Abstract
Unlike linear RNAs terminated with 5′ caps and 3′ tails, circular RNAs are characterized by covalently closed loop structures with neither 5′ to 3′ polarity nor polyadenylated tail. This intrinsic characteristic has led to the general under-estimation of the existence of circular RNAs in previous polyadenylated transcriptome analyses. With the advent of specific biochemical and computational approaches, a large number of circular RNAs from back-spliced exons (circRNAs) have been identified in various cell lines and across different species. Recent studies have uncovered that back-splicing requires canonical spliceosomal machinery and can be facilitated by both complementary sequences and specific protein factors. In this review, we highlight our current understanding of the regulation of circRNA biogenesis, including both the competition between splicing and back-splicing and the previously under-appreciated alternative circularization.
Keywords: alternative circularization, back-splicing, circularization, circular RNA, complementary sequence, splicing
Covalently closed circular RNAs were originally identified in plant viroids,1 yeast mitochondrial RNAs,2 and hepatitis δ virus.3 Later, circular RNAs were sporadically uncovered from either exons4-6 or introns,7,8 which were thought as by-products of spliceosome-mediated splicing errors (mis-splicing with scrambled exon orders)9,10 or intermediates escaped from intron lariat debranching,7,8 thus they are unlikely to play important roles in biological processes.
Being covalently closed, without polyadenylation at their 3′ ends,11,12 most circular RNAs have fallen below the radar of transcriptomic polyadenylated RNA profiling.13,14 Recently, with the depletion of both polyadenylated RNAs and rRNAs, non-polyadenylated human RNAs could be enriched for RNA sequencing (RNA-seq) analysis and unexpected RNA-seq signals were accumulated in certain exons (called excised exons) or introns (called excised introns).15 These non-polyadenylated signals were further confirmed as circular RNAs from back-spliced exons (circRNAs)16 or stabilized introns, having escaped debranching (circular intronic RNAs, ciRNAs).17 Nowadays, circRNAs have been extensively identified among a variety of transcriptomes.12,18/23 Although most of them are lowly expressed, some circRNAs have been proven to be more abundant than their linear counterparts.20 So far, endogenous circRNAs are not reported to associate with ribosomes for translation,18,21 indicating that they have a tendency to function as a new class of long noncoding RNAs (lncRNAs). However, exogenous circRNAs engineered with internal ribosome entry site (IRES) elements can be translated in vitro24 or in vivo,25 thus we can not exclude the possibility that some circRNAs are translatable.
The biogenesis of circRNAs via back-splicing is different from the canonical splicing of linear RNAs.12 Interestingly, it is also distinct from the formation of other types of circular RNAs, including but not limited to those generated by direct single strand RNA ligation,26 derived from intermediates of processed rRNAs27 or circularized introns escaped from debranching7,8,17 (reviewed by Lasda and Parker28). Here, we highlight recent advances in our understanding of circRNA biogenesis, with a particular focus on its regulations and the competition between back-splicing for circular RNAs and canonical splicing for linear ones.
