mutants of csd1, csd2, and ccs. The results of this study showed that the miR398-resistant mutants lost the ability to induce aberrant expression of heat-responsive genes and, therefore, were more sensitive to heat stress (Guan et al., 2013). Other experiments in Arabidopsis, as well as in common bean, have also demonstrated that miR398 is overexpressed during heat treatment, which supports other evidence showing that miR398 confers heat tolerance by promoting the cleavage of csd mRNAs (Guan et al., 2013; Naya et al., 2014). miR395 functions in abiotic stress response by regulating APS, an important enzyme catalysing the initial activation step of sulphate assimilation (Buchner et al., 2004). Several APS gene family members, including aps1 and aps2, are targeted by miR395 (Jones-Rhoades and Bartel, 2004). In addition to APS, miR395 also targets AST68 (AtSULTR2;1, At5g10180), which encodes a sulphate transporter (Allen et al., 2005).
One recent study has shown that trans-acting small interfering RNAs (tasiRNA) are generated through miRNA cleavage of TAS mRNAs, and that tasiRNAs are also involved in plant response to abiotic stress (S. Li et al., 2014). Currently, tasiRNAs have been identified in several plant species, including Arabidopsis (Peragine et al., 2004; Vazquez et al., 2004), rice (Allen et al., 2005), and cotton (Xie et al., 2014). tasiRNAs are generated from TAS mRNA cleavage by individual miRNAs and, after production, these tasiRNAs target ARF and MYB transcription factors, which are associated with different abiotic stresses. To date, there are at least three miRNAs (miR173, miR828, and miR390) that have been identified that target TAS mRNAs and produce tasiRNAs. miR173-cleaved tasiRNAs target HEAT-INDUCED TAS1 TARGET1 (HTT1) and HTT2 mRNAs, and have been shown to be involved in thermotolerance in Arabidopsis (S. Li et al., 2014), which suggests a new mechanism for miRNAs involved in plant response to abiotic stress.
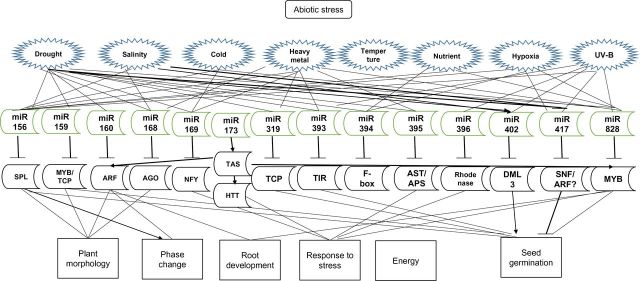
All this evidence shows that not only are miRNAs involved in plant response to abiotic stress, but that miRNAs function in plant stress response by targeting the transcripts of transcription factors and stress-responsive genes as well tasiRNAs. The entire miRNA gene network forms the basis of the regulatory mechanism (Fig. 2). This network is affected by plant hormones and different signalling pathways. miRNAs are also involved in the adaptation of roots to various environmental abiotic stresses by targeting genes within this network. Environmental conditions of the soil affect both primary root and lateral root development, and both of these are also partially affected by the miRNA-mediated gene network response. One of most important factors influencing root development is the supply of nutrients. Since nutrients are most probably located in the upper part of the soil, a deficiency in fertilizer usually causes an increase in lateral root development. This is evidenced by an increase in lateral root length and density, and also by a decrease in primary root extension. Using cell-sorting experiments, Gifford and colleagues (2008) found that Arabidopsis roots responded to nitrogen deficiency in a cell-specific manner. Similarly, after adding nitrate to nitrogen-depleted Arabidopsis plants, the expression of miR167 was decreased, which resulted in an increase of its targeted gene, ARF8, particularly in the pericycle cells (Gifford et al., 2008). These plants also displayed increased lateral root initiation, emergence, and development (Gifford et al., 2008). Drought and salinity stress are also known to affect root development, partially by way of miRNA-mediated gene regulation. There are many miRNAs that respond to different environmental stresses, and many of these stress-responsive miRNAs also modulate root development by targeting root development-related genes and transcription factors, such as NACs, ARFs, and HD-ZIPs.
Fig. 2.
The miRNA–target gene network is involved in plant response to environmental abiotic stresses. Different stresses induced and/or inhibited the expression of individual miRNAs that target transcription factors and/or stress-related genes. This network further regulates plant development as well as response to abiotic stress. Plant hormones are also involved in this process through directly/indirectly regulating the expression of miRNAs and their targets. (This figure is available in colour at JXB online.)
