Abstract
Background
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are characterized by chronic intestinal inflammation due to immunological, microbial and environmental factors in genetically predisposed individuals. Advances in the diagnosis, prognosis and treatment of IBD require the identification of robust biomarkers that can be used for molecular classification of diverse disease presentations. We previously identified five genes, RELA, TNFAIP3 (A20), PIGR, TNF and IL8, whose mRNA levels in colonic mucosal biopsies could be used in a multivariate analysis to classify patients with Crohn’s disease (CD) based on disease behavior and responses to therapy.
Aim
We compared expression of these 5 biomarkers in IBD patients classified as having CD or ulcerative colitis (UC), and in healthy controls.
Results
Patients with CD were characterized as having decreased median expression of TNFAIP3, PIGR and TNF in non-inflamed colonic mucosa as compared to healthy controls. By contrast, UC patients exhibited decreased expression of PIGR and elevated expression of IL8 in colonic mucosa compared to healthy controls. A multivariate analysis combining mRNA levels for all 5 genes resulted in segregation of individuals based on disease presentation (CD vs. UC) as well as severity, i.e., patients in remission vs. those with acute colitis at the time of biopsy.
Conclusion
We propose that this approach could be used as a model for molecular classification of IBD patients, which could further be enhanced by the inclusion of additional genes that are identified by functional studies, global gene expression analyses, and genome-wide association studies.
Keywords: Crohn’s disease, ulcerative colitis, colonic mucosa, mRNA biomarkers, multivariate analysis
Introduction
Inflammatory bowel diseases (IBD) represent a spectrum of disorders characterized by chronic gut inflammation in individuals with genetic and/or environmental risk factors [1, 2]. At one end of the clinical spectrum in IBD is Crohn’s disease (CD), defined by discontinuous, transmural, frequently granulomatous intestinal inflammation, which can involve the small and/or large bowel [3, 4]. At the other end of the spectrum is ulcerative colitis (UC), defined by continuous mucosal inflammation without granulomas, extending from the rectum to varying lengths into the colon [5, 6]. Individual presentations of IBD can vary along this spectrum, and can range in severity from mild to life-threatening. While differential diagnosis of CD, UC or IBD type unclassified is critical for clinical management of patients [7, 8], it has been estimated that up to 10% of patients presenting with IBD-associated colitis cannot be classified differentially using current diagnostic paradigms based on clinical, endoscopic and radiologic criteria [9]. It has been proposed that assessment of molecular markers in the evaluation of IBD patients may be a valuable complement to clinical criteria, and may better explain changes in disease behavior, particularly when several markers are combined [10, 11].
We previously reported that a multivariate analysis of mRNA levels in colonic mucosa for five “signature” genes, comprising RELA, TNFAIP3 (a/k/a A20), PIGR, TNF and IL8, was predictive of disease behavior and responses to therapy in CD patients [12]. These candidate genes were chosen based on their functional relevance to intestinal homeostasis and IBD. The RELA gene encodes the p65/RelA subunit of the nuclear factor (NF)-κB signaling complex that regulates epithelial barrier function but can also be associated with pathological inflammation in the intestine [13, 14]. NF-κB signaling induces transcription of the TNFAIP3 gene, encoding a ubiquitin-modifying enzyme (also known as A20) that negatively regulates the NF-κB signaling pathway [15–20]. The PIGR gene encodes the polymeric immunoglobulin receptor, which promotes mucosal homeostasis by transporting protective secretory (S)IgA antibodies across intestinal epithelial cells into the gut lumen [21–25]. The TNF gene encodes the pro-inflammatory cytokine tumor necrosis factor, which has been associated with mucosal inflammation and is the target of therapeutic antibodies for treatment of IBD [26–28]. However, TNF also has protective roles in innate immunity, including epithelial restitution, control of potentially pathogenic luminal bacteria and induction of pIgR expression [29–31]. The IL8 gene encodes interleukin (IL)−8 (also known as CXCL8), which is a potent chemoattractant for neutrophils, a major component of the cellular infiltrate in acute intestinal inflammation [32]. Polymorphisms in the RELA, TNFAIP3, PIGR, TNF and IL8 genes have been identified within genetic loci associated with increased risk for IBD [33, 34]. We hypothesize that genetic and/or environmental factors may influence expression of these genes in the colonic mucosa of IBD patients, and that altered levels of gene expression may influence the clinical course of IBD and responses to therapy.
In the current study, we expanded our study population to include individuals with UC as well as CD. The purpose of the present study was threefold: first, to evaluate the utility of these 5 mRNA biomarkers in colonic mucosa for predicting clinical outcome in IBD patients; second, to examine potential differences in biomarker expression between CD and UC patients; and third, to validate the predictive value of a multivariate approach with combined biomarkers as opposed to single biomarkers.
Materials and Methods
Study Individuals
Colonic mucosal biopsies were obtained from subjects undergoing colonoscopy at the University of Kentucky Medical Center, following written informed consent approved by the Institutional Review Board. The diagnosis of UC was based on clinical, endoscopic, radiological and histopathological criteria. The presence of active disease was based on endoscopic evidence of inflammation. Biopsies were collected from regions of the colon that were visibly inflamed and non-inflamed, as subsequently confirmed by histological examination. Control subjects underwent colonoscopies for evaluation of constipation or chronic abdominal pain or for routine screening for colon cancer. Patients were classified as normal when endoscopic, radiologic and/or histological evaluation detected no colonic disease.