Canonical Splicing and Back-Splicing
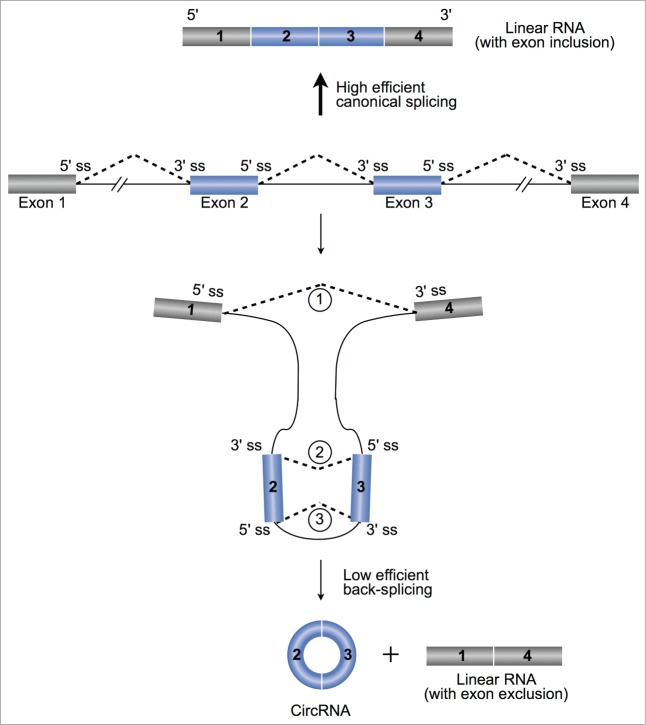
Canonical eukaryotic pre-mRNA splicing is catalyzed by the spliceosomal machinery to remove introns and join exons. Together with other co-/post-transcriptional processing, such as 5′ capping and 3′ polyadenylation, canonical splicing leads to the formation of a linear RNA transcript with 5′ to 3′ polarity (Top, Fig. 1). Different from canonical splicing that joins an upstream (5′) splice donor site with a downstream (3′) splice acceptor site, back-splicing ligates a downstream splice donor site reversely with an upstream splice acceptor site, resulting in a covalently closed circRNA transcript and an alternatively spliced linear RNA with skipped exons (Bottom, Fig. 1). However, evidence for co-expression of endogenous circRNAs and putative linear RNAs with exon exclusion is absent in most circRNAs,11,16,18 possibly due to their fast degradation. Nevertheless, it has been shown that both canonical splice signals and canonical spliceosomal machinery are required for back-spliced circularization.29,30 In addition, the majority of highly expressed circRNAs are usually processed from internal exons of pre-mRNAs and normally contain multiple exons, indicating that back-splicing is generally coupled with canonical splicing.12
Figure 1.
Back-splicing for circRNAs. Pre-mRNA can go through either high efficient canonical splicing to generate a linear RNA with exon inclusion (top) or low efficient back-splicing to produce both a circular RNA and an alternatively spliced linear RNA with exon exclusion (bottom). Canonical splicing signals and spliceosomal machinery are required for back-spliced exon circularization (blue bars). The coupling between splicing and back-splicing ( - ) are discussed in text. ss, splice site.
The coupling between splicing and back-splicing can be exemplified by a simple case illustrated in Fig. 1. A canonical alternative splicing event (canonical splicing ) joins 2 non-sequential exons together to form a linear RNA with skipped middle exons. This is coupled with the covalent circularization of the skipped middle exons by back-splicing (back-splicing ). Very often, these 2 skipped middle exons are further processed by another canonical splicing event (canonical splicing ) to remove the intron between them (discussed below). Two models have been proposed to explain the underlying mechanism of back-spliced circularization.11,16,18,20,28 Broadly speaking, the notable difference between these 2 models is which step, canonical splicing or back-splicing, happens first (Fig. 2).
Figure 2.