Phytohormones and their corresponding signalling pathways regulate miRNA–target gene networks and contribute to plant response to abiotic stress
Plant hormones play a central role in plant adaption to abiotic stress by regulating plant growth, development, nutrient allocation, and source/sink transitions (Peleg and Blumwald, 2011). Almost all plant hormones, including ABA, auxin, cytokinins, gibberellic acid (GA), ethylene, brassinosteroids, and jasmonic acid (JA), along with their corresponding signalling pathways, are involved in plant response to different environmental abiotic and biotic stresses. Cross-talk among different plant hormones, stress-related genes, and signalling pathways, as well as recently discovered miRNAs that are involved, results in synergetic or antagonistic interactions during plant response to abiotic stress (Peleg and Blumwald, 2011).
ABA is a stress-responsive plant hormone. Under stress conditions, ABA usually accumulates within the plant, which enhances the production of ROS. Together, ABA and ROS accumulation function as a stress signal that then induces the expression of mitogen-activated protein kinases (MAPKs) and antioxidant genes, ultimately enhancing plant tolerance to the stress condition. In maize seedlings, drought treatment inhibited the expression of miR168 and miR528, which resulted in overexpression of their target MAPK and peroxidase genes, and consequently overlapped with the ABA-involved signalling pathway (Wei et al., 2009). Drought conditions also induced stomatal movement and antioxidant defence in maize, which enhanced maize tolerance to drought stress (Wei et al., 2009).
miRNAs function in stress-related auxin signalling by modulating the expression of auxin response factor (ARF) transcription factors, which are involved in root development and stress response. At least five miRNAs (miR160, miR164, miR167, miR390, and miR393) have been shown to be involved in this network. For example, miR160 targets ARF10, ARF16, and ARF17, while miR167 interacts with ARF6 and ARF8, and miR390 targets ARF4. On the other hand, miR164 and miR393 are indirectly involved in the regulation of ARFs. All of these miRNAs and ARFs participate in root development and response to stress treatment (Meng et al., 2010). One study has suggested that auxin-induced miR164 functions to provide a homeostatic balance within the auxin signalling pathway by inhibiting NAC1, a transcription factor, and down-regulating auxin signalling (Guo et al., 2005).
The MAPK signalling pathway is also part of the miRNA–target gene network. MAPK signalling cascades are one of the most conserved pathways in plants and are known to regulate plant development as well as response to abiotic and biotic stresses (Raghuram et al., 2014). A recent study has shown that 98 out of 99 rice MAPK genes are potentially targeted by certain miRNAs, and the expression levels of these MAPKs were inversely correlated with the expression levels of their predicted miRNAs (Raghuram et al., 2014).
miRNAs, a new target for improving plant tolerance to abiotic stress
miRNAs play a significant role in plant growth and development and, recently, these molecules have emerged as important players in plant response to various environmental abiotic stresses. Further analysis has shown that miRNAs are located at the centre of complicated gene regulatory networks. Thus, miRNAs are becoming a novel target for plant improvement, including enhanced tolerance to different stresses (Zhang and Wang, 2015). To date, several miRNAs have been overexpressed in multiple plant species. Depending on their target genes, the miRNA-overexpressing transgenic plants exhibited either higher tolerance or sensitivity to different environmental abiotic stresses as compared with their wild type (Table 2). Several of these transgenic plants showed promise for using miRNA-based biotechnology for enhancing plant tolerance to harsh environments.
Table 2.
