Abstract
The Rb and Pten tumor suppressor genes are important regulators of bone development and both are frequently mutated in the bone cancer osteosarcoma (OS). To determine if Rb1 and Pten synergize as tumor suppressor genes for osteosarcoma, we co-deleted them in osteoprogenitor cells. Surprisingly, we observed rapid development of adipogenic but not osteosarcoma tumors in the ΔRb1/Pten mice. ΔPten solo deleted mice also developed lipoma tumors but at a much reduced frequency and later onset than those co-deleted for Rb1. Pten deletion also led to a marked increase in adipocytes in the bone marrow. To better understand the function of Pten in bone development in vivo, we conditionally deleted Pten in OSX+ osteoprogenitor cells using OSX-Cre mice. μCT analysis revealed a significant thickening of the calvaria and an increase in trabeculae volume and number in the femur, consistent with increased bone formation in these mice. To determine if Pten and Rb1 deletion actively promotes adipogenic differentiation, we isolated calvarial cells from Pten fl/fl and Pten fl/fl; Rb1 fl/fl mice, infected them with CRE or GFP expressing adenovirus, treated with differentiation media. We observed slightly increased adipogenic, and osteogenic differentiation in the ΔPten cells. Both phenotypes were greatly increased upon Rb1/Pten co-deletion. This was accompanied by an increase in expression of genes required for adipogenesis. These data indicate that Pten deletion in osteoblast precursors is sufficient to promote frequent adipogenic, but only rare osteogenic tumors. Rb1 hetero- or homo-zygous co-deletion greatly increases the incidence and the rapidity of onset of adipogenic tumors, again, with only rare osteosarcoma tumors.
Introduction
Mammalian bone formation occurs in a coordinated, sequential process that is characterized by the commitment of osteoprogenitor cells into pre- and mature osteoblasts whose function is to synthesize the bone matrix that later becomes mineralized. Osteoblasts originate from pluripotent mesenchymal stem cells that can also differentiate into other mesenchymal cell lineages such as adipocytes, chondrocytes, myoblasts, and fibroblasts depending on the nature of the activated transcriptional differentiation program, and if triggered prior to their commitment to the osteoblastic lineage. For example, MyoD transcription factors are critical for the maturation of muscle cell lineages [1], whereas the peroxisome proliferator-activated receptor γ (PPARγ) is necessary for the differentiation of adipocyte lineage cells [2]. Several transcription factors are critical for the commitment of mesenchymal stem cells into the osteoblast cell lineage, such as the runt-related genes Cbfa1 and Runx2 [3–5]. SP7, also known as Osterix and OSX, is a bone-specific transcription factor that regulates osteoblast development and is expressed in precursors at an early stage in the osteoblast lineage [6]. Understanding the regulation of mesenchymal stem cells into distinct lineages is critical because dysregulation or reactivation of these pathways have an important role in the pathogenesis of human disorders such as osteoporosis and osteosarcoma bone tumors.
Osteosarcoma (OS) is the most common primary bone cancer and is associated with excess osteoblastic proliferation. Mutations in several common human cancer genes such as TP53, RB1, CDKN2A, MYC, and PTEN drive OS development [7, 8]. OS incidence is increased several hundred-fold in patients with RB1 germline mutations and patients with hereditary retinoblastoma through loss of the Rb tumor suppressor protein, a key regulator of E2F transcription factors and cell cycle progression, and thus its deletion is often accompanied by defects in cell cycle exit and can result in an undifferentiated cellular phenotype [9–11]. In addition, pRB controls mesenchymal cell differentiation through interactions with RUNX2 to promote osteogenic differentiation [12], or through blocking PPAR-γ expression to suppress adipogenesis [13].
PTEN (phosphatase and tensin homolog) is a tumor suppressor gene that is mutated at high frequency in a wide variety of human cancers, such as glioblastoma, prostate, breast, osteosarcoma and was recently identified as a key driver of osteosarcoma in a murine forward genetic screen [8, 14, 15]. PTEN encodes a lipid phosphatase that dephosphorylates phosphatidylinositol (3,4,5)- triphosphate (PIP3) to oppose the activity of phosphatidylinositol 3-kinase (PI3K), which functions as a PIP3 kinase that is constitutively activated and functions as an oncogene in many cancer types [16]. Loss of PTEN causes persistent activation of the Akt serine-threonine kinase which promotes cell growth and proliferation, disables apoptosis, and controls metabolism by directly phosphorylating numerous downstream targets such as BAD, FOXO transcription factors, CASP9, MTOR, MDM2, and others [17].
