Abstract
MicroRNAs (miRNAs) emerge as important regulators of stem cell lineage commitment and bone development. MiRNA-26a (miR-26a) is one of the important miRNAs regulating osteogenic differentiation of both bone marrow-derived mesenchymal stem cells (BMSCs) and adipose tissue-derived mesenchymal stem cells (ADSCs). However, miR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs, suggesting distinct post-transcriptional regulation of tissue-specific MSC differentiation. However, the molecular basis is largely unknown. Here, we report that the function of miR-26a is largely depended on the intrinsic signaling regulation network of MSCs. Using bioinformatics and functional assay, we confirmed that miR-26a potentially targeted on GSK3β and Smad1 to regulate Wnt and BMP signaling pathway. Overall comparative analysis revealed that Wnt signaling was enhanced more potently and played a more important role than BMP signaling in osteogenic differentiation of BMSCs, whereas BMP pathway was more essential for promoting osteogenic differentiation of ADSCs. The distinct activation pattern and role of signaling pathways determined that miR-26a majorly targeted on GSK3β to activate Wnt signaling for promoting osteogenic differentiation of BMSCs, whereas it inhibited Smad1 to suppress BMP signaling for interfering with the osteogenic differentiation of ADSCs. Taken together, our study demonstrated that BMSCs and ADSCs applied different signaling pathway to facilitate their osteogenic differentiation, which determined the inverse function of miR-26a. The distinct transcriptional regulation and post-transcriptional regulation network suggested the intrinsic molecular differences between tissue-specific MSCs and the complexity in MSC research and MSC-based cell therapy.
Our understanding of the molecular mechanisms governing differentiation of mesenchymal stem cells (MSCs) developed rapidly in the past decades. A number of secretory molecules and transcription factors have been identified as regulators controlling osteoblastogenesis.1, 2, 3 Secreted molecules including bone morphogenetic proteins (BMPs), Wnt proteins, Indian hedgehog (IHH) and fibroblast growth factors (FGFs) are necessary for osteoblast differentiation and bone development.1, 3 These secreted molecules activate different signaling pathways through autocrine or paracrine signaling to regulate the expression of a set of transcription factors. Osteoblast-specific (Runx2, Osterix and ATF4) and nonspecific factors, expressing at distinct time points during the differentiation process, determine the lineage commitment of MSCs.1, 2
One recent breakthrough is that microRNAs (miRNAs), a class of 22–24 bp noncoding RNAs, emerge as important regulatory mechanism of MSC lineage commitment and bone development.4, 5 Several miRNAs have been identified as important regulators of osteogenesis.6 Among them, miR-26a is one of the important miRNAs regulating the osteogenic differentiation of both bone marrow-derived MSCs (BMSCs) and adipose tissue-derived MSCs (ADSCs). Expression of miR-26a is significantly increased in both BMSCs and ADSCs under osteogenic induction.7, 8, 9 Our recent work confirmed that miR-26a is a promoter of BMSC osteogenic differentiation.10 However, Luzi et al.11, 12 showed that miR-26a suppresses the osteogenic differentiation of ADSCs. These contradictory observations indicated that miR-26a plays distinct post-transcriptional regulatory function between BMSCs and ADSCs. However, the molecular basis is totally unknown.
BMSCs, which are capable of self-renewal and multipotent differentiation, play important role in bone homeostasis and regeneration.13 Among MSCs isolated from several connective tissues, ADSCs sharing many biological characteristics with BMSCs are attractive alternatives for cell therapy.14, 15, 16 Both BMSCs and ADSCs could potently differentiate into osteoblasts under similar induction conditions17 and have been applied to bone repair and regeneration in clinic trials.18, 19, 20 However, notwithstanding the similarity of cell behavior and function in vitro, BMSCs and ADSCs have distinct origin, location and physiological function in vivo. This raises the question of whether the molecular basis of osteogenic differentiation between BMSCs and ADSCs is identical. Indeed, a number of recent studies based on profiling strategy showed differences at transcriptional and proteomic levels between BMSCs and ADSCs.21, 22, 23, 24, 25, 26 Importantly, a number of differentially expressed genes are involved in Wnt signaling and other differentiation pathways,22, 26 suggesting a difference in signaling regulation programs between BMSCs and ADSCs. However, no conclusive functional study confirms that BMSCs and ADSCs adopt different sets of gene regulation programs for osteogenic differentiation.
The opposing function of miR-26a further suggests that the regulation program of BMSC and ADSC differentiation is different at post-transcriptional level. As one miRNA targets on numbers of mRNAs,27 the opposing function of miR-26a might be due to it targeting on different osteogenic regulators in BMSCs and ADSCs. In this study, we showed that the function of miR-26a is largely depended on the intrinsic signaling regulation network of MSCs. This finding indicates that there are specific signaling regulation networks between different MSCs, suggesting the complexity of tissue-specific MSC research and application in bone regeneration.
Results
MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs
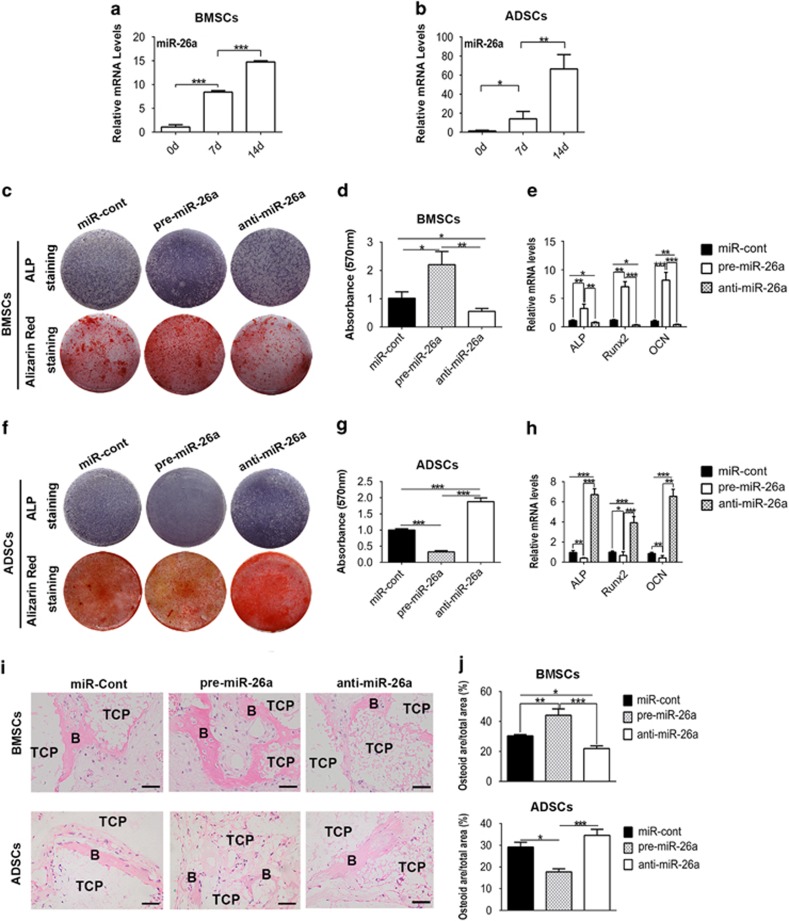
To investigate the function of miR-26a in osteogenic differentiation of BMSCs and ADSCs, we cultured BMSCs and ADSCs in a medium containing ascorbic acid, β-glycerophosphate and dexamethasone, widely used in in vitro osteogenesis model.28 After osteogenic induction, BMSCs and ADSCs highly expressed Alp (alkaline phosphatase, a marker of early osteogenic differentiation) and Ocn (osteocalcin, a critical marker of mature osteoblasts), and formed mineralized nodules (Supplementary Figure S1D–G). Notably, the expression of miR-26a was significantly increased during osteogenic differentiation of both BMSCs and ADSCs (Figures 1a and b).
