Substitutions in Vps13 suppress all measured phenotypic consequences of ERMES deficiency, and Vps13 dynamically localizes to vacuole–mitochondria and to vacuole–nucleus contact sites depending on growth conditions, suggesting that ERMES function can be bypassed by the activity of other contact sites, and that contact sites establish a growth condition–regulated organelle network.
Abstract
The endoplasmic reticulum–mitochondria encounter structure (ERMES) complex tethers the endoplasmic reticulum and the mitochondria. It is thought to facilitate interorganelle lipid exchange and influence mitochondrial dynamics and mitochondrial DNA maintenance. Despite this important role, ERMES is not found in metazoans. Here, we identified single amino acid substitutions in Vps13 (vacuolar protein sorting 13), a large universally conserved eukaryotic protein, which suppress all measured phenotypic consequences of ERMES deficiency. Combined loss of VPS13 and ERMES is lethal, indicating that Vps13 and ERMES function in redundant pathways. Vps13 dynamically localizes to vacuole–mitochondria and to vacuole–nucleus contact sites depending on growth conditions, suggesting that ERMES function can be bypassed by the activity of other contact sites, and that contact sites establish a growth condition–regulated organelle network.
Introduction
Eukaryotic cells are compartmentalized by membrane-bound organelles. Protein complexes tether various membranes at specific contact sites (Helle et al., 2013). In Saccharomyces cerevisiae, ER–plasma membrane contact sites, nuclear ER–vacuole junctions (NVJ; Pan et al., 2000), vacuole and mitochondria patches (vCLAMPs; Elbaz-Alon et al., 2014; Hönscher et al., 2014), and ER–mitochondria encounter structures (ERMES; Kornmann et al., 2009) have been described previously.
The ERMES complex is a well-characterized structure that tethers the ER to the outer mitochondrial membrane (OMM; Michel and Kornmann, 2012). The core–ERMES complex consists of the ER protein Mmm1 (mitochondria morphology maintenance 1), the OMM proteins Mdm10 and Mdm34, and the cytosolic protein Mdm12. ERMES localizes into foci (hereafter ERMES foci) at ER–mitochondria interfaces.
ERMES members were originally found in yeast genetic screens for mutants with altered mitochondrial distribution and morphology (MDM); while mitochondria form a tubular network in wild-type (WT) yeast cells, they collapse into swollen globules in strains defective for ERMES (hereafter ERMES mutants; Sogo and Yaffe, 1994; Berger et al., 1997). ERMES foci are often sites of mitochondrial fission, suggesting that the aberrant mitochondrial shape observed in ERMES mutants is caused by dysregulated fission (Murley et al., 2013). Moreover, ERMES foci are often found in proximity of replicating mitochondrial DNA (mtDNA), and ERMES mutants lose mtDNA, suggesting that ERMES functions in its maintenance (Aiken Hobbs et al., 2001; Meeusen and Nunnari, 2003).
We and others have suggested that ERMES has a role in ER–mitochondria lipid exchange, based on two lines of evidence: (1) three of four core ERMES components harbor lipid-binding domains (Kopec et al., 2010; Schauder et al., 2014) capable of sheltering lipid acyl chains from the aqueous cytosol during transfer from one membrane to another; and (2) a large-scale genetic interaction screen uncovered a connection between ERMES and the mitochondrial phosphatidylserine decarboxylase Psd1 (Kornmann et al., 2009). Since the substrate and the product of Psd1 come from and return to the ER, respectively, this connection suggests that ERMES plays a role in ER–mitochondria lipid exchange.
Yet, ERMES defects slow but do not block lipid exchange (Kornmann et al., 2009; Nguyen et al., 2012). A possible explanation for this conundrum is that ERMES is partially redundant with other structures. Indeed, two studies showed that the overexpression of the vacuole fusion factor Vps39 (vacuolar protein sorting 39) led to the formation of vacuole–mitochondria contact patches (vCLAMPs; Elbaz-Alon et al., 2014; Hönscher et al., 2014). Loss of ERMES and Vps39 are synthetic lethal, suggesting that they work in redundant pathways. Moreover, the growth and mitochondrial morphology defects of ERMES mutants can be rescued by overexpression of two mitochondrial proteins, Mcp1 and Mcp2, indicating that ERMES deficiency can be compensated for (Tan et al., 2013). Both Mcp1 and Mcp2 affect mitochondrial lipid composition, but how their overexpression leads to the rescue of ERMES mutants is unclear. An appealing hypothesis is that an up-regulation of ERMES-redundant pathways compensates for the loss of ERMES.
