Abstract
Biological robustness, the ability of an organism to maintain a steady-state output as genetic or environmental inputs change, is critical for proper development. MicroRNAs have been implicated in biological robustness mechanisms through their post-transcriptional regulation of genes and gene networks. Previous research has illustrated examples of microRNAs promoting robustness as part of feedback loops and genetic switches and by buffering noisy gene expression resulting from environmental and/or internal changes. Here we show that the evolutionarily conserved microRNAs mir-34 and mir-83 (homolog of mammalian mir-29) contribute to the robust migration pattern of the distal tip cells in Caenorhabditis elegans by specifically protecting against stress from temperature changes. Furthermore, our results indicate that mir-34 and mir-83 may modulate the integrin signaling involved in distal tip cell migration by potentially targeting the GTPase cdc-42 and the beta-integrin pat-3. Our findings suggest a role for mir-34 and mir-83 in integrin-controlled cell migrations that may be conserved through higher organisms. They also provide yet another example of microRNA-based developmental robustness in response to a specific environmental stress, rapid temperature fluctuations.
Keywords: mir-34, mir-83, mir-29, distal tip cell migrations, robustness
THE ability of a living system to maintain a steady-state output in the face of environmental and physiological stresses is referred to as biological robustness (Kitano 2004). Organisms must be able to compensate for adverse changes in gene expression caused by environmental stresses and internal gene expression noise if development is to proceed unchanged. The nematode Caenorhabditis elegans is a useful model for studying such robustness. C. elegans development has been mapped to a cell-by-cell level, such that we know exactly when cell divisions occur and what the fate of each cell is (Sulston 1976; Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). There is also extensive research on the worm’s responses to stress, including stress-induced alternative larval developmental choices such as stage-specific diapause (Baugh and Sternberg 2006; Fukuyama et al. 2006; Ruaud and Bessereau 2006; Schindler et al. 2014) and proceeding to the dauer larvae stage, an alternative third larval stage that allows C. elegans to lengthen their lifespan and survive food deprivation or heat stress (Cassada and Russell 1975; Liu et al. 1995).
MicroRNAs (miRNAs) are single-stranded RNAs of ∼22 nucleotides that negatively regulate the translation of their target messenger RNAs (mRNAs) by binding to their 3′ untranslated region (3′ UTR) as part of a protein–RNA complex called the miRNA-induced silencing complex (miRISC). Target recognition is determined by the miRNA’s seed sequence, nucleotides two through seven (reviewed in Ambros 2004; Bartel 2004). miRNAs with the same seed sequence can presumably regulate the same target mRNAs and are grouped together within a miRNA family (reviewed in Bartel 2009). In addition to being regulated by multiple members of a miRNA family, an mRNA can be cotargeted by multiple distinct miRNAs families, if it contains the corresponding distinct seed-complementary sites. Such interfamily and intrafamily cotargeting is considered one reason why most single miRNA gene deletion mutants in C. elegans do not display apparent phenotypes; although one negative regulator is deleted, the target mRNAs in question remain under the regulation of additional, functionally redundant miRNAs (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010). Findings reported by the Abbott lab exemplified this phenomenon (Brenner et al. 2010). They screened for developmental phenotypes resulting from miRNA deletions in genetically sensitized backgrounds, in which the miRNA-specific argonaute protein alg-1 was no longer functional. In the alg-1 mutant background, overall miRNA production is partially compromised, such that the additional deletion of an otherwise redundant single miRNA can result in target deregulation and visible phenotypes.
In their study, the Abbott lab members identified six miRNAs involved in gonad morphogenesis (Brenner et al. 2010), a normally robust developmental process that is dependent on the migration of two distal tip cells (DTCs) (reviewed in Wong and Schwarzbauer 2012). One of these miRNAs included mir-83, a miRNA that is highly conserved in animals, including mammals (known as mir-29, Supporting Information, Figure S1) (Lagos-Quintana et al. 2001, 2002; Lau 2001; Mourelatos et al. 2002; Dostie et al. 2003; Lim et al. 2003; Lim 2003; Michael et al. 2003; Suh et al. 2004; Poy et al. 2004; Landgraf et al. 2007; Lui et al. 2007). Mammalian mir-29 has been previously implicated in regulating cellular proliferation, differentiation, apoptosis, and the extracellular matrix (reviewed in Boominathan 2010; Kriegel et al. 2012). To better understand how mir-83 functions, we set out to determine its mRNA targets and miRNA coregulators in C. elegans. Using mirWIP (Hammell et al. 2008), we noticed an overlap between the predicted mRNA targets for mir-83 and mir-34, another miRNA that is conserved between nematodes and mammals (Figure S1, Table S1) (Lau 2001; Ambros et al. 2003; Grad et al. 2003; Houbaviy et al. 2003; Lim et al. 2003; Lim 2003; Landgraf et al. 2007). mir-34 has been shown to have tumor suppressor activity in mammalian systems. There, its transcription is activated by p53 and it functions to reinforce p53 negative regulation (reviewed in He et al. 2007; Yamakuchi and Lowenstein 2009; Hermeking 2009; Rokavec et al. 2014).
To further understand the roles of mir-83 and mir-34, we tested for genetic redundancy by creating the double mutant and looking at developmental phenotypes. We also identified potential targets of mir-34 and mir-83 by tests of genetic suppression. mir-83(n4638); mir-34(gk437) double mutants display a defect in gonad morphogenesis. In addition, this defect reflects a loss of developmental robustness; the mir-83(n4638); mir-34(gk437) phenotype is significantly enhanced in response to temperature changes but not by other environmental stresses that we tested. Based on our results, we conclude that mir-34 and mir-83 function together to help create or maintain robust function of the genetic network controlling gonad morphogenesis, such that the development of this important organ is protected from the temperature changes C. elegans may encounter, either the rapid oscillations tested here or subtler changes experienced in the wild. Furthermore, we observed that mir-83(n4638); mir-34(gk437) mutants have a decreased lifespan and decreased fecundity, suggesting that the loss of these two miRNAs has repercussions for the biological fitness of the animals.
Materials and Methods
C. elegans strains
The C. elegans Bristol N2 strain was used as wild type in the study (Brenner 1974). Additional strains are listed in Table S2. Both the n4638 and gk437 alleles were backcrossed to N2 four times upon receipt. The VT2595 strain was used as the mir-83(n4638); mir-34(gk437) double mutant except when VT3289 is explicitly discussed in Figure 2C.
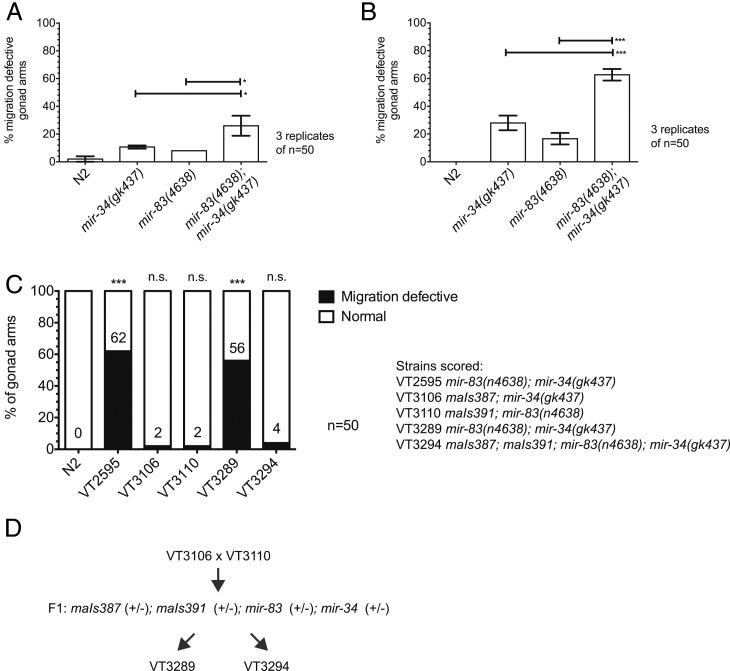
Figure 2.
The gonad migration defect is significantly enhanced in mir-83(n4638); mir-34(gk437) double mutants. Gonad arm morphology in young adult hermaphrodites was used to score for defects in DTC migrations during larval development at (A) 20° or (B) under an oscillating temperature regimen (15° for 15 min, 25° for 15 min, repeated from plating eggs until young adulthood). (C) Integrated transgenes expressing either mir-34, maIs387 or mir-83, maIs391, under their natural promoters rescue single mutants. Both VT3289 and VT3294 maIs387; maIs391; mir-83(n4638); mir-34(gk437) were produced by crossing VT3106 maIs387; mir-34(gk437) to VT3110 maIs391; mir-83(n4638), shown in D. As expected, the newly isolated mir-83(n4638); mir-34(gk437) double mutant, VT3289, displays the migration defect while the double mutant carrying both rescue transgenes, VT3294, appears normal. Significance asterisks compare to N2. Penetrance in VT2595 is not significantly different from penetrance in VT3289. Animals were cycled as described in B. ***P-value ≤ 0.005, *0.01 < P ≤ 0.05, not significant (n.s.) if P > 0.05.
C. elegans maintenance
Strains were maintained using standard procedures on nematode growth media (NGM) plates seeded with Escherichia coli strain HB101 (Brenner 1974), unless explicitly stated as being raised on OP50. Strains were raised at 20° unless otherwise stated when temperature was oscillated.
Sequence alignments, target prediction, and statistical significance
MicroRNA sequences were supplied by miRBase (Lagos-Quintana et al. 2001; Lau 2001; Lagos-Quintana et al. 2002; Mourelatos et al. 2002; Ambros et al. 2003; Aravin et al. 2003; Dostie et al. 2003; Grad et al. 2003; Houbaviy et al. 2003; Lim et al. 2003; Lim 2003; Michael et al. 2003; Sempere et al. 2003; Griffiths-Jones 2004; Poy et al. 2004; Suh et al. 2004; Griffiths-Jones et al. 2006, 2008; Landgraf et al. 2007; Lui et al. 2007; Kozomara and Griffiths-Jones 2011, 2014) and aligned by eye. Predicted targets were identified using mirWIP (Hammell et al. 2008). Throughout the manuscript, three significance asterisks (***) are used for a P-value of ≤0.005, two (**) for a P-value >0.005 and ≤0.01, and one (*) for a P-value >0.01 and ≤0.05.
Migration defective phenotype scoring
Hypochlorite treatment (Stiernagle 2006) was used to isolate embryos. Where noted, synchronized populations were created by allowing embryos to hatch in M9 buffer for ∼24 hr (Johnson et al. 1984). Embryos or starved L1’s were then plated on HB101-seeded NGM plates and raised to adulthood in the temperature scheme noted. Day one adults were paralyzed in 100 mM levamisole, mounted on 2% agarose pads, and scored using a Zeiss Axioskop differential interference contrast (DIC) microscope and a ×63 objective. A two-proportion z-test was used to determine significant differences between counts except where triplicates are presented. In such cases mean values were compared using an unpaired t-test performed by PRISM.