Figure 1.
MiR-26a inversely affects the osteogenic differentiation of BMSCs and ADSCs. (a and b) Expression of miR-26a in BMSCs (a) and ADSCs (b) during osteogenic induction. (c–h) BMSCs (c–e) and ADSCs (f–h) were transfected with miR-26a precursors (pre-miR-26a), miR-26a inhibitors (anti-miR-26a) and negative control (miR-cont) for 48 h before osteogenic induction. ALP staining and alizarin red staining were performed after 7 days or 14 days of induction separately (c and f). Alizarin red staining was extracted with cetylpyridinium chloride and quantified by spectrophotometer (d and g). Expression of Alp, Runx2 and Ocn (normalized to β-actin) was determined by real-time RT-PCR (e and h). (i) BMSCs and ADSCs transfected with miR-26a precursors, miR-26a inhibitors and negative control in vitro were transplanted with HA-TCP subcutaneously into immunocompromised mice for 8 weeks. The transplants were harvested and stained with H&E. B, bone; TCP, hydroxyaptite-tricalcium phosphate. (j) Osteoid formation in transplants was evaluated as osteoid area per total area in H&E staining photos with Image Pro software. Scale bar: 200 μm. Results are shown as mean±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=4)
To investigate the role of miR-26a in BMSC and ADSC differentiation, we performed gain- and loss-of-function assay. Synthesized pre-miR-26a or anti-miR-26a was transfected to efficiently overexpress or knock down miR-26a separately (Supplementary Figure S2). In BMSCs, overexpression of miR-26a increased ALP activity, Alp mRNA and Runx2 mRNA expression at day 7, and mineralized nodule formation and Ocn expression at day 14 (Figures 1c–e), mirroring what was seen in knockdown of miR-26a. In contrast, overexpression of miR-26a significantly inhibited osteogenic differentiation of ADSCs, whereas knockdown of miR-26a promoted ADSC osteogenic differentiation (Figures 1f–h).
To further confirm the function of miR-26a in vivo, we transplanted MSCs transfected with pre-miR-26a, anti-miR-26a or negative control subcutaneously into immunocompromised mice. H&E staining of implants showed that BMSC-overexpressed miR-26a (BMSC/pre-miR-26a) formed more bone tissue, whereas BMSC/anti-miR-26a formed less than control. Conversely, overexpression of miR-26a inhibited the in vivo bone formation of ADSCs, whereas knockdown of miR-26a promoted ADSC bone formation (Figures 1i and j). Consistent with previous studies,10, 11 our results indicated that miR-26 promotes BMSC osteogenic differentiation but inhibits ADSC osteogenic differentiation.
MiR-26a targets on both GSK3β and Smad1
To investigate the molecular basis of miR-26a, we employed three miRNA target prediction databases (TargetScan (http://www.targetscan.org/mmu_50/), PicTar (http://pictar.mdc-berlin.de/) and TargetRank (http://genes.mit.edu/targetrank/)) to predict the target mRNA. Among the predicted mRNAs that could regulate osteogenesis, Smad1 (Figure 2a) and GSK3β (Figure 2c) have been experimentally certified.11, 29, 30, 31 To confirm the direct binding of miR-26a on Smad1 and GSK3β mRNAs, we performed luciferase activity assay by co-transfecting pMIR reporter containing the binding sites of Smad1 or GSK3β 3′ UTR with pre-miR-26a or anti-miR-26a. Confirmative with the in silico prediction, overexpression of miR-26a inhibited the luciferase activity of reporters of Smad1 and GSK3β, whereas knockdown of miR-26a increased the luciferase activity of both reporters (Figures 2b and d). Gain- and loss-of-function assays further confirmed that miR-26a decreased both Smad1 and GSK3β protein accumulation in BMSCs (Figure 2f) and ADSCs (Figure 2h). Real-time reverse transcription-PCR (RT-PCR) showed that the mRNA levels of Smad1 and GSK3β were not significantly changed after overexpression or knockdown of miR-26a (Figures 2e and g), indicating that miR-26a functions through post-transcriptional regulation.
Figure 2.
MiR-26a targets on both Smad1 and GSK3β mRNA. (a) The diagram showed the binding region of miR-26a to 3′ UTR of Smad1 by complementary base paring. (b) Luciferase activity of pMIR-Reporter containing Smad1 3′ UTR was measured 48 h after co-transfection with pre-miR-26a, anti-miR-26a or miR-control. (c) The diagram showed the binding region of miR-26a to 3′ UTR of GSK3β. (d) Luciferase activity of pMIR-Reporter containing GSK3β 3′ UTR was measured 48 h after co-transfection. (e–h) BMSCs and ADSCs were transfected with pre-miR-26a, anti-miR-26a or miR-control for 48 h. Smad1 and GSK3β mRNA level in BMSCs (e) and ADSCs (g) was determined by real-time RT-PCR. Smad1 and GSK3β protein accumulation in BMSCs (f) and ADSCs (h) was determined by western blot. Relative protein abundance of each blots was normalized to the gray value of β-actin. Results are represented as mean±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=3)
MiR-26a regulates the osteogenic differentiation of BMSCs and ADSCs through different targets
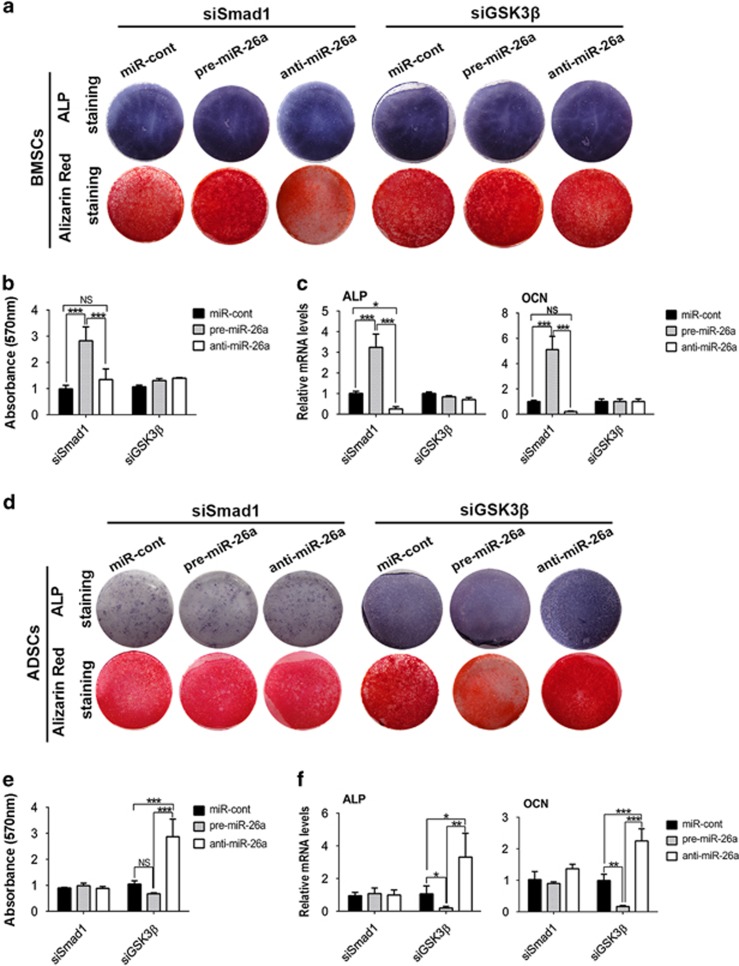
As Smad1 is the positive regulator but GSK3β is the negative regulator of BMSC osteogenesis,32, 33 we moved forward to explore which was the major target of miR-26a by knocking down GSK3β or Smad1 using specific siRNA (Supplementary Figure S3). After knocking down of GSK3β, miR-26a no longer obviously affected the osteogenic differentiation of BMSCs. However, miR-26a still potently promoted BMSC osteogenic differentiation after knocking down of Smad1 (Figures 3a–c). On the contrary, knockdown of Smad1 prohibited the function of miR-26a on osteogenic differentiation of ADSCs, but miR-26a still inhibited osteogenic differentiation of ADSCs after knocking down of GSK3β (Figures 3d–f). Taken together, these results indicated that miR-26a functions majorly by inhibiting GSK3β to promote osteogenesis of BMSCs, and restraining Smad1 to suppress ADSC differentiation.
