Abstract
N-methyl-d-aspartate receptors (NMDARs) belong to the large family of ionotropic glutamate receptors (iGluRs), which are critically involved in basic brain functions as well as multiple neurological diseases and disorders. The NMDARs are large heterotetrameric membrane protein complexes. The extensive extracellular domains recognize neurotransmitter ligands and allosteric compounds and translate the binding information to regulate activity of the transmembrane ion channel. Here, we review recent advances in the structural biology of NMDARs with a focus on pharmacology and function. Structural analysis of the isolated extracellular domains in combination with the intact heterotetrameric NMDAR structure provides important insights into how this sophisticated ligand-gated ion channel may function.
Introduction
NMDARs are pivotally involved in neurotransmission as well as neurological diseases and disorders. They belong to the iGluR family comprised of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR), kainate receptor, delta receptor and NMDAR. These different subclasses of iGluRs co-localize mainly at the postsynapses and regulate the excitatory postsynaptic potential (EPSP) that is a major component of neuronal plasticity. In general, non-NMDARs control amplitude and the early phase of the EPSP time course, while NMDARs contribute mostly in the late/slow phase. The NMDAR subclass is comprised of subunits from three groups of genes. The GluN1 subunit is expressed as eight different splice variants (GluN1-1a through GluN1-4a and GluN1-1b through GluN1-4b) where “a” and “b” notations refer to the exclusion (“a”) or inclusion (“b”) of a 21-residue polypeptide encoded by exon 5, while the numbering scheme denotes alternate splicing of exons in the long (~600 residues), largely unstructured Carboxyl Terminal Domain (CTD) [1]. Like the GluN1 subunits, the GluN3 (A–B) subunits bind the co-agonists glycine or d-serine, and GluN2, which is encoded by four different genes (A–D), binds glutamate or NMDA. Unlike non-NMDARs, the NMDARs cannot form functional homotetramers. The obligatory heterotetramers can consist of a wide variety of subunit combinations and confer functional diversity. Typically this includes two GluN1 subunits and either two GluN2 subunits of the same or different subtypes, or two GluN1 subunits and a GluN2 and a GluN3 subunit [1–3]. While the majority of functional studies have been done on NMDARs containing two copies of the same GluN2 subunits, evidence suggests that a large subset of receptors in vivo actually contain different combinations of GluN2 subunits [4]. Furthermore, NMDAR channels with different subunit compositions show spatio-temporal variation, with GluN2B and GluN2D expression highest in early development, shifting to increased, but not exclusive, GluN2A and GluN2C expression later in life, with expression levels varying across different regions of the brain as discussed in detail by Paoletti, et al. [2].
NMDARs show many more modes of regulation and functional diversity compared to non-NMDARs. In general, activation of NMDARs containing GluN1/GluN2 requires binding of both glycine and glutamate at the extracellular ligand-binding domains (LBDs) in addition to release of magnesium block by membrane depolarization at the transmembrane domain (TMD). Opening of the NMDAR ion channel results in significantly higher calcium permeation than that observed in calcium permeating non-NMDARs [1,2,5]. NMDARs also show slower, less-pronounced desensitization compared to non-NMDARs, but crucially, the specific subunit composition of the particular NMDAR in question imparts unique characteristics on channel activity [1,2,4]. For example, NMDARs containing GluN2D subunits show extremely slow deactivation and low open probability. Also, GluN2C/D receptors have similar low open probabilities, followed by GluN2B, and finally GluN2A with the highest open probability [2,5]. Over the years, concerted efforts have gradually uncovered the molecular elements responsible for the diverse functions of NMDARs described above. Here, we will describe recent advances based on structural studies on the isolated extracellular domains – the Amino Terminal Domain (ATD) and LBD – and the intact heterotetrameric NMDAR ion channel.
Structure and dynamics of the LBD
The first crystallographic studies on NMDARs focused on the isolated LBD due to its relative ease in recombinant expression and purification compared to ATD or the full length construct, and the field now has high-resolution crystal structures for LBDs from GluN1 (in the apo state, complexed with agonists, partial agonists, and antagonists), GluN2A (complexed with agonists and antagonists), GluN2D (complexed with agonists and partial agonists), GluN3A (apo, complexed with agonists, and partial agonists), and GluN3B (complexed with agonists) [6–8]. Furthermore, crystal structures of heterodimeric GluN1/GluN2A LBDs hinted at the pattern of subunit arrangement for the first time and reinforced the view that tetrameric NMDARs are a dimer of heterodimers [9]. An individual LBD adopts a bilobed architecture where agonists, partial agonists, and antagonists bind to the cleft between the two lobes (Figure 1). Crystal structures of GluN1 LBDs and GluN1/GluN2A LBDs complexed to agonists and antagonists revealed that the structures are stabilized in an open cleft or closed cleft conformation when bound to antagonists or agonists, respectively, for both GluN1 and GluN2 subunits [7,9–12]. This pattern of opening and closure of the bilobed structure is conserved among the other iGluR family members and is considered to be tied to ion channel gating since the LBD is directly linked to the TMD [13,14].