A list of studies that overexpressed miRNAs in order to alter plant tolerance to environmental stresses
| Targeted miRNAs | Transgenic plants | Targeted stress | References |
|---|---|---|---|
| 156 | Arabidopsis | Tolerance to heat stress | Stief et al. (2014) |
| 156 | Switchgrass | Increase of biomass | Fu et al. (2012) |
| 159 | Rice | Sensitive to heat stress | Wang et al. (2012) |
| 169 | Tomato | Enhancing plant tolerance to drought | Zhang et al. (2011) |
| 169 | Arabidopsis | Sensitivity to nitrogen deficiency | Zhao et al. (2011) |
| 173 | Arabidopsis | Thermotolerance | Li et al. (2014) |
| 319 | Bentgrass | Tolerance to salinity and drought | Zhou et al. (2013) |
| 319 | Rice | Tolerance to chilling temperature | Yang et al. (2013) |
| 393 | Arabidopsis | More sensitive to salinity and alkalinity | Gao et al. (2011) |
| 393 | Rice | More sensitive to salinity and alkalinity | Gao et al. (2011) |
| 394 | Arabidopsis | Tolerance to drought | Ni et al. (2012) |
| 395 | Arabidopsis | Drought and salinity stress | Kim et al. (2010a ) |
| 395 | Rapeseed | Enhanced tolerance to oxidative stress and heavy metal sterss | Zhang et al. (2013) |
| 396 | Arabidopsis | More sensitive to salinity and alkalinity | Gao et al. (2010) |
| 396 | Rice | More sensitive to salinity and alkalinity | Gao et al. (2010) |
| 402 | Arabidopsis | More tolerance to salinity, drought, and cold stress | Kim et al. (2010b ) |
| 417 | Arabidopsis | More sensitive to salinity and ABA | Jung and Kang (2007) |
| 828 | Sweet potato | Oxidative stress | Lin et al. (2012) |
miR156 was the first miRNA identified in plants and it has been shown to play a critical role in plant development and phase change. Recent studies demonstrated that miR156 was aberrantly expressed during plant exposure to various environmental stresses. Transgenic approaches have been used for validating miR156 function. miR156-overexpressing plants revealed that miR156 was required for plant heat stress memory, and these plants exhibited enhanced tolerance to heat stress (Stief et al., 2014). miR156 has also been used to improve plant biomass. Transgenic switchgrass that overexpressed the appropriate miR156 produced 58–101% more plant biomass in comparison with their control plants (Fu et al., 2012).
miR159 plays an important role in plant development. miR159 responded to various environmental stresses, and one study found that transgenic rice plants overexpressing miR159 were more sensitive to heat stress in comparison with the wild-type controls, suggesting that down-regulation of miR159 may contribute to heat stress tolerance (Wang et al., 2012).
miR169 is one of the largest miRNA families that is conserved in all plant species. miR169 is a significant contributor to proper plant development and also in plant response to environmental stress. Constitutive overexpression of miR169 in transgenic tomato significantly enhanced plant tolerance to drought stress after 7 d of drought treatment (Zhang et al., 2011). During drought treatment, non-transgenic wild-type tomato plants showed obvious dehydration symptoms, including wilting and turgor loss; however, the transgenic plants that overexpressed miR169 grew very well (Zhang et al., 2011). This study showed that transgenic tomato plants reduced their stomatal aperture index by 35–49% and their stomatal conductance by 33–45% (Zhang et al., 2011). Additionally, the transpiration rate of transgenic plants was reduced by 38–55% when compared with wild type non-transgenic tomato plants (Zhang et al., 2011). Thus, transgenic plants that overexpressed miR169 had reduced water loss through the leaves and required less water from the soil (Zhang et al., 2011). Overexpression of miR169 also caused transgenic Arabidopsis plants to be hypersensitive to nitrogen starvation, as evidenced by the yellowing of transgenic leaves under all tested nitrogen starvation conditions (Zhao et al., 2011). This suggests that miR169 also targets genes that function in nitrogen assimilation. Therefore, miR169 is a promising target for improving plant tolerance to drought stress and nitrogen deficiency.
miR319 has been identified to be associated with multiple abiotic stresses, and several studies have shown that miR319 is usually up-regulated during multiple stress conditions (Sunkar and Zhu, 2004; Zhou et al., 2010). A transgenic study found that constitutive expression of miR319 in creeping bentgrass significantly enhanced plant tolerance to salinity and drought stress, and also altered plant development (Zhou et al., 2013). This enhanced abiotic stress tolerance could be attributed to the regulation mechanism of miR319, since it is known to cleave the mRNAs of TCP transcription factors (Pieczynski et al., 2013). The transgenic plants that overexpressed miR319 exhibited increased water retention and cell membrane integrity when compared with their non-transgenic controls (Pieczynski et al., 2013). Under salinity stress conditions, the transgenic plants also accumulated less Na+ compared with the wild-type plants (Pieczynski et al., 2013). Overexpression of miR319 in transgenic rice was found to enhance rice tolerance to cold stress significantly (Yang et al., 2013). In this study, 7-day-old rice seedlings were acclimated to cold conditions by placing transgenic and wild-type seedlings at 12 °C for 2 d followed by 4 °C for 4 d (Yang et al., 2013). At the end of the experiment, transgenic rice that overexpressed miR319 had a higher survival rate (~50%) compared with the wild-type controls (13% of survival rate) (Yang et al., 2013). Taken together, the results of these studies suggest that overexpression of miR319 can be used to enhance plant tolerance to multiple environmental stresses.