Figure 3.
MiR-26a focuses on different targets to control BMSC and ADSC osteogenic differentiation. Smad1 or GSK3β siRNA was co-transfected with negative control, pre-miR-26a and anti-miR-26a into BMSCs (a–c) or ADSCs (d–f) for 48 h before osteogenic induction. (a, b, d and e) ALP staining and alizarin red staining were performed after osteogenic induction. (c and f) Alp and Ocn mRNA expression was analyzed by real-time RT-PCR after induction. Results are shown as mean±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=3)
GSK3β and Smad1 have distinct importance in osteogenic differentiation of BMSCs and ADSCs
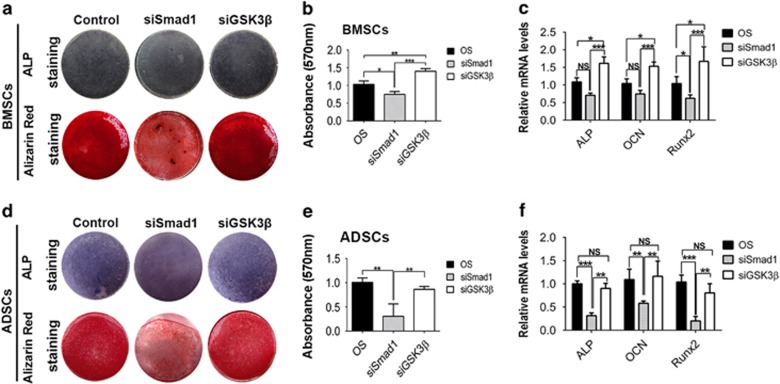
The observation that miR-26a targeted on both GSK3β and Smad1 but functioned majorly by regulating different targets in BMSCs and ADSCs raised a question. One possible answer is that GSK3β and Smad1 have different importance in the regulation of BMSC and ADSC differentiation. To verify this hypothesis, we compared the function of GSK3β and Smad1 between BMSCs and ADSCs through loss-of-function assay. As expected, GSK3β siRNA significantly promoted osteogenic differentiation of BMSCs (Figures 4a–c), whereas it caused no significant changes in ADSCs (Figures 4d–f). Smad1 siRNA significantly decreased osteogenic differentiation of ADSCs (Figures 4d–f), but only slightly affected the differentiation of BMSCs (Figures 4a–c).
Figure 4.
GSK3β and Smad1 have distinct importance in osteogenic differentiation of BMSCs and ADSCs. (a, b, d and e) ALP staining and alizarin red staining of BMSCs (a and b) and ADSCs (d and e) transfected with GSK3β siRNA, Smad1 siRNA or negative control after osteogenic induction. (c and f) Alp, Ocn and Runx2 mRNA expression was determined by real-time RT-PCR. Results are shown as mean±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=3)
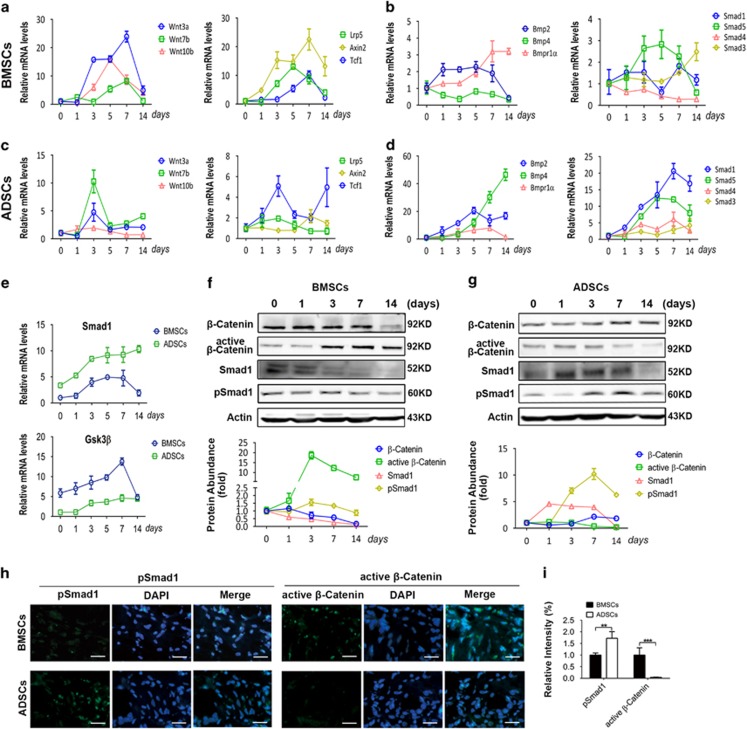
We moved to the next question of why GSK3β and Smad1 have distinct importance in osteogenic differentiation of different MSCs. The effectivity of one signaling molecule is largely dependent on the activation status of the signaling pathway. Real-time RT-PCR showed that the expression of Wnt ligands was increased over 10-fold during the early stage of BMSC osteogenic differentiation, and the expression of Wnt receptor and downstream genes was also significantly increased till day 7 (Figure 5a). Comparatively, only a few BMP pathway genes were slightly increased during BMSC differentiation (Figure 5b). In contrast, the expression of BMP pathway ligands, receptor and downstream regulators was significantly enhanced during osteogenic differentiation of ADSCs (Figure 5d), and only parts of the Wnt pathway genes were modestly increased (Figure 5c). During osteogenic differentiation process, BMSCs expressed more GSK3β but less Smad1 than ADSCs (Figure 5e).