Analysis of mRNA Levels in Colonic Mucosal Biopsies
Biopsied tissue samples were collected into an RNA-stabilizing solution (RNAlater, Qiagen, Germantown, MD) and stored at −80°C until analyzed. Total RNA was purified using the RNeasy Protect mini kit (Qiagen) and reverse-transcribed to generate cDNA templates using the TaqMan Gold RT-PCR kit (Applied Biosystems, Foster City, CA). Levels of individual mRNA transcripts were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR), using the ABI Prism 7700 Sequence Detection System (Applied Biosystems) as previously described [12]. The threshold cycles (CT) for test mRNAs were normalized to the CT for β2-microglobulin according to the formula: 2-(CTtest-CTβ2-microglobulin) × 100%.
Statistical Analyses
Human gene expression data were analyzed by non-parametric Mann-Whitney and Spearman correlation tests (http://www.winstat.com). Comparisons between gene expression in paired biopsies from non-inflamed and inflamed colonic mucosa were made using the Wilcoxon signed-rank test (http://vassarstats.net/wilcoxon.html). Multifactorial principal component analyses (PCA) of gene expression data were conducted using Multibase statistical analysis software (http://www.numericaldynamics.com).
Results
Biomarker expression in colonic mucosa of IBD patients and healthy controls
To determine whether mRNA levels for 5 candidate genes could be used to classify IBD patients into clinically relevant subsets, biopsies of colonic mucosa were collected during colonoscopy from 113 IBD patients (60 CD, 53 UC) and 44 healthy controls (Table 1). Diagnosis of CD or UC was based on clinical, radiological, and endoscopic criteria according to the Montreal classification [35], supported by histopathological findings. IBD patients were sub-classified into those with or without acute inflammation, as evidenced by areas of macroscopically visible tissue inflammation on colonoscopy. We hypothesized that changes in gene expression throughout the colonic epithelium of IBD patients could promote development of an inflammatory response, and would not necessarily be a consequence of localized inflammation. Accordingly, comparisons in gene expression between IBD patients and healthy controls were restricted to those regions of the colon that were macroscopically and microscopically non-inflamed. Although the median age of the cohort of healthy controls was higher than that of IBD patients (Table 1), there was no significant correlation between age at the time of biopsy and expression of RELA, TNFAIP3, PIGR or TNF (Table 2). However, younger subjects in general had significantly higher levels of IL8 expression in non-inflamed colonic mucosa, as evidenced by the negative correlation with age. Within individuals, levels of RELA, TNFAIP3 and PIGR mRNA were highly correlated, whereas expression of these genes was not correlated with IL8 (Table 2). Interestingly, expression of TNF was correlated with expression of all the other biomarkers within individuals. This finding is consistent with the hypothesis that elevated expression of RelA enhances transcription of the TNFAIP3, PIGR and TNF genes through the classical NF-κB signaling pathway. Other signaling pathways may also enhance transcription of the TNF and IL8 genes. While expression levels of individual biomarkers varied considerably among individuals, some differences were noted between clinical groups (Fig. 1). Median levels of TNFAIP3 and TNF mRNA were significantly lower in CD patients but not UC patients, compared to healthy controls. Interestingly, median levels of PIGR mRNA were reduced in both CD and UC patients compared to healthy controls. Although levels of RELA mRNA were correlated with levels of TNFAIP3, PIGR and TNF mRNA within all individuals (Table 2), the wide variation in RELA expression among individuals precluded the finding of significant differences among clinical groups. Although some individual CD and UC patients had elevated expression of IL8 mRNA, median levels for each category did not differ significantly from that of healthy controls. The finding that median IL8 mRNA levels were significantly elevated in UC patients with acute inflammation compared to UC patients in remission suggests that increased expression of IL8 may be associated with the inflammatory process in UC. This concept was supported by the finding that IL8 levels in UC patients, but not CD patients, were significantly elevated in biopsies from visibly inflamed colonic mucosa compared to paired samples from non-inflamed colonic mucosa from the same individuals (Table 3). By contrast, no significant differences were observed in the levels of RELA, TNFAIP3, PIGR or TNF between inflamed and non-inflamed colonic mucosa in either CD or UC patients.
Table 1.
Patient characteristics
| Diagnosis | Number of Subjects |
Age | Gender | ||
|---|---|---|---|---|---|
| Median | Range | Male | Female | ||
| Control | 44 | 54 | 17 – 78 | 34.1% | 65.9% |
| CD in remission | 42 | 40 | 20 – 64 | 36.4% | 63.6% |
| CD with acute inflammation | 18 | 31 | 19 – 72 | 50.0% | 50.0% |
| UC in remission | 34 | 39 | 24 – 65 | 45.5% | 54.5% |
| UC with acute inflammation | 19 | 38 | 26 – 61 | 71.4% | 28.6% |
Table 2.