Another conserved miRNA family that exists in both monocot and dicot plants and whose expression is altered by many environmental stresses is miR393. Thus, miR393 has become another target for improving plant tolerance to different stresses. Gao and colleagues (2011) transformed miR393 into rice and Arabidopsis under control of the Cauliflower mosaic virus (CaMV) 35S promoter. Transgenic and control plants were subjected to salinity (150mM NaCl) and alkalinity (75mM NaHCO3) treatments for 15 d (Gao et al., 2011). The results of this study showed that the T3 generation seedlings of miR393-overexpressing transgenic Arabidopsis were more sensitive to salinity and alkalinity treatment in comparison with wild-type plants. Likewise, the transgenic rice plants exhibited a similar phenotype (Gao et al., 2011). For both Arabidopsis and rice, overexpression of miR393 significantly inhibited seedling growth and root development (Gao et al., 2011).
miR394 is also evolutionarily conserved, and recent studies in Arabidopsis have demonstrated that the expression of miR394 is significantly induced by high salinity treatment and is also altered by iron and sulphate deficiency (Liu et al., 2008; Kong and Yang, 2010). Under drought conditions, transgenic Arabidopsis plants that overexpressed miR394 restricted water loss during leaf transpiration, which ultimately increased tolerance to drought stress (Ni et al., 2012).
miR395 has been shown to exhibit aberrant expression under different stress conditions. Transgenic approaches to elucidate miR395 function have found that overexpression of miR395 affects plant tolerance to salinity and drought stress, as evidenced by a decrease in seed germination and seedling growth (Kim et al., 2010a ). However, members of the miR395 family may regulate different targets during these processes due to a single nucleotide difference between their mature miRNA sequences. Hence, miR395 can also function in plant tolerance to heavy metals. Transgenic rapeseed that overexpressed miR395 showed a higher tolerance to cadmium stress (L.W. Zhang et al., 2013). This was evidenced by a lower degree of cadmium-induced oxidative stress in the transgenic plants compared with their wild-type controls (L.W. Zhang et al., 2013). The higher tolerance to cadmium stress could be attributed to the fact that the transgenic plants contained higher chlorophyll, glutathione, and non-protein thiols (L.W. Zhang et al., 2013).
miR396 is a known stress-responsive miRNA. During salinity and alkalinity stress treatments, transgenic Arabidopsis and rice plants that constitutively overexpressed miR396 had significantly stunted root growth as well as decreased plant growth and development (Gao et al., 2010). Interestingly, no phenotypic differences were observed between transgenic miR396-overexpressing Arabidopsis seedlings and wild-type plants under drought stress conditions. This suggests that miR396 is a negative regulator of plant response to salinity and alkalinity stresses.
miR402 has been shown to be up-regulated in Arabidopsis by salinity, dehydration, and cold stresses (Sunkar and Zhu, 2004; Kim et al., 2010b ). Overexpression of miR402 promoted seed germination in Arabidopsis under all three stress conditions; however, transgenic plants that overexpressed miR402 only exhibited enhanced plant growth under salinity stress but not under dehydration or cold stress conditions (Kim et al., 2010b ).
Additionally, salt and dehydration stresses altered the expression of miR417 (Jung and Kang, 2007). Constitutive overexpression of miR417 negatively impacted seed germination and survival of Arabidopsis seedlings under high salinity stress conditions and in the presence of ABA (Jung and Kang, 2007).
miR828 is a wound-induced miRNA. Interestingly, miR828 expression was induced by wounding but not by ethylene, hydrogen peroxide (H2O2), methyl jasmonate, or nitric oxide (NO) (Lin et al., 2012). Transgenic sweet potato overexpressing pre-miR828, however, exhibited an increase in lignin biosynthesis and the production of H2O2, two components that are generated to promote plant defence mechanisms (Lin et al., 2012).
The majority of transgenic technology studies have focused on the effects of stress conditions on seed germination and seedling growth. Since desirable crops may be allocated to grow on compromised lands, seed germination and seedling establishment will be critical for the survival, health, and overall yield of the crop. Therefore, all of the aforementioned transgenic studies, in which individual miRNAs were overexpressed, provide strong evidence for the use of a novel miRNA-based biotechnology for improving crop tolerance to various environmental stresses (Zhang and Wang, 2015).