Temperature oscillations
Temperature oscillations were performed using modified MJ Research Programable Thermocyclers. First, the Thermocyclers’ lids were removed. Next, a 0.25-inch-thick aluminum plate was attached to the heat block using silicone-based thermal conductive grease. Original lids were replaced by insulating covers constructed out of styrofoam. HB101-seeded NGM plates containing worms were placed inside the modified device, in contact with the aluminum plate. To assess the thermal dynamics of the system, a thermometer was embedded in an NGM plate and the temperature of the agar was monitored during programmed temperature cycles. We observed that the temperature of NGM plates did cycle in accordance with the Thermocycler program, although not as quickly as the heat block itself; we observed an ∼15- to 30-sec delay as the NGM cools or heats. Stated temperature cycles refer to the program run by the Thermocycler.
Electron microscopy
N2 and mir-83(n4638) IV; mir-34(gk437) X embryos were isolated using hypochlorite treatment, hatched in M9 buffer for ∼24 hr and plated as synchronized L1’s on HB101-seeded NGM plates. Worms were raised with temperature oscillations: 20° for 16 hr (15° for 15 min, 25° for 15 min, repeat three additional times), 20° until young adulthood. Worms were fixed and prepared for electron microscopy as previously described (Irazoqui et al. 2010) except they were cut below the pharynx rather than in half to preserve gonad morphology. Electron microscopy work was performed by the University of Massachusetts Medical School Electron Microscopy Core Facility.
Cloning and transgenics
Constructs were created using Life Technologies Gateway Cloning. Except where otherwise stated, gene fragments were first amplified from N2 genomic DNA using the primers listed in Table S3. PCR was next used to add the proper att sequences such that fragments could be moved into the appropriate Gateway DONR vector, as described in the Gateway Protocol. For the mir-34 promoter, the 5-kb promoter was digested from a separate plasmid and ligated to a modified 476p5Emcs (a gift from Nathan Lawson’s lab, modified to remove unnecessary SalI cut sites) to add the necessary att sites. For mutated 3′ UTRs, DNA fragments of the desired sequence were created de novo by GeneWiz. The constructed DONR vectors, along with two commercially available vectors, were then used for transgene construction as described in the Gateway Protocol (Table S4). The destination vectors were developed for the Mos1-mediated single-copy insertion technique (Frøkjaer-Jensen et al. 2008), which was used to generate transgenic strains (Table S2). Multicopy arrays were generated for cdc-42 and pat-3 GFP/mCherry reporters. mir-83(n4638) IV; mir-34(gk437) X males were first crossed to EG6701 to create a unc-119(ed3) III; mir-83(n4638) IV; mir-34(gk437) X strain (VT3087). VT3087 was subsequently injected with a mix of pBluescript SK+ (30 ng/μl), pIF9 (15 ng/μl), pCFJ150 (30 ng/μl), pCFJ210 (30 ng/μl), and either pSLB054 (3 ng/μl) and pSLB056 (3 ng/μl) or pSLB071 (3 ng/μl) and pSLB075 (3 ng/μl). Array maEx246 was generated from the pSLB054- and pSLB056-containing injection mix, while array maEx247 was made from the pSLB071 and pSLB075 mix. The array carrying strains, VT3118 (for maEx246) and VT3145 (for maEx247), were crossed to N2 males to cross out the mir-83(n4638) and mir-34(gk437) deletions, generating strains VT3136 and VT3178, respectively.
RNAi
RNA-mediated interference (RNAi) was performed by raising animals on dsRNA-producing E. coli as described in Kamath et al. (2001). Synchronized L1’s were placed on cdc-42 RNAi food, pat-3 RNAi food, or empty vector control food, and raised to young adulthood with the temperature scheme stated. Animals were scored as previously described.
Target reporter scoring
Hypochlorite treatment was used to isolate embryos, which were hatch in M9 to generate synchronized L1’s. Worms were plated on HB101-seeded NGM plates and raised with temperature oscillations: 20° for 16 hr (15° for 15 min, 25° for 15 min, repeat three additional times), 20° until time of scoring. L1 animals were scored starting 18 hr postplating, immediately following temperature oscillations. L3 animals were scored starting 43 hr postplating, shortly after the molt from L2 to L3. Adults were scored starting 67 hr postplating, after egg laying had commenced.
Worms were suspended in 100 mM levamisole, mounted on 2% agarose pads, and scored using a Leica TCS SPE confocal microscope. For cdc-42 reporters, DTCs were first identified in the DIC setting by eye using the ×63 objective. The confocal microscopy setting was subsequently used for imaging and quantifying the mean value florescence of each channel for a DTC using the Leica LAS AF software. For pat-3 reporters, the ×10 objective was used to observe whole animals. A 30-step z-series was taken per worm. The Leica AF software was used to quantify mean value fluorescence for each channel using the z-stack maximum projection. Datasets were compared using an unpaired t-test performed by PRISM.
Brood size counts
N2 and mir-83(n4638) IV; mir-34(gk437) X embryos were isolated using hypochlorite treatment and left in M9 buffer for ∼24 hr to hatch. Arrested L1’s were then plated on HB101-seeded NGM plates and either raised at 20° continuously or with temperature oscillations from hour 16 to hour 18 postplating [labeled “cycled”—20° for 16 hr (15° for 15 min, 25° for 15 min, repeat three additional times), 20° until death]. Worms were individually plated as L4’s, and the number of live offspring were counted from the start of egg laying until its end. Mean values were compared using an unpaired t-test performed by PRISM.
Mating assays
N2 and mir-83(n4638) IV; mir-34(gk437) X embryos were isolated using hypochlorite treatment and left in M9 buffer for ∼24 hr to hatch. Arrested L1’s were then plated on HB101-seeded NGM plates and either raised at 20° continuously or with temperature oscillations from hour 16 to hour 18 postplating [labeled “cycled”—20° for 16 hr (15° for 15 min, 25° for 15 min, repeat three additional times), 20° until death]. Once they reached adulthood, hermaphrodites were transferred daily away from progeny until the ability to lay eggs was exhausted (day four of adulthood). Each hermaphrodite was then moved to an individual plate, to which four 1-day-old adult N2 males were added. Animals were allowed to mate for 3 days, at which point the males were removed. The total number of offspring that hatched were counted for each hermaphrodite from the start of the assay until its death.
Lifespan assays
Survival assays were performed similarly to previously described (Kenyon et al. 1993). N2 and mir-83(n4638) IV; mir-34(gk437) X embryos were isolated using hypochlorite treatment and left in M9 buffer for ∼24 hr to hatch. Arrested L1’s were then plated on HB101-seeded NGM plates and either raised at 20° continuously or with temperature oscillations from hour 16 to hour 18 postplating [labeled “cycled”—20° for 16 hr (15° for 15 min, 25° for 15 min, repeat three additional times), 20° until death]. To track survival, 100 worms per replicate and three replicated per condition were moved to a new HB101-seeded NGM plate as L4’s. Worms were transferred to a new plate daily during the course of the assay to avoid overcrowding from the offspring. Worms were scored as alive or dead based on reaction to a gentle nose prod using a standard worm pick. Animals were tracked until their death. Any worms that died as a consequence of crawling up the side of the plate were removed from the assay. Replicates were used to calculate percent survival and the standard deviation for each day. Mean values for individual days were compared using a two sample t-test for means.
Additional stresses
Animals was raised on NGM plates containing 0.03% sodium arsenite to induce oxidative stress (Sahu et al. 2013). Incubation in 1% SDS for 30 min was used to isolate dauer animals from starved plates (Stiernagle 2006). Animals were raised on Pseudomonas auruginosa PA14 plates as previously described (Powell and Ausubel 2008).
Results
Improper distal tip cell migration paths in mir-83(n4638); mir-34(gk437) mutants
The shape of the C. elegans adult hermaphrodite gonad is determined by the migration of two DTCs, somatic gonadal cells at the tip of each gonad arm that drag the proliferative portion of the gonad with them as they migrate during the second through fourth larval stages of hermaphrodite development (Kimble and Hirsh 1979; Kimble and White 1981; Hedgecock et al. 1987). Both DTCs are born near the midbody during the first larval stage and begin to migrate along the ventral body wall muscles toward the head and tail during the second larval stage, referred to as phase 1 of migration (Hirsh et al. 1976; Kimble and Hirsh 1979; Kimble and White 1981). During phase 2, DTCs first turn dorsally to migrate from the ventral to the dorsal body wall muscle, crossing the hypodermis in the process (Hedgecock et al. 1987). A second turn is required to continue migration along the dorsal body wall muscles and return to the midbody (phase 3), creating two U-shaped gonad arms (Figure 1A) (Hirsh et al. 1976; Kimble and Hirsh 1979; Hedgecock et al. 1987). The final shape of the gonad arms can be used to deduce the path taken by each DTC. In a fraction of mir-83(n4638); mir-34(gk437) mutants (26% at 20°, quantified in Figure 2), anterior and posterior gonad arms appear displaced at various positions, improperly crossing the dorsal/ventral axis (Figure 1B). These misplaced gonad arms suggest DTCs wandered from the proper path during phase 1 and/or phase 3 of migration and often migrated too far and past the developing vulva. The penetrance of this overextension phenotype was highly variable and overextension was observed to occur either with or without wandering during phase 1 and/or phase 3. Unlike some classes of C. elegans migration mutants, DTCs in mir-83(n4638); mir-34(gk437) mutants execute both their first (dorsal) and second (anterior/posterior) turns at the proper positions in the animals and at the proper times in development. The migration defect of these mutants therefore reflects a defect in the precise pathfinding of the DTCs as they migrate longitudinally along the ventral or dorsal muscle. To score this phenotype we categorized gonad arms as either migration defective or normal. Gonad arms were categorized as migration defective if their abnormal shape indicated that the DTC had improperly crossed the dorsal/ventral axis during its migration, or that the DTC migrated completely past the vulva before stopping. Previously published findings indicated a low level of migration defects in the N2 strain (Peters et al. 2013). These included such phenotypes as DTCs stopping slightly short of the vulva and a slight ventralward “dip” of the DTCs when migration ceases. We observed such phenotypes as well, but due to their occurrence in N2 we classified such cases as within normal variation and did not score them as migration defects. It is also important to note that in the previously mentioned study (Peters et al. 2013), strains were raised at 23° rather than 20°.