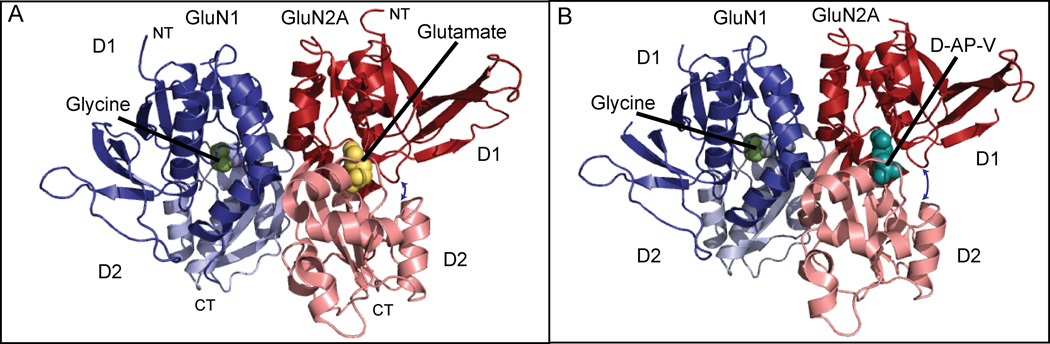
Figure 1.
Structure of the GluN1/ GluN2A LBD heterodimer. GluN1 is shown in blues and GluN2A is shown in reds. A) GluN1 in complex with glycine and GluN2A with glutamate (PDB: 4NF8). The clamshell is in a closed conformation (see arrow between GluN2A upper and lower lobes). B) GluN1 in complex with glycine and GluN2A with AP-V (PDB: 4NF5). Here, the clamshell is in an open state (see arrow between GluN2A upper and lower lobes). The d-AP-V site is shared among all competitive antagonists. Hypothetical binding sites for allosteric antagonists have not been structurally confirmed.
Conformational variations of the NMDAR and non-NMDAR LBDs have been further elaborated upon by single molecule Förster resonance energy transfer (FRET) and molecular dynamics [7,15,16]. While a number of studies indicated that the closure by agonists of the bilobed AMPAR LBD follows an induced fit mechanism, analysis of free energy landscapes suggests that NMDAR LBDs can fluctuate between open and closed conformations in the apo state, suggesting a conformational selection mechanism [7]. Similar studies showed greater probabilities of intermediate conformations upon the binding of partial agonists, suggesting that binding of partial agonists does not involve a fully-closed state which may explain the variety of responses in channel activation observed upon partial agonist binding rather than the full activation induced by an agonist [15].
One advantage of performing crystallographic studies on the isolated LBD is the high-resolution nature (often better than 2 Å) of the obtained structures, which facilitates clear identification of ligand-recognition elements necessary to create subtype-specific compounds targeting this domain; a handful of such compounds have been identified, but their utility and specificity has actually proven inconsistent in practice. For example, the compound PPDA was identified as a competitive antagonist with moderate selectivity for GluN2C/D over GluN2A/B [17], but it also inhibits non-NMDARs. Another antagonist, NVP-AAM077 (Box 1) was originally met with great enthusiasm because of its GluN2A-selectivity [18]; however, subsequent determination of Ki values showed this selectivity to be less than 10-fold [19].
Box 1.
Many of the small molecules which interact with the NMDAR are frequently presented as abbreviations in the interest of space. The full chemical names of those compounds which are relevant to this article are listed below.