We recently identified a functional connection between Rb1 and Pten in murine retinal progenitor cells, where Rb1 deletion deregulates E2F dependent cell cycle entry and cell death, and co-deleting Pten inactivates an E2F/FOXO transcriptional complex in vivo to prevent apoptosis and promote rapid retinal tumorigenesis in retinal progenitor cells [18]. We tested if co-deleting Rb1 and Pten in mouse osteo-progenitors resulted in osteosarcoma development. We surprisingly observed rapid lipoma formation and increased marrow adipocyte size and numbers in mice with Rb1 and Pten co-deleted in osteoprogenitor cells. Consistent with these observations, deleting Pten or co-deleting Rb1 and Pten in calvarial osteoblast cells significantly promoted adipocyte differentiation and adipogenic differentiation gene expression. These data indicate that Pten deletion in osteoblast precursors is sufficient to promote frequent adipogenic, but only rare osteogenic tumors. Rb1 hetero- or homo-zygous co-deletion greatly increases the incidence and the rapidity of onset of adipogenic tumors, again, with only rare osteosarcoma tumors.
Materials and Methods
Experimental Animals & Ethics Statement
Osx1-GFP::Cre mice [6], Rb1 lox/lox mice [19], p107 -/- mice [20], and Pten lox/lox mice [21] and associated PCR genotyping protocols have been described. Animals were housed in the University of Minnesota animal facility. All mouse experiments were performed in accordance with University of Minnesota Institutional Animal Care and Use Committee procedures and guidelines under IACUC protocols #1309-30948A (2013-present), and previously #0711A20882 (2010–2013). No analgesic or anesthetics were used. Mice were monitored at least three times weekly during the survival study for evidence of surface irritation, infection, inability to obtain feed or water, accompanying weight loss, but these symptoms were not observed. Mice were humanely euthanized with CO2 when tumors reached a size of 1cm3, or immediately if tumors began to ulcerate the skin, or if a general moribund status evidenced by a reluctance to stand or move around the cage was observed. The University of Minnesota Comparative Pathology Shared Resource assisted with H&E staining, Ki-67 & TUNEL immunohistochemistry (IHC) staining. Kaplan-Meier curves were calculated using GraphPad Prism software, and p-values were determined by Student’s t-test. Marrow fat percentage, adipocyte density and size were determined by histomorphometry of mouse tibial sections as described [22, 23].
Microcomputed tomography (μCT) analysis
Mouse femurs and calvaria were subjected to micro-CT scanning (model XT H 225, Nikon Metrology, Inc.) using the conditions of 90 kV, 90 μA, 0.5° steps with a pixel size at 12 μm and exposure time of 708 ms as described [24]. CT Pro 3D (Nikon metrology) software was used to reconstruct original three-dimensional images. VG Studio Max 2.1 (Volume Graphics GmbH) was used for visualization and three-dimensional rendering and creation of bmp files. Finally, histomorphometry analyses were performed using SkyScan CT-Analyzer Version 1.12 (Bruker micro-CT).
Calvarial osteoblast isolation, adenovirus infection and cell differentiation
Calvarial osteoblasts were harvested as described [10]. Briefly, calvaria were removed from newborn pups and rinsed in warmed MEM to remove excess tissue. Calvaria were shaken for 20 minutes at 37°C in collagenase digestion mixture and the first fraction was discarded. Calvaria were shaken with another 10 ml aliquot of digestion mix for 40 minutes, and a third aliquot for 90 minutes. Trypsin was quenched with 10% FBS after each digestion, and cells from fractions 2 and 3 were resuspended in 10% FBS MEM and plated on 60mm plates at an approximate density of 1 calvaria/1.5 plates. Adenovirus infections were performed as described [25]. Cells were infected 24 hours after isolation (60–80% confluence) with control, or CRE expressing adenovirus at a multiplicity of infection (MOI) of 100. Adipogenic Media (α-MEM containing 10% FBS, 1% anti-microbial, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 5 ug/ml h-insulin, and 0.1 μM dexamethasone) or osteogenic media (α-MEM containing 10% FBS, 1% anti-microbial, 100 mM L-ascorbic acid, 0.01M sodium phosphate) differentiation media was added five days after adenovirus infections. Cells were grown in differentiation media for 25 days, and media was changed every three days.
Oil Red O (ORO) and Alizarin Red staining of differentiated calvarial cells
ORO stock solution (3mg/mL in isopropanol) was prepared fresh each experiment and mixed at a 3/2 ratio to make the working solution, which was filtered and used within 2 hours. Cells were rinsed with PBS, fixed in 10% formalin for 30 minutes at room temperature, rinsed with water and treated with 60% isopropanol for 5 minutes. ORO working solution was added to cells for 5 minutes and then rinsed with water. Cells that had been treated with osteogenic differentiation media were fixed (10% formalin for 30 minutes). After rinsing fixed cells, alizarin red solution (2% Alizarin Red S in distilled water, pH adjusted to 4.2 with ammonium hydroxide) was added to cells for 15 seconds and then rinsed with water before imaging.