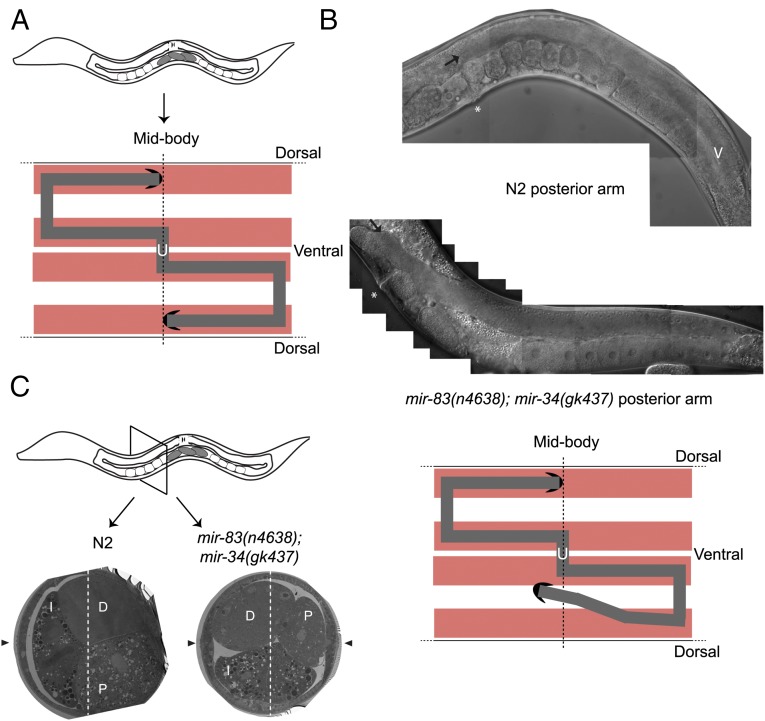
Figure 1.
mir-83(n4638); mir-34(gk437) mutants have gonad migration defects. (A) The C. elegans gonad consists of two U-shaped gonad arms. The shape of each arm is created by the migration path of the DTC. The two DTCs are initially located ventrally near the midbody. Each DTC migrates away from the midbody before turning dorsally, migrating to the dorsal body wall muscle, and then migrating back toward the midbody. The path of each DTC can be inferred by the location of the respective gonad arm. Body wall muscle in red, germline in gray, DTCs in black, uterus designated with “U.” (B) mir-83(n4638); mir-34(gk437) mutants have a gonad migration defect. The improper location of the mature gonad arms implies that DTCs did not migrate along the normal path. In the arm pictured (bottom), the DTC (black arrow) migrated too far, passing the vulva (white asterisk), and is displaced ventrally, as compared to N2 (top). Penetrance quantified in Figure 2. (C) Improper DTC migration causes a rearrangement of internal organs, due to space constraints. In N2 worms, the intestine is positioned laterally with respect to the dorsal/ventral axis (indicated by a white, dashed line), and the proximal and distal segments of the gonad are positioned ventrally and dorsally, respectively, on the other side from the intestine. In the mir-83(n4638); mir-34(gk437) worm shown here, the displaced gonad caused a displacement of the intestine. Black arrowheads point to the adult lateral alae, which are positioned on the left and right sides of the animal.
DTC wandering results in displaced gonad arms, which is reflected in a reorganization of the internal organs. To visualize this reorganization in mir-83(n4638); mir-34(gk437) mutants, we performed electron microscopy on cross-sections of adult worms. We first collected ∼20 mir-83(n4638); mir-34(gk437) worms whose anterior arm displayed a migration defect. These mutants and age-matched N2’s were fixed and processed for electron microscopy. As previously described, in N2 worms the intestine spans the dorsal/ventral axis (Figure 1C) (Hall and Altun 2008). The oocyte-containing proximal gonad is located ventrally while the distal gonad is dorsal (Hall and Altun 2008). For a DTC to wander from the appropriate migration path, it must displace the intestine. In addition, the somatic gonad itself will be displaced as it trails behind the migrating DTC. This rearrangement of internal organs, with the intestine, distal gonad arm, and proximal gonad arm all displaced, was visible in both of the two mir-83(n4638); mir-34(gk437) mutants sectioned and visualized (Figure 1C).
The penetrance of the migration defect is significantly enhanced by oscillating temperature
We first sought to quantify the penetrance of the migration defect in mir-83(n4638); mir-34(gk437) worms raised in standard laboratory conditions, namely on E. coli-seeded NGM plates kept at 20° (Brenner 1974). As expected, the DTCs migrate properly in N2 worms in the majority of worms (2 ± 2% migration defective, performed in triplicate, Figure 2A). Both mir-34(gk437) and mir-83(n4638) single mutant populations display migration defects at a low frequency, 10.67 ± 1.15% and 8 ± 0%, respectively. The phenotype’s penetrance is significantly enhanced to 26 ± 7.21% in the mir-83(n4638); mir-34(gk437) double mutant (P ≤ 0.05, unpaired t-test), suggesting that the two miRNAs contribute partially redundant functions in the context of DTC migration.
Due to the low penetrance of the phenotype in mir-83(n4638); mir-34(gk437) double mutants (Figure 2A), we reasoned that the functions of these miRNAs in DTC migration could be relatively unimportant under standard laboratory conditions, but perhaps more critical under stressful conditions. Therefore, we tested for enhancement of the DTC migration phenotype in worms exposed to various stresses. We observed no change in the penetrance of the phenotype when mutants were exposed continuously throughout development to high temperature (25°) or low temperature (15°) or to a diet of pathogenic P. aeruginosa, oxidative stress (0.03% arsenic), or starvation during early larval stages to induce dauer formation (Table S5). However, an enhanced phenotype was observed when worms developed under a changing temperature regimen (Figure 2B), a phenomenon previously observed for mir-7 Drosophila mutants in the context of Drosophila eye development (Li et al. 2009). Semisynchronized populations of developing larvae were exposed to a regimen of temperature changes every 15 min—first from 15° to 25°, and then back to 15°, and so on. The DTC migrations of N2 worms were unaffected by temperature oscillations (0 ± 0%). All three mutant strains [mir-34(gk437) and mir-83(n4638) single mutants and the mir-83(n4638); mir-34(gk437) double mutant] showed an enhanced penetrance of the DTC migration defective phenotype under oscillating temperature compared to constant temperature (20°); from 10.67 ± 1.15% to 28 ± 5.29% for mir-34(gk437), from 8 ± 0% to 16.67 ± 4.16% for mir-83(n4638), and from 26 ± 7.21% to 62.67 ± 4.16% for mir-83(n4638); mir-34(gk437). A similar pattern was observed for animals grown on OP50 rather than HB101 E. coli, suggesting that the migration defect and its enhancement by oscillating temperature are independent of food source (Figure S2). We also examined a second mir-34 null allele, n4276, and obtained similar results as those for gk437 (Figure S3).
To confirm that the DTC migration defects of mir-34 and mir-83 mutants are a consequence of the loss of the mir-34 and mir-83 genes, we generated single-copy integrated rescuing transgenes in their respective single mutant (Table S2). Both the mir-34 transgene, maIs387, and the mir-83 transgene, maIs391, rescue the migration defects of their respective single mutant (Figure 2C). Although this finding supports the conclusion that the migration defects can be explicitly attributed to the mir-34 and mir-83 loss of function, a caveat remained that the migration defect was due to background mutations, outside the mir-34 or mir-83 loci, that had been present in the single mutants and were segregated away during construction of the transgene-containing strains. In this scenario, the hypothetical background mutations would have been maintained during the construction of the mir-83(n4638); mir-34(gk437) double mutant, strain VT2595. To address this caveat, we crossed the maIs387; mir-34(gk437) and maIs391; mir-83(n4638) strains together to generate a new doubly mutant mir-83(n4638); mir-34(gk437) strain (VT3289) lacking both transgenes and a sibling doubly mutant strain carrying both transgenes (VT3294) (Figure 2C). We observed that VT3289 exhibited a 56% penetrance of migration defective, which is not statistically different from 62% migration defective exhibited by the original mir-83(n4638); mir-34(gk437) double mutant, VT2595 (Figure 2C, two-proportion z-test, P > 0.05). If the migration defect had been due to background mutations rather than the mir-34 and mir-83 deletions we would not expect the newly isolated double to have the defect. As expected, the double mutant carrying both rescue constructs, maIs387; maIs391; mir-83(n4638); mir-34(gk437), appears mostly normal (4% migration defective).
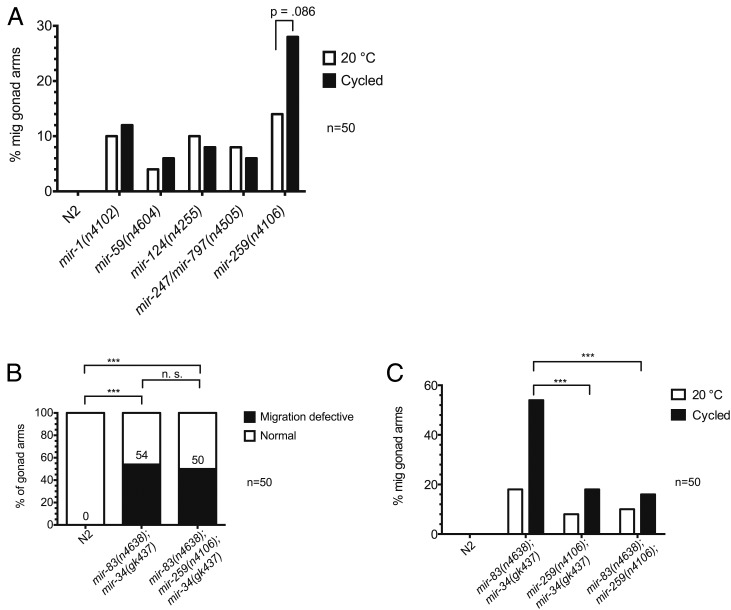
The above results suggest that DTC migrations may be inherently sensitive to unstable temperature, and that the activity of mir-34 and mir-83 helps protect the genetic network controlling this migration from environmental temperature changes. Although the penetrance of the phenotype was increased in mir-83(n4638); mir-34(gk437) mutants worms experiencing oscillating temperatures (from 26% at constant 20° to 62.67% in oscillating temperatures), the migration defect is still not fully penetrant in mir-83(n4638); mir-34(gk437) mutants, even with temperature oscillations. It was previously shown that for alg-1 dominant-negative alleles ma192 and ma202, respectively 71% and 73% of worms, have DTC migration defects, strongly suggesting that other miRNAs are involved in the regulation of DTC migrations (Zinovyeva et al. 2014). Therefore the additional miRNAs implicated in gonad migration by the Abbott lab (Brenner et al. 2010) were examined to see if being raised in changing temperatures enhanced any of the single mutants as it did for mir-34(gk437) and mir-83(n4638) mutants. We found that only the mir-259(n4106) migration defect appears to be enhanced by oscillating temperatures (Figure 3A), although the enhancement is not significant (P = 0.086, two-proportion z-test). Additionally, the migration defects exhibited by mir-259(n4106) mutants are similar to those of mir-34(gk437) and mir-83(n4638) single and double mutants; DTCs improperly cross the dorsal/ventral axis during phase 1 and phase 3 of migration and migrate past the vulva. However, the mir-259 deletion does not enhance the penetrance of these defects in a triple mutant mir-83(n4638); mir-259(n4106); mir-34(gk437) compared to the mir-83(n4638); mir-34(gk437) double mutant (Figure 3B, P ≥ 0.05, two-proportion z-test). This suggests that, although mir-259 regulates targets involved in DTC migration in a manner that is sensitive to temperature changes and could be involved in the same or a similar process as mir-34 and mir-83, mir-259 appears to not regulate the same targets as mir-34 and mir-83. If the three miRNAs were acting in parallel on the same set of mRNA targets, we would expect an elevated penetrance of the phenotype upon removal of mir-259 from the double mutant as the target mRNAs in question would be further derepressed. One caveat regarding this conclusion could be that the lack of enhancement seen in the mir-83(n4638); mir-259(n4106); mir-34(gk437) triple compared to the mir-83(n4638); mir-34(gk437) double (Figure 3B) might reflect common targets that were fully derepressed in the mir-83(n4638); mir-34(gk437) double, such that the loss of an additional regulator had no effect. In this regard, we note that the migration defect penetrance in both the mir-259(n4106); mir-34(gk437) and the mir-83(n4638); mir-259(n4106) double mutants are weaker than that of the mir-83(n4638); mir-34(gk437) mutant (Figure 3C, P ≤ 0.005, two-proportion z-test). This suggests that the relevant mRNA target sets for mir-34 and mir-83 may overlap more with each other than with mir-259. For these reasons, we chose to focus on the functionally interacting miRNAs mir-34 and mir-83 for the remainder of this study.