Correlation analysis of biomarker expression and age at biopsy1
| Variable | RELA | TNFAIP3 (A20) | PIGR | TNF | IL8 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | R | P | r | P | |
| - | ||||||||||
| Age at biopsy |
−0.009 | 0.468 | 0.091 | 0.206 | 0.153 | 0.082 | 0.021 | 0.424 | −0.274 | 0.006 |
| RELA | 0.618 | 7.80E-17 | 0.479 | 6.14E-10 | 0.392 | 6.05E-07 | 0.044 | 0.300 | ||
| TNFAIP3 | 0.594 | 2.20E-15 | 0.351 | 8.09E-06 | 0.119 | 0.079 | ||||
| PIGR | 0.246 | 0.001 | 0.036 | 0.336 | ||||||
| TNF | 0.362 | 4.12E-06 | ||||||||
Fig. 1.
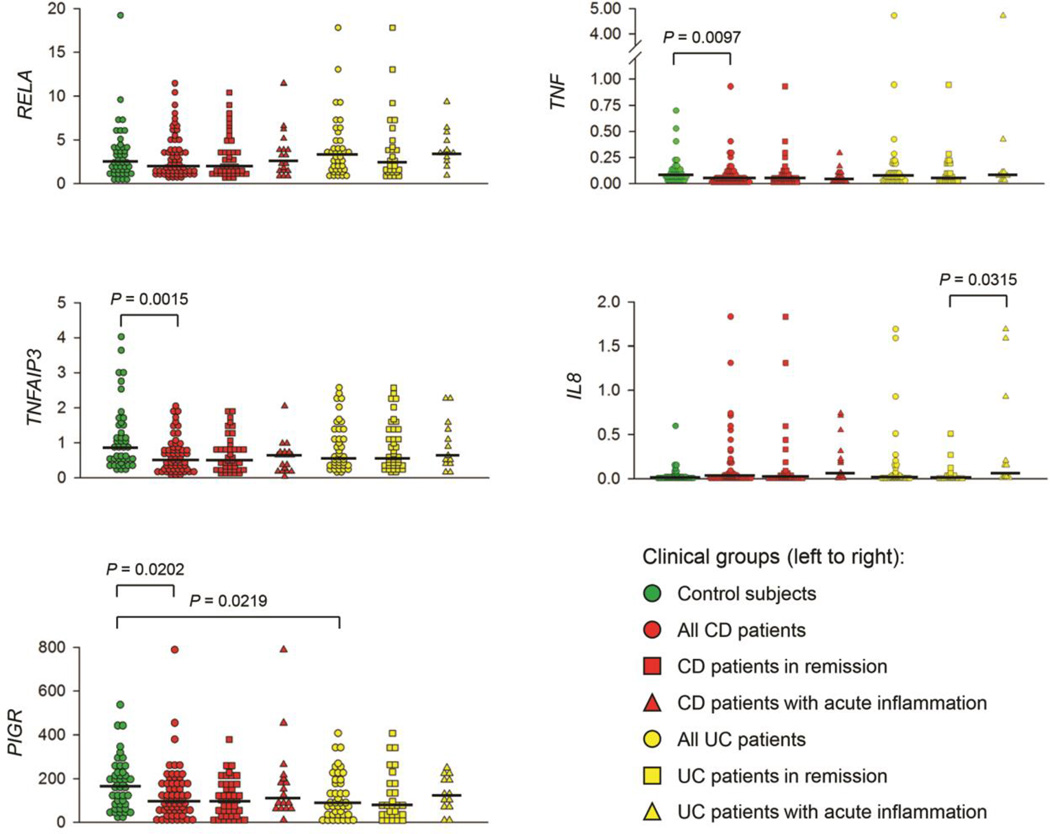
Expression of 5 biomarkers in colonic mucosa of UC patients and healthy controls. CD and UC patients were sub-classified as being in remission or with acute inflammation, based on macroscopic evidence of localized tissue inflammation during colonoscopy. For the purpose of this analysis, biopsies were obtained from regions of the colon of CD and UC patients that were not visibly inflamed, and absence of inflammation in these biopsies was confirmed microscopically. Levels of mRNA were measured by qRT-PCR and normalized to β2-microglobulin. Horizontal lines indicate the median level of mRNA for each group. Significant differences among groups were determined by the Mann-Whitney non-parametric test.
Table 3.
Comparison of biomarker expression in paired biopsies of non-inflamed and inflamed colonic mucosa from CD and UC patients1
| CD | UC | ||||
|---|---|---|---|---|---|
| Gene | Sample | Median | P-value | Median | P-value |
| RELA | Non-inflamed | 1.714 | 0.629 | 3.443 | 0.215 |
| Inflamed | 1.905 | 2.105 | |||
| TNFAIP3 | Non-inflamed | 0.441 | 0.119 | 0.643 | 0.889 |
| Inflamed | 0.382 | 0.450 | |||
| PIGR | Non-inflamed | 96.4 | 0.332 | 127.5 | 0.522 |
| Inflamed | 75.0 | 104.2 | |||
| TNF | Non-inflamed | 0.047 | 0.332 | 0.081 | 0.072 |
| Inflamed | 0.059 | 0.119 | |||
| IL8 | Non-inflamed | 0.038 | 0.791 | 0.025 | 0.049 |
| Inflamed | 0.045 | 0.757 | |||
Expression levels for biomarkers were analyzed as described in Fig. 1 in paired biopsies from regions of non-inflamed and inflamed colon from 17 CD and 15 UC patients. Statistical differences were determined by non-parametric Wilcoxon signed rank test; P-values < 0.05 were considered statistically significant and are noted in bold.