Quantitative real-time PCR
RNA was prepared from cells for quantitative real-time PCR using RNeasy, QIAshredder, and QuantiTect SYBR Green RT-PCR kits from QIAGEN. Each RT-PCR experiment was performed in triplicate and normalized against expression of GAPDH expression levels and fold changes were calculated using ΔΔCT method [26]. p-values were determined by Student’s t–test. p < 0.05 were considered significant. Primers are listed in S1 Table.
Results
Combined deletion of Pten and Rb1 in osteoblast precursor cells accelerates adipogenic tumor formation
Because the Rb1 and Pten tumor suppressor genes are frequently mutated in bone cancer, we predicted that their combined deletion might provoke a stronger tumor phenotype than individual deletion of either gene. The Osx-Cre transgenic mouse was chosen to conditionally delete Rb1 and Pten in these studies because Osterix is expressed in committed osteoprogenitor cells and because Osx-Cre has successfully been utilized to generate osteosarcoma mouse models that co-deleted Rb1 and the p53 tumor suppressor genes [6, 27, 28]. We generated Osx-Cre; Rb1 fl/fl; Pten fl/fl mice and aged them to determine if they develop OS.
We unexpectedly observed rapid development of large, bilateral adipogenic tumors in 100% (37/37) of the Osx-Cre; Rb1 fl/fl; Pten fl/fl mice within the subcutaneous space of the axillae and interscapular/dorsal thoracic region. These unencapsulated, non-invasive masses were detectable before one month of age, and grew rapidly until the tumor burden required mice to be euthanized. Tumors were composed of a multilobulated mass of well-differentiated white and brown fat that was interspersed and supported by multifocal to coalescing thin bands of fibrous connective tissue, stroma, and rare macrophages (Fig 1A). The mass was composed mainly of white fat (large vacuoles with eccentric nuclei) and rare, interspersed brown fat (small multi-vacuolated cells). The surrounding tissues were largely skeletal muscle and brown fat. These animals displayed little subcutaneous, retroperitoneal, or mesenteric fat stores, besides the large tumors. Detailed radiographic and histologic examination of the bones (skull, mandible, thoracic vertebrae and femur) from control and Osx-Cre; Rb1 fl/fl; Pten fl/fl mice revealed no bone lesions in any of these animals (data not shown).
Fig 1. Pten and Rb co-deletion in osteoblast precursor cells causes rapid lipoma but not osteosarcoma tumor onset.
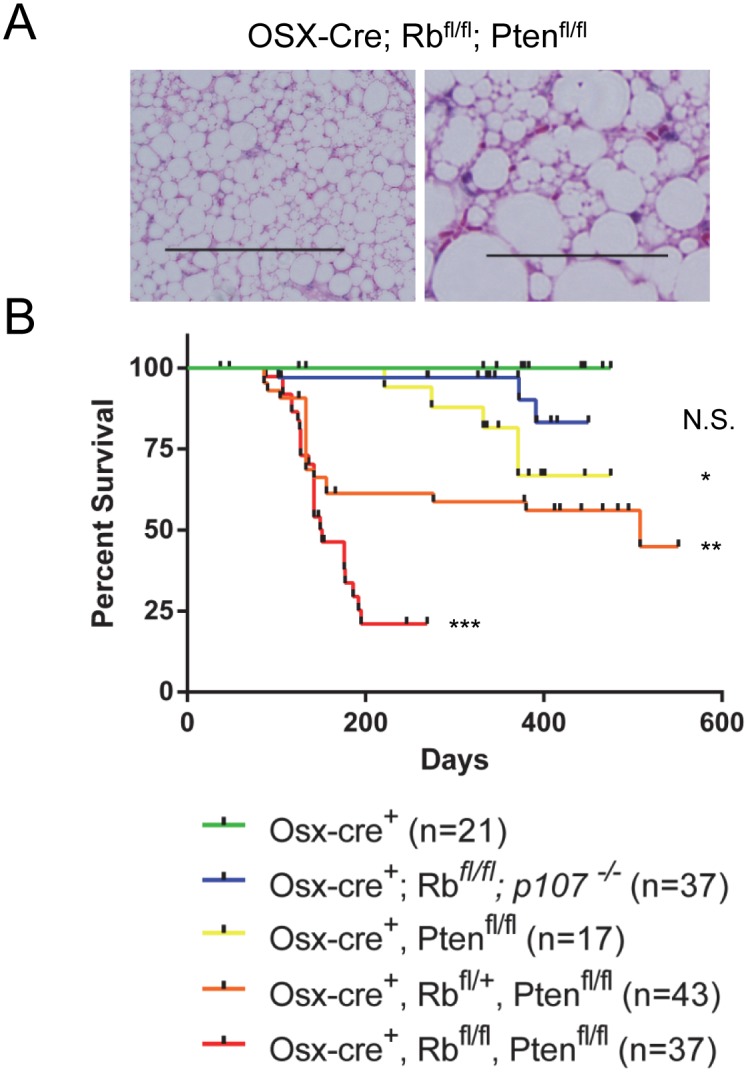
(A) H&E staining of sections from lipoma tumors derived from Osx-Cre; Rb1 fl/fl; Pten fl/fl mice. Scale bars, 200 μM (Left), 50 μM (Right). (B) Kaplan-Meier survival curve analysis for the indicated genotypes: Osx-Cre (n = 21), Osx-Cre; Rb1 fl/fl; p107 -/- (n = 37), Osx-Cre; Pten fl/fl (n = 17), Osx-Cre; Rb1 fl/+; Pten fl/fl (n = 43), and Osx-Cre; Rb1 fl/fl; Pten fl/fl (n = 37). p-values were determined by log rank test comparing a genotype with that of the control mice. *, p < 0.05; **, p < 0.01; ***, p < 0.005; N.S., not significant.
