Abstract
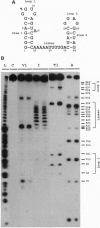
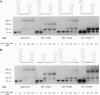
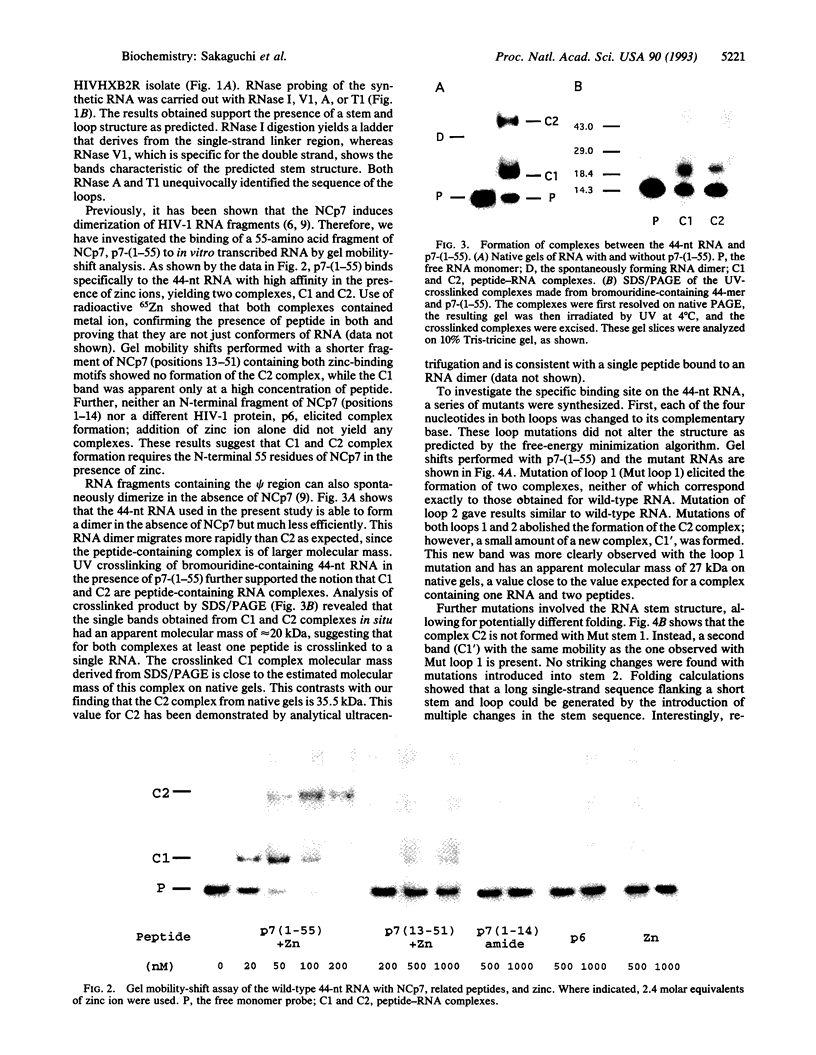
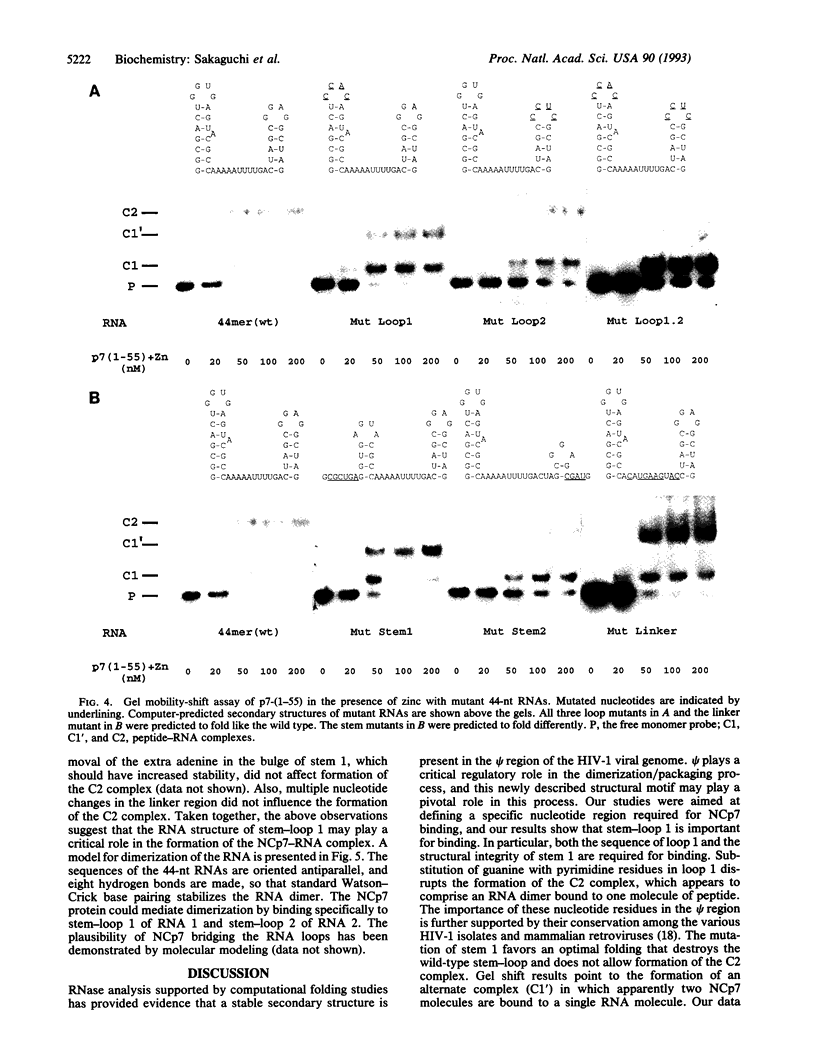
The nucleocapsid (NC) protein NCp7 of human immunodeficiency virus type 1 (HIV-1) is important for encapsidation of the virus genome, RNA dimerization, and primer tRNA annealing in vitro. Here we present evidence from gel mobility-shift experiments indicating that NCp7 binds specifically to an RNA sequence. Two complexes were identified in native gels. The more slowly migrating complex contained two RNA molecules and one peptide, while the more rapidly migrating one is composed of one RNA and one peptide. Further, mutational analysis of the RNA shows that the predicted stem and loop structure of stem-loop 1 plays a critical role. Our results show that NCp7 binds to a unique RNA structure within the psi region; in addition, this structure is necessary for RNA dimerization. We propose that NCp7 binds to the RNA via a direct interaction of one zinc-binding motif to stem-loop 1 followed by binding of the other zinc-binding motif to stem-loop 1, stem-loop 2, or the linker region of the second RNA molecule, forming a bridge between the two RNAs.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boone L. R., Skalka A. M. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. I. Kinetics of synthesis and size of minus- and plus-strand transcripts. J Virol. 1981 Jan;37(1):109–116. doi: 10.1128/jvi.37.1.109-116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille F., Mely Y., Ficheux D., Savignol I., Gerard D., Darlix J. L., Fournie-Zaluski M. C., Roques B. P. Solid phase synthesis of the retroviral nucleocapsid protein NCp10 of Moloney murine leukaemia virus and related "zinc-fingers" in free SH forms. Influence of zinc chelation on structural and biochemical properties. Int J Pept Protein Res. 1990 Dec;36(6):551–558. [PubMed] [Google Scholar]
- Darlix J. L., Gabus C., Nugeyre M. T., Clavel F., Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990 Dec 5;216(3):689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- De Rocquigny H., Gabus C., Vincent A., Fournié-Zaluski M. C., Roques B., Darlix J. L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R. J., Nigida S. M., Jr, Bess J. W., Jr, Arthur L. O., Henderson L. E., Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990 