Multivariate analysis of biomarker expression in colonic mucosa
Given the wide variability in expression of individual biomarkers within groups in our cohort, we hypothesized that a multivariate approach would be more useful for predicting clinical outcome in IBD patients. Accordingly, the composite data for expression of the 5 biomarkers were reduced to 2 “supervariables,” or principal components (PCs) for each individual (Table 4). The results of the PCA indicated that PC1 accounted for 40.7% of the overall variance in expression of the 5 biomarkers in this population of individuals, while PC2 accounted for 27.8%. For each individual, the value of PC1 and PC2 is calculated as the sum of normalized mRNA level for each gene, multiplied by the weighting coefficient (on a scale of −1 to +1) for that gene. As noted in Table 4, PC1 had strongly negative coefficients for RELA, TNFAIP3 and PIGR, and moderately negative coefficients for TNF and IL8. In other words, high expression levels for RELA, TNFAIP3 and/or PIGR (and to a lesser extent TNF and IL8) in an individual subject would result in a low score for PC1. By contrast, PC2 had strongly positive coefficients for TNF and IL8, a moderately negative coefficient for TNFAIP3, and weakly negative coefficients for RELA and PIGR. Therefore, high expression of TNF and/or IL8 in any individual subject would result in a high score for PC2, which could be lowered somewhat by high expression of TNFAIP3 and to a lesser extent RELA or PIGR.
Table 4.
Principal component analysis of biomarker expression in non-inflamed colonic mucosa of IBD patients and healthy controls1
| Principal Component Analysis | PC1 | PC2 |
|---|---|---|
| Contribution to overall variance | 40.7% | 27.8% |
| Eigenvalue | 1.64 | 1.12 |
| Weighting Coefficients | ||
| Genes | PC1 | PC2 |
| RELA | −0.544 | −0.050 |
| TNFAIP3 | −0.588 | −0.197 |
| PIGR | −0.563 | −0.089 |
| TNF | −0.166 | 0.682 |
| IL8 | −0.115 | 0.697 |
Expression levels for biomarkers were analyzed in biopsies from regions of non-inflamed colon as described in Fig. 1. Data for individual genes were combined for all study subjects and reduced to 2 principal components (PCs) as described in Methods. Weighting coefficients for each gene are listed for PC1 and PC2.
To examine the distribution of PC scores among clinical groups, a 2-dimensional scatter plot was constructed showing PC1 and PC2 scores for each individual (Fig. 2). This plot was divided into 4 quadrants based on positive or negative values for PC1 and PC2. By examining the distribution of individuals within each clinical group among the 4 quadrants, it can be determined whether levels of the composite variables PC1 and PC2 are useful for predicting clinical outcome. Chi-square analysis was used to determine whether individuals in different clinical groups were evenly distributed among the 4 quadrants, or whether there was a bias toward one or more quadrants (Table 5A). Healthy control subjects were distributed roughly equally between quadrant A (41%) and quadrant B (48%) and only minimally represented in quadrant C (7%) and quadrant D (5%). This pattern indicates that the majority of healthy controls had low values for PC2, but evenly distributed values for PC1. By contrast, CD patients were more heavily distributed in quadrant B (52%), followed by quadrant A (24%), quadrant C (18%) and quadrant D (8%). However, the subgroup of CD patients with clinical evidence of acute inflammation was more evenly distributed among the 4 quadrants (as noted by the non-significant chi-square value), with the greatest numbers found in quadrant B (44%) and quadrant C (33%). The distribution of all UC patients was similar to that of all CD patients in that there was a bias toward quadrant B (45%) followed by quadrant A (28%), but UC patients had a more even distribution between quadrants C (13%) and D (15%). As was the case with CD, UC patients with clinical evidence of acute inflammation were more evenly distributed among the 4 quadrants, as noted by the non-significant chi-square value.
Fig 2.