Results and discussion
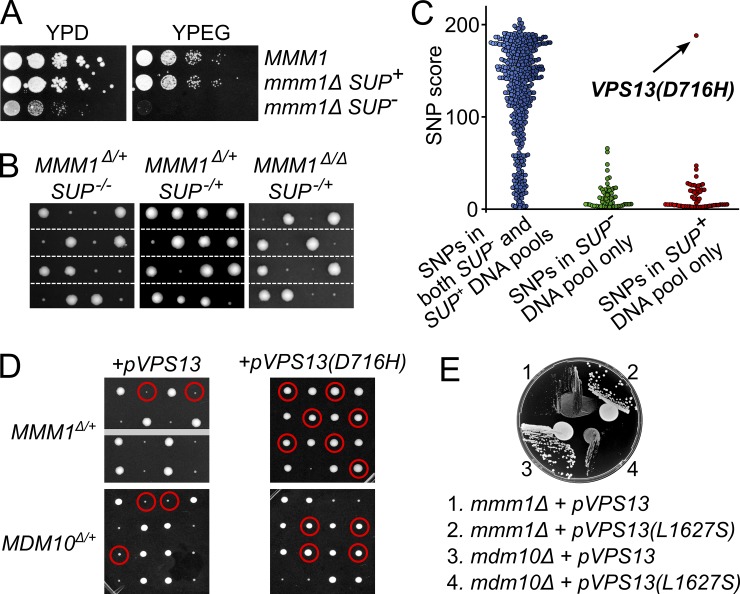
Yeasts deleted for any ERMES member grow slowly on fermentable media and cannot grow on a nonfermentable carbon source (Burgess et al., 1994; Berger et al., 1997; Youngman et al., 2004). Surprisingly, when we analyzed an mmm1Δ strain from a haploid deletion library (Giaever et al., 2002), we found that it grew indistinguishably from WT cells (Fig. 1 A). Backcrossing to an isogenic WT showed that this strain bore a Mendelian suppressor (SUP) mutation (Fig. 1 B). Crossing slow-growing (SUP−) mmm1Δ to fast-growing (SUP+) mmm1Δ cells gave rise to fast-growing diploids, indicating that the suppressor was dominant.
Figure 1.
A dominant mutation in VPS13 suppresses the growth defect of ERMES mutants. (A) Serial dilutions of strains of the indicated genotypes on fermentable (YPD: YP + 2% dextrose) or nonfermentable (YPEG: YP + 3% glycerol + 1.5% ethanol) media. Top: WT. Middle: isogenic mmm1Δ strain from a published deletion library (Giaever et al., 2002) that bears a suppressor mutation (SUP+). Bottom: isogenic mmm1Δ strain without suppressor mutation (SUP−). (B) Tetrad analysis of an MMM1/mmm1Δ heterozygote (left), a MMM1/mmm1Δ; SUP−/SUP+ heterozygote (middle), and a mmm1Δ homozygote, SUP−/SUP+ heterozygote (right). (C) Quality scores of SNPs were classified in three categories: SNPs found into both the SUP+ and SUP− DNA pools (left), SNPs found in the SUP− pool only (middle), and SNPs found in the SUP+ pool (right). Low-scoring SNPs represent sequencing errors while high scoring ones represent bona fide variants. (D) A diploid MMM1/mmm1Δ heterozygote was transformed with a plasmid encoding WT Vps13 (pVPS13, left) or the D716H allele (pVPS13(D716H), right). Spores circled in red bear both the deletion allele and the indicated plasmid. (E) A CEN/ARS plasmid bearing the VPS13(L1627S) allele (pVPS13(L1627S)) or its WT counterpart (pVPS13) were transformed into mmm1Δ and mdm10Δ cells. Transformants were spotted and streaked on YPD.
We devised a novel and potentially widely applicable whole-genome sequencing strategy that allowed us to distinguish the causative mutation from unlinked genetic variations. We first backcrossed mmm1Δ SUP+ cells to parental WT (SUP−) cells. We extracted and pooled genomic DNA from 20 mmm1Δ SUP− progenies and, in a separate pool, from 20 mmm1Δ SUP+ progenies. Unlinked mutations should segregate equally between these two pools, whereas the causal mutation should be found only in the SUP+ pool. We subjected the two pools to deep sequencing and classified the identified single-nucleotide polymorphisms (SNPs) into three categories (see Materials and methods): (1) SNPs present in both pools, (2) SNPs only present in the SUP− pool, and (3) SNPs only present in the SUP+ pool.
We found 463 SNPs between our strains and the reference genome, underscoring the substantial genetic heterogeneity between closely related laboratory strains (Fig. 1 C, left). In contrast, we found few, low-quality SNPs in the SUP− pool only, most likely representing sequencing errors (Fig. 1 C, middle). In the SUP+ pool, a single, high-scoring bona fide SNP stood out (Fig. 1 C, right). This C-to-G transversion caused a D716H substitution in Vps13 (vacuolar protein sorting 13). Engineered on a plasmid, this mutation conferred the suppression phenotype in a dominant manner (Fig. 1 D), proving that the VPS13(D716H) allele caused the suppression. Thus, our sequencing strategy allows unambiguous identification of mutations and promises to be widely applicable in forward genetics screens.
ERMES mutants lose their phenotype over time (Berger et al., 1997). We wondered if this phenomenon was due to the appearance of suppressor alleles in VPS13. We isolated four independent “adapted” strains from mmm1 or mdm34 mutant cells. In all cases, sequencing VPS13 identified nonsynonymous SNPs (Table 1). We engineered one of these (VPS13(L1627S)) on a plasmid, which, again, conferred the suppression phenotype (Fig. 1 E).