Figure 5.
Wnt and BMP signaling are differentially activated during osteogenic differentiation of BMSCs and ADSCs. (a–d) Expression of Wnt pathway-related genes and BMP pathway genes at different time points of BMSC (a and b) and ADSC (c and d) osteogenic differentiation was determined by real-time RT-PCR. (e) Smad1 and GSK3β mRNA expression in ADSCs and BMSCs during osteogenic differentiation was analyzed by real-time RT-PCR. (f and g) Western blot analysis of the markers of Wnt and BMP pathways during osteogenic differentiation of BMSCs (f) and ADSCs (g). β-Actin was used as the internal control. Relative protein abundance of each blots was normalized to the gray value of β-actin. (h and i) Immunofluorescence assay of p-Smad1 and active β-catenin in BMSC and ADSC transplants after 4 weeks of immunocompromised mice transplantation. The fluorescence intensity was quantified by Image Pro software (i). Scale bar: 200 μm. Results are represented as mean±S.D. **P<0.01, ***P<0.001 (n=3)
Moreover, western blot analysis showed that active β-catenin protein accumulation, a marker of Wnt pathway activation, was increased >10-fold during the middle stage of osteogenic differentiation of BMSCs (Figure 5f). Conversely, phosphorylated Smad1 (pSmad1) protein accumulation, a marker of BMP pathway activation, was slightly decreased at day 7 of BMSC osteogenic differentiation (Figure 5f). On the contrary, pSmad1 protein level was significantly increased whereas active β-catenin protein accumulation was decreased during osteogenic differentiation of ADSCs (Figure 5g).
To confirm the signaling activation status in vivo, we performed immunofluorescence assay in MSCs transplants harvested after 4 weeks. Consistent with the in vitro results, active β-catenin protein level in BMSCs was significantly higher than that in ADSCs, whereas pSmad1 protein accumulation in BMSCs was lower than that in ADSCs (Figures 5h and i). Taken together, these results indicated that Wnt signaling is dominantly activated during osteogenic differentiation of BMSCs, whereas BMP signaling is more active during ADSC differentiation.
BMSCs and ADSCs depend on different signaling pathways to promote osteogenic differentiation
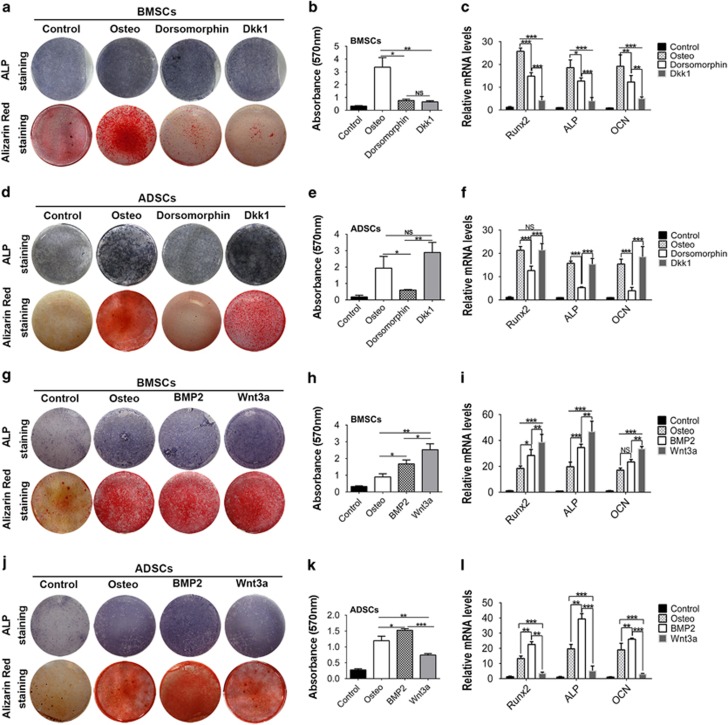
The differential activation of Wnt and BMP signaling suggested that BMSCs and ADSCs apply different signaling pathways to promote osteogenic differentiation. To certify the notion, we supplied Dorsomorphin to inhibit BMP pathway and Dickkopf-1 (DKK1) to inhibit Wnt pathway. Although both Dorsomorphin and DKK1 suppressed osteogenic differentiation of BMSCs, DKK1 was more effective than Dorsomorphin (Figures 6a–c). In ADSCs, Dorsomorphin significantly suppressed the osteogenic differentiation (Figures 6d–f). Interestingly, in contrast with its role in BMSC differentiation, DKK1 did not affect ADSC osteogenic differentiation (Figures 6d–f).
Figure 6.
Wnt and BMP signaling pathways play different role in BMSC and ADSC osteogenic differentiation. (a–c) BMSCs were cultured in osteogenic medium with Dorsomorphin or DKK1. Then, ALP and alizarin red staining were performed separately after 7 and 14 days of induction (a). Alizarin red staining was quantified by spectrophotometer (b). Expression of Runx2, Alp and Ocn was measured by real-time RT-PCR (c). (d–f) ALP staining (d), alizarin red staining (d and e) and real-time RT-PCR analysis (f) in ADSCs cultured in osteogenic medium containing Dorsomorphin or DKK1. (g–l) ALP staining, alizarin red staining and real-time RT-PCR were performed in BMSCs (g–i) and ADSCs (j–l) cultured in osteogenic medium containing recombinant BMP2 or Wnt3a. Results are represented as mean±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=3)
We next performed gain-of-function assay using recombinant Wnt3a to activate Wnt signaling and BMP2 to activate BMP signaling. Both BMP2 and Wnt3a enhanced the mineralized nodule formation and osteogenic markes expression in BMSCs, and Wnt3a was more effective than BMP2 (Figures 6g–i). In ADSCs, BMP2 markedly promoted the osteogenic differentiation of ADSCs, whereas Wnt3a slightly inhibited the osteogenic differentiation of ADSCs (Figures 6j–l). Taken together, these results indicated that Wnt pathway activation is indispensable for BMSC osteogenic differentiation, whereas BMP signaling activation is more important to promote ADSC osteogenic differentiation.
