SUMMARY
Epigenetic silencing of tumor suppressor genes is generally thought to involve DNA cytosine methylation, covalent modifications of histones, and chromatin compaction. Here, we show that silencing of the three transcription start sites in the bidirectional MLH1 promoter CpG island in cancer cells involves distinct changes in nucleosomal occupancy. Three nucleosomes, almost completely absent from the start sites in normal cells, are present on the methylated and silenced promoter, suggesting that epigenetic silencing may be accomplished by the stable placement of nucleosomes into previously vacant positions. Activation of the promoter by demethylation with 5-aza-2′-deoxycytidine involves nucleosome eviction. Epigenetic silencing of tumor suppressor genes may involve heritable changes in nucleosome occupancy enabled by cytosine methylation.
INTRODUCTION
CpG islands, which often occur at the transcription start sites of cancer-relevant genes, have been the focus of most studies on epigenetic silencing. CpG islands have an alternative structure to bulk chromatin in normal cells (Tazi and Bird, 1990). They are characterized by a lack of cytosine methylation, low levels of histone H1, high levels of histone acetylation, and hypersensitivity to DNaseI which has been equated with nucleosome-free regions (Mucha et al., 2000; Pfeifer and Riggs, 1991; Tazi and Bird, 1990). Genome-wide analysis has shown that DNaseI-sensitive sites are often present in both the expressing and nonexpressing states on many autosomal genes (Mito et al., 2007; Sabo et al., 2004). In a more specific case, the start site of the maternally imprinted and repressed Igf2 gene is unmethylated and DNaseI hypersensitive to the same extent as the paternal gene, which is strongly expressed (Sasaki et al., 1992a). These and other data (Davey et al., 2004) suggest that the “open” or permissive state of CpG islands for potential transcription is mitotically heritable. A promoter permissive for transcription may therefore be kept nucleosome free, allowing it to become activated by binding of appropriate transcriptional activators (Li et al., 2007). A central goal of tumor biology is to understand the mechanisms by which this permissive chromatin configuration is converted into a state that is permanently repressed and nonpermissive for expression.
The most well-characterized heritable covalent change at the start sites of CpG island promoters of tumor suppressor genes is abnormal promoter methylation (Herman and Baylin, 2003; Jones and Baylin, 2007). Much effort has been spent on investigating additional epigenetic changes such as covalent histone modifications responsible for aberrant gene regulation in cancer. Inactive, hypermethylated promoters are associated with a closed or repressive chromatin configuration, characterized by deacetylated histones and a variety of inactive histone marks. For examples, in colon and breast cancer cells, the silencing of MLH1 (Fahrner et al., 2002; McGarvey et al., 2006) and RASSF1A (Strunnikova et al., 2005) is associated with deacetylated histone H3 and increased H3-K9 methylation. The objective of this study was to explore additional determinants such as the role of nucleosomal occupancy in CpG island silencing in cancer because this has been implicated in transcriptional control in yeast, flies, and humans (Bernstein et al., 2004; Heintzman et al., 2007; Lee et al., 2004; Lieb and Clarke, 2005; Mito et al., 2005; Ozsolak et al., 2007; Yuan et al., 2005).
SIGNIFICANCE.
CpG islands are generally kept in an inherently “open” state independently of expression in normal cells and are methylation free and sensitive to nuclease cleavage in chromatin. Silencing in cancer involves DNA methylation and chromatin covalent and structural changes that are somatically heritable and contribute to the cancer phenotype. Using the MLH1 CpG island as an example, we show that the switch between the two heritable states involves the complete absence or presence of nucleosomes at the three start sites of the bidirectional promoter. Cytosine methylation may therefore ultimately lead to silencing, through the mediation of methylated DNA-binding proteins and other chromatin modifiers, by enabling the stable presence of nucleosomes at the start sites of cancer-related genes.
We focused on MLH1, a key player in the DNA mismatch repair system (Modrich and Lahue, 1996), which is frequently silenced by promoter hypermethylation in various cancers (Herman et al., 1998; Kanaya et al., 2003; Kane et al., 1997; Murata et al., 2002; Xiong et al., 2001). MLH1, which we determined to have two transcripts, 1a and 1b, is a member of a class of bidirectional gene pairs that are transcribed head-to-head on opposite strands, comprising ~10% of human genes (Adachi and Lieber, 2002; Lavia et al., 1987; Takai and Jones, 2004; Trinklein et al., 2004).
Until recently, methods for examining nucleosome positioning have relied on methodologies that reveal only the average state of a given promoter on all of the molecules in a cell population. However, we were able to analyze the scenario on individual molecules by using our high-resolution, methylase-based single-promoter analysis assay (MSPA), providing insights into the dynamics of chromatin remodeling (Fatemi et al., 2005; Gal-Yam et al., 2006). Our results, obtained in several cell lines and confirmed by traditional methods, show remarkable nucleosome depletion just upstream of each start site on the active MLH1 promoter, whereas the inactive promoter is associated with nucleosome occupancy in a mitotically heritable fashion.
We also show that nucleosomes are removed from promoter molecules upon gene reactivation by demethylation with 5-aza-2′-deoxycytidine (5-aza-CdR). Thus, our results provide strong evidence that epigenetic silencing of tumor suppressor genes may involve the insertion of nucleosomes into previously vacant positions.
RESULTS
Organization of the EPM2AIP1 and MLH1 Genes
The bidirectional genes EPM2AIP1 and MLH1 on human chromosome 3p22.1 are located within a CpG island that is 1.6 kb long and has a GC content of 57% and a CpG observed/expected ratio of 0.80 (Takai and Jones, 2003) (Figure 1A). The transcription start sites of the two genes were suggested to be 470 bp apart on the GenBank website. However, 5′-RACE analysis with mRNA isolated from LoVo colon cancer cells (data not shown) established that MLH1 has two transcription start sites (1a and 1b) spaced 309 bp apart, a distance that could accommodate almost two nucleosomes, whereas EPM2AIP1 initiates transcription at only one site, 321 bp upstream of the first initiation site, 1a, of MLH1 (Figure 1A). The start site of EPM2AIP1 was found to be ~150 bp closer to MLH1 (1a) than indicated by the GenBank website; therefore, the divergent transcription start sites are actually 321 bp apart, a distance that could also accommodate approximately two nucleosomes. Dual luciferase reporter assays confirmed that the promoter was indeed bidirectional (see Figure S1 in the Supplemental Data available with this article online).