Table 1. Potential suppressor alleles identified.
| Mutation | DNA | Protein | Found in | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | Position | Mutant | WT | Position | Mutant | ||||
| 1a | GAT | 2146 | CAT | D | 716 | H | mmm1Δ | ||
| 2 | TTA | 1814 | TCA | L | 605 | S | mdm34Δ | ||
| 3 | CCT | 3665 | CAT | P | 1222 | H | mdm34Δ | ||
| 4 | TTG | 2379 | TTC | L | 793 | F | mmm1Δ | ||
| 5a | TTG | 4880 | TCG | L | 1627 | S | mmm1-1 | ||
Alleles for which causality was confirmed (Fig. 1, D and E).
Vps13 is the fifth largest protein in yeast and has four orthologues in the human genome, two of which are linked to neurological disorders (Velayos-Baeza et al., 2004). VPS13 was originally found in screens for Golgi trafficking mutants (Bankaitis et al., 1986; Brickner and Fuller, 1997). Vps13 bears no apparent homology to other proteins, domains, or targeting sequences, but associates peripherally with the membrane of the endosomes (Huh et al., 2003) and with that of the prospore during meiosis (Park and Neiman, 2012). The molecular function of Vps13 has, however, remained obscure.
Vps13 acting as ERMES suppressor was surprising. Indeed, ERMES mutants are mostly defective in mitochondrial function, yet Vps13 is not known to affect mitochondria. We thus investigated whether, in addition to growth, the suppressor allele rescued other mitochondrial phenotypes of ERMES mutants.
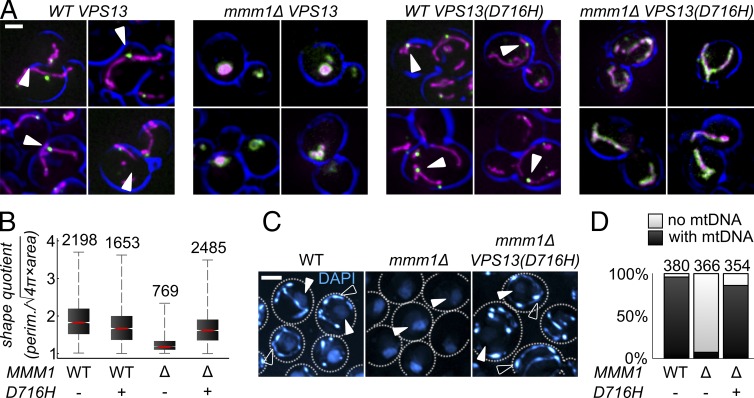
We imaged mmm1Δ cells bearing or not bearing the VPS13(D716H) allele, and expressing a mitochondria-targeted marker (mtDsRed) and a C-terminal GFP-tagged allele of Mdm34. As expected, Mdm34-GFP localized in foci in WT strains (Fig. 2 A) and had a diffuse mitochondrial staining in mmm1Δ strains. The suppressor allele did not restore the localization of Mdm34-GFP to foci in mmm1Δ cells (Fig. 2 A, right), indicating that the suppressor does not restore the assembly of ERMES complexes.
Figure 2.
The VPS13(D716H) allele suppresses pleiotropic consequences of ERMES deficiency. (A) Images of cells of the indicated genotype bearing GFP-tagged MDM34 (green) and a mitochondrial marker (mtDsRed, magenta). ERMES foci are indicated (arrowheads). The cell outline is shown in blue. Bar, 2 µm. (B) Cells of the indicated genotype were imaged as in A. A shape quotient was measured for each mitochondria. The number of mitochondria analyzed is indicated above each graph. (C) Cells of the indicated genotypes stained with DAPI. DAPI stains both nuclear DNA (solid arrowheads) and mtDNA (open arrowheads). Bar, 2 µm. (D) The number of cells in C displaying mitochondrial DAPI staining (with mtDNA) or not (no mtDNA) was counted for each genotype. The number of cells analyzed is indicated above. P < 10−100 for a t test and a Fisher’s exact test to compare mmm1Δ VPS13 and mmm1Δ VPS13(D716H) in B and D, respectively.
In contrast, while mmm1Δ VPS13 cells harbored round and swollen mitochondria, mmm1Δ VPS13(D716H) harbored a tubular mitochondrial network, comparable to WT cells. We quantified this effect using an algorithm (Script S1), which calculated a shape quotient [perimeter/√(4π × area)]. This quotient equals 1 for a sphere and increases with elongation. The shape quotient of mmm1Δ VPS13(D716H) mitochondria (1.67 ± 0.40, mean ± SD) was different from that of mmm1Δ (1.28 ± 0.22), and was comparable to isogenic WT cells (1.73 ± 0.46; Fig. 2 B).
Because ERMES mutants frequently lose mtDNA, we addressed mtDNA stability using DAPI staining. Although >90% of mmm1Δ cells lost their mtDNA (Aiken Hobbs et al., 2001; Fig. 2, C and D, middle), >85% of the mmm1Δ VPS13(D716H) cells retained it (Fig. 2, C and D, right), demonstrating that the suppressor restored mtDNA stability.