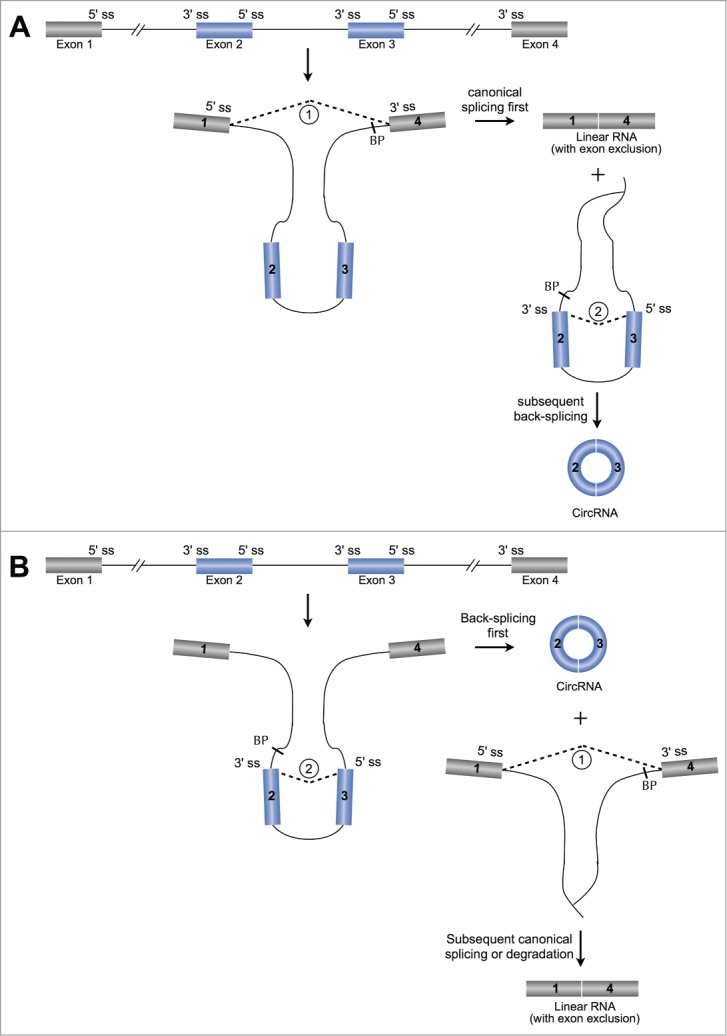
Two possible models for circRNA formation. (A) The “exon skipping” or “lariat intermediate” model for circRNA formation. The processing starts with canonical splicing for a linear RNA with skipped exons and a long intron lariat containing these skipped exons (blue bars), which is then further back-spliced to form a circRNA. (B) The “direct back-splicing” model for circRNA formation. The precessing starts with back-splicing for a circRNA together with an exon-intron(s)-exon intermediate, which can be further processed to produce a linear RNA with skipped exons or to be potentially degraded. ss, splice site. BP, branchpoint.
Which Comes First, Canonical Splicing or Back-Splicing
If canonical splicing happens first, it will initially generate a linear RNA with skipped exons and a long intron lariat containing these skipped exons, which are then back-spliced to generate a circRNA (Fig. 2A). Thus, this model is referred to as “exon skipping” or “lariat intermediate” model.11,16,18,20 On the other hand, if the back-splicing happens first, it will directly generate a circRNA together with an exon-intron(s)-exon intermediate, which can be further processed to produce a linear RNA with skipped exons or be potentially degraded (Fig. 2B). Accordingly, the second model has been highlighted as “direct back-splicing.”11,16,18,20 Although detailed lines of biochemical evidence are still required to evaluate these models, it appears that both proposed models can be effective in vivo.11,28
In both models, the expression of a circRNA is theoretically associated with an alternatively spliced linear RNA with exon exclusion (Fig. 2). However, although it has been exemplified in recapitulated assays,12 the correlation of endogenous circRNAs with alternatively spliced linear RNAs could only be identified in some, but not all, circRNA regions.11,12,18,28 Thus, to what extent the correlation of circularization with exon skipped splicing remains to be further investigated in endogenous circumstance. Furthermore, it is still unclear under which condition(s) the spliceosomal machinery chooses either canonical splicing (Fig. 2A) or back-splicing (Fig. 2B) to start with the production of a circRNA. These two steps may happen either stochastically or even synergistically.
Cis-Elements and Trans-Factors Regulate the Competition Between Canonical Splicing and Back-Splicing
Albeit more stable as covalently closed circles12,16,18,20 and catalyzed by canonical spliceosomal machinery,11,28,30 most circRNAs are less abundant than their linear mRNA counterparts.12,18,20 One possible explanation is that back-splicing is unfavorable for spliceosome assembly and thus less efficiently catalyzed by the spliceosomal machinery. To overcome this natural disadvantage, both cis-elements and trans-factors are required to bring the downstream donor and upstream acceptor sites close together to promote back-splicing. For examples, several recent studies have reported that the processing of circRNAs can be facilitated by either RNA pairing of reversely complementary sequences across their flanking introns (Fig. 3A)12,18,23,31 or protein factors binding to pre-mRNAs to bridge flanking introns together (Fig. 3B).29
Figure 3.