Figure 3.
Other miRNAs implicated in gonad migration function in separate pathways. (A) Previously implicated miRNA mutants were tested for enhancement of gonad migration defects by temperature oscillations. Worms were either maintained at a steady 20° throughout development or were subjected to oscillating temperature (15° for 15 min, 25° for 15 min) throughout development (“cycled”). (B) The mir-259(n4106) mutation was crossed into the mir-83(n4638); mir-34(gk437) strain to assess potential genetic interactions. Worms were subjected to an oscillating temperature regimen (15° for 15 min, 25° for 15 min, repeated until young adulthood). The difference in migration defective (mig) phenotype penetrance between mir-83(n4638); mir-34(gk437) and mir-83(n4638); mir-259(n4106); mir-34(gk437) is not significant. ***P-value ≤ 0.005. (C) Phenotype penetrance for the mir-259(n4106) mutation in combination with either the mir-34(gk437) mutation or the mir-83(n4638) mutation was compared to that in the mir-83(n4638); mir-34(gk437) double mutant. Worms were raised at 20° or cycled during the temperature-sensitive period discussed in Figure 4 [20° for 16 hr (15° for 15 min, 25° for 15 min, repeated three additional times), 20° until young adulthood]. ***P-value ≤ 0.005.
The temperature sensitivity of DTC migration is restricted to a 2-hr period during the L1 stage
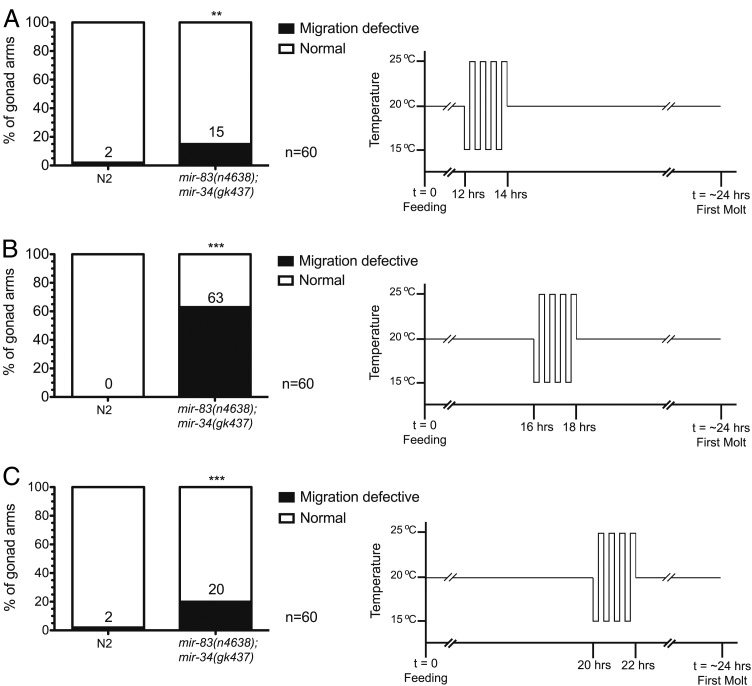
We next sought to determine if the entire DTC migration process was temperature sensitive in mir-83(n4638); mir-34(gk437) mutants, or if there was a more restricted developmental period during which DTCs are sensitive to changing temperature in these mutants. Synchronized populations of N2 and mutant worms were produced by hatching eggs in the absence of food. L1 larvae arrest in such conditions and will not proceed developing until the reintroduction of food (Johnson et al. 1984; Baugh 2013). Once food is reintroduced, it takes the larvae ∼24 hr at 20° to reach the first molt into the L2 larval stage. Using the DTC-specific reporter, Plag-2::GFP (Siegfried and Kimble 2002), we found that the DTCs are born ∼16 to 16.5 hr after the reintroduction of food (unpublished results) in both N2 and mir-83(n4638); mir-34(gk437) worms. This agrees with previous reports for the timing of DTC birth in N2’s (Sulston and Horvitz 1977; Kimble and Hirsh 1979). By examining the effects of 2-hr time windows during which the temperature oscillated between 15° and 25° every 15 min, we found that the temperature-sensitive period for DTCs overlapped with the approximate time of their births. When the 2-hr oscillating temperature regimen occurred before 14 hr of development (Figure 4A) or after 20 hr of development (Figure 4C), mir-83(n4638); mir-34(gk437) mutants displayed the migration phenotype at a penetrance similar to that seen in the nonenhanced 20° condition (see Figure 2A). However, when temperature oscillations occurred from hour 16 to hour 18 of development (Figure 4B) we observed an enhanced penetrance of the phenotype similar in magnitude to that of animals that had experienced temperature oscillations throughout larval development (see Figure 2B). This implies that the enhancement observed with temperature oscillations throughout development likely resulted from changing temperature in the interval between hour 16 to hour 18. Indeed, animals that experienced temperature oscillations throughout most of larval development but excluding the 2-hr period from hour 16 to hour 18 did not exhibit phenotypic enhancement (Figure S4).
Figure 4.
Temperature oscillations within a limited 2-hr window cause the migration defective phenotype enhancement in mir-83(n4638); mir-34(gk437) mutants. The temperature was oscillated every 15 min for a total of 2 hr (15° for 15 min, 25° for 15 min, repeated four times). (A) Temperature oscillations occurred prior to the birth of the DTCs (12 to 14 hr after plating starved L1’s on HB101-seeded NGM plates). (B) Temperature oscillations occurred over an interval (16 to 18 hr after plating starved L1’s on HB101) corresponding to the time of DTC birth. (C) Temperature oscillations occurred after the birth of the DTCs (20 to 22 hr after plating starved L1’s on HB101). ***P-value ≤ 0.005, **0.005 < P ≤ 0.01, significance asterisks compare N2 to mir-83(n4638); mir-34(gk437) mutants.
Our results indicate that cycling between 15° and 25° captures the full enhancement of the migration phenotype under the conditions we have tested. Increasing the upper temperature to 37°, thereby heat shocking the worms in 15-min increments, did not further enhance the penetrance of the phenotype (Figure S5). We also found that the phenotypic enhancement occurred whether the first temperature change was a decrease (Figure 4) or an increase (Figure S6) in temperature and was largely independent of the magnitude of the temperature change; 10-degree or 5-degree changes in temperature resulted in similar phenotype enhancement (Figure S7). We observed that a single oscillation cycle during the hour-16 to hour-18 period was insufficient to enhance the phenotype (Figure S8), suggesting that multiple cycles are necessary to elicit the stress that compromises the integrity of gonad morphogenesis in the absence of mir-34 and mir-83.
Tissue specificity of mir-34 and mir-83
To determine the anatomical site of action of mir-34 and mir-83 in regulating the integrity of DTC migrations we generated constructs expressing each of these miRNAs driven by its natural promoter or by promoters from genes expressed in the hypodermis (dpy-7), DTCs (lag-2, also expressed in some vulval cells), gonadal sheath cells (lim-7), or muscle (myo-3, also expressed in muscle-like sheath cells 3–5) (Johnstone et al. 1992; Henderson et al. 1994; Hall et al. 1999; Dupuy et al. 2007; Fox et al. 2007; Ono et al. 2007). These constructs were built in Mos1-mediated single-copy insertion (MosSCI) destination vectors and were used to generate integrated transgenic lines (Frøkjaer-Jensen et al. 2008) (Table S2).
As expected, both mir-34 or mir-83 expressed under their cognate natural promoters rescued the DTC migration defect of the respective single mutants (Table 1 and Table 2, P ≤ 0.005, two-proportion z-test). mir-34(gk437) and mir-83(n4638) phenotypes were also rescued to varying degrees by tissue-specific heterologous promoters. mir-34 expressed in the DTCs (driven by the lag-2 promoter; Plag-2::mir-34) partially rescued the mir-34(gk437) defect, from 28.3 to 11.7% (Table 1, P ≤ 0.05, two-proportion z-test). The lack of full rescue could be due to a difference in expression levels and/or a requirement for mir-34 in more than one tissue. mir-83 expressed in either the DTCs (Plag-2::mir-83) or muscle (Pmyo-3::mir-83), significantly rescued the migration defect (from 30% in the mutant to 5% in each rescue line, Table 2, P ≤ 0.005, two-proportion z-test). Potential explanations for this result, where mir-83 appears to function in either the DTCs or the muscle, are discussed in the Discussion section.
Table 1. Tissue-specific mir-34 rescue.
| Strain | Migration defective (%) (N = 60) |
|---|---|
| N2 | 0 |
| mir-34(gk437) | 28.3 |
| Pmir-34::mir-34; mir-34(gk437) | 0*** |
| Pdpy-7::mir-34; mir-34(gk437) | 20 |
| Plag-2::mir-34; mir-34(gk437) | 11.7* |
| Plim-7::mir-34; mir-34(gk437) | 31.7 |
| Pmyo-3::mir-34; mir-34(gk437) | 26.7 |
Synchronized L1’s were raised with oscillating temperatures: 20° for 16 hr (15° for 15 min, 25° for 15 min, repeated three additional times), 20° until young adulthood. ***P-value ≤ 0.005, *0.01 < P ≤ 0.05, significance asterisks compare rescue strains to mir-34(gk437) mutants.
Table 2. Tissue-specific mir-83 rescue.
| Strain | Migration defective (%) (N = 60) |
|---|---|
| N2 | 0 |
| mir-83(n4638) | 30 |
| Pmir-83::mir-83; mir-83(n4638) | 1.7*** |
| Pdpy-7::mir-83; mir-83(n4638) | 25 |
| Plag-2::mir-83; mir-83(n4638) | 5*** |
| Plim-7::mir-83; mir-83(n4638) | 41.7 |
| Pmyo-3::mir-83; mir-83(n4638) | 5*** |
Synchronized L1’s were raised with oscillating temperatures: 20° for 16 hr (15° for 15 min, 25° for 15 min, repeated three additional times), 20° until young adulthood. ***P-value ≤ 0.005, significance asterisks compare rescue strains to mir-83(n4638) mutants.