DCKA: 5,7-dichlorokynurenic acid
D-AP-V: (2R)-amino-5-phosphonovaleric acid
PPDA: 1-(phenanthrene-2-carbonyl)-piperazine-2,3-dicarboxylic acid
NVP-AAM077: (R)-[(S)-1-(4-bromo-phenyl)-ethylamino]-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-5-yl)-methyl]-phosphonic acid
TCN-201: 3-Chloro-4-fluoro-N-[4-[[2-phenylcarbonyl)hydrazino]carbonyl]benzyl]benzenesulfonamide
Ifenprodil: 4-[2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol
Eliprodil: 1-(4-chlorophenyl)-2-[4-[(4-fluorophenyl)methyl]piperidin-1-yl]ethanol
PYD-106: Methyl 4-(3-acetyl-4-hydroxy-1-(2-(2-methyl-1H-indol-3-yl)ethyl)-5-oxo-2,5-dihydro-1H-pyrrol-2-yl)benzoate
QNZ46: 4-[6-Methoxy-2-[(1E)-2-(3-nitrophenyl)ethenyl]-4-oxo-3(4H)quinazolinyl]benzoic acid
CIQ: (3-Chlorophenyl) [3,4-dihydro-6,7-dimethoxy-1-[(4-methoxyphenoxy)methyl]-2(1H)-isoquinolinyl]methanone
MK-801: [5R,10S]-[+]-5-methyl-10,11- dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine
Memantine: 3,5-dimethyladamantan-1-amine
Structures of the LBD bound to d-AP-V and PPDA (Figure 1B) provided clear evidence that a compound must interact with a conserved tyrosine in the binding pocket (Tyr735 in GluN2A) to be specific to NMDARs and showed a GluN2A-specific element (Lys738) that can be targeted for potential GluN2A-specific compound design [10]. Fluorometric high-throughput screening led to the discovery of an allosteric modulator that was later shown to target the LBD of GluN2A-containing NMDARs at the subunit interface. The compound, TCN-201 (Box 1), shows 275-fold GluN2A-selectivity over GluN2B and GluN2D subtypes [20] through a putative binding site at the LBD dimer interface [21]. One potential complication, though, is that TCN-201 potency is diminished in the presence of GluN1-agonists such as glycine [21]. Thus, TCN-201 can be useful in well-controlled in vitro experimental conditions, specifically at low glycine concentration; however, this compound may not be straightforward to use in more complicated in vivo studies. Undoubtedly, it will be important to understand the precise mode of binding for the above allosteric compounds in order to improve their specificity, efficacy, and potency.
ATD and allosteric regulation
Much work has been done in recent years on the ATD of the NMDAR resulting in an improved understanding of the structure and function of this domain. Unlike AMPARs and kainate receptors, a large component of NMDAR function is mediated through the ATD [22,23]. A high degree of divergence can be found among the various iGluR ATDs as well as among the different subunits of NMDARs. One consequence of this dissimilarity is that a wide range of subtype-specific functional effects can be conferred by the ATD and several electrophysiology and structural biology studies have identified small molecule binding sites in this domain which could exploit these differences [3].
The ATD allosterically regulates channel open probability and deactivation kinetics through its binding of modulators such as zinc and the phenylethanolamines mentioned above [24]. As might be expected due to the relatively low sequence identity among NMDAR ATDs, the different subtypes exert different effects on channel activity as a result of modulator binding; indeed, these results can be experimentally transferred between different subtypes of NMDARs by interchanging ATDs [25].
A structural study of the isolated GluN2B ATD [26] illustrated a bilobed clamshell-like architecture distinct from the LBD (Figure 2A); later, the structure of the GluN1b/GluN2B ATD dimer [27] revealed an offset in the orientation of the two domains in the dimer. While the upper (R1) lobes of the clamshells make extensive contacts with one another, the lower (R2) lobes are nearly completely separated (Figure 2B). This is in stark contrast to the ATDs of the AMPAR [28] and kainate receptor [29], in which both the R1 and R2 lobes interact (Figure 2C). Interestingly, molecular dynamics predict that the bilobed structure of both the AMPAR and NMDAR ATDs would be capable of largely similar motions, except for a limited (and most likely crucial) set of restrictions imposed by dimerization [30]. The nature of this motion is thought to be composed of two major components: an opening and closing of the clamshell, in addition to a rotation around the hinge region or twisting between the two lobes [30,31], and several lines of experimental evidence support this model. A modified cysteine in the hinge region of the clamshell allowed Gielen, et al. to demonstrate that perturbing closure of the clamshell potentiates NMDAR activity [25]. FRET data recently reported by the Jayaraman group suggests that the binding of zinc promotes closure of the GluN2A ATD [32], and while the GluN2B ATD is on average more closed than the GluN2A ATD, the presence of spermine leads to the opening of the GluN2B ATD [33]. The precise structural details of the changes in the ATD clamshell and how it correlates with ion channel function is as yet unknown due to a lack of high resolution structures and remains a crucial question.
