Abstract
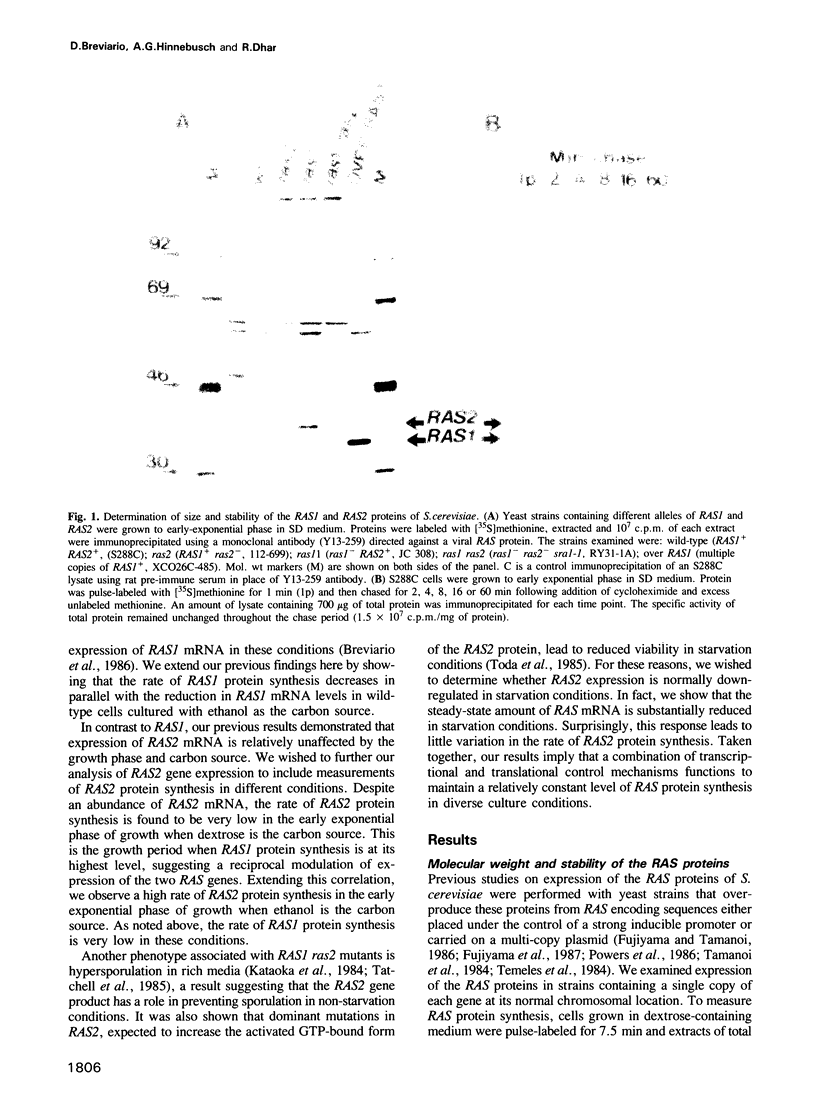
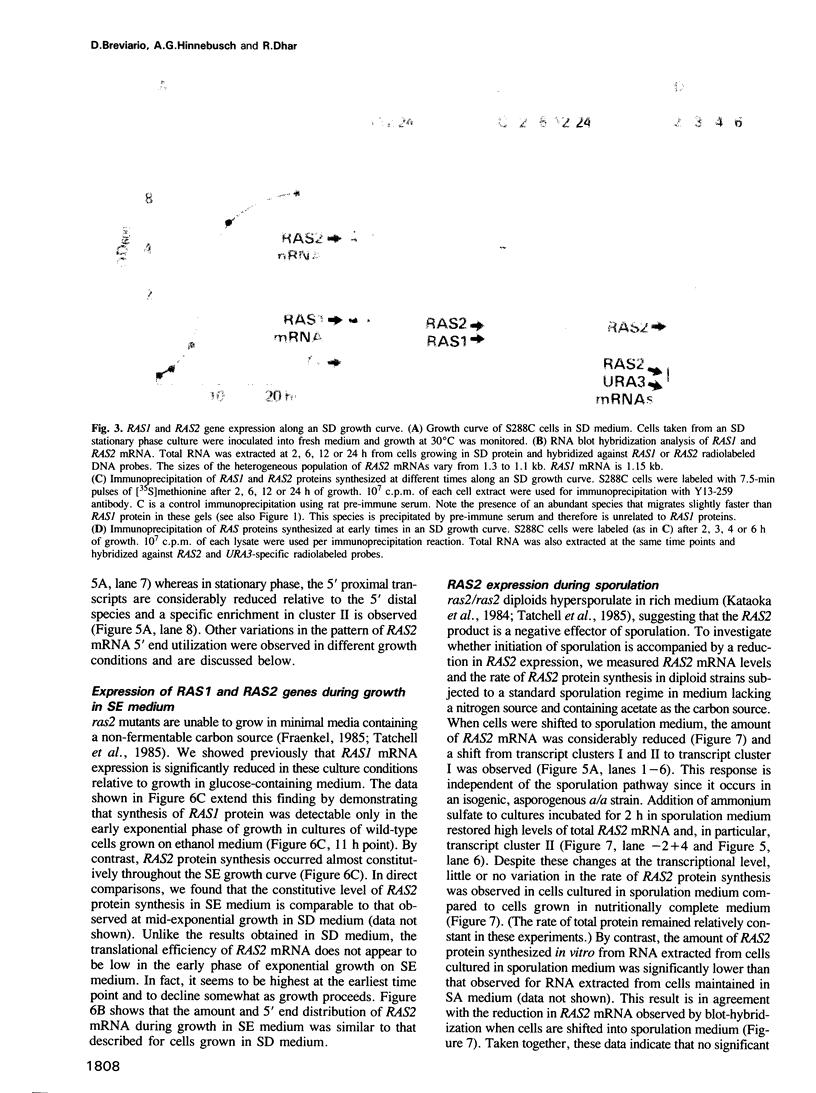
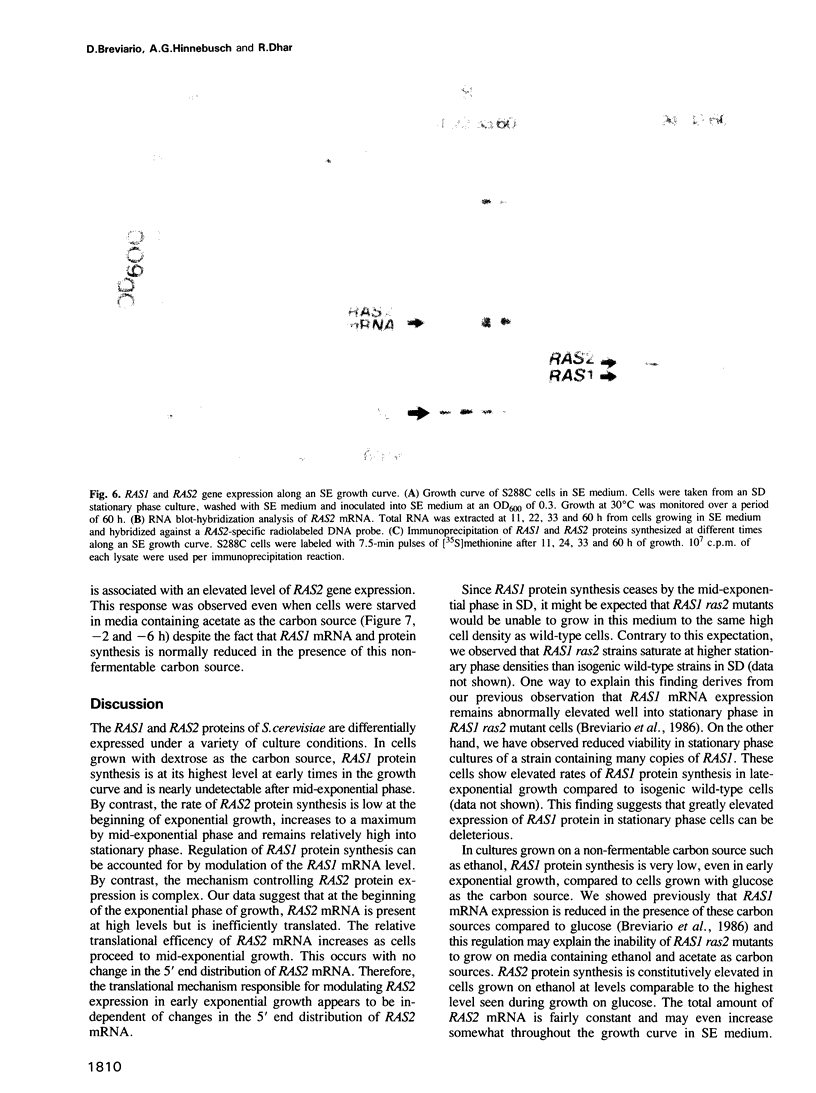
Expression of the RAS1 and RAS2 genes of Saccharomyces cerevisiae has been examined at the transcriptional and translational levels. When dextrose is the carbon source, the steady-state amount of RAS1 mRNA and the rate of RAS1 protein synthesis are reduced in parallel as cells approach the mid-exponential phase of growth. RAS1 mRNA levels and protein synthesis are very low at all stages of growth when ethanol rather than dextrose is provided as the sole carbon source. The rate of RAS2 protein synthesis is regulated differently. In cells cultured on dextrose, it is lowest in the early exponential phase, increases approximately 10-fold and remains nearly constant as cells approach stationary phase. By contrast, RAS2 mRNA is found at uniformly high levels at all phases of exponential growth, suggesting that the translational efficiency of RAS2 mRNA is repressed during the early exponential phase. This repression is not observed when ethanol is the sole carbon source. Nutrient starvation, resulting in G1 arrest and sporulation in diploids, leads to greatly decreased amounts of RAS2 mRNA, accomplished in part by selective repression of RAS2 transcripts with particular 5' ends. However, this reduction in RAS2 mRNA levels has little effect on the rate of RAS2 protein synthesis, suggesting that the translational efficiency of RAS2 mRNA is stimulated by nutrient starvation. The combination of transcriptional and translational controls which regulate yeast RAS gene expression seems to ensure that one or the other RAS proteins will be produced over a wide range of physiological states.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradley M. O., Kraynak A. R., Storer R. D., Gibbs J. B. Experimental metastasis in nude mice of NIH 3T3 cells containing various ras genes. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5277–5281. doi: 10.1073/pnas.83.14.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breviario D., Hinnebusch A., Cannon J., Tatchell K., Dhar R. Carbon source regulation of RAS1 expression in Saccharomyces cerevisiae and the phenotypes of ras2- cells. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4152–4156. doi: 10.1073/pnas.83.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J. E., Sefton B. M. Direct identification of palmitic acid as the lipid attached to p21ras. Mol Cell Biol. 1986 Jan;6(1):116–122. doi: 10.1128/mcb.6.