Therefore, the VPS13(D716H)-mediated suppression is pleiotropic.
Cytosolic Vps13(D716H) is unlikely to play a direct role in mtDNA maintenance. The fact that it suppresses ERMES mtDNA defects indicates that these are secondary to the loss of a primary ERMES function. By uncoupling ERMES primary function from indirect downstream phenotypes, suppressors will be instrumental to address the role of ERMES in other processes, like mitochondrial fission, protein import, and mitophagy (Meisinger et al., 2007; Murley et al., 2013; Böckler and Westermann, 2014).
The fact that the VPS13(D716H) and VPS13(L1627S) alleles were dominant indicated that they caused a gain of function in Vps13. Surprisingly, deletions of both MMM1 and VPS13 were synthetic lethal (Fig. S1 A; Costanzo et al., 2010; Hoppins et al., 2011). This paradox indicates that Vps13 function is redundant with that of ERMES; enhancing Vps13-enabled processes by expression of gain-of-function alleles compensates for the loss of ERMES, whereas a loss of Vps13 renders ERMES indispensable.
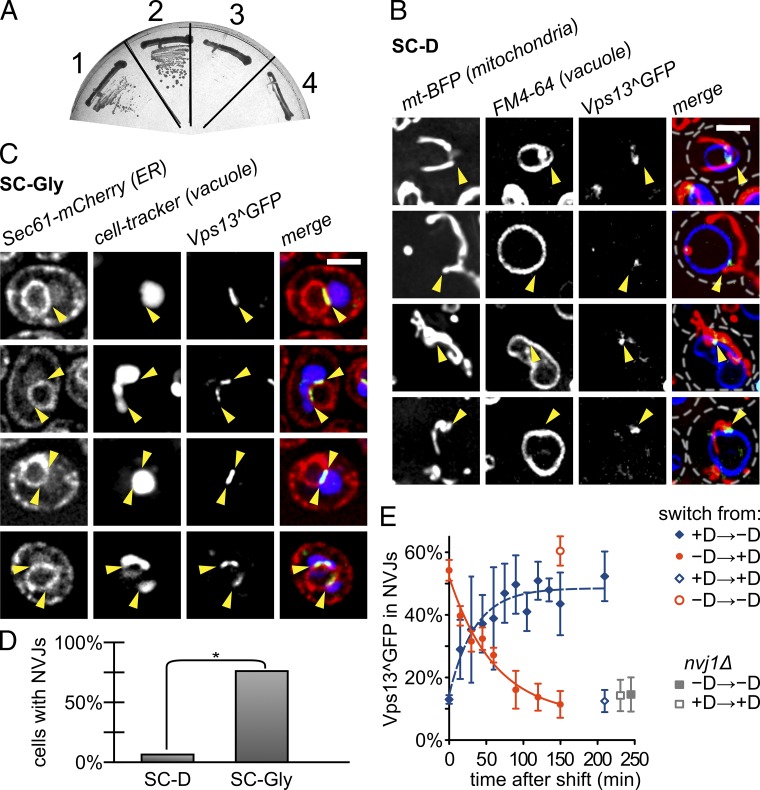
To define this redundant pathway, we sought to GFP-tag Vps13. A previous study suggested that GFP-tagged Vps13 localized to endosomes (Huh et al., 2003), though without addressing whether the fusion protein was functional. We tested the functionality of GFP-tagged constructs using the synthetic lethality between VPS13 and MMM1. Both C- and N-terminal GFP fusions yielded nonfunctional proteins (Fig. S1, B and C). In the absence of structural information, alignment between fungal Vps13 orthologues hinted toward variable loops that were potentially tolerant to GFP insertion. We transformed a temperature-sensitive mmm1-1 strain (Burgess et al., 1994) and looked for viability upon shifting to the nonpermissive temperature. Of the 15 internal locations tested, one yielded a functional fusion protein, referred to hereafter as Vps13^GFP (Fig. 3 A and Tables S1 and S2).
Figure 3.
Vps13 localizes to alternative membrane contact sites. (A) Assessment of the functionality of GFP-tagged Vps13 using synthetic lethality with MMM1. mmm1 thermo-sensitive strains harboring a GFP inserted in Vps13 after amino acid 499 (1), 446 (3), 473 (4), or untagged Vps13 (2) were streaked onto YPD and grown at the nonpermissive temperature (37°C). Only GFP inserted after amino acid 499 yields a functional protein. (B) Intracellular localization of Vps13^GFP in otherwise WT cells grown on SC-Dextrose, expressing a mitochondrial marker (mtBFP) and stained with a vacuole dye (FM4-64). Vps13^GFP is often found in foci colocalizing with the vacuole and mitochondria. Bar, 2 µm. (C) Intracellular localization of Vps13^GFP in otherwise Sec61-mCherry cells grown on SC-Glycerol, and stained with the vacuole marker CellTracker Blue CMAC. Vps13^GFP relocalizes to NVJs (yellow arrowheads). (D) Quantification of the number of cells in which Vps13^GFP shows NVJ localization in dextrose- (SC-D) and glycerol- (SC-Gly) containing medium. *, P < 10−100 from a Fisher’s exact test. (E) Time course showing the percentage of Vps13 found in NVJs upon carbon source shifting. At time 0, cells grown to exponential phase in dextrose-replete (+D) or depleted (-D) medium were washed and resuspended in indicated medium. Percentages were calculated automatically with ImageJ Script S3.