Conclusions and perspectives
Over the course of time, numerous scientists have tried to elucidate the mechanisms underlying plant response to abiotic stress. In order to support the world’s growing human population, future endeavours will be needed that will focus on breeding new crop cultivars with high tolerance to different environmental stress conditions. Although great progress has been made over the past 20 years, including the identification of both protein-coding genes and small RNAs responsive to abiotic stress, all of these studies are still in their infancy and, therefore, more time is needed before miRNAs become a real target for improving crop tolerance.
miRNA-related research needs to focus on functional analysis instead of miRNA identification
In the past decade, particularly with the development of computational programs and the advancement of deep sequencing technologies, the majority of plant miRNA-related research has focused on the identification of miRNAs from different plant species; however, there are only a few studies that have aimed to elucidate the functions of these miRNAs. Therefore, future studies in this field should switch from identifying miRNAs that are responsive to abiotic stresses to validating the roles of individual miRNAs in plant tolerance to stress conditions. In order to achieve this goal, new technologies must be developed that are able to screen and test multiple genes, including miRNAs, at the same time. This is especially important for miRNAs because there are many miRNAs that have been identified to be responsive to abiotic stress. One new approach may include virus-induced gene silencing (VIGS). Recently, one study successfully overexpressed miR156 in cotton using Cotton leaf crumple virus (CLCrV)-induced gene silencing, which resulted in abnormal leaf development in the transgenic plants (Gu et al., 2014). This study also used VIGS technology to transform a miR156 small tandem target mimic (STTM) into cotton in order to knock down a target of miR156 (Gu et al., 2014). Thus, VIGS can be an efficient approach for studying the function of individual miRNAs.
Using both miRNAs and artificial miRNAs (amiRNAs) to develop transgenic plants with high tolerance to abiotic stress
More and more studies are showing that abiotic stresses induce the aberrant expression of miRNAs and that miRNAs sit at the hub of gene networks that regulate plant response to abiotic stress. This leads to the promise of utilizing miRNAs as a new target for genetically improving agriculturally important traits, including plant response to environmental stress. However, only a few studies have been performed that transformed targeted miRNAs into crops with the goal of improving crop stress tolerance. Additionally, only a few reports have focused on validating miRNA function. One reason why this type of research may be hindered is because we have limited knowledge of the miRNA-regulated gene network that is involved in plant response to abiotic stress. The use of artificial miRNAs (amiRNAs), which can target stress-responsive mRNAs, may solve this problem. In a recent study, amiRNAs were designed to knock down nuclear cap-binding protein 80 [CBP80; also known as Abscisic Acid Hypersensitive 1 (ABH1)] in potato (Pieczynski et al. 2013). The results of this study showed that transgenic potato plants exhibited a higher tolerance to drought stress, particularly due to an increase in leaf stomata and trichome density, ABA-hypersensitive stomatal closing, and compact cuticle structures containing a lower number of microchannels (Pieczynski et al., 2013).
Is miRNA response to abiotic stress a common mechanism or a genotype/stress-dependent mechanism?
It is obvious that different kinds of environmental stress induce the aberrant expression of certain miRNAs in all plant species. Although many studies have been performed in this field, particularly on individual plant species, no study has systemically investigated the common and non-common miRNA-regulated mechanisms that occur during various stresses across a variety of plant species. Therefore, miRNA-mediated reactions may or may not be genotype/stress dependent. To test this, a large-scale study should be performed that compares miRNA expression profiles among several plant species during environmental stress conditions. Ultimately, it will be better to compare major agricultural crops (e.g. maize, soybean, and cotton) with model plant species (e.g. Arabidopsis) so that knowledge gathered from the model plants can be directly translated into functional studies for improving crop tolerance to environmental stress.
Is miRNA modification involved in plant response to abiotic stress?
During miRNA biogenesis, miRNAs may go through several modifications, including truncation, addition, and nucleotide substitution. Several recent reports have shown that miRNA modification widely exists in plants (Xie et al., 2015c; Zhai et al., 2013; J. Zhang et al., 2013), including agriculturally important crops (Xie et al., 2015c). Although one study has proposed that miRNA modification may play an important role in miRNA regulation of plant growth and development, no experimental study has been performed to elucidate the function of individually modified plant miRNAs. Functional analysis of miRNA modifications may provide new insights into plant miRNA biogenesis and ultimately may aid in generating plants with enhanced tolerance to abiotic stress.