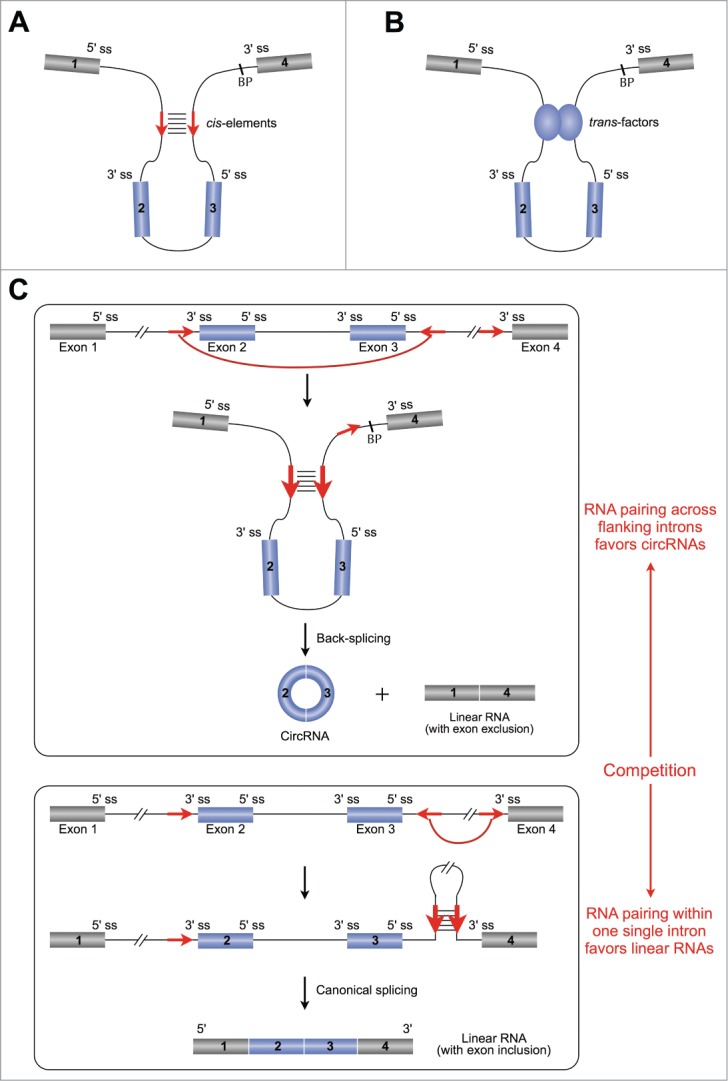
The competition of RNA pairing for splicing or back-splicing. (A) (B) Both cis-elements (A) and trans-factors (B) can affect back-splicing efficiency by taking the downstream splice donor and upstream acceptor sites close together. (C) The competition model of RNA pairing. Top, RNA pairing formed across flanking introns promotes back-splicing, leading to the formation of a circRNA and a linear RNA with exon exclusion. Bottom, the RNA pairing formed within one individual intron promotes the canonical splicing, resulting in a linear RNA with exon inclusion, but no back-splicing. Red arrows, complementary sequences. ss, splice site. BP, branchpoint.
RNA pairing across flanking introns is an efficient way to promote exon circularization. RNA pairing can be formed by either repetitive elements, such as the very abundant Alu elements32 in human, or non-repetitive but complementary sequences.12 Although short sequences (as small as 30 to 40 nucleotides) were observed to be able to sufficiently facilitate circRNA biogenesis,31 a strong pairing capacity could dramatically enhance the production of circRNAs.12
However, it is worth noting that RNA pairing is not always formed across flanking introns to facilitate back-splicing for circRNAs (top, Fig. 3C), but can also be formed within an individual intron to promote canonical splicing for linear RNA formation (bottom, Fig. 3C).12 For example, a conserved RNA pairing within a long intron was reported to bridge the upstream splice donor site close to the downstream splice acceptor site, and thus could promote a canonical splicing for Rbfox-mediated exon inclusion.33 Thus, the selection of RNA pairing across flanking introns or within a single individual intron leads to competition between back-splicing for circRNAs and canonical splicing for linear RNAs, which has been predicted to occur genome-wide and demonstrated in recapitulated assays.12 It should be noted that being under regularoty control, the competition can be very dynamic, leading to different expression patterns of circRNAs and linear RNAs.