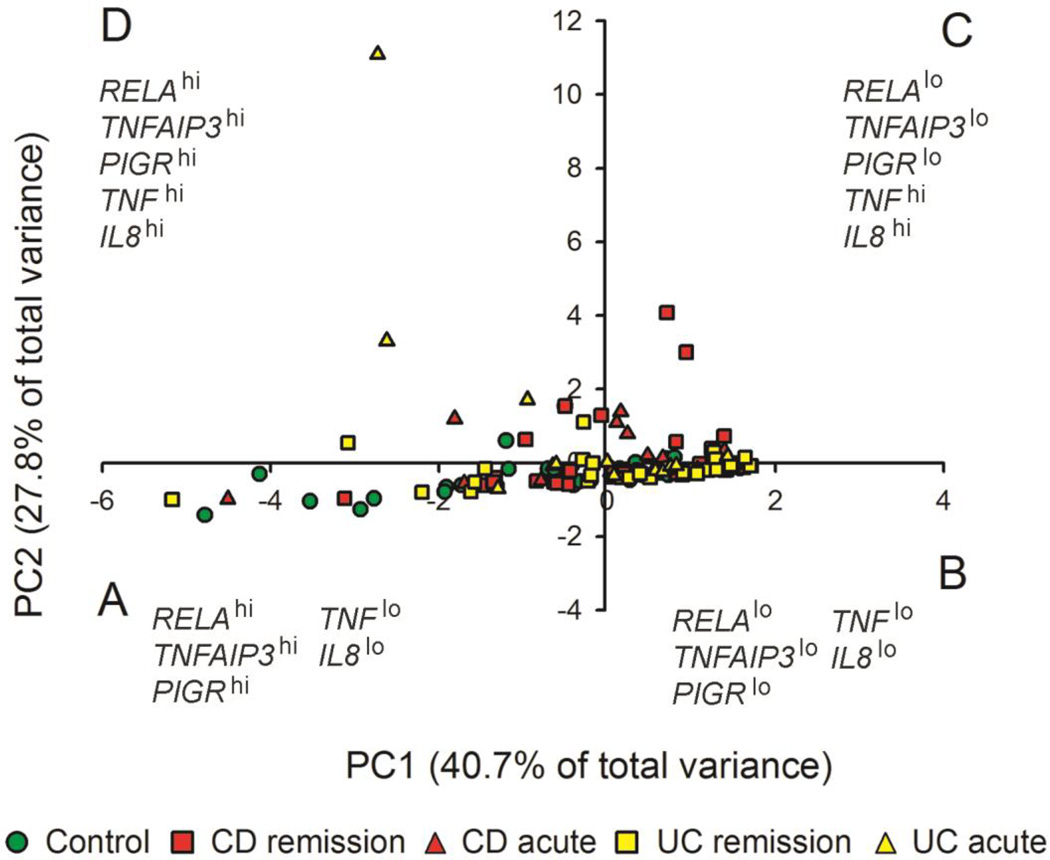
Principal component analysis of gene expression in colonic mucosa. Levels of mRNA for 5 biomarkers were analyzed as described for Fig. 1, combined for all subjects, then reduced to 2 principal components (PCs) for each individual based on the sum of the normalized expression level multiplied by the weighting coefficient for each gene (Table 4). Symbols denote individual subjects, including healthy controls (green), CD patients (red) and UC patients (yellow). CD or UC patients in remission are denoted by squares, and patients with acute inflammation are denoted by triangles. Individuals were distributed into 4 quadrants based on their scores for PC1 and PC2: A, PC1 < 0, PC2 < 0; B, PC1 > 0, PC2 < 0; C, PC1 > 0, PC2 > 0; D, PC1 < 0, PC2 > 0.
Table 5.
Chi-square analysis of distribution of study subjects based on principal component analysis of biomarker expression in non-inflamed colonic mucosa1
| A. Distribution of clinical groups into PCA quadrants23 | ||||||
|---|---|---|---|---|---|---|
| Fraction within each quadrant | ||||||
| Clinical group | A | B | C | D | Chi-square | P-value |
| Control subjects | 0.41 | 0.48 | 0.07 | 0.04 | 26.73 | 6.72E-06 |
| All CD patients | 0.22 | 0.52 | 0.18 | 0.08 | 25.07 | 1.50E-05 |
| CD remission | 0.24 | 0.55 | 0.12 | 0.09 | 21.81 | 7.15E-05 |
| CD acute | 0.17 | 0.44 | 0.33 | 0.06 | 6.44 | 0.092 |
| All UC patients | 0.28 | 0.45 | 0.12 | 0.15 | 10.60 | 0.014 |
| UC remission | 0.30 | 0.48 | 0.11 | 0.11 | 10.19 | 0.017 |
| UC acute | 0.23 | 0.39 | 0.15 | 0.23 | 1.46 | 0.691 |
| B. Comparison of clinical groups within each PCA quadrant4 | |||||
|---|---|---|---|---|---|
| Quadrant | |||||
| Comparison | A | B | C | D | |
| Control vs. CD | 5.73 | 0.16 | 4.84 | 0.69 | Chi-square |
| 0.017 | 0.689 | 0.028 | 0.405 | P-value | |
| CD in remission | 1.20 | 1.22 | 9.80 | 1.00 | Chi-square |
| vs. CD acute | 0.274 | 0.269 | 0.002 | 0.317 | P-value |
| Control vs. UC | 2.45 | 0.10 | 1.80 | 5.00 | Chi-square |
| 0.118 | 0.756 | 0.180 | 0.025 | P-value | |
| UC in remission | 0.92 | 1.16 | 0.62 | 4.24 | Chi-square |
| vs. UC acute | 0.336 | 0.281 | 0.433 | 0.040 | P-value |
Levels of mRNA for RELA, TNFAIP3, PIGR, TNF and IL8 were analyzed as described in Fig. 1, and reduced to 2 principal components (PCs) as described in Table 4.
A 2-dimensional ordination plot was used to categorize study subjects into 4 quadrants based on individual scores for PC1 and PC2, as described in Fig. 2: A, PC1 < 0, PC2 < 0; B, PC1 > 0, PC2 < 0; C, PC1 > 0, PC2 > 0; D, PC1 < 0, PC2 > 0.
This chi-square analysis tests the null hypothesis that subjects within each clinical group are evenly distributed among the 4 quadrants. Values shown in bold are statistically significant (p < 0.05).