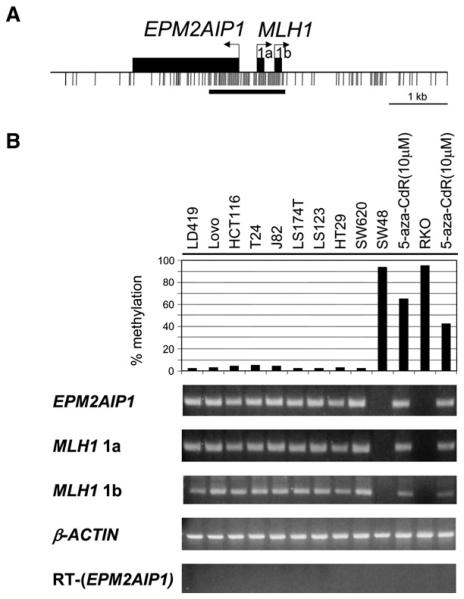
Figure 1. Correlation of the Methylation Status of the Bidirectional Promoter and the Expression Patterns of EPM2AIP1 and MLH1.
(A) The bidirectional promoter and the CpG island. Horizontal arrows show the transcription start sites as established by 5′-RACE analysis, and black boxes show the respective first exons. Black tick marks indicate CpG dinucleotides. The horizontal bar underneath the tick marks represents the CpG island.
(B) The average methylation levels of three CpGs in the EPM2AIP1/MLH1 promoter region were analyzed by Ms-SNuPE, and the expression of both genes was determined by RT-PCR in a normal LD419 human bladder fibroblast cell line and various human cancer cell lines. β-ACTIN expression served as a control for the input amount of cDNA. RT served as a negative control for the intronless gene, EPM2AIP1.
Correlation of the Endogenous Methylation Status of the Bidirectional Promoter and the Expression Patterns of EPM2AIP1 and MLH1
To determine the relationship between the methylation status of the bidirectional promoter and the expression of the two genes, we used the quantitative methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) assay (Bender et al., 1999) and RT-PCR in normal human fibroblast cells and various colon and bladder cancer cell lines. Normal human fibroblast LD419 cells and cell lines with low methylation levels (LoVo, HCT116, T24, J82, LS174T, LS123, HT29, and SW620) expressed EPM2AIP1 and both transcripts of MLH1 (Figure 1B). Conversely, none of the transcripts was produced in the colon cell lines SW48 and RKO, displaying promoter hypermethylation (93% and 95%, respectively, Figure 1B). Treatment of both cell lines (SW48 and RKO) with the demethylating agent 5-aza-CdR for 24 hr (Jones and Taylor, 1980) caused a decrease in the level of CpG methylation, with a concordant reactivation of all three transcripts (Figure 1B). Thus, the endogenous methylation status of the bidirectional promoter correlates quite well with the expression patterns of both genes, as was also shown by others (Shu et al., 2006).
The MLH1 Promoter Has Only One Highly Positioned Nucleosome in Expressing Cells, Whereas It Is Occupied by Nucleosomes in Nonexpressing Cells
Because nucleosomal positioning and histone tail modifications play essential roles in transcriptional regulations (Bernstein and Allis, 2005; Rice and Allis, 2001), we first investigated chromatin structure at all three transcription start sites by using the DNaseI hypersensitivity assay. Nuclei from LD419 and RKO cells were treated with increasing concentrations of DNaseI to obtain suitable levels of digestion (Figure 2A).
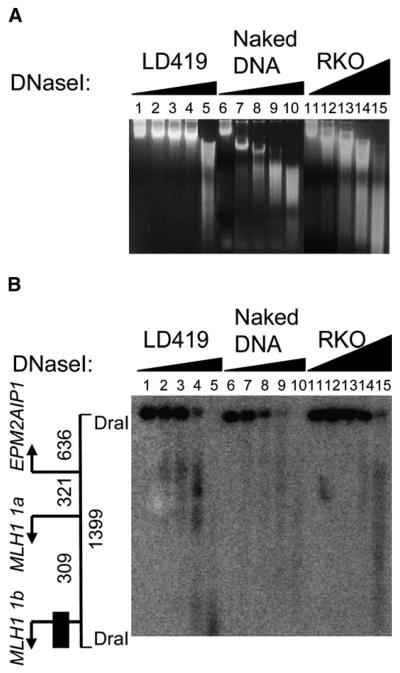
Figure 2. Detection of Hypersensitive Sites by DNaseI Digestion.
(A) Genomic naked DNA and nuclei from LD419 and RKO cells were treated with increasing concentrations of DNaseI, DNA from each sample was then purified and treated with DraI, and the digestion products were analyzed by Southern blot. Naked DNA, as a control, was used to confirm the lack of sequence specificity of the enzyme. Digested samples prior to DraI treatment were resolved by gel electrophoresis as shown.
(B) Southern blot analysis revealed DNaseI hypersensitivity in the EPM2AIP1/MLH1 promoter. On the left, drawn to scale, the 1399 bp DNA fragment generated by DraI digestion, transcription start sites (arrows), and the probe fragment (black box) are indicated. Numbers show the distance in bp.
Southern blot images revealed a region of hypersensitivity in the lanes of minimally digested LD419 chromatin samples (Figure 2B, lanes 2–4), which was mapped to the EPM2AIP1/MLH1 1a promoter region. Interestingly, another hypersensitive region corresponding to the region upstream of MLH1 1b was also detected at higher enzyme concentrations (Figure 2B, lane 4). These patterns were not detected in control naked DNA, suggesting the lack of sequence preference of DNaseI for that region. At the genomic level, compared to LD419 nuclei, RKO nuclei were digested to a greater extent with higher concentrations of DNaseI (Figure 2A, lanes 11–14). However, even under such digestion conditions, at the specific loci examined, the 1399 bp DraI fragment persisted in lanes 11–14 (Figure 2B) without showing any discrete DNaseI hypersensitivity. The region in RKO cells is therefore more compact and inaccessible to DNaseI. Thus, the regions just upstream of all three start sites are highly accessible in expressing LD419 cells, but not in nonexpressing RKO cells. The hypersensitive sites in LD419 cells may suggest the lack of nucleosomes, as a result of changes in chromatin structure correlated with the transcriptional activity of the genes.
To confirm the nucleosome depletion suggested by DNaseI footprinting, we examined the presence of nucleosomes in the promoter regions at the mononucleosomal level. Nuclei from LD419 and RKO cells were partially digested with MNase, yielding molecules of various nucleosomal repeats. Nucleosomes from the digested nuclei were then run on sucrose gradients to isolate fractions enriched in mononucleosomes, and the DNA, derived from the mononucleosomes, was analyzed by quantitative PCR by using eight primer sets positioned across the region (Figure 3A). Consistent with the DNaseI results, in expressing LD419 cells, the region between the EPM2AIP1 and MLH1 1a transcription initiation sites (TISs) (Figure 3A, b–d) and the region of ~150 bp just upstream of MLH1 1b TIS (Figure 3A, f) showed virtually no signals after PCR. Interestingly, there is one, and only one, highly positioned nucleosome in the MLH1 promoter (1a+1b), just downstream of the MLH1 1a TIS (Figure 3A, e), corresponding to the region between two hypersensitive sites (Figure 2B, lane 4). However, no analogous nucleosomal depletion was observed in nonexpressing RKO cells (Figure 3A). Taken together, these results argue that the promoter regions just upstream of all three start sites are depleted of nucleosomes in expressing LD419 cells, but are occupied by nucleosomes in nonexpressing RKO cells.