So far, 2 RNA binding proteins, muscleblind (MBNL1) and Adenosine deaminase 1 acting on RNA (ADAR1), have been reported to play a role in circRNA biogenesis, doing so by different modes of action. MBNL1 was shown to bind to its own pre-mRNA and bridge 2 flanking introns close together (Fig. 3B) to induce back-splicing, resulting in up-regulated circRNA formation from its own RNA.29 ADAR1 knockdown specifically up-regulates some circRNA expression, implying a role of ADAR1 in suppression of circRNA biogenesis.23 Mechanistically, it was suggested that such regulation is associated with Adenosine-to-Inosine (A-to-I) editing.23 Double strand RNA (dsRNA) pairing structures are known as A-to-I RNA editing targets by ADARs.34,35 In the normal condition, highly enriched A-to-I editing in dsRNA regions could diminish RNA pairing structures, resulting in reduced RNA pairing and thus less efficient back-splicing for circRNA formation. Whereas, with the reduced level of A-to-I editing after ADAR1 knockdown, RNA pairing across flanking introns is more stable and favors back-splicing for circRNA production.23 Notably, ADAR1 was recently shown to act as a dsRNA binding protein to interfere with microRNA processing,36 thus it is also possible that ADAR1 may regulate circRNA formation directly through its dsRNA binding activity, independent of RNA editing. As there are hundreds (and maybe thousands) of RNA binding proteins,37 it will be of interest to identify additional trans-factors involved in circRNA formation.
Alternative Circularization: Multiple Circular RNAs from a Single Precursor RNA
It is becoming increasingly evident that circRNAs are not just by-products from mis-splicing or splicing errors, but are processed from regulated back-splicing with distinct sets of cis-elements and/or trans-factors. Accordingly, some circRNAs were reported to be more abundant than their linear counterparts in certain cell lines20 or upregulated during neural development in flies.22 Remarkably, the production of circRNAs is even more complex with the discovery that multiple circular RNAs can be identified from a single gene locus,12,16,18,22,38 a phenomenon referred to as alternative circularization.12
There are several ways to generate different alternative circularization events. First of all, multiple circRNAs can be processed from a single gene locus with different numbers of exons included,12,16,18,22,38 possibly due to the competition of RNA pairing across different intron sets. For instance, the existence of multiple human circRNAs from a single gene locus is associated with a variety of Alu pairing in human introns (red arcs, Fig. 4A), suggesting a role of competition between Alu pairing in the formation of alternative circularization.12 In fact, the competition of RNA pairing across different flanking introns can be very complicated, due to the fact that the competition of RNA pairing happens either within repetitive or non-repetitive elements,12 and that the RNA pairing can also be regulated by additional factors, such as RNA binding proteins.23
Figure 4.