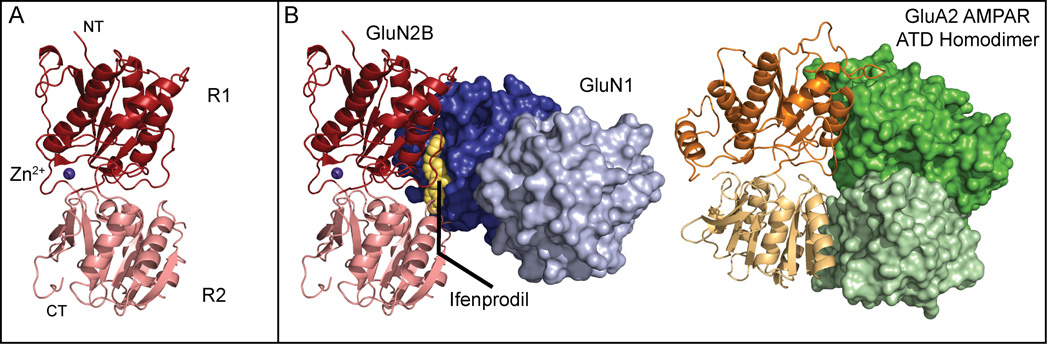
Figure 2.
Composite structure and ligand binding of the NMDAR ATD. (A) The isolated GluN2B ATD (PDB: 3JPW) adopts a clamshell-like structure consisting of an upper (R1, dark red) and lower (R2, pink) lobe. Zinc (purple) binds near the hinge between the two lobes and allosterically inhibits GluN2A, and to a lesser extent GluN2B, receptors. (B) Contrary to early work, the allosteric antagonist ifenprodil (yellow) binds between the GluN1b/GluN2B ATD subunits (blues, R1 and R2 lobes, and reds, R1 and R2 lobes, respectively) stabilizing dimer formation (PDB: 3QEL). While the R1 lobes of the GluN1b and GluN2B ATDs are closely apposed, there is practically no interaction between the R2 lobes in the dimer. Conversely, the AMPAR ATD homodimer (PDB: 3H5V) exhibits extensive interactions between both R1 and R2 lobes.
Due to the different protomer arrangements in the ATD dimers of NMDARs and AMPARs and the effect that dimerization has on these domains’ activities, it is possible that small molecule binding to the ATDs is more favored in one set of iGluRs than others. Indeed, to date, no small molecule has been found that binds specifically to the ATDs of non-NMDAR ATDs, while a number of ligands have been identified that bind the NMDAR ATD. One class of allosteric inhibitor compounds in particular, the phenylethanolamines, includes the neuroprotectants ifenprodil [34] and eliprodil [35] (Box 1) in addition to a multitude of variants. This class of small molecules was shown to bind with high specificity to NMDAR ATDs containing the GluN2B subunit [5,36] yet the binding site differs in only one residue versus GluN2A [27]. Perplexingly, swapping this residue into GluN2A does not “induce” ifenprodil sensitivity, suggesting that differences in the association of the subunits themselves is a key factor in compound selectivity [27]. Recently, another phenylethanolamine variant known as 93-31 was identified that shows promise in the treatment of stroke through reduced allosteric inhibitory activity at physiological pH as compared to the lower pH observed in ischemic tissue [37]. Notably, the phenylethanolamines all appear to bind not between the lobes of a particular clamshell, but at the dimer interface between the GluN1 and GluN2B R1 lobes (Figure 2B). This is in stark contrast to the binding site of zinc, a physiological allosteric inhibitor, which binds within the clamshell cleft of GluN2A and GluN2B.
The endogenous polyamines spermine and spermidine have been known to act on the ATD and potentiate function of the GluN1/GluN2B NMDARs. The binding site of polyamines is predicted to lie between the R2 lobes of the GluN1/GluN2B heterodimer [33,38], but the precise mode of binding and activity is largely unknown. Both potentiation by polyamines and inhibition by phenylethanolamines or zinc involve alteration of sensitivity to protons (or pH) that inhibits NMDAR function [39–41]. That is, polyamines and phenylethanolamines potentiate and attenuate the NMDAR activity by respectively alleviating and enhancing sensitivity to protons. Determination of the structural elements responsible for proton sensitivity will likely provide an in-depth understanding of allosteric modulation in general.
Structure of intact heterotetrameric NMDARs
Clearly, a thorough understanding of the details of small molecule binding to the ATD and LBD and how this effects differential channel activity is critical to the development of pharmacologically-active agents targeting the NMDAR (Figure 3A). A significant step forward on this path was the recent elucidation of the crystal structure of the intact NMDAR. Two groups solved highly similar structures of heterotetrameric NMDARs from Rattus norvegicus [42] and Xenopus laevis [43]. Speaking to the difficulty of this task, neither model was fully complete to high resolution: various small portions of different domains were lacking in electron density. Taken in aggregate, however, the combined models provide a fairly comprehensive view of the NMDAR structure. Consistent with previous predictions [1,9,44], the NMDAR assembles with GluN1/GluN2 subunits assuming a dimer of heterodimers arrangement at the extracellular domains and a pseudo-fourfold symmetry in the TMD. One interesting feature of this arrangement is a domain-swap in subunits between the ATD and LBD, in which the first GluN1/GluN2 dimer in the ATD (Fig. 3B, dark blue and pink, respectively) associates instead with the complementary subunit from the second dimer at the LBD (dark blue and dark red) [42], as is also the case in homotetrameric AMPARs and kainate receptors [45–47]. A comparison of the structure and function of NMDARs and non-NMDARs can be found elsewhere in more detail [48].