Mir-34 and mir-83 regulate two key proteins involved in DTC migrations
Our findings that DTC-specific expression of mir-34 or mir-83 can rescue the gonad migration defect of mir-34(gk437) or mir-83(n4638) mutants suggests that mir-34 and mir-83 may regulate a gene or genes whose activity impacts the fidelity of the DTC migration process, and that the migration defect reflects abnormal pathfinding by the DTCs during larval development. An alternative hypothesis, that the displaced gonad arms observed in mutant adults resulted from the shifting of internal organs after otherwise proper migrations, was tested by examining whether the animals’ movement impacted their gonad morphology. We observed that the migration-defective phenotype was not affected in genetically paralyzed worms compared to fully active worms, arguing against a contribution of movement-derived structural damage as a cause of the migration defect (Figure S9).
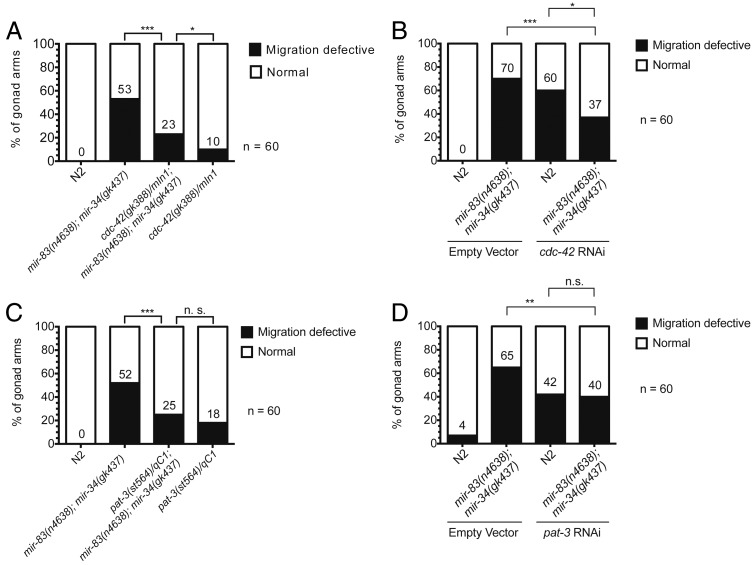
To identify potential targets of mir-34 and mir-83 in the regulation of DTC migration, we tested for suppression of the phenotype in mir-83(n4638); mir-34(gk437) mutants by RNAi knockdown of genes predicted by mirWIP (Hammell et al. 2008) to be targets of both mir-34 and mir-83 (Table S1), prioritizing genes known to be involved in cell migrations, larval development, or miRNA function. cdc-42 and pat-3, both expressed in the DTCs and body wall muscle and known to be involved in their migration (Lee et al. 2001; Cram et al. 2006; Lucanic and Cheng 2008), are predicted targets of mir-34 and mir-83 and were subsequently examined.
cdc-42 is a GTPase shown to be downstream of integrin signaling. Upon activation by integrin, cdc-42 acts to bring about the actin cytoskeleton rearrangements associated with cell migration (Van Aelst and D’Souza-Schorey 1997; Price et al. 1998; Ren et al. 1999). Previously it has been shown that when a constitutively active allele of cdc-42 (unable to hydrolyze GTP to GDP) is expressed in the DTCs, those DTCs display pathfinding defects (Peters et al. 2013). If cdc-42 is a direct target of mir-34 and mir-83, we would expect its expression to be elevated in animals mutant for these two miRNAs. Therefore, we hypothesized that by lowering the level of cdc-42 in mir-83(n4638); mir-34(gk437) mutants we could suppress the gonad migration defect. This is in fact the case. The cdc-42 null allele gk388 deletes a portion of cdc-42’s 5′ UTR, the first exon, and a portion of the first intron (Kimata et al. 2012). mir-83(n4638); mir-34(gk437) mutants heterozygous for the cdc-42(gk388) allele were significantly rescued, from 53% of the population displaying the gonad migration-defective phenotype to 23% (Figure 5A, P ≤ 0.005, two-proportion z-test). Mutants containing one copy of gk388 and wild-type copies of both mir-34 and mir-83 also are migration defective (wandering in phase 1 and/or phase 3) at a low penetrance (10% of the population). This is expected as cdc-42 is a critical regulator of the integrin signaling network (Van Aelst and D’Souza-Schorey 1997; Price et al. 1998; Ren et al. 1999). We confirmed this suppression using RNAi-induced knockdown of cdc-42 (Figure 5B); cdc-42(RNAi) in a mir-83(n4638); mir-34(gk437) mutant significantly suppressed the penetrance of the migration defect from 70 to 37% (P ≤ 0.005, two-proportion z-test). N2 worms on cdc-42 RNAi food exhibit wandering phenotypes similar to that observed in the balanced heterozygote. This result supports the idea that the reduction of cdc-42 is responsible for the suppression seen in mir-83(n4638); mir-34(gk437) rather than being the result of the inclusion of the balancer chromosome mIn1.
Figure 5.
The mir-83(n4638); mir-34(gk437) migration defect is suppressed in cdc-42 and pat-3 heterozygotes. The penetrance of the migration defective phenotype is significantly reduced in mir-83(n4638); mir-34(gk437) mutants when (A) carrying one copy of cdc-42, (B) raised on cdc-42 RNAi food as compared to empty vector control, (C) carrying one copy of pat-3, or (D) raised on pat-3 RNAi food as compared to empty vector control. Arrested L1’s were plated on HB101-seeded NGM plates to restart development. Once plated, temperature was held at 20° for 16 hr, then cycled as follows: 15° for 15 min, 25° for 15 min, four times, then held at 20° until young adulthood. ***P-value ≤ 0.005, **0.005 < P ≤ 0.01, *0.01 < P ≤ 0.05, P > 0.05 not significant (n.s.).
C. elegans has one β-integrin gene, pat-3 (Williams and Waterston 1994; Gettner et al. 1995). Integrin signaling is the major regulatory network involved in phase 1 and phase 3 of DTC migration (Baum and Garriga 1997; Lee et al. 2001; Cram et al. 2006). Using the pat-3 null allele st564 (Williams and Waterston 1994), we created a strain homozygous for deletions of both mir-34 and mir-83 and carrying only one functional copy of pat-3. In this case, partial loss of pat-3 reduced the migration defect from 52 to 25% (Figure 5C, P ≤ 0.005, two-proportion z-test), supporting the conclusion that pat-3 overexpression contributes to the migration defect in mir-83(n4638); mir-34(gk437) mutants. Note that 18% of worms with one functional copy of pat-3 are migration defective (we observed wandering during phase 1 and phase 3 and overextension, but no defects in phase 2 turns), indicating that the fidelity of DTC migration may depend critically on pat-3 dosage. RNAi against pat-3 also significantly suppressed the migration defect in a mir-83(n4638); mir-34(gk437) mutant from 65 to 40% (Figure 5D, P ≤ 0.01, two-proportion z-test), confirming that the reduction of pat-3 is responsible for the suppression.
The suppression in mir-83(n4638); mir-34(gk437) mutant phenotype by genetic reduction of either cdc-42 or pat-3 activity supports the supposition that cdc-42 and pat-3 function downstream of these miRNAs in the regulation of DTC migration. The fact that both cdc-42 and pat-3 are predicted to contain sites for both mir-34 and mir-83 in their 3′ UTR sequences strongly suggests direct targeting by these miRNAs. In an attempt to gather additional data in support of direct regulation, we constructed a set of nuclear localized mCherry and GFP reporters using the promoters and 3′ UTR sequences of each target (see Methods). mCherry was paired with the wild-type target 3′ UTR while GFP was fused to a 3′ UTR where the predicted mir-34 and mir-83 binding sites were mutated. Specifically, the 3′ UTR bases predicted to be part of the mRNA–miRNA seed base pairing were mutated to the complementary base, A to T, C to G, G to C, and U to A, such that mir-34 and mir-83 loaded miRISCs would no longer to be able to bind the mutated 3′ UTRs. For each wild-type 3′ UTR reporter, the corresponding GFP construct with a mutated 3′ UTR served as an internal control, as it should not be subjected to regulation by either miRNA. The mCherry construct, however, should be regulated by both miRNAs and therefore, should be expressed differently in their presence or absence. We generated multicopy arrays (single-copy transgenes were unsuccessful, see Discussion) containing both constructs in mir-83(n4638); mir-34(gk437) mutants and crossed these arrays into N2 worms for comparison to the mutant background. GFP and mCherry fluorescence was subsequently quantified within DTCs or for the whole animal. We did not observe a difference in the ratio of mCherry to GFP fluorescence expressed in DTCs for the cdc-42 reporters, in any of the stages observed (Figure S10A, P ≥ 0.05, unpaired t-test).
As for the pat-3 reporters, the results were indicative but not definitive. Expression of the pat-3 reporters was not detectable in DTCs in larval stages. In adults, pat-3 reporter expression was very dim and bleached rapidly, necessitating scoring whole animals. In whole adult animals, there was a slight increase in the ratio of mCherry to GFP fluorescence for the pat-3 reporters when mir-34 and mir-83 were deleted (Figure S10B, P ≤ 0.05, unpaired t-test), suggesting that the two miRNAs do directly regulate pat-3. Although these reporter experiments could not confirm direct regulation of both cdc-42 and pat-3 by mir-34 or mir-83, there were numerous technical issues that limited the sensitivity and fidelity of the reporter assays (see Discussion). It is possible that cdc-42 and pat-3 could be acting in parallel pathways that indirectly oppose the activities of mir-34 and mir-83, such that the observed suppression is the result of decreasing the activity of an opposing pathway. However, based on the computational prediction of cotargeting by mir-34 and mir-83, combined with the fact that both cdc-42 and pat-3 are known to be required for proper pathfinding, we propose that these predicted mir-34 and mir-83 common targets function downstream of the two miRNAs in conferring robustness to DTC migration in the face of temperature changes.