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. F., Gibbs J. B., Tatchell K. Suppressors of the ras2 mutation of Saccharomyces cerevisiae. Genetics. 1986 Jun;113(2):247–264. doi: 10.1093/genetics/113.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- De Vendittis E., Vitelli A., Zahn R., Fasano O. Suppression of defective RAS1 and RAS2 functions in yeast by an adenylate cyclase activated by a single amino acid change. EMBO J. 1986 Dec 20;5(13):3657–3663. doi: 10.1002/j.1460-2075.1986.tb04696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D., Scolnick E. M., Koller R., Dhar R. ras-Related gene sequences identified and isolated from Saccharomyces cerevisiae. Nature. 1983 Dec 15;306(5944):707–709. doi: 10.1038/306707a0. [DOI] [PubMed] [Google Scholar]
- DeFeo-Jones D., Tatchell K., Robinson L. C., Sigal I. S., Vass W. C., Lowy D. R., Scolnick E. M. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985 Apr 12;228(4696):179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- Dhar R., Nieto A., Koller R., DeFeo-Jones D., Scolnick E. M. Nucleotide sequence of two rasH related-genes isolated from the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Apr 25;12(8):3611–3618. doi: 10.1093/nar/12.8.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel D. G. On ras gene function in yeast. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4740–4744. doi: 10.1073/pnas.82.14.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A., Matsumoto K., Tamanoi F. A novel yeast mutant defective in the processing of ras proteins: assessment of the effect of the mutation on processing steps. EMBO J. 1987 Jan;6(1):223–228. doi: 10.1002/j.1460-2075.1987.tb04742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A., Tamanoi F. Processing and fatty acid acylation of RAS1 and RAS2 proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1266–1270. doi: 10.1073/pnas.83.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth M. E., Davis L. J., Fleurdelys B., Scolnick E. M. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family. J Virol. 1982 Jul;43(1):294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Broek D., Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985 Dec;43(2 Pt 1):493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Changing patterns of gene expression during sporulation in yeast. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7323–7327. doi: 10.1073/pnas.81.23.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980 Sep;143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Axel R., Myers A. M. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Feb;84(3):779–783. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGrath J. P., Capon D. J., Goeddel D. V., Levinson A. D. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984 Aug 23;310(5979):644–649. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- Papageorge A., Lowy D., Scolnick E. M. Comparative biochemical properties of p21 ras molecules coded for by viral and cellular ras genes. J Virol. 1982 Nov;44(2):509–519. doi: 10.1128/jvi.44.2.509-519.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., Kataoka T., Fasano O., Goldfarb M., Strathern J., Broach J., Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell. 1984 Mar;36(3):607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- Powers S., Michaelis S., Broek D., Santa Anna S., Field J., Herskowitz I., Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986 Nov 7;47(3):413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Tamanoi F., Walsh M., Kataoka T., Wigler M. A product of yeast RAS2 gene is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6924–6928. doi: 10.1073/pnas.81.22.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Tatchell K. RAS genes and growth control in Saccharomyces cerevisiae. J Bacteriol. 1986 May;166(2):364–367. doi: 10.1128/jb.166.2.364-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Robinson L. C., Breitenbach M. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3785–3789. doi: 10.1073/pnas.82.11.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeles G. L., DeFeo-Jones D., Tatchell K., Ellinger M. S., Scolnick E. M. Expression and characterization of ras mRNAs from Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2298–2305. doi: 10.1128/mcb.4.11.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeles G. L., Gibbs J. B., D'Alonzo J. S., Sigal I. S., Scolnick E. M. Yeast and mammalian ras proteins have conserved biochemical properties. Nature. 1985 Feb 21;313(6004):700–703. doi: 10.1038/313700a0. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Christensen A., Hubbert N. L., Papageorge A. G., Lowy D. R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984 Aug 16;310(5978):583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]