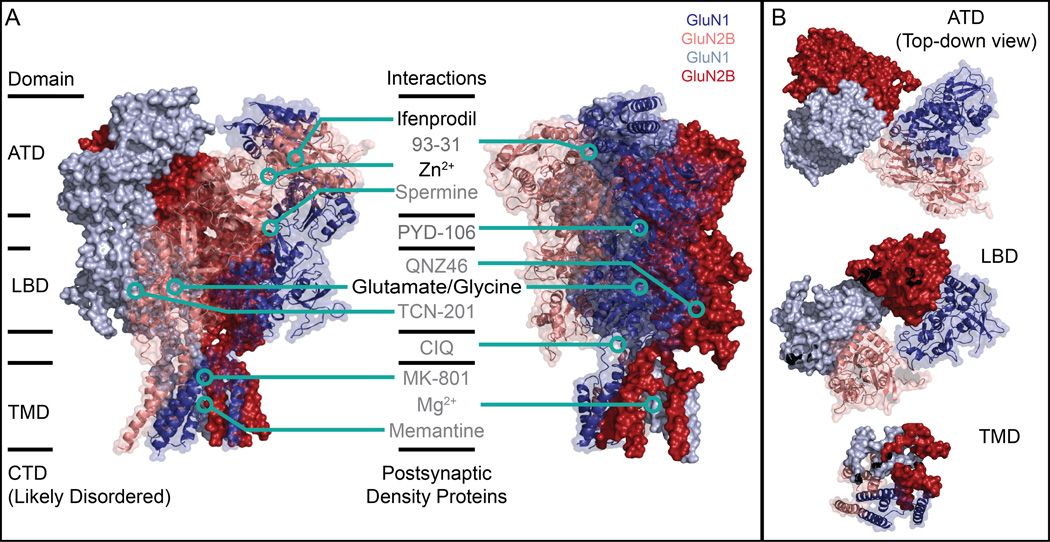
Figure 3.
Architecture of the intact NMDAR and selected interactions. (A) The heterotetrameric GluN1a/GluN2B NMDAR (PDB: 4PE5) consists of four domains: the extracellular ATD and LBD, the transmembrane TMD, and the disordered intracellular CTD. A number of small molecule and protein interaction partners have been identified; the structure of the intact receptor provides a framework to visualize where these interactions occur in relation to each other in addition to potential mechanical and pharmacological modulatory mechanisms. Critically, the binding sites of many of these small molecules are as yet putative at best. Several binding sites have been visualized at high-resolution by x-ray crystallography, while for others point mutagenesis studies suggest that the different small molecules bind at distinct sites on the NMDAR. In this figure, small molecules listed in black have been verified by x-ray crystallography while those in grey have not. (B) The domain swap between ATD and LBD is a general feature of iGluRs. In this 1-2-1-2 arrangement, two pairs of GluN1a (blues) /GluN2B (reds) ATD dimers form, shown here as a dark blue/pink dimer and a light blue/dark red dimer. At the LBD, however, the dimers are instead composed of the dark blue/dark red pair and the pink/light blue pair. The TMD shows quasi-fourfold symmetry, with the ion channel pore in the center of this structure in what is believed to be the closed state.
Most importantly, the intact NMDAR structure provides a context in which to understand the structures and interactions of the individual domains. Notably, this structure revealed an intimate association between the ATDs and LBDs, which stands in contrast to structures of non-NMDARs [45–47,49]. In addition to the dynamics of the individual domains, it is highly likely that this ATD/LBD interaction is fundamental to the ability of ligand binding at the ATD to be propagated and thus affect channel activity, which is not the case in the separated ATD/LBD domains in AMPARs or kainate receptors. Similarly, conformational changes at the LBD related to ligand binding appear to be directly linked to opening or closing of the LBD clamshell, which applies tension to the M3-S2 peptide linker connecting this domain to the TMD [14]. Single amino acid additions or deletions to this linker have been shown to dramatically alter the NMDAR’s response to agonist binding [13], indicating a sensitive and efficient mechanism for communicating conformational alterations across the macromolecule.