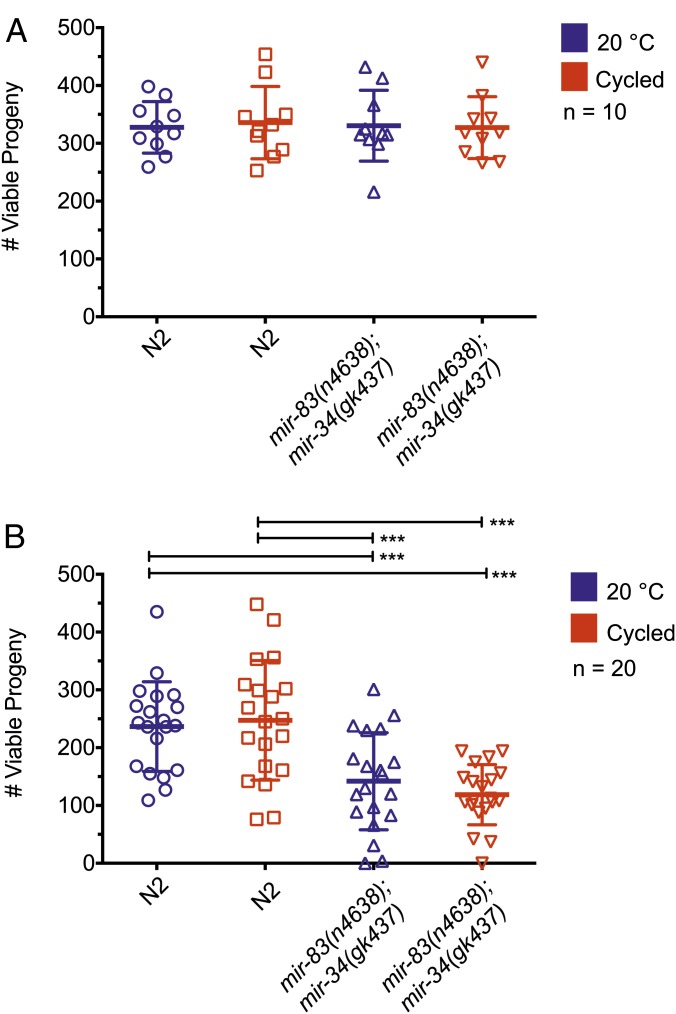
mir-83(n4638); mir-34(gk437) mutants display reduced cross-progeny fecundity
In addition to guiding the morphology of the adult gonad, DTCs are also required to signal to the germline and regulate the production of germ cells (reviewed in Hubbard and Greenstein 2000). We therefore tested whether the deletion of mir-34 and mir-83 affected the function of the hermaphrodite germline by quantifying offspring. We scored the total number of viable progeny, animals that hatched from laid eggs. We did not observe a noticeable difference in the number of dead eggs laid by mir-83(n4638); mir-34(gk437) mutants vs. N2 worms; therefore, we did not include them in our quantification. When raised at 20° or with temperature oscillations occurring during the previously described 2-hr temperature-sensitive period (referred to as cycled), there was no statistical difference in the number of viable self-progeny produced by mir-83(n4638); mir-34(gk437) mutants compared to wild type (Figure 6A, P ≥ 0.05, unpaired t-test).
Figure 6.
mir-83(n4638); mir-34(gk437) mutants produce normal sized self-broods and reduced cross-broods. (A) The total number of living offspring was counted to determine brood size for N2 and mir-83(n4638); mir-34(gk437) worms raised at 20° throughout development or subjected to temperature cycles (15° for 15 min, 25° for 15 min, repeated four times) from 16 to 18 hr of development after plating starved L1’s on HB101. There is no statistically significant difference between any of the groups. (B) Individual 4-day-old adult hermaphrodites that had exhausted their self-progeny were crossed to four 1-day-old N2 males and number of cross-brood progeny was counted. The average cross-brood size was reduced for mir-83(n4638); mir-34(gk437) hermaphrodites whether they were raised at 20° or subjected to temperature cycles (15° for 15 min, 25° for 15 min, repeated four times) from 16 to 18 hr of development. Averages were compared using an unpaired t-test, performed by PRISM. ***P-value ≤ 0.005. Each group was compared to the three others. The absence of significance asterisks denotes a lack of statistical significance.
The self-fertility of a C. elegans hermaphrodite is limited by the number of sperm that it produces. Once self-sperm are exhausted, the hermaphrodite’s total reproductive capacity is defined by the number of additional oocytes it can produce that are competent to produce viable cross-progeny upon mating to males (Brenner 1974). To investigate cross-progeny production, N2 and mir-83(n4638); mir-34(gk437) mutants were first either raised at 20° or cycled. After day 4 of adulthood, animals had exhausted their own supply of sperm and could no longer produce fertilized embryos (Byerly et al. 1976). At this point, young adult N2 males were mated to aged animals to assess their fertility (Figure 6B). Temperature oscillations did not affect the number of cross-progeny produced by wild-type hermaphrodites crossed to wild-type males (247.25 ± 77.52 vs. 236.5 ± 103.44 at 20°, P ≥ 0.05, unpaired t-test). Additionally, there was no significant difference between the number of cross-progeny produced by mir-83(n4638); mir-34(gk437) worms raised in either constant temperature or with temperature oscillations when mated to wild-type males (141.85 ± 84.01 at 20° vs. 118.65 ± 51.99 cycled, P ≥ 0.05, unpaired t-test). There were, however, significant differences between strains in total cross-progeny produced. Specifically, the number of cross-progeny produced by mir-83(n4638); mir-34(gk437) mutants when mated to wild-type males was significantly less than the number of cross-progeny produced by wild-type hermaphrodites mated to wild-type males regardless of the temperature regime during development (P ≤ 0.005, unpaired t-test).
If the fecundity defect in mir-83(n4638); mir-34(gk437) hermaphrodites was a direct consequence of the gonad migration defect, we would expect to see a greater number of cross-progeny produced by mir-83(n4638); mir-34(gk437) mutants when raised at 20° compared to temperature cycled, as temperature cycling dramatically decreases the percentage of animals with normal gonad morphology. Moreover, we would expect to see two populations of animals within the cycled conditioned, a lower number of viable progeny for the ∼60% of the population expected to have a gonad migration defect, and a higher number of viable progeny for the 40% of the population expected to lack any defect. However, the reduced fecundity of mir-83(n4638); mir-34(gk437) hermaphrodites was unaffected by temperature regimen and the population distribution of fecundity for mir-83(n4638); mir-34(gk437) worms was not consistent with a 60/40% split. Thus it appears that the fecundity defect in mir-83(n4638); mir-34(gk437) mutants is likely independent of the gonad migration defect. It is possible that mir-34 and mir-83 may regulate targets other than cdc-42 and pat-3 within the DTCs that affect their ability to signal to the germline for the regulation of oocyte production. Alternatively, there may be subtle changes in the mating behavior of mir-83(n4638); mir-34(gk437) hermaphrodites that could be detected with closer study.
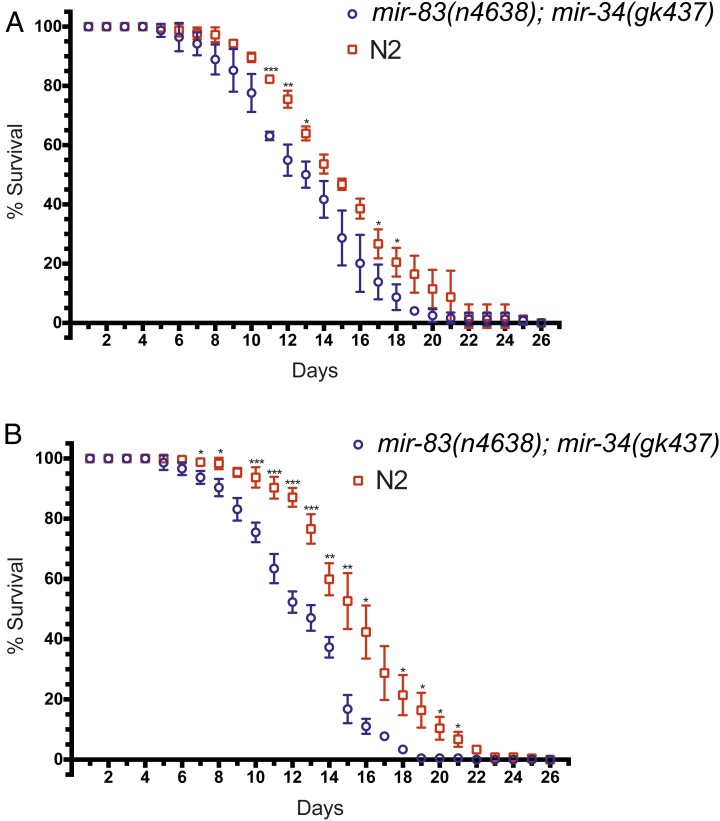
mir-83(n4638); mir-34(gk437) mutants have a decreased lifespan
A rearrangement of internal organs (as seen in Figure 1C) might be expected to have negative consequences for the overall fitness and viability of the affected worm. Moreover, it has been shown that an animal’s fecundity can correlate with its longevity (Hsin and Kenyon 1999; Berman and Kenyon 2006; Kenyon 2010). To explore this possibility, we measured the lifespan of N2 and mir-83(n4638); mir-34(gk437) mutants raised at either 20° continuously (Figure 7A) or with temperature oscillations during the time-sensitive period (Figure 7B). Although mir-83(n4638); mir-34(gk437) worms raised at 20° appeared to have a slightly shortened lifespan compared to N2, the difference was only marginally statistically significant (Figure 7A, ***P ≤ 0.005, **0.005 < P ≤ 0.01, *0.01 < P ≤ 0.05, unpaired t-test). However, for animals subjected to oscillating temperatures during the temperature-sensitive period (from 16 to 18 hr postplating) the difference between N2 and mir-83(n4638); mir-34(gk437) mutant worms was more pronounced and statistically significant from day 7 to day 21 (with the exception of days 9 and 17, Figure 7B, ***P ≤ 0.005, **0.005 < P ≤ 0.01, *0.01 < P ≤ 0.05, unpaired t-test). The difference in lifespans may be related to the DTC migration phenotype. A relation between the two would explain the slight, less significant difference in lifespan at 20°, when only ∼20% of the mir-83(n4638); mir-34(gk437) population is migration defective, vs. the significant difference in lifespan when animals undergo temperature changes, as now ∼60% of the population is migration defective. This hypothesis has yet to be explored as we have not scored individual worms for both the migration phenotype and lifespan due to the fact that scoring for the migration phenotype involves paralyzing the worms and is potentially detrimental to their lifespan. It is possible that the more pronounced difference in lifespan between mir-83(n4638); mir-34(gk437) and N2 worms when raised with temperature oscillations does not reflect a direct relationship between lifespan and gonadal migration but instead could reflect a general decrease in the fitness of mir-83(n4638); mir-34 (gk437) worms when experiencing temperature changes.
Figure 7.
mir-83(n4638); mir-34(gk437) mutants have a decreased lifespan compared to wild type. Wild-type (N2) and mir-83(n4638); mir-34(gk437) embryos were harvested by hypochlorite treatment and synchronized L1 larvae were obtained by hatching overnight in M9. Larvae were plated on HB101-seeded NGM plates and raised (A) continuously at 20° or (B) subjected to temperature cycles (15° for 15 min, 25° for 15 min, repeated four times) from 16 to 18 hr of development after plating starved L1’s on HB101. For each longevity assay shown in panels A and B, 100 animals per replicate, three replicates per condition, were plated as L4’s (day 0) and tracked until their death. Alive or dead was determined by prodding worms on their nose and looking for a reaction. Daily averages were compared using a two sample t-test for means. ***P-value ≤ 0.005, **0.005 < P ≤ 0.01, *0.01 < P ≤ 0.05.