We imaged Vps13^GFP together with mitochondria and the vacuole. Vps13^GFP localized in foci, which overlapped often with both organelles (Fig. 3 B), suggesting that Vps13 was a part of vCLAMPs. Some of the Vps13 foci did not overlap with either mitochondria or vacuoles, and may mark endosomal structures. Using an automated counting algorithm (Script S2), we quantified the fraction of the foci colocalized with mitochondria at 22.3 ± 0.6% (mean ± SEM). This number is likely an underestimate, due to the conservative automatic segmentation of the mitochondria. To validate that this percentage was different than expected by chance, we picked random points on the vacuole and quantified the percentage of those colocalizing with mitochondria. This number was significantly lower (16.0 ± 0.63%, P < 10−5 from a paired t test). A reciprocal analysis detected 56.3 ± 4.0% of Vps13 on the vacuole. This proportion dropped to 20.7 ± 0.76% for random points on mitochondria (P < 10−5). Thus, the localization of Vps13^GFP foci is consistent with a function at vCLAMPs.
To further test the idea that Vps13 might localize to vCLAMPs, we assessed Vps13 localization in medium containing glycerol as carbon source (SC-Gly). In these conditions, vCLAMP formation is repressed (Hönscher et al., 2014) and Vps13^GFP relocalized from dotted structures to streaks (Fig. 3, C and D). This relocalization was fast and reversible (Fig. 3 E). We identified these streaks as nuclear–vacuole junctions (NVJs; Fig. 3 C, arrowheads). Indeed, Vps13^GFP relocalization was abrogated in mutants lacking NVJs (nvj1Δ; Fig. 3 E). NVJs are well-known structures, which play a role in piecemeal microautophagy of the nucleus, a process of unclear function (Roberts et al., 2003). NVJs also harbor lipid-transport proteins like Osh1 (Levine and Munro, 2001) and Nvj2 (Toulmay and Prinz, 2012).
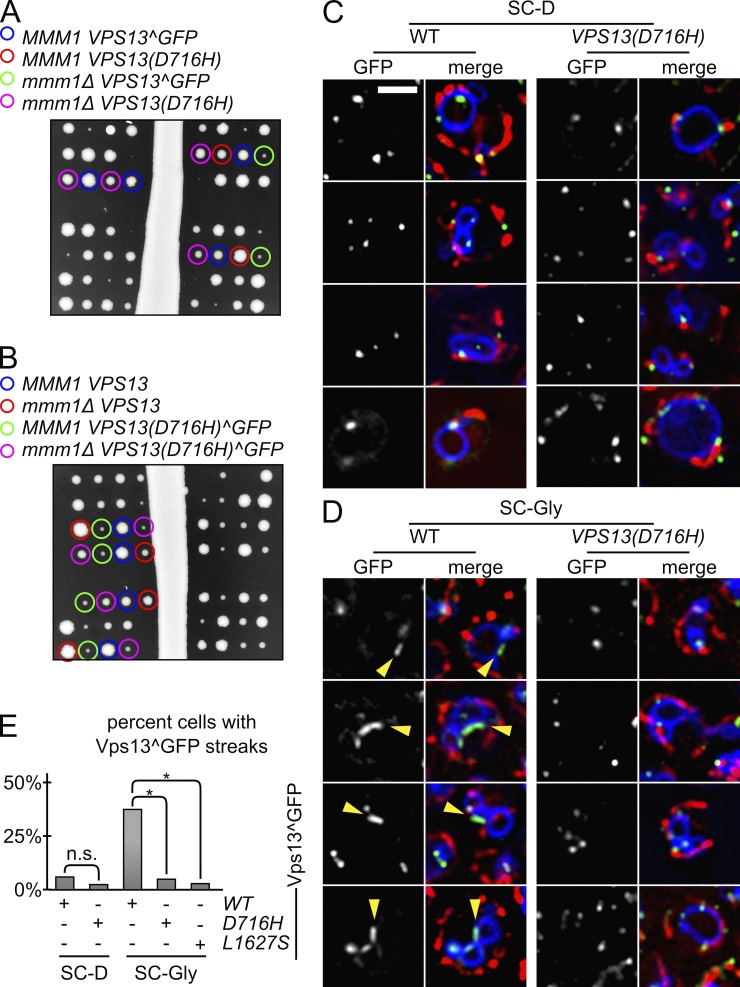
We wondered if the suppressor mutations altered the localization of the protein. We thus engineered the Vps13^GFP fusion protein in a MMM1/mmm1Δ VPS13/VPS13(D716H) double heterozygous strain. Because the GFP fusion construct could integrate either into the WT or the D716H allele of VPS13, we sporulated several strains, two of which are shown here. In the first strain, the GFP fusion and the D716H allele segregated away from each other (Fig. 4 A), while in the second strain they segregated together (Fig. 4 B). mmm1Δ strains harboring the VPS13(D716H)^GFP allele grew better than those harboring a WT VPS13 allele, but slightly less than strains harboring untagged VPS13(D716H). Thus, the suppressing ability is only slightly impaired upon GFP tagging. Vps13(D716H)^GFP localized to dotted structures, like its WT counterpart (Fig. 4 C), but failed to relocalize to NVJs upon shift to glycerol-containing medium (Fig. 4, C–E). The same was true for another suppressor allele (Vps13(L1627S); Fig. 4 E).