Pharmacologically, these first NMDAR structures couple our nascent mechanistic understanding of receptor function with an as-yet incomplete picture of how small molecules modulate NMDAR activity. In particular, a considerable number of modulators (Box 1) are thought to bind at the interfaces of different domains or subunits which cannot be feasibly explored in studies using the isolated domains. In addition to compounds such as ifenprodil and TCN-201 that bind, respectively, to the ATD dimer interface and the LBD dimer interface, the positive allosteric modulator PYD-106 was reported to bind at the ATD/LBD interface of GluN2C-containing receptors [50]. The GluN2D-specific inhibitor QNZ46 is thought to bind at the bottom of the LBD, near the interface with the TMD [51], which, interestingly, is also near the putative binding site of the GluN2C/D allosteric potentiator CIQ [51]. The channel blocker MK-801, frequently used in both electrophysiological and structural studies [3,36], is believed to bind to the assembled helices of the TMD [43], although specific structural data verifying this is still somewhat sparse. Other channel blockers, including memantine, one of the few compounds approved for treating Alzheimer’s disease, and ketamine, show limited subtype-specificity in the presence of magnesium [52]. Additional work is required to rigorously define binding modes for these compounds, and the means to crystallize the fully-assembled NMDAR will provide a powerful template for characterizing these complex interactions. While recent functional experiments have provided clues as to the binding sites of all of these compounds, it is only through further structural experiments that these sites can be confirmed.
Conclusion and future directions
The structural analyses of recent years have revealed the molecular basis for some aspects of the complicated pharmacology and function of NMDARs. With the completion of the structure of intact NMDARs, the field has a basis from which to extrapolate conformational alterations at each individual domain in the context of the heterotetrameric ion channel. The structures of intact NMDARs also provide an opportunity to predict compound binding sites, which are frequently at the inter-subunit and inter-domain interfaces [53]. We anticipate that the combination of the recent structures of NMDARs at low resolution along with the high-resolution structures of the extracellular regions will serve as a blueprint for compound development.
Structural insights into ligand-gating of NMDAR ion channels are limited since the only available structure at this point is the GluN1/GluN2B NMDAR with a closed channel, bound to agonists and allosteric inhibitors. While some evidence of conformational alterations have been observed in the extracellular domains [10,16,31,33,49], how this ensemble of movements translates into gating regulation of the channel remains to be seen. Furthermore, the intact NMDAR structures were solved using constructs that lack the CTD. The CTD interacts with a number of postsynaptic density proteins, some of which are known to regulate ion channel activity via direct interaction or phosphorylation [54–57]. Since the CTD is predicted to be unstructured [58], alternative approaches such as single molecule FRET could be relevant to solving the issue of how interactions at this domain alter channel activity [57]. Other molecular factors that could regulate NMDAR activities are transmembrane auxiliary proteins; whether or not such auxiliary proteins exist for NMDARs continues to be an intriguing question, although NETO1 [59], EphB [60], and the zinc transporter ZNT1 [61] have been suggested to at least indirectly affect NMDAR activity. Ultimately, the combination of structural and dynamics research into NMDAR function, together with advances in neuropharmacology, will not only fill the current knowledge gaps in the field, but will also be the driving force for the design of compounds with better affinity, potency, and selectivity.
Highlights.
The NMDA receptor is a membrane protein critical to mammalian learning and memory.
Heterotetrameric NMDARs consist of three structured domains that regulate ion flux.
Small molecule binding at different sites can dramatically affect channel activity.
Recent structural work provides a framework for understanding NMDAR regulation.
Acknowledgements
This work was supported by the National Institutes of Health (MH085926 and GM105730 to HF), the Stanley Institute of Cognitive Genomics, and the Robertson Research Fund of Cold Spring Harbor Laboratory (to HF). ARH is funded by the Genentech Foundation Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Traynelis SF, Wollmuth LP, Mcbain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr. Opin. Pharmacol. 2015;20:14–23. doi: 10.1016/j.coph.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasgow NG, Siegler Retchless B, Johnson JW. Molecular bases of NMDA receptor subtype-dependent properties. J. Physiol. 2014;593:83–95. doi: 10.1113/jphysiol.2014.273763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF, et al. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol. Pharmacol. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure. 2013;21:1788–1799. doi: 10.1016/j.str.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. • In an early study on the isolated NMDAR LBD, the authors provide structural evidence that the NMDAR assembles as a dimer of heterodimers.
- 10. Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural insights into competitive antagonism in NMDA receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. • The NMDAR antagonists D-AP-V and PPDA were shown here to have distinct binding modes to the LBD, resulting in subtle conformational differences which could potentially be extrapolated to advance the development of subtype-specific NMDAR inhibitors.