Discussion
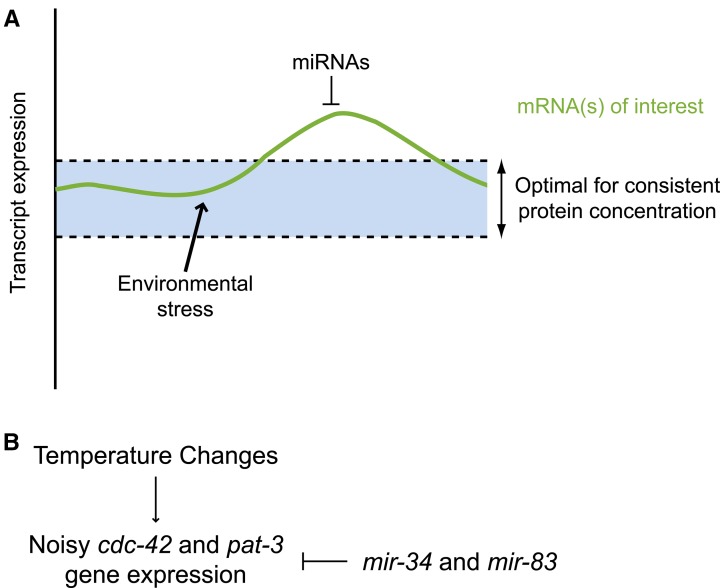
There is an obvious benefit for building biological robustness into genetic networks; the fitness of an organism critically depends on the fidelity of developmental processes and reproductive capacity in the face of environmental changes. Previous research across numerous model organisms has described evolutionarily conserved responses to stresses such as food deprivation, heat shock, and various forms of toxicity (reviewed in Lant and Storey 2010). miRNAs are thought to contribute to ensuring the fidelity of gene expression programs in the face of both external stresses and internal transcriptional noise (Li et al. 2009) (reviewed in Hornstein and Shomron 2006; Ebert and Sharp 2012; Posadas and Carthew 2014). Previous studies have shown how miRNAs can participate in regulatory loops to precisely regulate gene expression levels (reviewed in Tsang et al. 2007; Ebert and Sharp 2012) or establish genetic switches. miRNAs have also been shown to repress leaky transcription, thereby dampening noise within genetic networks (reviewed in Hornstein and Shomron 2006; Posadas and Carthew 2014).
The precise spatiotemporal program of DTC migration is controlled by a complex genetic network including genes pat-3 and cdc-42 (Cram et al. 2006), which encode, respectively, β-integrin and a GTPase downstream of integrin signaling. Genetic networks need to be robust so as to compensate for adverse changes in gene expression that can result from stress (reviewed in Lant and Storey 2010; Zhou et al. 2011). Cells can compensate for stress-induced positive fluctuations in mRNA levels through the post-transcriptional repressive action of miRNAs (Figure 8A). Likewise, negative fluctuations in mRNA levels can be compensated for by activating transcription and/or translation, including releasing inhibition imparted by miRNAs. Thus miRNAs can contribute to the robustness of a genetic network by fine tuning the expression levels of networked genes and buffering their expression from environmentally driven adverse perturbations.
Figure 8.
A model proposing robustness functions for mir-34 and mir-83 through dampening noisy cdc-42 and pat-3 expression in the face of temperature changes. (A) Environmental stresses, such as changing temperatures, may lead to fluctuations in the expression levels of various transcripts, which could challenge cellular proteomic homeostasis. Repression of protein production by miRNAs can provide a means of stabilizing protein output from fluctuating target transcripts. (B) In the case of mir-34 and mir-83, the fidelity of DTC migration is proposed to be maintained in part by inhibiting noisy cdc-42 and pat-3 expression incited by unstable environmental temperature.
For DTCs, phase 1 and phase 3 of their migration is regulated by cell-intrinsic integrin signaling (Baum and Garriga 1997; Lee et al. 2001; Cram et al. 2006; Meighan and Schwarzbauer 2007). We propose that both mir-34 and mir-83 help protect the robustness of this genetic network by regulating cdc-42 and pat-3 (Figure 8B), particularly when the network is stressed by temperature changes. Our finding that DTC expression of mir-34 and mir-83 can rescue the migration defect of their respective mutants supports the hypothesis that the two miRNAs regulate cdc-42 and pat-3 within the DTCs. Although these rescue data strongly suggest that mir-34 and mir-83 function within the DTCs, we were not able to confirm expression of these miRNAs in DTCs. Transgenes with either the mir-34 or mir-83 promoter driving GFP did not produce detectable expression of GFP in the DTCs (although expression in body wall muscle cells was detected for mir-34 driven GFP). It is possible that the endogenous mir-34 and mir-83 genes are expressed in the DTCs, but these transgenic constructs may express GFP at levels below limits of detection.
Interestingly, we also observed rescue of the migration defect in mir-83(n4638) mutants by expression of mir-83 in muscle cells using the myo-3 promoter. This could reflect a low level of activity of the myo-3 promoter in the DTCs. Alternatively, it is possible that mir-83 may perform functions within both the DTCs and muscle, and that function in either cell type can rescue DTC migration. A third possibility is that mir-83 may function only in the DTCs, but that the mir-83 miRNA can be supplied either cell intrinsically or cell extrinsically from muscle-expressed mir-83. Intriguingly, another known component of the DTC migration gene network, the metalloprotease mig-17, is secreted by body wall muscle cells and localizes to the gonadal basement membrane (Nishiwaki et al. 2000). It is also possible that mir-83 synthesized in the muscle could be transported to the DTCs to function, explaining why mir-83 expression in either of these two tissues can result in similar levels of rescue.
For technical reasons, our fluorescent reporter transgene approach did not permit us to confirm 3′ UTR-dependent regulation of cdc-42 or pat-3 by mir-34 or mir-83. Both genes are widely expressed; cdc-42 is most highly expressed in the intestine and muscles, while pat-3 expression is particularly high in muscles (unpublished results and Plenefisch et al. 2000). Since the two DTCs make up such a small fraction of the worm, protein expression changes within the DTCs are not easily analyzed by Western blotting as expression throughout the entire worm will mask any small cell-specific changes. We used fluorescent reporters due to the difficulty of seeing cell-specific changes. We first attempted to generate single copy insertions of the four reporters. For both the pat-3 GFP reporter and the cdc-42 mCherry reporter, we were not able to generate single copy integrated transgenes that expressed at detectable levels. Therefore we were forced to use multicopy arrays, which most certainly reflect an overexpression of mRNA. We suspect that the absence of measurable response of these reporters to mir-34 and mir-83 activity in our experiments likely reflects an overexpression of the reporters, in excess to the endogenous miRNA levels. It is also possible that mir-34 and mir-83 may regulate their targets in a fashion too dynamic to visualize using fluorescent reporters.
Here we showed that mir-34 and mir-83 both contribute to keeping DTC migrations robust and protecting the fidelity of DTC migratory behavior from changes in temperature. In addition to organizing the morphogenic processes that shape the mature gonad, DTCs are also responsible for regulating meiosis in the germline (Hubbard and Greenstein 2000). Therefore DTCs have a direct impact on C. elegans reproductive capacity, and there is a direct benefit for the worm to protect these cells from external stresses. Interestingly, although mir-83(n4638); mir-34(gk437) mutants produced the normal number of self-progeny in our experiments, they nevertheless exhibited reduced numbers of cross-progeny when mated to wild-type males. Thus, the overall maximum reproductive capacity of mir-83(n4638); mir-34(gk437) hermaphrodites is compromised. Our results suggest that this maximum fecundity defect is not a direct consequence of the gonad migration defect, as the penetrance of the fecundity defect is higher than that of the migration defect and is not enhanced by temperature oscillations. Additionally the fecundity defect may reflect a role for mir-34 and mir-83 in regulating fertility that is entirely unrelated to their role in regulating DTC migrations. We do not yet know the pathway or pathways through which mir-34 and mir-83 affect fertility or whether the fertility phenotypes represent functions within the germline, DTCs, or both.
We identified a 2-hr temperature-sensitive period, during which temperature oscillations can induce the enhanced DTC migration phenotype of mir-83(n4638); mir-34(gk437) hermaphrodites. This temperature-sensitive period overlaps with the birth of the DTCs during the L1 larval stage and is well before the DTCs begin their migration (Sulston 1976; Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). We have not determined what aspect of DTC specification and/or differentiation may be inherently sensitive to temperature changes such that in the absence of mir-34 and mir-83, DTCs display a stronger defect in pathfinding fidelity than when under constant temperature. It is intriguing that this defect is enhanced by fluctuating temperature specifically during the L1 stage, suggesting that DTCs are particularly sensitive to the stress of unstable temperature prior to the execution of their migratory program. It is known that the rate of development is closely tied to the environmental temperature in C. elegans. Temperature sensitivity during the L1 stage may reflect a requirement to buffer noise within the integrin genetic network early in development. A lack of proper initial buffering may sensitize DTCs such that their subsequent migration is no longer robust. Alternatively, early temperature fluctuations may lead to changes in gene expression that are subsequently buffered by mir-34 and mir-83 as the DTCs actively migrate during later larval stages.
In mammals, both mir-34 and mir-29 (the mammalian mir-83 homolog) have been implicated in cancer, and mir-29 has also been implicated in regulating fibrosis and cell–cell interactions (reviewed in Boominathan 2010; Kriegel et al. 2012). Mammalian mir-34 homologs are transcriptionally activated by p53 and mediate post-transcriptional regulatory processes downstream of p53 (reviewed in He et al. 2007; Hermeking 2009; Boominathan 2010). mir-34a overexpression has been shown to reduce lung cancer tumor cell proliferation and tumor volume (Xue et al. 2014). Human cells contain four paralogs of mir-29/mir-83: namely, mir-29a, mir-29b-1, mir-29b-2, and mir-29c (reviewed in Kriegel et al. 2012). Expression of the mir-29 miRNAs are regulated by both c-Myc and NF-κB and have been shown to regulate genes involved in apoptosis, cell proliferation, differentiation, and extracellular matrix components (reviewed in Kriegel et al. 2012). The mir-29 miRNAs have been shown to be downregulated in a variety of cancers, including cervical, colon, liver, leukemia, lung, and melanoma (Calin et al. 2005; Yanaihara et al. 2006; Cummins et al. 2006; Pekarsky et al. 2006; Garzon et al. 2008, 2009; Xiong et al. 2010; Li et al. 2011; Nguyen et al. 2011). They have also been shown to be up-regulated in certain breast cancers. Human CDC42 was shown to be a direct target of the mir-29 miRNAs in work from the Kim lab (Park et al. 2008). Additionally, they showed that the suppression of CDC42 by mir-29 led to p53 up-regulation and increased apoptosis. Taken together with the work in mammals showing that mir-34 reinforces p53 negative regulation, this suggests that the coregulation of a genetic network by mir-34 and mir-83/mir-29 is evolutionarily ancient and conserved. Furthermore, TargetScan (Lewis et al. 2005; Grimson et al. 2007; Friedman et al. 2009; Garcia et al. 2011) predicts conserved targeting of human ITGB1 (integrin beta-1), ITGA11 (integrin alpha-11), and ITGA6 (integrin alpha-6) by the mir-29 family and conserved targeting for human ITGB8 (integrin beta-8) and ITGA10 (integrin alpha-10) by the mir-34 family.