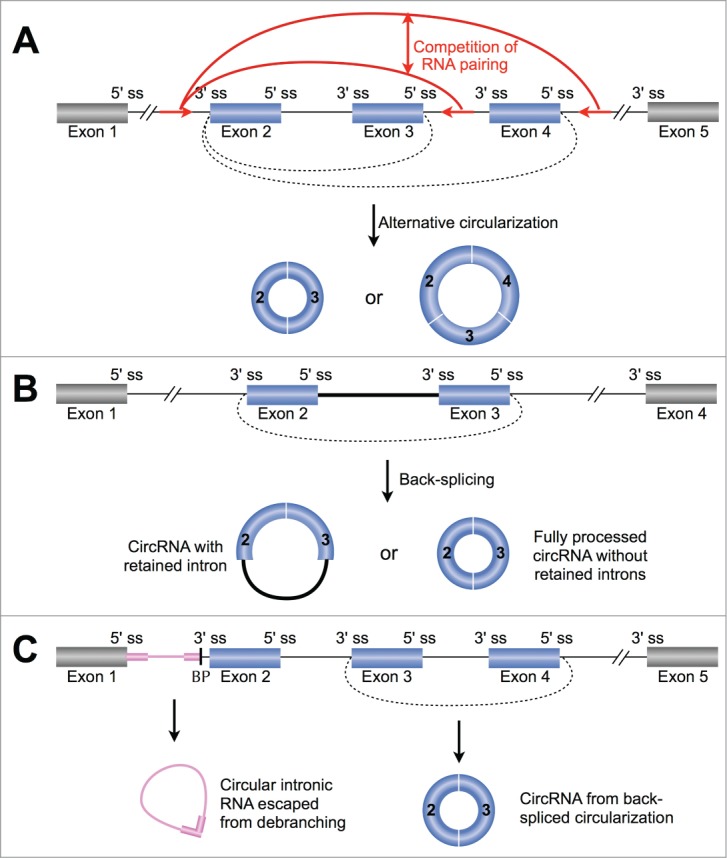
Possible mechanisms for alternative circularization. (A) Multiple circRNAs can be processed from a single gene locus with different numbers of exons regulated by the competition of RNA pairing across different introns (red arcs). (B) Multiple circRNAs can be produced from a single gene locus with the internal intron included or excluded with unknown mechanisms. (C) Multiple circular RNAs from either exons or introns can be generated from a single gene locus through distinct circular RNA formation pathways. See text for details. Red arrows, complementary sequences. ss, splice site. BP, branchpoint.
In addition, multiple circRNAs can be produced from a single gene locus with the internal intron included or excluded (Fig. 4B).12,20,39 Interestingly, the level of internal intron retention within a circRNA can be very different when examined from various cell lines, suggesting that the inclusion of internal introns is under regulatory control.20 Finally, multiple circular RNAs can also be generated from either exons or introns in a single gene locus (Fig. 4C, and data not shown) by distinct circular RNA formation pathways. For instance, a circular intronic RNA is produced from intron lariats that escape debranching (pink circle, Fig. 4C) and the production of such circular intronic RNAs is highly associated with consensus motifs near to 5′ splice sites and branchpoint sites (pink bars, Fig. 4C).17 While in the same locus, an additional circular RNA from back-spliced exons can be simultaneously generated (blue circle, Fig. 4C), resulting in multiple circular RNAs from a single gene locus.
Although endogenous conditions and regulations for alternative circularization are complex and require further investigation, the identification of alternative circularization definitely expands our understanding of back-splicing and its regulation.
Future Remarks
Genome-wide annotations have revealed that circRNAs are widely expressed in different cell lines and across various species,12,18,19,22,23,31 greatly expanding our understanding of the ever growing list of lncRNAs. Recent studies have uncovered that the processing of circRNAs is not only catalyzed by canonical spliceosomal machinery,23,30 but also regulated as canonical splicing by both cis-elements12,18,23,31 and trans-factors.23,29 Given the discrete expression of circRNAs, additional cis-/trans-regulators involved in circRNA biogenesis are required to be further defined.
A challenging unanswered question is the function of circRNAs. Recently, a handful of circRNAs have been shown to be involved in gene expression regulation as miRNA sponges.19,21,40,41 Strikingly, some circular intronic RNAs and circRNAs with retained introns were also reported to function as positive regulators of RNA Pol II transcription in the nucleus.17,39 In addition, it has been proposed that some circRNAs may “sponge” other factors, such as RNA binding proteins.42 The processing of circRNA biogenesis itself might also regulate the formation of linear mRNAs through the competition between splicing and back-splicing.28 Since dozens of thousands of circRNAs have been identified, in the future, it will be of great interest to study what the vast majority of circRNAs can do in both physiological and pathological conditions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Andrew E Teschendorff for critical reading of this manuscript. We apologize to authors whose work could not cite here owing to space/content limitations.
Funding
Our work is supported by 31471241, 31322018 and 91440202 from NSFC.