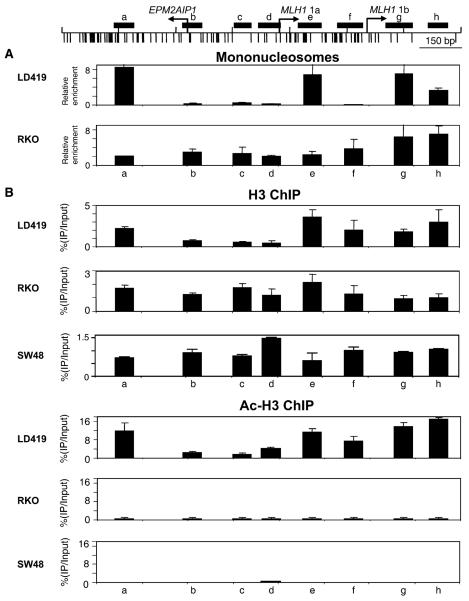
Figure 3. Nucleosomal Depletion by Mononucleosomal DNA and ChIP Analyses.
(A) Mononucleosomal DNA analysis. Nuclei from LD419 and RKO were digested partially with MNase, and the reaction mixture was run on a sucrose gradient to isolate mononucleosomal DNA. Enrichment of mononucleosomal DNA was analyzed by real-time PCR by using primers specific for eight regions (a–h), shown as black rectangles on the top of the figure.
(B) Distinct chromatin structures at the EPM2AIP1/MLH1 promoter in expressing LD419 and nonexpressing RKO and SW48 cells. ChIP analysis performed with antibodies against histone H3 and acetylated histone H3. Immunoprecipitated DNA was analyzed by real-time PCR as described in (A). The fraction of immunoprecipitated DNA was calculated as a percentage of input DNA. Results are shown as the mean (bar) ± SD of two or three experiments from two independent chromatin preparations.
Distinct Chromatin Structures at the EPM2AIP1/MLH1 Promoter in Expressing and Nonexpressing Cells
Next, we used the chromatin immunoprecipitation assay (ChIP) to validate the above-described results. Because we wanted as high a resolution map as possible over the relatively small region between the start sites, particular attention was paid to ensuring that the fragments generated by chromatin sonication were of the order of 200 bp.
Consistent with the DNaseI and the mononucleosomal DNA analyses, H3 occupancy was clearly lower in the region between the EPM2AIP1/MLH1 1a start site in expressing LD419 cells and was much higher in nonex-pressing RKO and SW48 cells (Figure 3B, b–d). A zone of nucleosomal depletion or a “dip” in histone H3 occupancy was seen in the EPM2AIP1/MLH1 1a promoter region of the expressing LD419 cells (Figure 3B, b-d). No prominent dips were present in nonexpressing RKO and SW48 cells, and H3 occupancy was higher in the EPM2AIP1/MLH1 1a promoter region compared to LD419 (Figure 3B, b–d). However, ChIP analysis did not detect the depletion of the nucleosome upstream of the start site of MLH1 1b in LD419 (Figure 3B, f), probably as a result of the limited resolution of sonication. Overall, histone H3 acetylation was high in expressing LD419 and virtually none in RKO and SW48 (Figure 3B, a–h). Remarkably, the H3 acetylation pattern in LD419 recapitulated the patterns seen in the H3 and mononucleosomal DNA analyses, showing a prominent dip in the EPM2AIP1/MLH1 1a promoter and a lesser dip in the region just upstream of MLH1 1b (Figure 3B, LD419, regions b–d and f). Although many studies have shown that promoter regions are often associated with increased acetylation in active promoters without specific references to H3 occupancy, our results indicate that the increased acetylation is actually coupled with H3 enrichment in regions surrounding the promoter (Figure 3B, a and e).
To test the generality of these findings, the chromatin structure of the BRCA1/NBR2 CpG island promoter was also investigated. Consistent with the MLH1 promoter, nucleosomal depletion was again seen in the unmethylated promoter in expressing T47D and Caov3 cells (see Figures S2, S3A, and S3B, regions R5 and R4).
We next used a high-resolution MSPA to footprint individual DNA molecules in the MLH1 promoter (Fatemi et al., 2005; Gal-Yam et al., 2006; Kladde and Simpson, 1996). Nuclei from expressing, unmethylated LD419 cells were treated with M.SssI, which methylates all accessible CpG sites in purified DNA, but is unable to methylate CpG sites that are found within a nucleosome, or bound by tight-binding transcription factors (Fatemi et al., 2005; Gal-Yam et al., 2006; Kladde and Simpson, 1996). After M.SssI treatment, bisulfite conversion of extracted DNA, and PCR amplification of the promoter region, single PCR products were cloned and sequenced to show the accessibilities of individual DNA strands to the methylase.
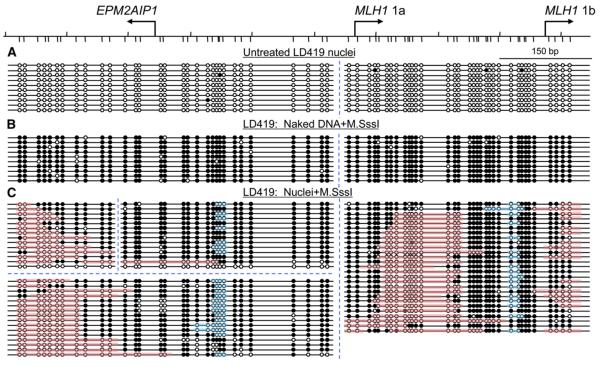
As expected, the regions analyzed were virtually unmethylated before M.SssI treatment in untreated LD419 nuclei (Figure 4A). Control experiments showed that naked DNA extracted from the same cells was almost completely methylated by M.SssI treatment at the region under the same experimental conditions used for nuclei, with no preferential sites of methylation (Figure 4B). Analysis of the M.SssI-treated nuclei revealed that the 321 bp region between the two start sites (EPM2AIP1 and MLH1 1a) as well as a region of ~150 bp upstream of MLH1 1b were largely accessible to M.SssI, as shown by the extensive acquired methylation, indicating the absence of nucleosomes in these regions (Figure 4C). No nucleosome footprint was seen in 28 out of 30 promoter replicas analyzed in the 321 bp region, and none were seen in any of the 27 molecules analyzed in the 150 bp upstream of MLH1 1b (Figure 4C). Furthermore, there were two clear patches of substantial inaccessibility, suggesting the presence of nucleosomes downstream of the MLH1 1a and EPM2AIP1 start sites (Figure 4C, pink). Patches are defined as at least two consecutively unmethylated sites flanked on each side by at least two consecutively methylated CpG sites (Fatemi et al., 2005). Patches at both the 5′ and 3′ ends were also considered as nucleosomes on the basis of the ChIP and mononucleosome experiments (Figures 3A and 3B). In fact, a well-defined nucleosome was precisely positioned in 93% (25/27) of molecules examined in the region downstream of MLH1 1a (Figure 4C, pink).