Acknowledgements
Mr Jun Ma and Ms Runrun Sun helped to collect some data on miRNA expression in different plant species under different abiotic stresses. We appreciate Ms Taylor Frazier of Virginia Polytechnic Institute and State University for critically proofreading this manuscript. The related project has been partially supported by the USDA NIFA, Cotton Incorporated, and the North Carolina Biotechnology Center.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. 2004. Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Al Abdallat AM, Ayad JY, Abu Elenein JM, Al Ajlouni Z, Harwood WA. 2014. Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Molecular Breeding 33, 401–414. [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. 2005. Antiquity of microRNAs and their targets in land plants. The Plant Cell 17, 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Figueroa B, Gao L, Diop N, Wu Z, Ehlers J, Roberts P, Close T, Zhu J-K, Liu R. 2011. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biology 11, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. 2004. Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. Journal of Experimental Botany 55, 1765–1773. [DOI] [PubMed] [Google Scholar]
- Budak H, Akpinar A. 2011. Dehydration stress-responsive miRNA in Brachypodium distachyon: evident by genome-wide screening of microRNAs expression. Omics 15, 791–799. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L. 2010. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. Journal of Integrative Plant Biology 52, 946–951. [DOI] [PubMed] [Google Scholar]
- Diaz-Vivancos P, Faize M, Barba-Espin G, Faize L, Petri C, Antonio Hernandez J, Burgos L. 2013. Ectopic expression of cytosolic superoxide dismutase and ascorbate peroxidase leads to salt stress tolerance in transgenic plums. Plant Biotechnology Journal 11, 976–985. [DOI] [PubMed] [Google Scholar]
- Ding D, Zhang LF, Wang H, Liu ZJ, Zhang ZX, Zheng YL. 2009. Differential expression of miRNAs in response to salt stress in maize roots. Annals of Botany 103, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldem V, Çelikkol Akçay U, Ozhuner E, Bakır Y, Uranbey S, Unver T. 2012. Genome-wide identification of miRNAs responsive to drought in peach Prunus persica by high-throughput deep sequencing. PLoS One 7, e50298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. 2014. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. Journal of Experimental Botany 65, 2119–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J-E. 2014. Understanding olive adaptation to abiotic stresses as a tool to increase crop performance. Environmental and Experimental Botany 103, 158–179. [Google Scholar]
- Frazier T, Sun G, Burklew C, Zhang B. 2011. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Molecular Biotechnology 49, 159–165. [DOI] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C, et al. 2012. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnology Journal 10, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ashwin G, Parida A. 2012. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. Journal of Experimental Botany 63, 4549–4561. [DOI] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y. 2010. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231, 991–1001. [DOI] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y. 2011. osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Molecular Biology Reports 38, 237–242. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA 105, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N. 2014. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Frontiers in Plant Science 5, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Li X, VandenLangenberg KM, Wen D, Sun S, Wei M, Li Y, Yang F, Shi Q, Wang X. 2014. Overexpression of S-adenosyl- l -methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnology Journal 12, 694–708. [DOI] [PubMed] [Google Scholar]
- Gu Z, Huang C, Li F, Zhou X. 2014. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnology Journal 12, 638–649. [DOI] [PubMed] [Google Scholar]
- Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. 2013. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. The Plant Journal 74, 840–851. [DOI] [PubMed] [Google Scholar]
- Guo H-S, Xie Q, Fei J-F, Chua N-H. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. The Plant Cell 17, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Gustafson P, Langridge P, Shi B-J. 2015. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnology Journal 13, 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H-o, Tomari Y. 2013. Molecular insights into microRNA-mediated translational repression in plants. Molecular Cell 52, 591–601. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. 2009. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Jia X, Wang W-X, Ren L, Chen Q-J, Mendu V, Willcut B, Dinkins R, Tang X, Tang G. 2009. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Molecular Biology 71, 51–59. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhang C, Lu P, Jiang G, Liu X, Dai F, Gao J. 2014. RhNAC3, a stress- associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress- related genes in rose petals. Plant Biotechnology Journal 12, 38–48. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Jung HJ, Kang H. 2007. Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiology and Biochemistry 45, 805–811. [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas S, Budak H. 2011. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233, 471 – –484. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Jung H, Maruyama K, Suzuki N, Kang H. 2010. a . Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kwak KJ, Jung HJ, Lee HJ, Kang H. 2010. b . MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant and Cell Physiology 51, 1079–1083. [DOI] [PubMed] [Google Scholar]