MiR-26a regulates different signaling pathways in BMSCs and ADSCs
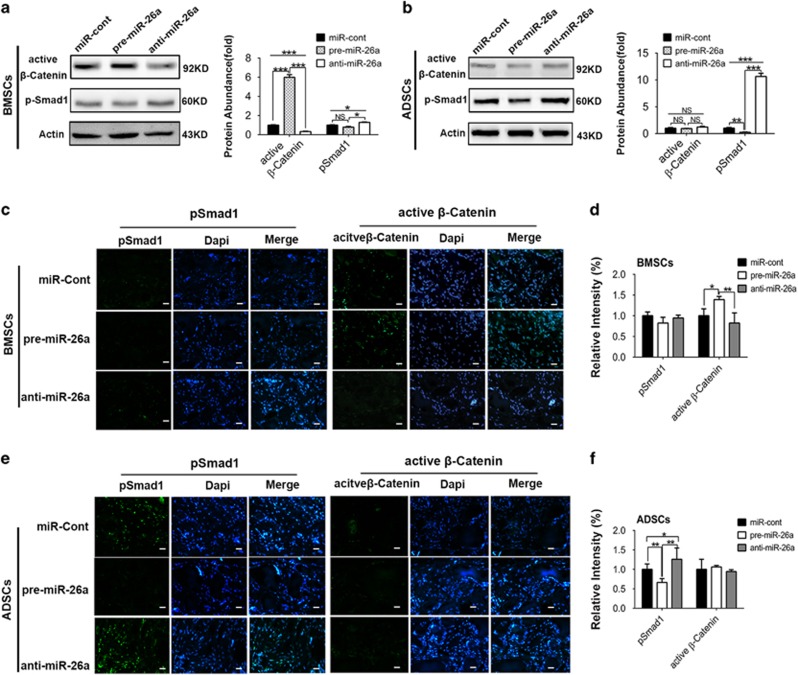
Our results above showed that differential activation of Wnt and BMP pathways determined the distinct importance of GSK3β and Smad1 in BMSC and ADSC differentiation. To further verify the notion, we checked the effects of miR-26a on activation of Wnt and BMP pathways in different MSCs after 14 days of osteogenic induction. Overexpression of miR-26a increased active β-catenin expression, whereas knockdown of miR-26a decreased active β-catenin expression in BMSCs (Figure 7a). MiR-26a also inhibited pSmad1 protein slightly in BMSCs. As pSmad1 protein rarely expressed in BMSCs after osteogenic differentiation, the absolute quantity change of pSmad1 was much lower than that of active β-catenin (Figure 7a). Immunofluorescence assay of in vivo transplants revealed that miR-26a significantly inhibited the expression of GSK3β (Supplementary Figure S4A and B) and increased the level of active β-catenin (Figures 7c and d). As pSmad1 expression in BMSC implants was scarce, we did not detect the effect of miR-26a on pSmad1 level in vivo (Figures 7c and d).
Figure 7.
MiR-26a potently regulates different signaling pathways in BMSC and ADSC osteogenic differentiation. (a and b) BMSCs and ADSCs were transfected with negative control (miR-cont), miR-26a precursors (pre-miR-26a) and inhibitors (anti-miR-26a) for 48 h before osteogenic induction. Western blot was performed to analyze the protein level of active β-catenin or pSmad1 in BMSCs (a) and ADSCs (b) after 14 days of induction in vitro. Relative protein abundance of each blots was normalized to the gray value of β-actin. (c–f) BMSCs and ADSCs transfected with negative control (miR-cont), miR-26a precursors (pre-miR-26a) and inhibitors (anti-miR-26a) were implanted into nude mice for 4 weeks. Expression of pSmad1 and active β-catenin in transplants of BMSCs (c and d) and ADSCs (e and f) was detected by immunofluorescence assay, and quantified by Image Pro software. Scale bar: 200 μm. Results represent means±S.D. *P<0.05, **P<0.01, ***P<0.001 (n=3)
In ADSCs, miR-26a markedly effected pSmad1 protein expression (Figure 7b). Although miR-26a slightly inhibited GSK3β, miR-26a knockdown or overexpression did not significantly affect the active β-catenin protein level in ADSCs. Immunofluorescence assay also confirmed that miR-26a inhibited the activation of Smad1 (Supplementary Figure S4C and D) and pSmad1 (Figures 7e and f) in vivo. We did not detect the regulation of miR-26a on active β-catenin of ADSC transplants (Figures 7e and f).
In conclusion, we found that BMP and Wnt signaling pathways were differentially activated and played distinct role in osteogenic differentiation of BMSCs and ADSCs. The different gene regulation program determined the opposite function of miR-26a in BMSC and ADSC osteogenic differentiation. Our findings uncovered that tissue-specific MSCs applied differential signaling regulation programs for cell-fate commitment, suggesting the complexity in tissue-specific stem cell research and application.
Discussion
Both Wnt and BMP are critical pathways controlling MSC fate. Notably, WNT and BMP pathways are reported to be closely related with each other during osteogenic differentiation of MSCs. BMPs repress Wnt signaling in skeletal progenitor cells to control osteoblastic differentiation.34, 35 Studies using pluripotent mesenchymal cell lines show that BMP upregulates Wnt signaling to promote osteogenic differentiation.36, 37 Wnt3a, in turn, regulates BMP signaling in osteoblast.38 The interplay of BMP and Wnt signaling seems to be cell specific. In this study, we confirmed that Wnt and BMP signaling are antagonistically activated during osteogenic differentiation of BMSCs and ADSCs. Wnt ligand expressions were potently activated, but BMP expressions were slightly elevated during BMSC osteogenesis. In contrast, BMP expressions were enhanced much more potently than the expression of Wnt ligands during ADSC osteogenic differentiation, suggesting that only one signaling plays the dominate role in each cell. However, it remains elusive whether Wnt or BMP signaling directly inhibits each other in MSCs. Further gain- and loss-of-function studies are necessary to uncover the mechanism of BMP and Wnt signaling interplay in tissue-specific MSCs.
A number of comparative studies demonstrated a difference in the differentiation capacity of MSCs from specific tissues.39, 40, 41, 42, 43 However, there remains a question of whether tissue-specific MSCs apply different gene regulation programs for osteogenic differentiation. Some studies showed that many genes, including genes involving in important signaling pathways, are differentially expressed between different MSCs.21, 22, 23, 24, 43 Here, through study of the opposite function of miR-26a in BMSCs and ADSCs, we confirmed distinct signaling regulation networks between osteogenic differentiation of BMSCs and ADSCs. We thoroughly compared the activation states and function of Wnt and BMP pathways during BMSC and ADSC differentiation. Interestingly, we found that Wnt and BMP signaling were differentially activated during osteogenic differentiation of BMSCs and ADSCs. Function assay further confirmed that BMSCs majorly applied Wnt signaling to promote osteogenic differentiation, but ADSCs majorly used BMP signaling to facilitate osteogenic differentiation. In accordance with previous studies showing that WNT and BMP are differentially activated in MSCs derived from different tissues, our findings supported that tissue-specific MSCs apply distinct signaling pathways for differentiation. Further works are needed to uncover the physiological significance and the underlying mechanism of this phenomenon.
Notably, our results showed that Wnt signaling pathway potently promotes osteogenic differentiation of BMSCs, whereas it inhibits ADSC osteogenic differentiation, and this is supported by previous reports.32, 44 Very little is known about the molecular basis of this phenomenon. One possibility is that the tissue-specific epigenetic signatures determine transcription factors of Wnt and BMP pathways to activate different cohorts of genes. A deeper study of epigenetic signature between tissue-specific MSCs would be necessary to understand the gene regulation of signaling pathways.
MiR-26a regulates the differentiation of several cells, such as BMSCs, adipose progenitors, smooth muscle cells, monocytes and so on. MiR-26a targeted E2F7 to inhibit monocytic differentiation.45 MiR-26a was upregulated and targeted ten-eleven translocation (TET) enzymes during pancreatic cell differentiation.46 The transcription factors Smad1 and Smad4 were regulated by miR-26a to promote the differentiation of myoblasts.47 MiR-26 family and its downstream effector ADAM17 functioned in human adipocyte differentiation by promoting characteristics of energy-dissipating thermogenic adipocytes.48 These results suggested that miR-26a functions as an important regulator of cell lineage commitment. In our study, we identified miR-26a as a positive regulator of BMSC osteogenic differentiation but a negative regulator of ADSC osteogenic differentiation. Interestingly, it was reported that miR-26a is upregulated during adipogenesis of BMSCs,9 suggesting the more complicated role of miR-26a in MSCs osteogenic–adipogenic differentiation balance. As BMSCs are inclined to differentiate into osteoblasts and ADSCs prefer to differentiate into adipocytes in physiological conditions, miR-26a might be a crucial regulator to ensure normal lineage commitment of tissue-specific MSCs.