- 11.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Kazi R, Dai J, Sweeney C, Zhou H-X, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 2014;17:914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous movements prior to pore opening in NMDA receptors. J. Neurosci. 2013;33:12052–12066. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Zhou H-X. Reduced curvature of ligand-binding domain free-energy surface underlies partial agonism at NMDA receptors. Structure. 2015;23:228–236. doi: 10.1016/j.str.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolino DM, Cooper D, Ramaswamy S, Jaurich H, Landes CF, Jayaraman V. Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. J. Biol. Chem. 2015;290:797–804. doi: 10.1074/jbc.M114.605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng B, Tse HW, Skifter Da, Morley R, Jane DE, Monaghan DT. Structure-activity analysis of a novel NR2C/NR2D-preferring NMDA receptor antagonist: 1-(phenanthrene-2-carbonyl) piperazine-2,3-dicarboxylic acid. Br. J. Pharmacol. 2004;141:508–516. doi: 10.1038/sj.bjp.0705644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M. 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg. Med. Chem. Lett. 2002;12:1099–1102. doi: 10.1016/s0960-894x(02)00074-4. [DOI] [PubMed] [Google Scholar]
- 19.Frizelle PA, Chen PE, Wyllie DJA. Equilibrium constants for (R) - [(S) -1- (4-bromophenyl) - methyl] -phosphonic acid (NVP-AAM077) acting at recombinant NMDA receptors: implications for studies of synaptic transmission. Mol. Pharmacol. 2006;70:1022–1032. doi: 10.1124/mol.106.024042. [DOI] [PubMed] [Google Scholar]
- 20.Bettini E, Sava A, Griffante C, Carignani C, Buson A, Capelli AM, Negri M, Andreetta F, Senar-sancho SA, Guiral L, et al. Identification and characterization of novel NMDA receptor antagonists selective for NR2A- over NR2B-containing receptors. J. Pharmacol. Exp. Ther. 2010;335:636–644. doi: 10.1124/jpet.110.172544. [DOI] [PubMed] [Google Scholar]
- 21.Hansen KB, Ogden KK, Traynelis SF. Subunit-Selective Allosteric Inhibition of Glycine Binding to NMDA Receptors. J. Neurochem. 2012;32:6197–6208. doi: 10.1523/JNEUROSCI.5757-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa H. Structure and function of glutamate receptor amino terminal domains. J. Physiol. 2012;590:63–72. doi: 10.1113/jphysiol.2011.213850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J. Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. • With Ref. 25, this work showed that a number of subtype-dependent characteristics, including channel open probability and deactivation kinetics, are affected by the NMDAR ATD. Specifically, by swapping ATDs of various GluN2 subtypes, the functional traits associated with that subtype of NMDAR can be transferred to a different subtype.
- 24.Sobolevsky AI. Structure and gating of tetrameric glutamate receptors. J. Physiol. 2013;593:29–38. doi: 10.1113/jphysiol.2013.264911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. • The authors show that channel open probability is not only influenced allosterically by the NMDAR ATD and the linker between the ATD and LBD, but that characteristic responsiveness of a particular NMDAR subtype can be conferred on a different subtype by swapping the ATDs, which complements data from Ref. 23.
- 26.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar J, Schuck P, Mayer ML. Structure and Assembly Mechanism for Heteromeric Kainate Receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta A, Shrivastava IH, Sukumaran M, Greger IH, Bahar I. Comparative dynamics of NMDA- and AMPA-glutamate receptor N-terminal domains. Structure. 2012;20:1838–1849. doi: 10.1016/j.str.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat. Struct. Mol. Biol. 2013;20:477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
- 32.Sirrieh RE, Maclean DM, Jayaraman V. Amino-terminal Domain Tetramer Organization and Structural Effects of Zinc Binding in the N-Methyl-D-aspartate (NMDA) Receptor. J. Biol. Chem. 2013;288:22555–22564. doi: 10.1074/jbc.M113.482356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sirrieh RE, MacLean DM, Jayaraman V. Subtype dependent NMDA receptor amino-terminal domain conformations and modulation by spermine. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.649723. [no volume]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avenet P, Lonardon J, Besnard F, Graham D, Frost J, Depoortere H, Langer SZ, Scatton B. Antagonist properties of the stereoisomers of ifenprodil at NR1A/NR2A and NR1A/NR2B subtypes of the NMDA receptor expressed in Xenopus oocytes. Eur. J. Pharmacol. 1996;296:209–213. doi: 10.1016/0014-2999(95)00700-8. [DOI] [PubMed] [Google Scholar]