This chi-square analysis tests the null hypothesis that subjects within each quadrant are evenly distributed between the indicated clinical groups. Values shown in bold are statistically significant (p < 0.05).
Another approach for evaluation of the 2-dimensional PCA data was to compare the distribution of individuals in different clinical groups within each quadrant (Table 5B). Within quadrant A (PC1 < 0, PC2 < 0), there was a significantly higher proportion of healthy controls compared to CD patients (p = 0.017). However, this did not hold true for UC patients compared to healthy controls, nor were there significant differences within quadrant A between CD or UC patients with or without acute inflammation. Within quadrant B (PC1 > 0, PC2 < 0), there were no significant differences among any of the groups. Interestingly, a bias toward active CD was seen in quadrant C (PC1 > 0, PC2 > 0), as demonstrated by a significantly higher proportion of CD patients compared to healthy controls (p = 0.028), and an even greater proportion of CD patients with acute inflammation compared to CD patients in remission (p = 0.002). By contrast, a bias toward UC was seen in quadrant D (PC1 < 0, PC2 > 0), as demonstrated by a significantly higher proportion of UC patients compared to healthy controls (p = 0.025), as well as a significantly higher proportion of UC patients with acute inflammation compared to UC patients in remission (p = 0.04). In summary, a multivariate approach combining 5 biomarkers was more predictive of clinical outcome than was any single biomarker. A characteristic of both CD and UC was a shift toward elevated levels of PC2 (quadrants C and D), which could result from elevated expression of TNF and/or IL8, and could be mitigated somewhat by increased expression of TNFAIP3. The finding that healthy controls were more highly represented than were CD patients in quadrant A (PC1 < 0, PC2 < 0) suggested that lower levels of PC1, resulting from elevated expression of RELA, TNFAIP3 and/or PIGR, could be protective against inflammation in this disease. This conclusion is consistent with the greater representation of CD patients with acute inflammation than those in remission in quadrant C (PC1 > 0, PC2 > 0). However, low PC1 values were not associated with improved outcome in UC patients, as evidenced by no significant difference from healthy controls in quadrant A (PC1 < 0, PC2 < 0), and the greater representation of UC patients with acute inflammation compared to those in remission in quadrant D (PC1 < 0, PC2 > 0). These differences between CD and UC patients in overall patterns of biomarker expression, which were not revealed by analysis of single biomarkers, may represent inherent differences in the underlying pathology of the two forms of IBD.
Discussion
The findings from the present study extend our earlier findings on gene expression in CD patients [12], and reveal interesting differences in gene expression between patients with CD and UC (Fig. 1). In contrast to our previous finding with a smaller study population, we did not find a significant reduction in RELA mRNA in CD patients compared to healthy controls. However, consistent with our earlier study, we found that median levels of TNFAIP3, PIGR and TNF mRNA in non-inflamed colonic mucosa were significantly lower in CD patients compared to healthy controls. In this context it is important to note that TNFAIP3, PIGR and TNF are target genes of the RelA-dependent NF-κB signaling pathway in intestinal epithelial cells [36]. Therefore, decreased transcription of the TNFAIP3, PIGR and TNF genes could result either from decreased levels of RelA protein or from suppression of the RelA signaling pathway, and these regulatory events could differ from patient to patient. Interestingly, median levels of PIGR mRNA were decreased in UC patients, but levels of RELA, TNFAIP3 and TNF mRNA were not significantly different between UC patients and healthy controls. The finding of decreased PIGR mRNA in the colonic mucosa of both CD and UC patients is consistent with important role of protective SIgA antibodies in protection against intestinal inflammation [24]. We and others have reported that targeted deletion of the Pigr gene in mice is associated with alterations in the gut microbiota and increased susceptibility to chemically-induced colitis [25, 37, 38]. Surprisingly, median levels of TNFAIP3 were not reduced in the colonic mucosa of UC patients. Furthermore, median levels of mRNA encoding IL8, a pro-inflammatory factor that we observed not to be correlated with RELA, was elevated in colonic mucosa of UC patients but not CD patients. Taken together, these findings suggest that some signaling pathways controlling target gene expression may be differentially regulated in CD vs. UC, whereas other pathways, including some that regulate PIGR gene transcription, could be dysregulated in both CD and UC. Further studies of these signaling pathways in IBD patients could reveal unique features of CD and UC that could have important implications for targeted molecular therapies.
Consistent with our earlier study with CD patients [12], findings from the present study highlight the benefits of a multivariate approach in analyzing patterns of mucosal gene expression in both CD and UC patients (Table 5). When all 5 biomarkers were considered together, unique distributions were observed for patients with CD, UC and healthy controls. In contrast to the analysis with individual biomarkers, the multivariate approach distinguished patients in remission from those with acute colonic inflammation. Another useful outcome of a multivariate biomarker analysis could be the identification of specific individuals who segregate outside the range of the majority of patients within each category. Recognition of differences in patterns of gene expression among individual IBD patients could be important for development of personalized treatment strategies, and potentially for monitoring the efficacy of medical interventions. We did not observe a significant effect of medications on expression of these 5 biomarkers in our previous study of CD patients, but this could have been due to variable drug combinations and duration of treatments in our cohort [12]. In the present study, we did not observe significant effects of medications on biomarker expression in those individuals for whom medical histories were available (data not shown). However, it could prove valuable to monitor changes in biomarker expression in a prospective study under conditions where combinations, doses and duration of medications could be controlled.