Figure 4.
Suppressor alleles of Vps13 fail to relocalize to NVJs. (A and B) Tetrad dissection of sporulated MMM1/mmm1Δ VPS13/VPS13(D716H) diploids in which the VPS13^GFP construct has been engineered in the VPS13 (A) or VPS13(D716H) (B) alleles. (C) Cells harboring the Vps13^GFP construct in the indicated VPS13 allele were grown in dextrose- (SC-D; C) or glycerol-containing (SC-gly) medium (D), and stained with DAPI (mitochondria, red) and FM4-64 (vacuole, blue). The formation of streaks (arrowheads) denotes the relocalization of Vps13^GFP to NVJs. Bar, 2 µm. (E) Quantification of the number of cells showing streaks in the indicated conditions. *, P < 10−20; n.s., P > 0.05 from a Fisher’s exact test.
Mitochondria–vacuole contacts have been recently described as extensive patches, appearing under strong Vps39 overexpression (Hönscher et al., 2014; Elbaz-Alon et al., 2014). We show that under physiological conditions, Vps13 localizes to diffraction-limited foci, which may reflect the natural morphology of vCLAMPs. Vps13 was found among a list of proteins interacting with overexpressed Vps39 (Elbaz-Alon et al., 2014), the hitherto only known component of vCLAMPs. We have not been able to detect endogenous Vps39 in pull-downs of Vps13^GFP (unpublished data), perhaps because physiological levels of Vps39 at vCLAMPs are too low for proper detection. However, we found strong genetic interaction between VPS13(D716H) and VPS39; the synthetic lethality between VPS39 and MMM1 was not alleviated by VPS13(D716H) (Fig. S2 A), indicating that Vps13(D716H) needs Vps39 for its suppression function. As our understanding of vCLAMP biology remains sparse, Vps13, as the first marker of natural vCLAMPs, may prove an invaluable tool for future characterization.
Because Mcp protein overexpression can suppress ERMES mutant phenotypes, we addressed whether they were required for Vps13 suppressor function by crossing VPS13(D716H) to mcp1Δ and mcp2Δ. Whereas Mcp2 deletion did not impair Vps13-mediated suppression, Mcp1 behaved as Vps39; mcp1Δ and mmm1Δ were synthetic lethal and this was not alleviated by VPS13(D716H) (Fig. S2, B and C). Thus, while Mcp2 is clearly part of another pathway, Mcp1 may function in the same pathway as Vps13 and Vps39.
It is surprising that gain-of-function mutations in VPS13 arise frequently (Table 1), as it is generally easier to destroy a function than to create a new one. One way to generate gain-of-function mutations is through loss of a regulatory module (Wilkie, 1994). This could explain the appearance of suppressors; the loss of a regulatory module may reassign Vps13 to serve primarily in the vCLAMP pathway, thereby increasing the pathway’s output and rendering ERMES dispensable. This conjecture is supported by the fact that Vps13(D716H)^GFP and Vps13(L1627S)^GFP fail to relocalize to NVJs. However, preventing relocalization by deleting NVJ1 does not confer a suppressor phenotype. This indicates that failure to relocalize to NVJ might not be the cause but rather a consequence of the suppression phenomenon. Thus, the link between relocalization and suppression is complex and needs to be clarified.
We show here that Vps13 links vCLAMPs, NVJs, and ERMES. These unsuspected connections suggest that membrane contact sites act in a networked fashion involving redundancy and cross-regulation. Severing one interorganelle link may reinforce another, and conversely reinforcing an interorganelle link may render another link dispensable, as we have shown here. It is plausible that such redundant interconnectivity is a widespread principle currently under-appreciated, which complicates the phenotypical characterization of contact site mutants.
Because the ERMES complex bears lipid transporters (Kopec et al., 2010; Schauder et al., 2014) and because vCLAMPs are partially redundant with ERMES for mitochondrial lipid supply (Elbaz-Alon et al., 2014), it is tempting to speculate that suppression occurs by restoring lipid homeostasis in mitochondria. We have performed lipid measurement assays in strains bearing a deletion of VPS13 and a thermosensitive allele of mmm1, either assessing steady-state lipid levels or phosphatidylcholine biosynthesis rates using standard assays. We have not been able to evidence any gross abnormality (Fig. S3). Although these are, in essence, negative results, they may indicate that, rather than playing a direct role in lipid exchange, Vps13 may serve as a regulator of contact sites; by fine-tuning their lipid transport activity, Vps13 may compensate for the loss of ERMES, without necessarily causing measurable lipid changes. More research on the biochemical activity of ERMES and of its backup pathways is needed to clarify this point.