One microRNA might play different roles in different tissues. For example, miR-21 promoted the proliferation of squamous cell carcinoma and breast tumor cells,49, 50 but it inhibited the proliferation of ADSCs.51 MiR-214 promoted the differentiation of myocytes,52 whereas it inhibited the differentiation of osteoblasts.53 This could be explained as one miRNA potentially targets on a number of mRNAs. As gene expression varies in different types of cells, microRNA function is likely to be determined by which genes are highly expressed and functional. In our study, we find that Smad1 and GSK3β mRNA expression is quite different between BMSCs and ADSCs. Furthermore, Wnt signaling and BMP signaling is differentially activated and plays different roles to regulate osteogenic differentiation of MSCs. Thus, these differences might determine the opposing function of miR-26a in different MSCs. Therefore, further systemic research of signaling regulation program is necessary for understanding miRNA-mediated post-transcriptional regulation of tissue-specific MSCs.
Materials and Methods
Animals
C57BL/6J mice and BALB/c nude mice were obtained from the Laboratory Animal Research Centre of the Fourth Military Medical University (Xi'an, China). All procedures that involved animals were approved by the animal use and care committee of the Fourth Military Medical University (License Number: 2014 KQ-005). All mice were housed under specific pathogen-free conditions (22 °C, 12-h light/12-h dark cycle, and 50–55% humidity) with free access to food pellets and tap water.
Materials
Recombinant mouse BMP2 (R&D, Minneapolis, MN, USA), recombinant mouse Wnt3a (R&D), recombinant mouse Dkk1 (PeproTech, Rocky Hill, NJ, USA) and dorsomorphin (Santa Cruz Biotechnology, Santa Cruz, TX, USA) were obtained commercially.
Cell culture and identification
The 4- to 6-week-old C57BL/6J mice were used for cell culture. BMSCs were isolated and cultured as previously described.54 Mouse were killed and the hindlimbs were aseptically removed and bones were dissected free of soft tissues. Marrow cavities of femur and tibia were flushed with culture medium as α-MEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS, 1% penicillin and streptomycin and 2-mercaptoethanol (2-ME). 1.5 × 107 cells were seeded in 10 cm tissue culture flasks and incubated in humidified atmosphere of 5% CO2 at 37 °C. Nonadherent cells were removed by frequent medium change during 72 h. The remaining adherent colonies were cultured for 14 days until confluent, and passaged after digestion with 0.25% trypsin for 3 min.
ADSCs were isolated from scraps of subcutaneous adipose tissues as previously described.55 In brief, the adipose tissues were minced and incubated in 1% collagenase type I for 1.5 h in a 37 °C water bath shaker. The digested solution was filtered through a 200-μM and a 100-μm cell strainer to separate undigested tissue fragments. Adipocytes and aqueous supernatant were separated by centrifugation at 800 × g for 5 min. Then, the deposit was resuspended in culture medium and plated in 10 cm tissue culture flasks and maintained in humid incubator at 37 °C and 5% CO2. Hematopoietic lineage cells attached to Petri dish after 1 h were removed, and the nonadherent cells containing ADSC populations were fed with α-MEM supplemented with 10% FBS, 1% penicillin and streptomycin until colony forming and reaching 70–80% confluence.
Cells at passage 3 were used for all experiments. Both BMSCs and ADSCs highly expressed MSC markers (Sca-1, CD90, CD106) and did not express hematopoietic cell marker (CD34), and had self-renewal and multipotent differentiation potential (Supplementary Figures S1A–K).
Osteogenic differentiation and mineralization assay
After the cells reached 70–80% confluence, differentiation medium containing α-MEM, 10% FBS, 100 μg/ml ascorbic acid, 2 mM β-glycerophosphate and 10 nM dexamethasone was used to induce osteogenic differentiation of both MSCs. The medium was changed every 3 days. After induction for 7 days, ALP staining was performed with the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime Co., Shanghai, China) following the standard protocol. After 14 days of osteogenic differentiation, 1% alizarin red (Sigma, St. Louis, MO, USA) was used to detect calcium accumulation according to the manufacturer's suggested protocol. Then, alizarin red was incubated with 100 mM cetylpyridinium chloride (Sigma) for 30 min at room temperature and quantified by spectrophotometer at 540 nm. The final calcium levels in each group were normalized with the total protein concentration obtained from the duplicate plates.
Real-time RT-PCR of mRNA and miRNA
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's standard instructions. For reverse transcription of mRNA, random-primed cDNA was synthesized from 2 μg of total RNA using a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). Real-time RT-PCR analysis was performed using the SYBR Premix Ex Taq II Kit (TaKaRa) and detected on the ABI Prism 7500 HT Sequence Detection System (Applied Bio-Systems, Foster City, CA, USA). β-Actin and U6 were used as loading controls for quantitation of mRNA and microRNA. The optimized microRNA-specific primers for miR-26a and the endogenous control U6 were commercially obtained (RiboBio, Guangzhou, China). Primers of PCR are listed in Supplementary Table S1.
Western blot analysis
Cells were harvested in RIPA lysis buffer (Beyotime Co.). Whole-cell protein extracts were quantified by the BCA assay, separated on 12% SDS-PAGE and then transferred to PVDF membranes (Millipore, Billerica, MA, USA) and blocked with 5% nonfat milk powder in PBST for 3 h. The membranes were probed overnight with the primary antibodies and then 4 h with peroxidase-conjugated secondary antibodies (Boster, Wuhan, China). Antibodies used in this study included the mouse β-Actin, rabbit Smad1, rabbit phospho-Smad1, mouse GSK3β, rabbit phospho-GSK3β (Cell Signaling, Beverly, MA, USA), rabbit β-catenin (Abcam, Cambridge, MA, USA) and mouse active β-catenin (Millipore). The blots were visualized using an ECL Kit (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's recommended instructions. To measure the protein abundance, gray value of the blots in scanned images was measured with ImageJ Plus software (National Institutes of Health, Bethesda, MD, USA). Gray value of each target protein was normalized to that of β-Actin before comparison.
The in vivo bone formation assay
BALB/c nude mice, 6 weeks old, were used as hosts for implantation. BMSCs and ADSCs were first transfected with miR-26a precursors, inhibitors or negative controls as described before, and then cultured in osteogenic differentiation medium for 3 days. Approximately 4 × 106 cells were mixed with 40 mg hydroxyaptite-tricalcium phosphate (HA-TCP) powders (Sigma-Aldrich, St. Louis, MO, USA) and subcutaneously implanted into the dorsal surface of immunocompromised mice. Implants were harvested after 4 or 8 weeks for immunofluorescence and histological assay separately.