References
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73: 3852-6; PMID:1069269; http://dx.doi.org/ 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell 1980; 19: 313–9; PMID:6986989; http://dx.doi.org/ 10.1016/0092-8674(80)90505-X [DOI] [PubMed] [Google Scholar]
- 3.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558–60; PMID:2429192; http://dx.doi.org/ 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- 4.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607–13; PMID:1991322; http://dx.doi.org/ 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- 5.Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 1992; 11: 1095–8; PMID:1339341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019–30; PMID:7684656; http://dx.doi.org/ 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- 7.Kopczynski CC, Muskavitch MA. Introns excised from the Delta primary transcript are localized near sites of Delta transcription. J Cell Biol 1992; 119: 503–12; PMID:1383233; http://dx.doi.org/ 10.1083/jcb.119.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res 1992; 20: 5345–50; PMID:1437551; http://dx.doi.org/ 10.1093/nar/20.20.5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155–60; PMID:7678559 [DOI] [PubMed] [Google Scholar]
- 10.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA 1996; 2: 603–10; PMID:8718689 [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature Biotechnol 2014; 32: 453–61; http://dx.doi.org/ 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159: 134–47; PMID:25242744; http://dx.doi.org/ 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Graveley BR. Molecular biology: power sequencing. Nature 2008; 453: 1197–8; PMID:18580940; http://dx.doi.org/ 10.1038/4531197b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 2008; 453: 1239–43; PMID:18488015; http://dx.doi.org/ 10.1038/nature07002 [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 2011; 12:R16; PMID:21324177; http://dx.doi.org/ 10.1186/gb-2011-12-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7: e30733; PMID:22319583; http://dx.doi.org/ 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51: 792–806; PMID:24035497; http://dx.doi.org/ 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 18.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19: 141–57; PMID:23249747; http://dx.doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 20.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; http://dx.doi.org/ 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15: 409; PMID:25070500; http://dx.doi.org/ 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9: 1966–80; PMID:25544350; http://dx.doi.org/ 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al.. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10: 170–7; PMID:25558066; http://dx.doi.org/ 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 24.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268: 415–7; PMID:7536344; http://dx.doi.org/ 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21: 172–9; PMID:25449546; http://dx.doi.org/ 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores R, Navarro JA, de la Pena M, Navarro B, Ambros S, Vera A. Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group. Biol Chem 1999; 380: 849–54; PMID:10494833; http://dx.doi.org/ 10.1515/BC.1999.104 [DOI] [PubMed] [Google Scholar]
- 27.Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Hüttenhofer A. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 2002; 30: 921–30; PMID:11842103; http://dx.doi.org/ 10.1093/nar/30.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20: 1829–42; PMID:25404635; http://dx.doi.org/ 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56: 55–66; PMID:25242144; http://dx.doi.org/ 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 30.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015; 10: 103–11; PMID:25543144; http://dx.doi.org/ 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 31.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28: 2233–47; PMID:25281217; http://dx.doi.org/ 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle 2008; 7: 3294–301; PMID:18948735; http://dx.doi.org/ 10.4161/cc.7.21.6927 [DOI] [PubMed] [Google Scholar]
- 33.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al.. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 2013; 20: 1434–42; PMID:24213538; http://dx.doi.org/ 10.1038/nsmb.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2004; 2: e391; PMID:15534692; http://dx.doi.org/ 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 2004; 14: 1719–25; PMID:15342557; http://dx.doi.org/ 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Xiang JF, Zhu S, Chen S, Yin QF, Zhang XO, Zhang J, Feng H, Dong R, Li XJ, et al.. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res 2015; in press; PMID:25708366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012; 149: 1393–406; PMID:22658674; http://dx.doi.org/ 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 38.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6: e1001233; PMID:21151960; http://dx.doi.org/ 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Kjems J. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; in press; PMID:25664725 [DOI] [PubMed] [Google Scholar]
- 40.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30: 4414–22; PMID:21964070; http://dx.doi.org/ 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 42.Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J 2013; 32: 923–5; PMID:23463100; http://dx.doi.org/ 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]