Figure 4. Accessibility of Native Chromatin to M.SssI at the EPM2AIP1/MLH1 Promoter Region in Expressing LD419 Cells.
(A–C) Nuclei were extracted from expressing unmethylated LD419 cells and were then treated with M.SssI for 15 min, followed by bisulfite genomic sequencing. Two independent bisulfite-sequencing reactions were done to avoid introducing a bias in the analyses. Four PCR products of different sizes, indicated by the blue, dotted lines, were included in the analysis. (A) Untreated nuclei. (B) Naked DNA treated with M.SssI. (C) Nuclei treated with M.SssI. Horizontal lines with circles indicate individual molecules that were sequenced after PCR amplification and cloning of bisulfite-treated DNA. Solid circles, methylated CpG dinucleotides; open circles, unmethylated CpG dinucleotides. Pink bars indicate areas or patches that are inaccessible to M.SssI, suggesting the presence of nucleosomes. Patches are defined as at least two consecutively unmethylated sites flanked on each side by at least two consecutively methylated CpG sites (Fatemi et al., 2005). Blue bars show the putative protein-binding regions.
Two inaccessible patches whose sizes were too small to qualify as nucleosomes were also detected. One patch of ~30 bp was observed in the region between EPM2AIP1 and MLH1 1a, and another patch of ~20 bp was also detected in the region between MLH1 1a and 1b, which may be unidentified protein-binding sites (Figure 4C, blue). In addition, there was another region of substantial inaccessibility downstream of MLH1 1b that could indicate another nucleosome, as also suggested by the high enrichment in region g in Figure 3. Together, these results suggest that, in expressing LD419 cells, the region between EPM2AIP1 and MLH1 1a is devoid of nucleosomes, whereas there is precisely one nucleosome positioned downstream of MLH1 1a, followed by another nucleosome-free region upstream of the MLH1 1b start site. The nucleosome-free regions correspond to the hypersensitive regions seen in the DNaseI assay (Figure 2B, lane 4) and also to precisely the same regions with the lowest enrichment seen in the mononucleosomal DNA analysis (Figure 3A, regions b–d and f).
In addition, to verify that the nucleosome depletion observed is not just cell line specific, the MSPA assay was also applied to expressing T24 cells. Consistently, the region between EPM2AIP1 and MLH1 1a was virtually nucleosome free in 17 of 20 promoter replicas, as shown by high levels of accessibility to M.SssI (see Figure S4B).
Nucleosome Eviction after 5-aza-CdR Treatment
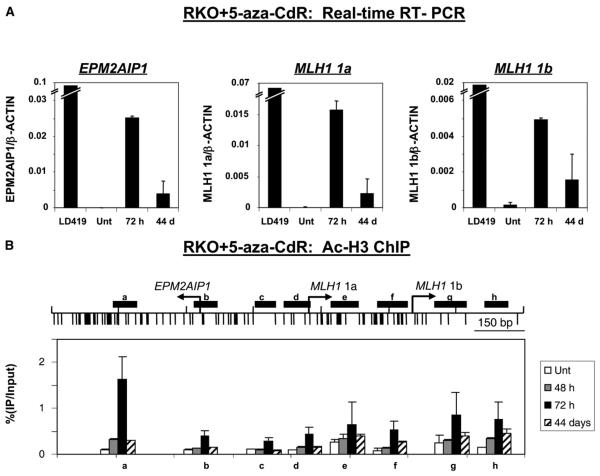
Because the promoter in nonexpressing RKO cells was occupied by nucleosomes, we were interested to see the chromatin structural changes accompanying gene activation. First, we quantified the level of expression of the silenced genes after 5-aza-CdR treatment by using quantitative RT-PCR. As found previously (Figure 1B), 24 hr of 5-aza-CdR treatment caused a concordant reactivation of all three transcripts at 72 hr after addition of the drug (Figure 5A). Expression for all three transcripts was sustained even 44 days after drug treatment, although with a 3- to 7-fold decrease in the level of expression (Figure 5A). The decreased expression as a function of time after treatment has been observed with other genes activated by 5-aza-CdR treatment and is due to a gradual resilencing effect (Bender et al., 1999).
Figure 5. Chromatin Structural Changes upon Gene Reactivation by 5-Aza-CdR.
RKO cells were treated with 5-aza-CdR and then harvested 72 hr and 44 days after drug addition for RT-PCR and ChIP analyses. (A) Quantitative RT-PCR. Expression levels were normalized with β-ACTIN, which served as a control for the input cDNA. A minus-RT control served as a negative control for the intronless gene, EPM2AIP1 (data not shown). Results are shown as the mean (bar) ± SD of two PCRs from two independent cDNA preparations. (B) Histone H3 acetylation patterns by ChIP. Immunoprecipitated DNA was analyzed by real-time PCR as described (Figure 3B). The fraction of immunoprecipitated DNA was calculated as a percentage of input DNA. Results are shown as the mean (bar) ± SD of two or three experiments from two independent chromatin preparations.
We then analyzed the chromatin changes upon gene activation by ChIP analysis of RKO cells 48 hr, 72 hr, and 44 days after 5-aza-CdR treatment. H3 acetylation was minimal in untreated RKO cells, increased slightly by 48 hr, and rose substantially by several fold in every region examined 72 hr after drug treatment (Figure 5B, a–h), as seen by others (Fahrner et al., 2002). Even 44 days after drug treatment, the H3 acetylation was still maintained at levels intermediate to those seen in untreated cells and after 72 hr in virtually all regions. Strikingly, after gene activation, acetylation patterns recapitulated the scenario seen in the mononucleosomal DNA analysis, showing a dip in the EPM2AIP1/MLH1 1a promoter and a lesser dip upstream of the MLH1 1b at all time points after treatment. Qualitatively, the changes in H3 acetylation levels correlate well with the changes in gene expression levels.