Jul;64(7):3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison G. P., Lever A. M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992 Jul;66(7):4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L. E., Copeland T. D., Sowder R. C., Smythers G. W., Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J Biol Chem. 1981 Aug 25;256(16):8400–8406. [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings D. A., Nash M. A., Maizel J. V., Arlinghaus R. B. Novel GACG-hairpin pair motif in the 5' untranslated region of type C retroviruses related to murine leukemia virus. J Virol. 1992 Feb;66(2):632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985 May;54(2):401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Prats A. C., Roy C., Wang P. A., Erard M., Housset V., Gabus C., Paoletti C., Darlix J. L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990 Feb;64(2):774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts W. J., Elliott J. I., McMurray W. J., Williams K. R. Synthesis of the p10 single-stranded nucleic acid binding protein from murine leukemia virus. Pept Res. 1988 Nov-Dec;1(2):74–80. [PubMed] [Google Scholar]
- Roy C., Tounekti N., Mougel M., Darlix J. L., Paoletti C., Ehresmann C., Ehresmann B., Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990 Dec 25;18(24):7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Appella E., Omichinski J. G., Clore G. M., Gronenborn A. M. Specific DNA binding to a major histocompatibility complex enhancer sequence by a synthetic 57-residue double zinc finger peptide from a human enhancer binding protein. J Biol Chem. 1991 Apr 15;266(11):7306–7311. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shapiro B. A., Zhang K. Z. Comparing multiple RNA secondary structures using tree comparisons. Comput Appl Biosci. 1990 Oct;6(4):309–318. doi: 10.1093/bioinformatics/6.4.309. [DOI] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Summers M. F., South T. L., Kim B., Hare D. R. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990 Jan 16;29(2):329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- Tsurumi T., Lehman I. R. Release of RNA polymerase from vero cell mitochondria after herpes simplex virus type 1 infection. J Virol. 1990 Jan;64(1):450–452. doi: 10.1128/jvi.64.1.450-452.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]