Maximizing the benefit of multivariate gene expression analysis as a diagnostic, prognostic and therapeutic tool for the management of IBD patients will require the identification of sensitive and specific biomarkers. Our candidate gene approach, based on functional relevance to intestinal inflammation, has identified 5 potential biomarkers. This approach could be extended by including additional genes that are identified by functional studies, global gene expression analyses, and genome-wide association studies. Technological advances in multiplex analysis of mRNA levels in small tissue samples, accompanied by robust statistical analysis, should allow multivariate analysis of mucosal gene expression to become an important component of personalized medicine for IBD patients.
Acknowledgments
This work was supported by NIH/NIDCR grant 5P20RR020145 to M.E.C.B. and R.I.A.; and NIH grant AI069027 (and an associated American Recovery and Reinvestment Act supplement) and a Senior Research Award from the Crohn’s and Colitis Foundation of America (CCFA) to C.S.K.
Footnotes
The authors state that they have no conflicts of interest relevant to this manuscript.
Contributor Information
Maria E. C. Bruno, Email: mebrun2@uky.edu, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Lexington, Kentucky 40536. Current address: Markey Cancer Center, University of Kentucky, Lexington, Kentucky 40536.
Eric W. Rogier, Email: erogier@cdc.gov, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Lexington, Kentucky 40536. Current address: Centers for Disease Control and Prevention, Division of Parasitic Diseases and Malaria, Atlanta, GA 30333.
Razvan I. Arsenescu, Email: arsenescu.1@osu.edu, Department of Internal Medicine, University of Kentucky, Lexington, Kentucky 40536. Current address: Department of Internal Medicine, The Ohio State University, Columbus, Ohio.
Deborah R. Flomenhoft, Email: deborah.flomenhoft@uky.edu, Department of Internal Medicine, University of Kentucky, Lexington, Kentucky 40536.
Cathryn J. Kurkjian, Email: ckurkjia@email.unc.edu, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Lexington, Kentucky 40536. Current address: Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, 450 West Drive, Chapel Hill, NC 27599.
Gavin I. Ellis, Email: gavellis@mail.med.upenn.edu, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Lexington, Kentucky 40536. Current Address: Department of Microbiology, Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, 3400 Civic Center Blvd, Philadelphia, PA 19104.
Charlotte S. Kaetzel, Email: cskaet@uky.edu, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Lexington, Kentucky 40536.
References
- 1.Maher MM. Inflammatory bowel disease: review and future view. Frontiers in Bioscience (Elite Edition) 2012;4:1638–1647. doi: 10.2741/485. 1638–1647. [DOI] [PubMed] [Google Scholar]
- 2.Foersch S, Waldner MJ, Neurath MF. Innate and adaptive immunity in inflammatory bowel diseases. Dig Dis. 2013;31:317–320. doi: 10.1159/000354685. [DOI] [PubMed] [Google Scholar]
- 3.Van Assche G, Dignass A, Panes J, Beaugerie L, et al. The second European evidence-based Consensus on the diagnosis and management of Crohn’s disease: Definitions and diagnosis. Journal of Crohn’s & Colitis. 2010;4:7–27. doi: 10.1016/j.crohns.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Laass MW, Roggenbuck D, Conrad K. Diagnosis and classification of Crohn’s disease. Autoimmun Rev. 2014;13:467–471. doi: 10.1016/j.autrev.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 5.Dignass A, Eliakim R, Magro F, Maaser C, et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 1: definitions and diagnosis. Journal of Crohn’s & Colitis. 2012;6:965–990. doi: 10.1016/j.crohns.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Conrad K, Roggenbuck D, Laass MW. Diagnosis and classification of ulcerative colitis. Autoimmun Rev. 2014;13:463–466. doi: 10.1016/j.autrev.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 7.Burisch J, Munkholm P. Inflammatory bowel disease epidemiology. Curr Opin Gastroenterol. 2013;29:357–362. doi: 10.1097/MOG.0b013e32836229fb. [DOI] [PubMed] [Google Scholar]
- 8.Feakins RM. Ulcerative colitis or Crohn’s disease? Pitfalls and problems. Histopathology. 2014;64:317–335. doi: 10.1111/his.12263. [DOI] [PubMed] [Google Scholar]
- 9.Tontini GE, Vecchi M, Pastorelli L, Neurath MF, et al. Differential diagnosis in inflammatory bowel disease colitis: State of the art and future perspectives. World J Gastroenterol. 2015;21:21–46. doi: 10.3748/wjg.v21.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scaldaferri F, Correale C, Gasbarrini A, Danese S. Mucosal biomarkers in inflammatory bowel disease: key pathogenic players or disease predictors? World J Gastroenterol. 2010;16:2616–2625. doi: 10.3748/wjg.v16.i21.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeire S, Van Assche G, Rutgeerts P. Classification of inflammatory bowel disease: the old and the new. Curr Opin Gastroenterol. 2012;28:321–326. doi: 10.1097/MOG.0b013e328354be1e. [DOI] [PubMed] [Google Scholar]