Most ERMES components, though present in the common ancestor of metazoans and fungi, have been lost in metazoans (Flinner et al., 2013; Wideman et al., 2013), indicating that, at one point in evolution, ERMES has been rendered dispensable. Our data show how ERMES can be bypassed and suggest that in metazoans, Vps13 participates in functions performed by ERMES complexes in fungi. This conjecture may advance our understanding of human VPS13 orthologues, some of which are linked to familial neurological diseases. In the light of our data, it is possible that, akin to many other neurological diseases (Kwong et al., 2006), the molecular causes of disease could result from mitochondrial defects.
Materials and methods
Yeast strains and plasmids
Yeast genetics were performed using classical methods. To keep the strain background clean of any unwanted spontaneous suppressors, we kept ERMES mutant strains as heterozygous diploids, thereby avoiding any selective pressure on the appearance of suppressors. All genetic manipulations were performed in the heterozygous diploid, which was subsequently sporulated to generate haploid strains of the appropriate genotypes. In every case, multiple spores of the scored phenotype were analyzed. Deletions were made using the Longtine toolbox (Longtine et al., 1998), while internal GFP-tagging of VPS13 was done using the Gauss toolbox (Gauss et al., 2005). Yeast strains, oligonucleotides, and plasmids used in this work are listed in Tables S1, S2, and S3, respectively. The pVPS13 plasmid (originally named pSOI1-1), containing the full VPS13 gene cloned in a CEN/ARS LEU2 plasmid, was a gift from R.S. Fuller and M. De (University of Michigan, Ann Arbor, MI). The pVPS13(D716H) and pVPS13(L1627S) plasmids were generated by introducing the relevant alleles into pVPS13 by gap-repair cloning. The mtBFP plasmid was a gift of C. Osman (University of California, San Francisco, San Francisco, CA). The mmm1-1 thermosensitive strain was provided by R.E. Jensen and H. Sesaki (John Hopkins School of Medicine, Baltimore, MD), and the GAL1-VPS13 strain was provided by J. Svejstrup (Francis Crick Institute, London, England, UK).
Whole genome resequencing
DNA from 20 SUP− and 20 SUP+ strains originating from the same cross was extracted according to standard protocol (Hoffman and Winston, 1987) and pooled in equal proportions. DNA sequencing was performed by paired-end Illumina sequencing according to the manufacturer’s protocol. Reads were aligned to the reference yeast genome (S288C_reference_genome_R64-1-1_20110203) using Bowtie (Langmead et al., 2009), and SNPs were identified using SAMtools (Li et al., 2009) with default parameters settings. The resulting variant call format (VCF) file for the SUP+ and SUP− pools was imported into a spreadsheet program for comparison and plotting.
Microscopy
Yeast cells were grown in exponential phase in SD complete SD-Uracil (for selection of the pVTU-mtDsRed or pVTU-mtBFP plasmids) or SD-Leu (for selection of pVPS13 derivatives), and imaged on a microscope (DeltaVision MPX; Applied Precision) equipped with a 60× 1.42 NA oil Plan-ApoN or a 100× 1.40 NA oil UplanS-Apo objective lens (Olympus), a multicolor solid state illumination light source, a CoolSNAP HQ2 camera (Roper Scientific), and an incubator set at 30°C. Acquisition was performed with the SoftWoRx software and deconvoluted using the manufacturer’s parameters. Subsequent analysis was performed using the Fiji ImageJ bundle. Vacuolar and DNA staining was performed by incubating exponentially growing cells in SD medium containing 1.7 ng/µl FM4-64 (Life Technologies), 0.1 mM CellTracker Blue CMAC (Life Technologies), or 2 µg/ml DAPI at 30°C for 20 min in the dark. Cells were then washed once in SD medium. For FM4-64 staining, cells were incubated for another 20 min in dye-free medium before imaging. Cells were then mounted between a slide and a coverslip for direct live imaging. For quantification of mitochondrial shape, z sections of the whole cell were stacked using a maximal intensity projection. Mitochondria were detected using ImageJ particle analysis, which outputs mitochondrial perimeters and areas used to compute the data in Fig. 2 B. The procedure was performed by Script S1. All relevant parameters are contained therein. For the quantification of the amount of mtDNA per cell, DAPI fluorescence was imaged as z sections of the whole cell, which were then stacked using a maximal intensity projection. Colocalization analysis between Vps13^GFP, the vacuoles and the mitochondria was performed using the ImageJ Script S2.
Online supplemental material
Fig. S1 shows the synthetic lethality between mmm1Δ and C-terminal GFP fusion, N-terminal GFP fusion, or deletion of VPS13. Fig. S2 shows the genetic interaction between mmm1Δ VPS13(D716H) and vps39Δ, mcp1Δ, or mcp2Δ. Fig. S3 shows the kinetics of phosphatidylcholine biosynthesis. Table S1 summarizes the yeast strains used in this study. Table S2 summarizes the primers used in this study. Table S3 summarizes the plasmids used in this study. Script S1 is an ImageJ macro used to calculate mitochondrial shape quotient. Script S2 is an ImageJ macro used to quantify Vps13 localization at the mitochondria and vacuole. Script S3 is an ImageJ macro used to measure the percentage of Vps13 at NVJs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502105/DC1.