H&E staining of MSC implants
MSC implants separated from surrounding fibrous capsule were fixed in 4% neutral buffered formalin (Sigma) overnight and decalcified with EDTA (10%) (Sigma) for 2 weeks. Then, the implants were embedded in paraffin and sectioned at 5 mm. H&E staining was performed according to standard protocols. Photos were taken under magnification of × 40 on four consecutive microscopic fields of the sections of implants. Osteoid formation in transplants was semiquantified as osteoid area per total area in H&E staining photos with Image Pro software (Media Cybernetics, Silver Spring, MD, USA).
Immunofluorescence assay of MSC implants
Decalcified MSC implants were dehydrated in 30% sucrose solutions until submerged in bottom. Samples were embedded into optimal cutting temperature compound (OCT) for frozen sections, and analyzed by immunofluorescence assay following the standard protocol. Antibodies used in this study including goat Smad1 (Santa Cruz Biotechnology), mouse active β-catenin (Millipore), rabbit phospho-Smad1 and rabbit GSK3β (Cell Signaling). The samples were treated with fluorescence-labeled secondary antibodies and the nuclei were stained with 100 ng/ml of 4',6-diamidno-2-phenylinde (DAPI) (Beyotime Co.). All samples were examined under a confocal microscope (Olympus Optical, Tokyo, Japan).
Transfection of miRNA precursors and inhibitors
Cells were seeded into 12-well or 6-well plates and transfection was performed shortly before or at 70% confluence. The microRNA precursors, inhibitors and negative controls of miR-26a were commercially purchased (Ambion, Austin, TX, USA). The cells were transfected with pre-miR-26a and negative control at a final concentration of 50 nM, and anti-miR-26a of 100 nM. Cell suspensions were overlaid onto the transfection complexes and incubated at 37 °C and 5% CO2 for 24 h until further study.
Transfection of siRNA
Cells were seeded into 12-well or 6-well plates and transfected at 40–60% confluence. The Smad1 siRNA sequence (5′-GAACUGAAGCCUCUGGAAU-3′, 3′-CUUGACUUCGGAGACCUUA-5′), Gsk3β siRNA sequence (5′-GGUAUAUCAAGCCAAACUU-3′, 3′-CCAUAUAGUUCGGUUUGAA-5′) and scramble RNA were commercially synthesized (RiboBio). The cells were transfected with miRNAs together with Smad1 siRNA or Gsk3β siRNA by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and transected with scramble RNA as a negative control at the same time. Final concentration of microRNA precursors, negative control and siRNA were 50 nM, and the concentration of inhibitor was 100 nM. The cells were incubated at 37 °C in the incubator for 24 h before further study.
Luciferase reporter assay
To construct a vector of mouse Smad1 and GSK3β, the Smad1 oligonucleotide sequence were amplified using primers (forward: 5′- GGCTCCTTCGTCAGGTCTCCA-3′ reverse: 5′-ACGGATGAAATAGGATTGTGGG-3′) with Hindlll and Spel sites at their extremities to insert the pMIR-Report vector (Ambion). GSK3β oligonucleotide sequences were amplified using primers (forward: 5′- CCACCATCCTTATCCCTCCAC-3′ reverse: 5′-GGAGGAGCAGAGCATTAAACACA-3′). HEK293T cells were seeded in a 96-well plate at 50 to 60% confluence. After 24 h, cells were co-transfected with reporter constructs as pMIR-Report (pMIR-Cont), pMIR-Smad1 or pMIR-GSK3β plasmids (125ng), and microRNA controls, precursors or inhibitors (20 pmol). All transfections were conducted using Lipofectamine 2000 (Invitrogen). Luciferase assays were performed 48 h after transfection using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well. Each transfected well was assayed in triplicate.
Statistical analysis
Data are presented as mean±S.D. Comparisons were analyzed using Student's two-tailed t-test or one-way ANOVA for experiments with more than three groups. All experiments were repeated at least three times, and representative experiments are shown. Differences were considered to be significant when P<0.05.
Acknowledgments
This work was supported by grants from the National Major Scientific Research Program of China (2011CB964700) and the Nature Science Foundation of China (31030033, 81470710).
Author contributions
Xiaoxia Su and Li Liao: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; Yi Shuai and Huan Jing: collection and assembly of data; Shiyu Liu and Hong Zhou: data analysis and interpretation; Yong Liu: conception and design, data analysis and interpretation; Yan Jin: conception and design, data analysis and interpretation, manuscript writing, financial support, financial approval of manuscript.
Glossary
- ADSC
adipose tissue-derived mesenchymal stem cell
- ALP
alkaline phosphatase
- BMP
bone morphogenetic protein
- BMSC
bone marrow-derived mesenchymal stem cell
- DKK1
Dickkopf-1
- HA-TCP
hydroxyaptite-tricalcium phosphate
- miRNA
microRNA
- MSC
mesenchymal stem cell
- OCN
osteocalcin
- pSmad1
phosphorylated Smad1
- RT-PCR
reverse transcription PCR
- WNT
wingless-type MMTV integration site family
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks PB. MicroRNAs and parallel stem cell lives. Cell. 2009;138:423–424. doi: 10.1016/j.cell.2009.07.025. [DOI] [PubMed] [Google Scholar]
- Clark EA, Kalomoiris S, Nolta JA, Fierro FA. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32:1074–1082. doi: 10.1002/stem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan L, Liu S, Liu W, Zhang H, Zhou T, et al. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34:5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- Luzi E, Marini F, Tognarini I, Galli G, Falchetti A, Brandi ML. The regulatory network menin-microRNA 26a as a possible target for RNA-based therapy of bone diseases. Nucleic Acid Ther. 2012;22:103–108. doi: 10.1089/nat.2012.0344. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Fraser JK, Wulur I, Alfonso Z, Hedrick MH. Fat tissue: an underappreciated source of stem cells for biotechnology. Trends Biotechnol. 2006;24:150–154. doi: 10.1016/j.tibtech.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Tobita M, Uysal AC. Concise review: Adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells. 2012;30:804–810. doi: 10.1002/stem.1076. [DOI] [PubMed] [Google Scholar]
- cDeschaseaux F, Pontikoglou C, Sensebe L. Bone regeneration: the stem/progenitor cells point of view. J Cell Mol Med. 2010;14:103–115. doi: 10.1111/j.1582-4934.2009.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Nixon AJ. Stem cell therapy in bone repair and regeneration. Ann NY Acad Sci. 2007;1117:62–72. doi: 10.1196/annals.1402.074. [DOI] [PubMed] [Google Scholar]
- Gomez-Barrena E, Rosset P, Muller I, Giordano R, Bunu C, Layrolle P, et al. Bone regeneration: stem cell therapies and clinical studies in orthopaedics and traumatology. J Cell Mol Med. 2011;15:1266–1286. doi: 10.1111/j.1582-4934.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CC, Lin AB, Hung SC. Mesenchymal stem cells in regenerative medicine for musculoskeletal diseases: bench, bedside, and industry. Cell Transplant. 2014;23:505–512. doi: 10.3727/096368914X678328. [DOI] [PubMed] [Google Scholar]