- 12.Arsenescu R, Bruno ME, Rogier EW, Stefka AT, et al. Signature biomarkers in Crohn’s disease: toward a molecular classification. Mucosal Immunol. 2008;1:399–411. doi: 10.1038/mi.2008.32. [DOI] [PubMed] [Google Scholar]
- 13.Eckmann l, Neish AS. NF-κB and mucosal homeostasis. Curr Top Microbiol Immunol. 2011;349:145–58. doi: 10.1007/82_2010_103. [DOI] [PubMed] [Google Scholar]
- 14.Pasparakis M. Role of NF-kappaB in epithelial biology. Immunol Rev. 2012;246:346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 15.Vereecke L, Sze M, Mc GC, Rogiers B, et al. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolodziej LE, Lodolce JP, Chang JE, Schneider JR, et al. TNFAIP3 maintains intestinal barrier function and supports epithelial cell tight junctions. PLoS ONE. 2011;6:26352. doi: 10.1371/journal.pone.0026352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee L, Murphy SF, Kolodziej LE, Grimm WA, et al. Expression of TNFAIP3 in intestinal epithelial cells protects from DSS- but not TNBS-induced colitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G220–G227. doi: 10.1152/ajpgi.00077.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shembade N, Harhaj EW. Regulation of NF-κB signaling by the A20 deubiquitinase. Cell Mol Immunol. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4:598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frantz AL, Bruno ME, Rogier EW, Tuna H, et al. Multifactorial patterns of gene expression in colonic epithelial cells predict disease phenotypes in experimental colitis. Inflamm Bowel Dis. 2012;18:2138–2148. doi: 10.1002/ibd.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frantz AL, Rogier EW, Weber CR, Shen L, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–512. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaetzel CS. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism. Immunol Lett. 2014:10. doi: 10.1016/j.imlet.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogier EW, Frantz AL, Bruno MEC, Wedlund L, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slebioda TJ, Kmiec Z. Tumour necrosis factor superfamily members in the pathogenesis of inflammatory bowel disease. Mediators Inflamm. 2014;2014:325129. doi: 10.1155/2014/325129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danese S, Colombel JF, Peyrin-Biroulet L, Rutgeerts P, et al. Review article: the role of anti-TNF in the management of ulcerative colitis -- past, present and future. Aliment Pharmacol Ther. 2013;37:855–866. doi: 10.1111/apt.12284. [DOI] [PubMed] [Google Scholar]
- 28.Oikonomopoulos A, van Deen WK, Hommes DW. Anti-TNF antibodies in inflammatory bowel disease: do we finally know how it works? Curr Drug Targets. 2013;14:1421–1432. doi: 10.2174/13894501113149990164. [DOI] [PubMed] [Google Scholar]
- 29.Goncalves NS, Ghaem-Maghami M, Monteleone G, Frankel G, et al. Critical role for tumor necrosis factor alpha in controlling the number of lumenal pathogenic bacteria and immunopathology in infectious colitis. Infect Immun. 2001;69:6651–6659. doi: 10.1128/IAI.69.11.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corredor J, Yan F, Shen CC, Tong W, et al. Tumor necrosis factor regulates intestinal epithelial cell migration by receptor-dependent mechanisms. Am J Physiol Cell Physiol. 2003;284:C953–C961. doi: 10.1152/ajpcell.00309.2002. [DOI] [PubMed] [Google Scholar]
- 31.Bruno MEC, Kaetzel CS. Long-term exposure of the HT-29 human intestinal epithelial cell-line to TNF causes sustained up-regulation of the polymeric Ig receptor and pro-inflammatory genes through transcriptional and post-transcriptional mechanisms. J Immunol. 2005;174:7278–7284. doi: 10.4049/jimmunol.174.11.7278. [DOI] [PubMed] [Google Scholar]
- 32.Dent G, Loweth SC, Hasan AM, Leslie FM. Synergic production of neutrophil chemotactic activity by colonic epithelial cells and eosinophils. Immunobiology. 2014;219:793–797. doi: 10.1016/j.imbio.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Franke A, McGovern DP, Barrett JC, Wang K, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jostins L, Ripke S, Weersma RK, Duerr RH, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverberg MS, Satsangi J, Ahmad T, Arnott ID, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of aWorking Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 36.Bruno ME, Frantz AL, Rogier EW, Johansen FE, et al. Regulation of the polymeric immunoglobulin receptor by the classical and alternative NF-κB pathways in intestinal epithelial cells. Mucosal Immunol. 2011;4:468–478. doi: 10.1038/mi.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murthy AK, Dubose CN, Banas JA, Coalson JJ, et al. Contribution of polymeric immunoglobulin receptor to regulation of intestinal inflammation in dextran sulfate sodium-induced colitis. J Gastroenterol Hepatol. 2006;21:1372–1380. doi: 10.1111/j.1440-1746.2006.04312.x. [DOI] [PubMed] [Google Scholar]
- 38.Reikvam DH, Derrien M, Islam R, Erofeev A, et al. Epithelial-microbial cross-talk in polymeric Ig receptor deficient mice. Eur J Immunol. 2012;42:2959–2970. doi: 10.1002/eji.201242543. [DOI] [PubMed] [Google Scholar]