Here we show a link between mir-34, mir-83/mir-29, and integrin-controlled cell migration. Integrin and extracellular matrix misregulation is a key factor in epithelial to mesenchymal transition (EMT) (reviewed in Seguin et al. 2015). The implication of mir-34 and mir-83/mir-29 in various cancers may reflect a conserved involvement in metastasis through altered EMT that has yet to be explored. The wandering DTC defect in C. elegans mir-34 and mir-83/mir-29 mutants, which we have shown results from misregulation of pat-3 and cdc-42, may reflect a homologous miRNA–integrin axis in tumor formation, proliferation, and metastasis in higher animals, including humans.
Supplementary Material
Acknowledgments
We thank the members of the Ambros lab for helpful discussion, the labs of Nathan Lawson and Oliver Hobert for plasmids, and the lab of Amy Walker for use of their confocal microscope. We also thank the Electron Microscopy Core Facility at the University of Massachusetts for electron microscopy data. The Electron Microscopy Core Facility is supported by award no. S10RR027897 from the National Center For Research Resources. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). The authors are solely responsible for the content of this paper and it does not necessarily represent the official views of the National Center for Research Resources of the NIH. This research was supported by NIH R01GM34028 and by a grant from the Ellison Medical Foundation.
Author contributions: S.L.B. performed the experiments and analyzed the data. S.L.B., M.H., and V.A. designed experiments; S.L.B. and V.A. wrote the paper.
Footnotes
Communicating editor: D. I. Greenstein
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179184/-/DC1.
Literature Cited
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R. C., Lavanway A., Williams P. T., Jewell D., 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., et al. , 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Bartel D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16: 780–785. [DOI] [PubMed] [Google Scholar]
- Baum P. D., Garriga G., 1997. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron 19: 51–62. [DOI] [PubMed] [Google Scholar]
- Berman J. R., Kenyon C., 2006. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124: 1055–1068. [DOI] [PubMed] [Google Scholar]
- Boominathan L., 2010. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 29: 613–639. [DOI] [PubMed] [Google Scholar]
- Brenner J. L., Jasiewicz K. L., Fahley A. F., Kemp B. J., Abbott A. L., 2010. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 20: 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L., Cassada R. C., Russell R. L., 1976. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev. Biol. 51: 23–33. [DOI] [PubMed] [Google Scholar]
- Calin G. A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., et al. , 2005. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. The dauer larva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- Cram E. J., Shang H., Schwarzbauer J. E., 2006. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J. Cell Sci. 119: 4811–4818. [DOI] [PubMed] [Google Scholar]
- Cummins J. M., He Y., Leary R. J., Pagliarini R., Diaz L. A., et al. , 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 103: 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Mourelatos Z., Yang M., Sharma A., Dreyfuss G., 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D., Bertin N., Hidalgo C. A., Venkatesan K., Tu D., et al. , 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25: 663–668. [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Sharp P. A., 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. M., Watson J. D., Stetina Von S. E., McDermott J., Brodigan T. M., et al. , 2007. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol. 8: R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K.-H., Burge C. B., Bartel D. P., 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Rougvie A. E., Rothman J. H., 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16: 773–779. [DOI] [PubMed] [Google Scholar]
- Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., et al. , 2011. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Garofalo M., Martelli M. P., Briesewitz R., Wang L., et al. , 2008. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA 105: 3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Heaphy C. E. A., Havelange V., Fabbri M., Volinia S. et al, 2009. MicroRNA 29b functions in acute myeloid leukemia. Blood 114: 5331–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner S. N., Kenyon C., Reichardt L. F., 1995. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 129: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y., Aach J., Hayes G. D., Reinhart B. J., Church G. M., et al. , 2003. Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11: 1253–1263. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., 2004. The microRNA Registry. Nucleic Acids Res. 32: D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J., 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34: D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J., 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K. K.-H., Johnston W. K., Garrett-Engele P., Lim L. P., et al. , 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D. H., and Z. F. Altun, 2008 C. Elegans Atlas, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. [DOI] [PubMed] [Google Scholar]
- Hammell M., Long D., Zhang L., Lee A., Carmack C. S., et al. , 2008. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods 5: 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., He L., Hannon G. J., 2007. The guardian’s little helper: microRNAS in the p53 tumor suppressor network. Cancer Res. 67: 11099–11101. [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D., 1987. Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100: 365–382. [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Hermeking H., 2009. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17: 193–199. [DOI] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N., 2006. Canalization of development by microRNAs. Nat. Genet. 38(Suppl): S20–S24. [DOI] [PubMed] [Google Scholar]
- Houbaviy H. B., Murray M. F., Sharp P. A., 2003. Embryonic stem cell-specific MicroRNAs. Dev. Cell 5: 351–358. [DOI] [PubMed] [Google Scholar]
- Hsin H., Kenyon C., 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Greenstein D., 2000. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev. Dyn. 218: 2–22. [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E., Troemel E. R., Feinbaum R. L., Luhachack L. G., Cezairliyan B. O., et al. , 2010. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6: e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. E., Mitchell D. H., Kline S., Kemal R., Foy J., 1984. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 28: 23–40. [DOI] [PubMed] [Google Scholar]
- Johnstone I. L., Shafi Y., Barry J. D., 1992. Molecular analysis of mutations in the Caenorhabditis elegans collagen gene dpy-7. EMBO J. 11: 3857–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., 2010. The genetics of ageing. Nature 464: 504–512. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kimata T., Tanizawa Y., Can Y., Ikeda S., Kuhara A., et al. , 2012. Synaptic polarity depends on phosphatidylinositol signaling regulated by myo-inositol monophosphatase in Caenorhabditis elegans. Genetics 191: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. [DOI] [PubMed] [Google Scholar]
- Kitano H., 2004. Biological robustness. Nat. Rev. Genet. 5: 826–837. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A. J., Liu Y., Fang Y., Ding X., Liang M., 2012. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genomics 44: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., et al. , 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., et al. , 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant B., Storey K. B., 2010. An overview of stress response and hypometabolic strategies in Caenorhabditis elegans: conserved and contrasting signals with the mammalian system. Int. J. Biol. Sci. 6: 9–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N. C., 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee M., Cram E. J., Shen B., Schwarzbauer J. E., 2001. Roles for pat-3 Integrins in Development and Function of Caenorhabditis elegans Muscles and Gonads. J. Biol. Chem. 276: 36404–36410. [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P., 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W., 2009. A microRNA imparts robustness against environmental fluctuation during development. Dev. Cell 137: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang F., Xu J., Ye F., Shen Y., et al. , 2011. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J. Pathol. 224: 484–495. [DOI] [PubMed] [Google Scholar]
- Lim L. P., 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Glasner M. E., Yekta S., Burge C. B., Bartel D. P., 2003. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Liu Z., Kirch S., Ambros V., 1995. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121: 2471–2478. [DOI] [PubMed] [Google Scholar]
- Lucanic M., Cheng H.-J., 2008. A RAC/CDC-42–independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 4: e1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui W.-O., Pourmand N., Patterson B. K., Fire A., 2007. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 67: 6031–6043. [DOI] [PubMed] [Google Scholar]
- Meighan C. M., Schwarzbauer J. E., 2007. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 21: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael M. Z., O’ Connor S. M., van Holst Pellekaan N. G., Young G. P., James R. J., 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1: 882–891. [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., et al. , 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3: e215–e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., et al. , 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Kuo C., Nicholl M. B., Sim M.-S., Turner R. R., et al. , 2011. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics 6: 388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K., Hisamoto N., Matsumoto K., 2000. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science 288: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Ono K., Yu R., Ono S., 2007. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev. Dyn. 236: 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Lee J. H., Ha M., Nam J.-W., Kim V. N., 2008. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat. Struct. Mol. Biol. 16: 23–29. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y., Santanam U., Cimmino A., Palamarchuk A., Efanov A., et al. , 2006. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 66: 11590–11593. [DOI] [PubMed] [Google Scholar]
- Powell J. R., Ausubel F. M., 2008. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol. Biol. 415: 403–427. [DOI] [PubMed] [Google Scholar]
- Plenefisch J. D., Zhu X., Hedgecock E. M., 2000. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development 127: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Posadas D. M., Carthew R. W., 2014. MicroRNAs and their roles in developmental canalization. Curr. Opin. Genet. Dev. 27: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., Ausubel F. M., 2008. Models of Caenorhabditis elegans Infection by Bacterial and Fungal Pathogens. Methods Mol. Biol. 415: 403–427. [DOI] [PubMed] [Google Scholar]
- Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., et al. , 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230. [DOI] [PubMed] [Google Scholar]
- Price L. S., Leng J., Schwartz M. A., Bokoch G. M., 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. D., Kiosses W. B., Schwartz M. A., 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18: 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokavec M., Li H., Jiang L., Hermeking H., 2014. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 6: 214–230. [DOI] [PubMed] [Google Scholar]
- Ruaud A.-F., Bessereau J.-L., 2006. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development 133: 2211–2222. [DOI] [PubMed] [Google Scholar]
- Sahu S. N., Lewis J., Patel I., Bozdag S., Lee J. H., et al. , 2013. Genomic Analysis of Stress Response against Arsenic in Caenorhabditis elegans. PLoS ONE 8: e66431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler A. J., Baugh L. R., Sherwood D. R., 2014. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10: e1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin L., Desgrosellier J. S., Weis S. M., Cheresh D. A., 2015. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 25: 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., Ambros V., 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259: 9–18. [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Kimble J., 2002. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development 129: 443–453. [DOI] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook. 1.101. 1. 10.1895/wormbook [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M.-R., Lee Y., Kim J. Y., Kim S.-K., Moon S.-H., et al. , 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270: 488–498. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., 1976. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275: 287–297. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A., 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L., D’Souza-Schorey C., 1997. Rho GTPases and signaling networks. Genes Dev. 11: 2295–2322. [DOI] [PubMed] [Google Scholar]
- Williams B. D., Waterston R. H., 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124: 475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.-C., Schwarzbauer J. E., 2012. Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite. Wiley Interdiscip. Rev. Dev. Biol. 1: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Fang J.-H., Yun J.-P., Yang J., Zhang Y., et al. , 2010. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51: 836–845. [DOI] [PubMed] [Google Scholar]
- Xue W., Dahlman J. E., Tammela T., Khan O. F., Sood S., et al. , 2014. Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. USA 111: E3553–E3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M., Lowenstein C. J., 2009. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle 8: 712–715. [DOI] [PubMed] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., et al. , 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198. [DOI] [PubMed] [Google Scholar]
- Zhou K. I., Pincus Z., Slack F. J., 2011. Longevity and stress in Caenorhabditis elegans. Aging (Albany, N.Y. Online) 3: 733–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Bouasker S., Simard M. J., Hammell C. M., Ambros V., 2014. Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genet. 10: e1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.