- Kong WW, Yang ZM. 2010. Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiology and Biochemistry 48, 153–159. [DOI] [PubMed] [Google Scholar]
- Kong X, Zhang M, Xu X, Li X, Li C, Ding Z. 2014. System analysis of microRNAs in the development and aluminium stress responses of the maize root system. Plant Biotechnology Journal 12, 1108–1121. [DOI] [PubMed] [Google Scholar]
- Kulcheski FR, de Oliveira LFV, Molina LG, et al. 2011. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Li C-W, Koh KW, Chuang H-Y, Chen Y-R, Lin C-S, Chan M-T. 2014. MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. Journal of Experimental Botany 65, 5049–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BS, Qin YR, Duan H, Yin WL, Xia XL. 2011. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. Journal of Experimental Botany 62, 3765–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu J, Liu Z, Li X, Wu F, He Y. 2014. EAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. The Plant Cell 26, 1764–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Oono Y, Zhu J, He X, Wu J, Iida K, Lu X, Cui X, Jin H, Zhu J. 2008. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell 20, 2238 – –2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo C, Gu J, Duan W, Zhao M, Ma C, Du X, Lu W, Xiao K. 2014. Overexpression of VP, a vacuolar H-pyrophosphatase gene in wheat (Triticum aestivum L.), improves tobacco plant growth under Pi and N deprivation, high salinity, and drought. Journal of Experimental Botany 65, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liang M, Haroldsen V, Cai X, Wu Y. 2006. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant, Cell and Environment 29, 746–753. [DOI] [PubMed] [Google Scholar]
- Lin J-S, Lin C-C, Lin H-H, Chen Y-C, Jeng S-T. 2012. MicroR828 regulates lignin and H2O2 accumulation in sweet potato on wounding. New Phytologist 196, 427–440. [DOI] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang L, Zhou X, Zhou M, Lu Y, Ma L, Ma H, Zhang Z. 2013. Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. Journal of Experimental Botany 64, 2243–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Li Q, Wei H, et al. 2013. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proceedings of the National Academy of Sciences, USA 110, 10848–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SF, Sun Y H, Shi R, Clark C, Li LG, Chiang VL. 2005. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. The Plant Cell 17, 2186–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q-J, Mittal A, Jia F, Rock C. 2012. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Molecular Biology 80, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Chen S, Li A, Zhai C, Jing R. 2014. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One 9, e84359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. 2012. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. Journal of Experimental Botany 63, 2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Agrawal D, Jajoo A. 2014. Photosynthesis: response to high temperature stress. Journal of Photochemistry and Photobiology B Biology 137, 116–126. [DOI] [PubMed] [Google Scholar]
- Meng Y, Ma X, Chen D, Wu P, Chen M. 2010. MicroRNA-mediated signaling involved in plant root development. Biochemical and Biophysical Research Communications 393, 345–349. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends in Plant Science 11, 15–19. [DOI] [PubMed] [Google Scholar]
- Mondal TK, Ganie SA. 2014. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 535, 204–209. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2012. NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta 1819, 97–103. [DOI] [PubMed] [Google Scholar]
- Naya L, Paul S, Valdés-López O, Mendoza-Soto AB, Nova-Franco B, Sosa-Valencia G, Reyes JL, Hernández G. 2014. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS One 9, e84416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. 2012. Overexpression of gma-MIR394a confers tolerance to drought in transgenic Arabidopsis thaliana. Biochemical and Biophysical Research Communications 427, 330–335. [DOI] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. 2013. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Molecular Biology 82, 113–129. [DOI] [PubMed] [Google Scholar]
- Pandurangaiah M, Rao GL, Sudhakarbabu O, Nareshkumar A, Kiranmai K, Lokesh U, Thapa G, Sudhakar C. 2014. Overexpression of horsegram (Macrotyloma uniflorum Lam.Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Molecular Biotechnology 56, 758–769. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E. 2011. Hormone balance and abiotic stress tolerance in crop plants. Current Opinion in Plant Biology 14, 290–295. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes and Development 18, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczynski M, Marczewski W, Hennig J, et al. 2013. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnology Journal 11, 459–469. [DOI] [PubMed] [Google Scholar]
- Qin YR, Duan ZX, Xia XL, Yin WL. 2011. Expression profiles of precursor and mature microRNAs under dehydration and high salinity shock in Populus euphratica. Plant Cell Reports 30, 1893–1907. [DOI] [PubMed] [Google Scholar]