- Lee RH, Kim B, Choi I, Kim H, Choi HS, Suh K, et al. Characterization and expression analysis of mesenchymal stem cells from human bone marrow and adipose tissue. Cell Physiol Biochem. 2004;14:311–324. doi: 10.1159/000080341. [DOI] [PubMed] [Google Scholar]
- Monaco E, Bionaz M, Rodriguez-Zas S, Hurley WL, Wheeler MB. Transcriptomics comparison between porcine adipose and bone marrow mesenchymal stem cells during in vitro osteogenic and adipogenic differentiation. PLoS One. 2012;7:e32481. doi: 10.1371/journal.pone.0032481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Sim WY, Min BH, Yang SS, Khademhosseini A, Kaplan DL. Chip-based comparison of the osteogenesis of human bone marrow- and adipose tissue-derived mesenchymal stem cells under mechanical stimulation. PLoS One. 2012;7:e46689. doi: 10.1371/journal.pone.0046689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strioga M, Viswanathan S, Darinskas A, Slaby O, Michalek J. Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells Dev. 2012;21:2724–2752. doi: 10.1089/scd.2011.0722. [DOI] [PubMed] [Google Scholar]
- Izadpanah R, Trygg C, Patel B, Kriedt C, Dufour J, Gimble JM, et al. Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue. J Cell Biochem. 2006;99:1285–1297. doi: 10.1002/jcb.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel D, Caton D, Roche S, Bony C, Lehmann S, Casteilla L, et al. Cell specific differences between human adipose-derived and mesenchymal-stromal cells despite similar differentiation potentials. Exp Cell Res. 2008;314:1575–1584. doi: 10.1016/j.yexcr.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MJ, Fernandes MH. Human bone cell cultures in biocompatibility testing. Part II: effect of ascorbic acid, beta-glycerophosphate and dexamethasone on osteoblastic differentiation. Biomaterials. 2000;21:1095–1102. doi: 10.1016/s0142-9612(99)00192-1. [DOI] [PubMed] [Google Scholar]
- Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res. 2013;113:1231–1241. doi: 10.1161/CIRCRESAHA.113.301780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J Biol Chem. 2010;285:29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JH, Choi E, Cha MJ, Song BW, Ham O, Lee SY, et al. Up-regulation of miR-26a promotes apoptosis of hypoxic rat neonatal cardiomyocytes by repressing GSK-3beta protein expression. Biochem Biophys Res Commun. 2012;423:404–410. doi: 10.1016/j.bbrc.2012.05.138. [DOI] [PubMed] [Google Scholar]
- Krause U, Harris S, Green A, Ylostalo J, Zeitouni S, Lee N, et al. Pharmaceutical modulation of canonical Wnt signaling in multipotent stromal cells for improved osteoinductive therapy. Proc Natl Acad Sci USA. 2010;107:4147–4152. doi: 10.1073/pnas.0914360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ji X, Shi X, Cao X. Smad1 domains interacting with Hoxc-8 induce osteoblast differentiation. J Biol Chem. 2000;275:1065–1072. doi: 10.1074/jbc.275.2.1065. [DOI] [PubMed] [Google Scholar]
- Minear S, Leucht P, Miller S, Helms JA. rBMP represses Wnt signaling and influences skeletal progenitor cell fate specification during bone repair. J Bone Miner Res. 2010;25:1196–1207. doi: 10.1002/jbmr.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Kobayashi T, Mochida Y, Yu PB, Yamauchi M, Kronenberg HM, et al. Wnt inhibitors Dkk1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J Bone Miner Res. 2010;25:200–210. doi: 10.1359/jbmr.090806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbalaviele G, Sheikh S, Stains JP, Salazar VS, Cheng SL, Chen D, et al. Beta-catenin and BMP-2 synergize to promote osteoblast differentiation and new bone formation. J Cell Biochem. 2005;94:403–418. doi: 10.1002/jcb.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Whetstone HC, Youn A, Nadesan P, Chow EC, Lin AC, et al. Beta-catenin signaling pathway is crucial for bone morphogenetic protein 2 to induce new bone formation. J Biol Chem. 2007;282:526–533. doi: 10.1074/jbc.M602700200. [DOI] [PubMed] [Google Scholar]
- Zhang R, Oyajobi BO, Harris SE, Chen D, Tsao C, Deng HW, et al. Wnt/beta-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone. 2013;52:145–156. doi: 10.1016/j.bone.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A, Breit S, Parsch D, Benz K, Steck E, Hauner H, et al. Cartilage-like gene expression in differentiated human stem cell spheroids: a comparison of bone marrow-derived and adipose tissue-derived stromal cells. Arthritis Rheum. 2003;48:418–429. doi: 10.1002/art.10767. [DOI] [PubMed] [Google Scholar]
- Toupadakis CA, Wong A, Genetos DC, Cheung WK, Borjesson DL, Ferraro GL, et al. Comparison of the osteogenic potential of equine mesenchymal stem cells from bone marrow, adipose tissue, umbilical cord blood, and umbilical cord tissue. Am J Vet Res. 2010;71:1237–1245. doi: 10.2460/ajvr.71.10.1237. [DOI] [PubMed] [Google Scholar]
- Sakaguchi Y, Sekiya I, Yagishita K, Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52:2521–2529. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- Niemeyer P, Fechner K, Milz S, Richter W, Suedkamp NP, Mehlhorn AT, et al. Comparison of mesenchymal stem cells from bone marrow and adipose tissue for bone regeneration in a critical size defect of the sheep tibia and the influence of platelet-rich plasma. Biomaterials. 2010;31:3572–3579. doi: 10.1016/j.biomaterials.2010.01.085. [DOI] [PubMed] [Google Scholar]
- Shafiee A, Seyedjafari E, Soleimani M, Ahmadbeigi N, Dinarvand P, Ghaemi N. A comparison between osteogenic differentiation of human unrestricted somatic stem cells and mesenchymal stem cells from bone marrow and adipose tissue. Biotechnol Lett. 2011;33:1257–1264. doi: 10.1007/s10529-011-0541-8. [DOI] [PubMed] [Google Scholar]
- Lagathu C, Christodoulides C, Virtue S, Cawthorn WP, Franzin C, Kimber WA, et al. Dact1, a nutritionally regulated preadipocyte gene, controls adipogenesis by coordinating the Wnt/beta-catenin signaling network. Diabetes. 2009;58:609–619. doi: 10.2337/db08-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori B, Iosue I, Mangiavacchi A, Loddo G, Padula F, Chiaretti S, et al. The microRNA-26a target E2F7 sustains cell proliferation and inhibits monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2012;3:e413. doi: 10.1038/cddis.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci USA. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbiener M, Pisani DF, Frontini A, Oberreiter LM, Lang E, Vegiopoulos A, et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells. 2014;32:1578–1590. doi: 10.1002/stem.1603. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Hwang SH, Cho HH, Shin KK, Bae YC, Jung JS. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J Cell Physiol. 2012;227:183–193. doi: 10.1002/jcp.22716. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc. 2009;4:102–106. doi: 10.1038/nprot.2008.221. [DOI] [PubMed] [Google Scholar]
- Sugii S, Kida Y, Berggren WT, Evans RM. Feeder-dependent and feeder-independent iPS cell derivation from human and mouse adipose stem cells. Nat Protoc. 2011;6:346–358. doi: 10.1038/nprot.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.