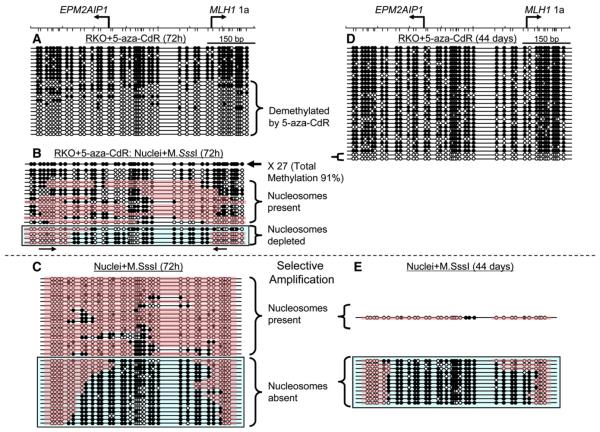
We next applied the MSPA assay to detect chromatin structural changes in RKO cells after gene activation upon 5-aza-CdR treatment. We focused on the region between the EPM2AIP1/MLH1 1a start sites to simplify the analysis, and almost all of the CpG sites in the promoter were endogenously methylated in untreated RKO cells (see Figure S5A). In accordance with the expected mechanism of action of the drug (Egger et al., 2004), ~62% (15/24) of the molecules were methylation free or had sporadic residual methylation 72 hr after treatment (Figure 6A). These molecules could potentially serve as substrates for M.SssI, thus allowing us to search for new footprints by using the MSPA mapping technology.
Figure 6. Eviction of Nucleosomes by 5-Aza-CdR Treatment.
(A–E) RKO cells were treated with 5-aza-CdR for 24 hr and were harvested 72 hr and 44 days after drug treatment started. Both DNA and nuclei from drug-treated RKO cells were extracted and then subjected to M.SssI treatment, followed by bisulfite genomic sequencing. (A and D) Demethylation of the promoter in RKO cells (A) 72 hr and (D) 44 days after addition of 5-aza-CdR. (B) Accessibility of native chromatin to M.SssI at the EPM2AIP1/MLH1 1a promoter region in RKO cells 72 hr after drug addition. To filter out extensively methylated molecules that were not suitable for M.SssI treatment, PCR analyses were done with selective primers that only anneal to unmethylated molecules. See the main text for rationale. (C and E) M.SssI treatments on nuclei from drug-treated RKO cells (C) 72 hr and (E) 44 days after drug addition, followed by bisulfite genomic sequencing with selective primers. Please refer to Figure 4 for descriptions of molecules and patches. The DNA molecules with nucleosome depletion in the promoter region are boxed in blue.
We first verified that naked DNA extracted from 5-aza-CdR-treated cells could be fully converted to methylated molecules by M.SssI. This was indeed the case, because 95% (20/21) of the sequenced molecules were extensively methylated after M.SssI treatment (see Figure S5B). Therefore, residual 5-aza-cytosine in DNA did not complicate the analysis by the generation of spurious patterns.
Nuclei prepared from 5-aza-CdR-treated cells were exposed to M.SssI, and 45 molecules were sequenced and analyzed (Figure 6B). A total of 27 clones (60%) were almost completely methylated (total methylation level of 91%), but these were noninformative because we could not distinguish whether they represented parent DNA molecules that had not been demethylated by the drug or whether they had been remethylated by M.SssI. However, ~40% (18 of 45 clones) of the molecules had probably become modified by M.SssI, because none of patterns resembled any of the molecules in RKO cells after 5-aza-CdR treatment (Figure 6A). In addition, there were partially methylated molecules, which had modification patterns not present before M.SssI treatment. Of these, ~61% (11 of 18 clones) had protected patches (as previously defined), large enough to accommodate nucleosomes, and many of these patches had the diagnostic ~150 or multiples of 150 bp size footprints of a nucleosome (Fatemi et al., 2005)(Figure 6B). These represent demethylated molecules that still harbored nucleosomes in the MLH1 promoter, whereas others strongly resembled the patterns observed in expressing LD419 cells (Figure 4C), with high levels of M.SssI accessibility between the transcription start sites (Figure 6B, blue box).
These data were obtained by amplification of the region including parent methylated molecules that had not been demethyated by 5-aza-CdR treatment. Thus, to filter out such noninformative molecules and further refine the analysis, selective primers were designed, and the reverse primer was annealed only to unmethylated molecules in order to obtain a better representation of informative promoter replicas (Figure 6B, arrows underneath the blue box). As controls, no PCR products were produced from the amplification of the DNA from untreated RKO cells (data not shown), whereas after 5-aza-CdR treatment, only substantially unmethylated molecules were amplified and cloned, confirming the specificity of the primers (see Figure S5C).
Nuclei prepared from 5-aza-CdR-treated cells 72 hr after drug addition were then exposed to M.SssI, and promoter molecules were sequenced and analyzed by selective amplification (Figure 6C). Approximately 54% (20 of 37 molecules) had nucleosomal patches (as defined above) (Figure 6C). These represent demethylated molecules that still harbored nucleosomes in the MLH1 promoter. In contrast, a second subset of promoter had high levels of M.SssI accessibility between the transcription start sites, flanked by inaccessible areas, indicative of a nucleosome-depleted region reminiscent of the patterns seen in the expressing cells (Figure 6C, blue box, compared to Figure 4C). Thus, the data strongly suggest that, at least in a subset of promoters, the nucleosomes are evicted when the genes become reactivated after drug treatment. As for the demethylated promoters that seemed to be occupied by nucleosomes, these could possibly reflect hemimethylated promoter molecules that are still present 72 hr after drug addition according to the drug mechanism (Bender et al., 1999; Egger et al., 2004); thus, nucleosomes might be trapped on the hemimethylated DNA (Figure 6C).
To eliminate this variable, identical analyses were done on RKO cells treated with 5-aza-CdR, followed by 44 days of culture without the drug, at which time no or very little hemimethylated DNA would be expected to be present. Bisulfite genomic sequencing revealed that ~7% (2/29) of the sequenced molecules were still completely unmethylated, whereas the rest were extensively methylated (Figure 6D), probably due to remethylation after removal of the drug (Bender et al., 1999).
Again, only substantially unmethylated molecules were detected by selective amplification from 44 days after 5-aza-CdR treatment (see Figure S5D). Among them, ~92% (12/13) had extensive accessibility to M.SssI between the transcription start sites, indicative of a nucleosome-depleted region, recapitulating the patterns seen in expressing cells (Figure 6E, blue box). These results show that nucleosomes are evicted from a subset of promoters upon gene reactivation by drug-induced methylation. Not only that, data from 44 days indicate that nearly all of the demethylated promoter molecules lack nucleosomes, establishing the heritability of nucleosomal eviction on the demethylated, active promoter.
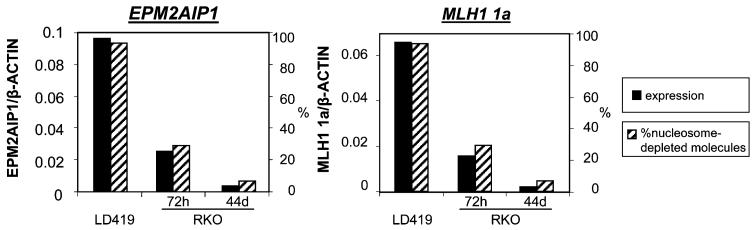
By combining the data from quantitative RT-PCR and the MSPA analyses (Figures 4-6), we found a remarkable correlation between the expression level and the percentage of nucleosome-depleted molecules for MLH1 1a and EPM2AIP1, where the decrease in expression is associated with a reduced nucleosome-depleted population (Figure 7). Thus, it seems very likely that reactivation of such an epigenetically silenced gene requires nucleosome depletion in addition to demethylation of DNA and the application of positive histone marks.