Supplementary Material
Acknowledgements
We are grateful to A.H. Michel, M. Schuldiner, M. Peter, and the Kornmann laboratory for helpful discussions; and to R.S. Fuller, M. De, C. Osman, R. Jensen, H. Sesaki, and J. Svejstrup for strains and plasmids. Sequencing was done at the Functional Genomics Center Zürich. Imaging was performed at the ETH Zürich ScopeM facility.
This work was supported by the Swiss National Science Foundation (grant #PP00P3_133651) and the European Research Council (ERC-2013-StG 337906-OrgaNet) to B. Kornmann. P. Walter is an Investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ERMES
- ER–mitochondria encounter structure
- MDM
- mitochondrial distribution and morphology
- MMM
- mitochondria morphology maintenance
- mtDNA
- mitochondrial DNA
- NVJ
- nuclear–vacuole junction
- SNP
- single-nucleotide polymorphism
- SUP
- suppressor
- vCLAMP
- vacuole and mitochondria patch
- VPS
- vacuolar protein sorting
- WT
- wild type
References
- Aiken Hobbs A.E., Srinivasan M., McCaffery J.M., and Jensen R.E.. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410. 10.1083/jcb.152.2.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V.A., Johnson L.M., and Emr S.D.. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA. 83:9075–9079. 10.1073/pnas.83.23.9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.H., Sogo L.F., and Yaffe M.P.. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136:545–553. 10.1083/jcb.136.3.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckler S., and Westermann B.. 2014. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell. 28:450–458. 10.1016/j.devcel.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Brickner J.H., and Fuller R.S.. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36. 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.M., Delannoy M., and Jensen R.E.. 1994. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126:1375–1391. 10.1083/jcb.126.6.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L.Y., Toufighi K., Mostafavi S., et al. 2010. The genetic landscape of a cell. Science. 327:425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Flinner N., Ellenrieder L., Stiller S.B., Becker T., Schleiff E., and Mirus O.. 2013. Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim. Biophys. Acta. 1833:3314–3325. 10.1016/j.bbamcr.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Gauss R., Trautwein M., Sommer T., and Spang A.. 2005. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 22:1–12. 10.1002/yea.1187 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Helle S.C.J., Kanfer G., Kolar K., Lang A., Michel A.H., and Kornmann B.. 2013. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 1833:2526–2541. 10.1016/j.bbamcr.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Hoffman C.S., and Winston F.. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 57:267–272. 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., and Nunnari J.. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195:323–340. 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Kopec K.O., Alva V., and Lupas A.N.. 2010. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 26:1927–1931. 10.1093/bioinformatics/btq326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong J.Q., Beal M.F., and Manfredi G.. 2006. The role of mitochondria in inherited neurodegenerative diseases. J. Neurochem. 97:1659–1675. 10.1111/j.1471-4159.2006.03990.x [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T.P., and Munro S.. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 12:1633–1644. 10.1091/mbc.12.6.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Meeusen S., and Nunnari J.. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510. 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Pfannschmidt S., Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M.J., Jensen R.E., Chacinska A., Guiard B., et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26:2229–2239. 10.1038/sj.emboj.7601673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A.H., and Kornmann B.. 2012. The ERMES complex and ER-mitochondria connections. Biochem. Soc. Trans. 40:445–450. 10.1042/BST20110758 [DOI] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Lewandowska A., Choi J.-Y., Markgraf D.F., Junker M., Bilgin M., Ejsing C.S., Voelker D.R., Rapoport T.A., and Shaw J.M.. 2012. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 13:880–890. 10.1111/j.1600-0854.2012.01352.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., and Neiman A.M.. 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125:3004–3011. 10.1242/jcs.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., and Goldfarb D.S.. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141. 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., and Reinisch K.M.. 2014. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 510:552–555. 10.1038/nature13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo L.F., and Yaffe M.P.. 1994. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126:1361–1373. 10.1083/jcb.126.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T., Ozbalci C., Brügger B., Rapaport D., and Dimmer K.S.. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126:3563–3574. 10.1242/jcs.121244 [DOI] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2012. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125:49–58. 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza A., Vettori A., Copley R.R., Dobson-Stone C., and Monaco A.P.. 2004. Analysis of the human VPS13 gene family. Genomics. 84:536–549. 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Wideman J.G., Gawryluk R.M.R., Gray M.W., and Dacks J.B.. 2013. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol. Biol. Evol. 30:2044–2049. 10.1093/molbev/mst120 [DOI] [PubMed] [Google Scholar]
- Wilkie A.O. 1994. The molecular basis of genetic dominance. J. Med. Genet. 31:89–98. 10.1136/jmg.31.2.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman M.J., Hobbs A.E.A., Burgess S.M., Srinivasan M., and Jensen R.E.. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164:677–688. 10.1083/jcb.200308012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.