- Raghuram B, Sheikh AH, Sinha AK. 2014. Regulation of MAP kinase signaling cascade by microRNAs in Oryza sativa . Plant Signaling and Behavior 9, e29804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua N-H. 2007. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal 49, 592–606. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Rong W, Qi L, Wang A, Ye X, Du L, Liang H, Xin Z, Zhang Z. 2014. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnology Journal 12, 468–479. [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I. 2014. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell 26, 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. 2006. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell 18, 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu J. 2004. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Tamirisa S, Vudem DR, Khareedu VR. 2014. Overexpression of pigeonpea stress-induced cold and drought regulatory gene (CcCDR) confers drought, salt, and cold tolerance in Arabidopsis. Journal of Experimental Botany 65, 4769–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I., Capitão C, Dalmay T, Fevereiro M, Santos DM. 2010. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231, 705–716. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crété P. 2004. Endogenous trans-Acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular Cell 16, 69–79. [DOI] [PubMed] [Google Scholar]
- Wang J-W, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang Q, Zhang B. 2013. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530, 26–32. [DOI] [PubMed] [Google Scholar]
- Wang TZ, Chen L, Zhao MG, Tian QY, Zhang WH. 2011. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. 2003. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun F, Cao H, Peng H, Ni Z, Sun Q, Yao Y. 2012. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One 7, e48445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhang D, Xiang F, Zhang Z. 2009. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. International Journal of Plant Sciences 170, 979–989. [Google Scholar]
- Winter J, Diederichs S. 2011. Argonaute proteins regulate microRNA stability. RNA Biology 8, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Xie FL, Jones DC, Wang QL, Sun RR, Zhang BH. 2015. a . Small RNA sequencing identifies miRNA roles in ovule and fiber development. Plant Biotechnology Journal (in press). [DOI] [PubMed] [Google Scholar]
- Xie F, Stewart CN, Taki FA, He Q, Liu H, Zhang B. 2014. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnology Journal 12, 354–366. [DOI] [PubMed] [Google Scholar]
- Xie F, Wang Q, Sun R, Zhang B. 2015. b . Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. Journal of Experimental Botany 66, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie FL, Wang QL, Zhang BH. 2015. c . Global microRNA modification in cotton (Gossypium hirsutum L.). Plant Biotechnology Journal (in press). [DOI] [PubMed] [Google Scholar]
- Xu MY, Zhang L, Li WW, Hu XL, Wang M-B, Fan YL, Zhang CY, Wang L. 2014. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana . Journal of Experimental Botany 65, 89–101. [DOI] [PubMed] [Google Scholar]
- Xu Q, He Q, Li S, Tian Z. 2014. Molecular characterization of StNAC2 in potato and its overexpression confers drought and salt tolerance. Acta Physiologiae Plantarum 36, 1841–1851. [Google Scholar]
- Yang C, Li D, Mao D, Liu XUE, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L. 2013. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant, Cell and Environment 36, 2207–2218. [DOI] [PubMed] [Google Scholar]
- Yin Z, Li Y, Yu J, Liu Y, Li C, Han X, Shen F. 2012. Difference in miRNA expression profiles between two cotton cultivars with distinct salt sensitivity. Molecular Biology Reports 39, 4961–4970. [DOI] [PubMed] [Google Scholar]
- Zhai J, Zhao Y, Simon SA, et al. 2013. Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. The Plant Cell 25, 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. 2006. Conservation and divergence of plant microRNA genes. The Plant Journal 46, 243–259. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang Q. 2015. MicroRNA-based biotechnology for plant improvement. Journal of Cellular Physiology 230, 1–15. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. 2007. MicroRNAs and their regulatory roles in animals and plants. Journal of Cellular Physiology 210, 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. 2005. Identification and characterization of new plant microRNAs using EST analysis. Cell Research 15, 336–360. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang S, Li S, Han S, Wu T, Li X, Qi L. 2013. A genome-wide survey of microRNA truncation and 3′ nucleotide addition events in larch (Larix leptolepis). Planta 237, 1047–1056. [DOI] [PubMed] [Google Scholar]
- Zhang LW, Song JB, Shu XX, Zhang Y, Yang ZM. 2013. miR395 is involved in detoxification of cadmium in Brassica napus. Journal of Hazardous Materials 250–251, 204–211. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z. 2011. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnology Letters 33, 403–409. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y. 2009. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Molecular Biology 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BT, Liang RQ, Ge LF, Li W, Xiao HS, Lin HX, Ruan KC, Jin YX. 2007. Identification of drought-induced microRNAs in rice. Biochemical and Biophysical Research Communications 354, 585–590. [DOI] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu J-K, Zhang F, Li W-X. 2011. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytologist 190, 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. 2010. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. Journal of Experimental Botany 61, 4157–4168. [DOI] [PubMed] [Google Scholar]
- Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H. 2013. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiology 161, 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W. 2007. UV-B responsive microRNA genes in Arabidopsis thaliana. Molecular Systems Biology 3, 103–103. [DOI] [PMC free article] [PubMed] [Google Scholar]