Figure 7. Correlation between the Expression Level and the Percentage of Nucleosome-Depleted Molecules.
Data were graphed based on the results from Figures 5, 6, and 7. A decrease in expression is associated with a reduced nucleosome-depleted population.
DISCUSSION
We set out to determine how the permissive chromatin configuration of CpG islands, which is inherited independently of transcriptional activity, is converted into a somatically heritable silent or nonpermissive state. Depletion of nucleosomes just upstream of transcription start sites has recently been observed in genome-wide screens of yeast, flies, and humans (Heintzman et al., 2007; Lee et al., 2004; Mito et al., 2005; Ozsolak et al., 2007). This strongly suggests that lack of nucleosomes is needed to allow access of the transcriptional machinery to the permissive promoter (Li et al., 2007). Indeed, nucleosomal remodeling factors have been shown to be causally linked to transcription activation; for example, nucleosomes in the PHO5 promoter are completely disassembled upon transcriptional activation in yeast (Boeger et al., 2004). It therefore seems likely that the absence of at least one nucleosome is necessary for gene transcription. However, we should again emphasize that many autosomal CpG islands in human cells are in an apparently permissive configuration, implying that nucleosomes are constitutively depleted at start sites (Sabo et al., 2004; Sasaki et al., 1992a).
Our results, obtained with a complex human promoter, build on these studies in several important ways. First, the high-resolution MSPA method, validated by traditional approaches, shows that there is a constitutive complete absence (i.e., total depletion) of a single nucleosome upstream of each start site. This is similar to what we saw in the GRP78 promoter, in which no trace of a nucleosome was seen in 356 promoter replicas we examined (Gal-Yam et al., 2006). While it is possible that this might be due to a rapid and reversible nucleosomal occupancy, we think this is unlikely because fixation with formaldehyde before ChIP analysis confirmed very low occupancy in the regions examined. With regard to the generality of our findings, our results at the BRCA1/NBR2 promoter were consistent with those at the MLH1 promoter, suggesting that nucleosome depletion in the CpG islands is quite common.
Second, we find it remarkable that the entire 630 bp promoter, which generates three transcripts, acts as a unit in the sense that either all transcripts were present or none were present; thus, the start sites seem to be coordinately controlled. Equally remarkable is the presence of a highly positioned, acetylated nucleosome in almost all of the promoter replicas in expressing cells. CpG islands often do not have TATA boxes and thus initiate transcription in quite a heterogeneous fashion. Our finding might help explain the existence of multiple transcription start sites in many mammalian CpG island promoters. Perhaps nucleosome occupancy near the start sites is responsible for defining the transcripts generated in a given CpG island. Support for this idea comes from our observations of a high level of positional variation and heterogeneous start sites in the p16(INK4A) gene (Fatemi et al., 2005) in contrast to the three “sharply defined” sites observed by 5′-RACE in MLH1. Further work will clearly be necessary to resolve these issues.
The third implication of our work relates to the role of occupancy in epigenetic silencing, which is, by definition, mitotically heritable. Most work to date has focused on constitutively active genes or on the chromatin changes associated with gene induction. As an extension of a previous study on p16(INK4A) shows that DNA methylation may serve to lock in the repressed state after H3-K9 methylation (Bachman et al., 2003), our data suggest that heritable DNA methylation patterns may maintain silencing by contributing to heritable changes in nucleosome occupancy via positioning nucleosomes at the start sites. The almost “digital” quality of this process was seen in our experiments with 5-aza-CdR. Here, the erasure of DNA methylation by drug treatment led to the reactivation of all three start sites, application of the covalent activating acetylated marks, and removal of the nucleosomes from the region examined. Interestingly, the MLH1 promoter had already adopted the “dip” conformation for histone H3 acetylation 48 hr after drug treatment started. However, we have not as yet determined whether histone modifications precede the nucleosomal rearrangement, or whether the active marks are applied to the rearranged nucleosomes. Nonetheless, even though the gene was resilenced as a function of time after treatment, the 7% of promoters that remained unmethylated in the mass population were also nucleosome free. Indeed, the level of expression of all three transcripts in reactivated RKO cells was highly correlated with the level of nucleosome deficiency.
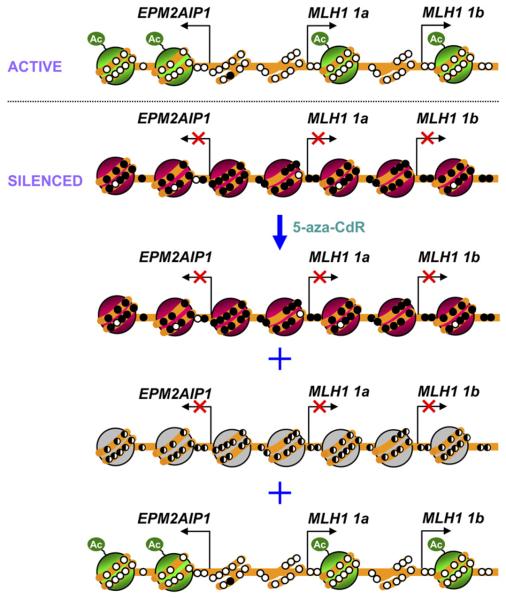
Thus, we suggest a model of the epigenetic silencing of the MLH1 gene in which the unmethylated and active promoters of EPM2AIP1 and MLH1 (1a and 1b) are depleted of one nucleosome just upstream of each of the transcription start sites (Figure 8). In the silenced state, the methylated and inactive promoters are occupied by nucleosomes, and treatment with 5-aza-CdR causes considerable DNA demethylation of the promoter region, although some molecules remain methylated. Remarkably, while some hemimethylated promoter molecules are occupied by nucleosomes, the nucleosome-free zone is established in a substantial portion of the promoter molecules. Because these promoter molecules show strikingly similar patterns to those seen in expressing cells (Figure 4C), they are probably associated with the reexpression of genes (Figure 8). Conversely, the genes are most likely silenced in the hemimethylated promoter molecules that have nucleosomes in the promoters because hemimethylation has been shown to block transcription by inhibiting transcription factor binding (Sasaki et al., 1992b).
Figure 8. A Simplified Model for the Epigenetic Silencing of the MLH1 Gene.