- 35.Lengyel C, Dézsi L, Biliczki P, Horváth C, Virág L, Iost N, Németh M, Tálosi L, Papp JG, Varró A. Effect of a neuroprotective drug, eliprodil on cardiac repolarisation: importance of the decreased repolarisation reserve in the development of proarrhythmic risk. Br. J. Pharmacol. 2004;143:152–158. doi: 10.1038/sj.bjp.0705901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogden KK, Traynelis SF. New advances in NMDA receptor pharmacology. Trends Pharmacol. Sci. 2011;32:726–733. doi: 10.1016/j.tips.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan H, Myers SJ, Wells G, Nicholson KL, Swanger SA, Lyuboslavsky P, Tahirovic YA, Menaldino DS, Ganesh T, Wilson LJ, et al. Context-dependent GluN2B-selective inhibitors of NMDA receptor function are neuroprotective with minimal side effects. Neuron. 2015;85:1–14. doi: 10.1016/j.neuron.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traynelis SF, Hartley M, Heinemann SF. Control of Proton Sensitivity of the NMDA Receptor by RNA Splicing and Polyamines. Science (80-.) 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- 40.Mott DD, Doherty JJ, Zhang S, Washburn MS, Fendley MJ, Lyuboslavsky P, Traynelis SF, Dingledine R. Phenylethanolamines inhibit NMDA receptors by enhancing proton inhibition. Nat. Neurosci. 1998;1:659–667. doi: 10.1038/3661. [DOI] [PubMed] [Google Scholar]
- 41.Low C, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl- D -aspartate NR1 / NR2A receptors. Proc. Nat. Acad. Sci. U.S.A. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. •• The first image of the assembled heterotetrameric NMDAR, as observed through x-ray crystallographic studies of the rat GluN1/GluN2B intact receptor, followed within months by the structure of the frog NMDAR in Ref. 43.
- 43. Lee C-H, Lü W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. • See Ref. 38.
- 44.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J. Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dürr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, Mchaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. • Through a series of crystal structures of the AMPAR in different states, the authors illustrate a range of conformations accessible to iGluRs at different points during their activity cycle. Even here, however, the structure of a receptor in a true open state was elusive.
- 47.Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karakas E, Regan MC, Furukawa H. Emerging structural insights into the function of ionotropic glutamate receptors. Trends Biochem. Sci. 2015;40:328–337. doi: 10.1016/j.tibs.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Durr KL, Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science (80-.) 2014;354:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khatri A, Burger PB, Swanger Sa, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol. Pharmacol. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acker TM, Khatri A, Vance KM, Slabber C, Bacsa J, Snyder JP, Traynelis SF, Liotta DC. Structure-activity relationships and pharmacophore model of a noncompetitive pyrazoline containing class of GluN2C/GluN2D selective antagonists. J. Med. Chem. 2013;56:6434–6456. doi: 10.1021/jm400652r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotermanski SE, Johnson JW. Mg2+ Imparts NMDA Receptor Subtype Selectivity to the Alzheimer’s Drug Memantine. J. Neurosci. 2009;29:2774–2779. doi: 10.1523/JNEUROSCI.3703-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green T, Nayeem N. The multifaceted subunit interfaces of ionotropic glutamate receptors. J. Physiol. 2014;593:73–81. doi: 10.1113/jphysiol.2014.273409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea Ma. The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains. Structure. 2007;15:1603–1617. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S-JR, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicoll Ra, Roche KW. Long-term potentiation: peeling the onion. Neuropharmacol. 2013;74:18–22. doi: 10.1016/j.neuropharm.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi UB, Xiao S, Wollmuth LP, Bowen ME. Effect of Src kinase phosphorylation on disordered C-terminal domain of N-methyl-D-aspartic acid (NMDA) receptor subunit GluN2B protein. J. Biol. Chem. 2011;286:29904–29912. doi: 10.1074/jbc.M111.258897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi UB, Kazi R, Stenzoski N, Wollmuth LP, Uversky VN, Bowen ME. Modulating the intrinsic disorder in the cytoplasmic domain alters the biological activity of the N-methyl-D-aspartate-sensitive glutamate receptor. J. Biol. Chem. 2013;288:22506–22515. doi: 10.1074/jbc.M113.477810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cousins SL, Innocent N, Stephenson FA. Neto1 associates with the NMDA receptor/amyloid precursor protein complex. J. Neurochem. 2013;126:554–564. doi: 10.1111/jnc.12280. [DOI] [PubMed] [Google Scholar]
- 60.Dalva MB, Takasu Ma, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 61.Mellone M, Pelucchi S, Alberti L, Genazzani Aa, Di Luca M, Gardoni F. Zinc transporter-1: a novel NMDA receptor-binding protein at the postsynaptic density. J. Neurochem. 2015;132:159–168. doi: 10.1111/jnc.12968. [DOI] [PubMed] [Google Scholar]