The active promoters of 1a and 1b are depleted of at least one nucleosome just upstream of each of the transcription start sites. In the silenced state, the inactive promoters of 1a and 1b are occupied by nucleosomes. Treatment with 5-aza-CdR causes substantial DNA demethylation of the promoter region, although some molecules remain methylated. While some hemimethylated promoter molecules may still be occupied by nucleosomes, the nucleo-some-free zone is established in some of the promoter molecules. The activation of genes is probably derived from these molecules with a nucleosome-free zone in the promoter, although we could not determine this definitively. Green represents nucleosomes bearing active marks. Red represents nucleosomes bearing repressive marks. Gray represents nucleosomes occupying the region of hemimethylation. Open circles indicate unmethylated CpG dinucleotides, and filled circles represent methylated CpG dinucleotides. Hemimethylated DNA is shown as half-filled circles. Ac refers to acetylated histone tails.
Our data are consistent with earlier studies showing that unmethylated CpG islands are nuclease accessible, whereas methylated counterparts are occupied by randomly positioned nucleosomes (Patel et al., 1997; Pfeifer and Riggs, 1991). However, these studies relied on relatively low-resolution nuclease digestions or in vivo footprinting, whereas we were able to show this digitally with our high-resolution assay. The transition from the inactive to the active state, after gene reactivation by 5-aza-CdR, seems to involve nucleosome eviction. Our model is supported by a study by Kass et al. (1997) that showed mechanistically that a methylated CpG island promoter, but not its unmethylated counterpart, forms nucleosomal arrays after injection into Xenopus oocytes. This study, done in an artificial system, complements our findings and potentially provides a mechanistic basis for what we see in cancer cells.
Methyl-binding proteins such as MBD1, MBD2, or MeCP2 are likely to be involved in silencing of methylated promoters (Klose and Bird, 2006), and we have documented the presence of MeCP2 at methylated CpG islands associated with tumor suppressor genes (Nguyen et al., 2001, 2002). Thus, it is conceivable that MeCP2 and other methylated binding proteins stabilize the presence of nucleosomes in methylated promoters. Direct structural changes in the properties of the DNA helix elicited by methylation might also directly alter occupancy (Pennings et al., 2005). Which mechanism predominates is unclear; nevertheless, our data suggest that the core mechanism responsible for permanent silencing may be the insertion of a nucleosome into a previously unoccupied site. Although the exact chromatin remodeling complex regulating the eviction of nucleosomes that we observed has not been identified, nucleosomal occupancy might be the essential outcome of a chromatin remodeling process involving covalent modification of DNA, histones, and other chromosomal proteins.
EXPERIMENTAL PROCEDURES
Cell Culture
Colorectal cancer cell lines (LoVo, HCT116, LS174T, LS123, HT29, SW620, SW48, RKO), bladder cancer cell lines (T24, J82), and a cervical cancer cell line (HeLa) were obtained from the American Type Culture Collection (ATCC). All cells were cultured as recommended by the ATCC. A normal LD419 human bladder fibroblast cell line was generated in our laboratory and cultured in McCoy's 5A supplemented with 20% FBS.
5-Aza-2′-Deoxycytidine Treatment
Cells were plated (4 × 105 cells/100 mm dish or 2 × 106 cells/150 mm dish), and 24 hr later, they were treated with 10−5 M 5-aza-2′-deoxycytidine (Sigma) for 24 hr. The culture was then replenished with fresh medium without the drug for 2 more days, and then nuclei, DNA, and RNA were isolated from the drug-treated culture.
Methylation-Sensitive Single-Nucleotide Primer Extension
Genomic DNA was treated with sodium bisulfite as previously described (Frommer et al., 1992). The mean cytosine methylation levels of CpG sites were determined by the methylation-sensitive single-nucleotide primer extension assay as described previously (Nguyen et al., 2001). All primer sequences are available upon request.
RT-PCR
Total RNA was isolated from cells with the Trizol reagent (Invitrogen). Reverse transcription was performed with random primers. The 5′-3′ sequences of the primers used in the PCR are: β-ACTIN forward, TTTGAGACCTTCAACACCCCAG; β-ACTIN reverse, TTTCGTGGATGCCACAGGA; MLH1 forward, CAGCTAATGCTATCAAAGAGATGAT TG (1a) and GAGACCTTTTAAGGGTTGTTTGG (1b); MLH1 reverse, GTTGTAAGAGTAACATGAGCCACATG; EPM2AIP1 forward, TTTGTGGCCTATGAGAACTACC; EPM2AIP1 reverse, GCTCTGATTCAGATTCCGTTAG. The PCR conditions were 95° C for 9 min, 30 cycles (25 cycles for β-ACTIN) of 95° C for 30 s, 62° C for 1 min, 72° C for 1 min, and 72° C for 10 min. PCR products were resolved on 2% agarose gels. Quantitative RT-PCR was performed with an Opticon light cycler with SYBR green I (Sigma), by using the primers listed above. All values were normalized to β-ACTIN expression ratios, and a set of known amounts of standards was used for quantitation.
Bisulfite Genomic Sequencing
DNA extracted from cells were treated with bisulfite, were PCR amplified with primers specific to the bisulfite-converted DNA, and then ligated into the pGEM-T easy vector (Promega). For MLH1 1a, three PCR amplifications of DNA sizes of 250, 536, and 708 bp were produced. For MLH1 1b, a PCR amplification of DNA size of 416 bp was produced. Primer sequences and PCR conditions are available upon request. In addition, the selective amplification in the MSPA assay on RKO cells was performed with the reverse primer only annealing to the unmethylated molecules to eliminate the amplification of extensively methylated clones that could not be analyzed (see the main text for rationale). To increase the specificity of the primer to unmethylated DNA, a mismatch was incorporated at the 3′ end of the reverse primer, which will destabilize unspecific duplex formation. For selective amplification, the 5′-3′ sequences of the primers were as follows: forward, TGGGTTGGAAAATTAGAGTTTTGTT; reverse, ACCAAATAACCCCT ACCACAAATA. Individual plasmid molecules were sequenced by an automated DNA sequencer at Laragen (LA) and at the microchemical core laboratory at the University of Southern California (USC).
Rapid Amplification of cDNA Ends, 5′-RACE
Total RNA was extracted from LoVo cells as described above, and the 5′ ends of mRNA were determined by using the RLM-RACE Kit (Ambion) according to the manufacturer's instructions. 5′-RACE reaction products were cloned into a pGEM-T easy vector (Promega) and sequenced.
M.SssI Treatment
Nuclei preparation and M.SssI reactions were performed as described previously (Fatemi et al., 2005). Briefly, purified genomic DNA and freshly extracted nuclei were treated with M.SssI for 15 min at 37° C. Reactions were stopped by the addition of an equal volume of stop solution (20 nM Tris-HCl [pH 7.9], 600 mM NaCl, 1% SDS, 10 mM EDTA, 400 μg/ml proteinase K), incubated at 55° C overnight, and DNA was purified by phenol/chloroform extraction and ethanol precipitation.
DNaseI Footprinting
Nuclei from LD419 cells were extracted as described above. Nuclei were resuspended in RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2) plus 0.25 M sucrose and were then incubated with various concentrations of DNaseI (Worthington) at 37° C for 15 min to obtain a suitable range of digestion of genomic DNA revealed by EtBr staining. Digested DNA, purified as described above, was cut again by the DraI restriction enzyme to be resolved on a 1.5% agarose gel, which was then Southern blotted. The blot was hybridized with a 163 bp PCR-amplified DraI probe spanning −29 to 134 bp relative to MLH1 1b. The probe was labeled with [α-32P]dCTP by using High Prime (Roche #0) and was hybridized by ExpressHyb Hybridization Solution (BD Biosciences). The 5′-3′ PCR primer sequences used for the probe amplification were as follows: forward, GTTCCCTGACGTGCCAGTCA; reverse, AAATTAAGTGGCTTCCTTACTTAGTTAACG. The blot was visualized by Molecular Dynamics PhosphorImager.
Mononucleosomal DNA Preparation and Analysis
Detailed protocols were published previously (Gal-Yam et al., 2006). Quantitative PCR was performed by using AmpliTaq Gold DNA polymerase (Applied Biosystems) and SYBR green I with the DNA Engine Opticon System (MJ Research, Cambridge, MA). The primer sequences are available upon request. The following PCR conditions were used: 95° C for 9 min, and 45 cycles of 95° C for 30 s, 62° C for 1 min, and 72° C for 1 min, followed by 72° C for 10 min. The values were normalized with naked DNA.
ChIP Assays
ChIP analyses were performed as described previously (Nguyen et al., 2001). The following antibodies were used: 10 μl of either anti-Histone H3 (Abcam) or anti-acetylated Histone H3 (Upstate) and 1 μl rabbit IgG (Upstate) as a nonspecific antibody control.
Real-Time PCR Amplification of Immunoprecipitated DNA
Quantitative PCR was performed by using AmpliTaq Gold DNA polymerase (Applied Biosystems) and TaqMan probes (Biosearch) with the DNA Engine Opticon System (MJ Research, Cambridge, MA). The primers and probe sequences are available upon request. The following PCR conditions were used: 95° C for 10 min, and 45 cycles of 95° C for 15 s and 59° C for 1 min. For each PCR, a set of known amounts of DNA was included as a quantitation standard, and immunoprecipitated samples with nonspecific antibody were also included. The fraction of immunoprecipitated DNA was calculated as a percentage of input DNA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Susan J. Clark for her suggestions. This work was supported by National Institutes of Health grant R01 CA82422. The figures for bisulfite sequencing were generated by using the CpG Bubble Chart Generator software created by Mark A. Miranda (markmir@ucla.edu).
Footnotes
Supplemental Data: The Supplemental Data include the promoter deletion analysis of the EPM2AIP1/MLH1 1a promoter and additional data on the BRCA1/NBR2 promoter, as well as five supplemental figures, and can be found with this article online at http://www.cancercell.org/cgi/content/full/12/5/432/DC1/.
REFERENCES
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- Bender CM, Gonzalgo ML, Gonzales FA, Nguyen CT, Robertson KD, Jones PA. Roles of cell division and gene transcription in the methylation of CpG islands. Mol. Cell. Biol. 1999;19:6690–6698. doi: 10.1128/mcb.19.10.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Davey CS, Pennings S, Reilly C, Meehan RR, Allan J. A determining influence for CpG dinucleotides on nucleosome positioning in vitro. Nucleic Acids Res. 2004;32:4322–4331. doi: 10.1093/nar/gkh749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T, Kyo S, Maida Y, Yatabe N, Tanaka M, Nakamura M, Inoue M. Frequent hypermethylation of MLH1 promoter in normal endometrium of patients with endometrial cancers. Oncogene. 2003;22:2352–2360. doi: 10.1038/sj.onc.1206365. [DOI] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- Kladde MP, Simpson RT. Chromatin structure mapping in vivo using methyltransferases. Methods Enzymol. 1996;274:214–233. doi: 10.1016/s0076-6879(96)74019-3. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lavia P, Macleod D, Bird A. Coincident start sites for divergent transcripts at a randomly selected CpG-rich island of mouse. EMBO J. 1987;6:2773–2779. doi: 10.1002/j.1460-2075.1987.tb02572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Clarke ND. Control of transcription through intragenic patterns of nucleosome composition. Cell. 2005;123:1187–1190. doi: 10.1016/j.cell.2005.12.010. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Mucha M, Lisowska K, Goc A, Filipski J. Nucleasehypersensitive chromatin formed by a CpG island in human DNA cloned as an artificial chromosome in yeast. J. Biol. Chem. 2000;275:1275–1278. doi: 10.1074/jbc.275.2.1275. [DOI] [PubMed] [Google Scholar]
- Murata H, Khattar NH, Kang Y, Gu L, Li GM. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002;21:5696–5703. doi: 10.1038/sj.onc.1205683. [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Gonzales FA, Jones PA. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 2001;29:4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Patel SA, Graunke DM, Pieper RO. Aberrant silencing of the CpG island-containing human O6-methylguanine DNA methyltransferase gene is associated with the loss of nucleosome-like positioning. Mol. Cell. Biol. 1997;17:5813–5822. doi: 10.1128/mcb.17.10.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings S, Allan J, Davey CS. DNA methylation, nucleosome formation and positioning. Brief. Funct. Genomic. Proteomic. 2005;3:351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Riggs AD. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. USA. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Jones PA, Chaillet JR, Ferguson-Smith AC, Barton SC, Reik W, Surani MA. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 1992a;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Hansen RS, Gartler SM. Hemimethylation and hypersensitivity are early events in transcriptional reactivation of human inactive X-linked genes in a hamster x human somatic cell hybrid. Mol. Cell. Biol. 1992b;12:3819–3826. doi: 10.1128/mcb.12.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J, Jelinek J, Chang H, Shen L, Qin T, Chung W, Oki Y, Issa JP. Silencing of bidirectional promoters by DNA methylation in tumorigenesis. Cancer Res. 2006;66:5077–5084. doi: 10.1158/0008-5472.CAN-05-2629. [DOI] [PubMed] [Google Scholar]
- Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell. Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Takai D, Jones PA. Origins of bidirectional promoters: computational analyses of intergenic distance in the human genome. Mol. Biol. Evol. 2004;21:463–467. doi: 10.1093/molbev/msh040. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Wu AH, Bender CM, Tsao JL, Blake C, Shibata D, Jones PA, Yu MC, Ross RK, Laird PW. Mismatch repair deficiency and CpG island hypermethylation in sporadic colon adenocarcinomas. Cancer Epidemiol. Biomarkers Prev. 2001;10:799–803. [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.