Because the RB protein is a reported positive regulator of osteogenic differentiation, we predicted that retaining one copy of the Rb1 gene in the Osx-Cre Rb1 fl/+; Pten fl/fl mice might shift the tumor formation away from adipogenic and towards osteogenic tumors. Cohorts of control, Osx-Cre; Pten fl/fl, and Osx-Cre; Rb1 fl/+; Pten fl/fl mice were generated and aged to around 18 months to assess tumor incidence. We also generated Osx-Cre; Rb1 fl/fl; p107 -/- mice to test the effect of co-deleting the Rbl1 (p107) gene, a pRB pocket protein family member, because its deletion augments other Rb driven tumors like retinoblastoma in mice [29]. However, we found that the Osx-Cre Rb1 fl/+; Pten fl/fl mice still developed adipogenic tumors, albeit at a lower incidence and reduced rapidity compared to those arising in the Osx-Cre; Rb1 fl/fl; Pten fl/fl mice (Fig 1B). Retention of both copies of Rb1 (Osx-Cre; Pten fl/fl) reduced the formation and delayed onset of lipoma formation even further, and only 4 of 17 mice developed them at around 9–12 months old. We detected a single sternal OS on the Osx-Cre; Pten fl/fl (1/7 mice analyzed) and a single OS, also sternal, on one of the Osx-Cre Rb1 fl/+; Pten fl/fl mice (S1 Fig). No detectable OS or adipogenic tumors or other abnormal phenotype were detected on the control or Osx-Cre; Rb1 fl/fl; p107 -/- mice. These data indicate that Pten deletion in osteoblast precursors strongly promotes lipoma tumor formation, that Rb1 hetero- or homo-zygous co-deletion greatly increases the incidence and the rapidity of onset of adipogenic tumors, with only a modest increase in detectable osteosarcoma tumors compared to control mice.
Pten loss in osteoprogenitor cells leads to accumulation of bone marrow adipocytes
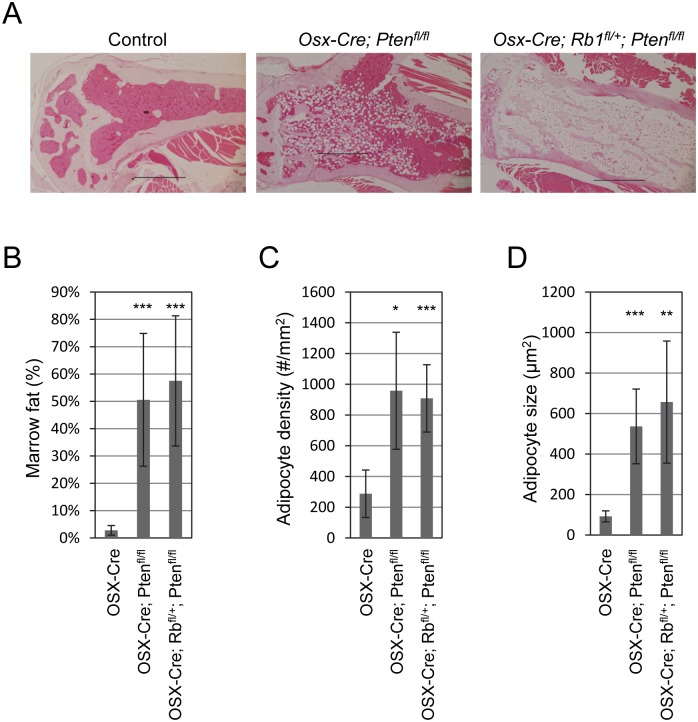
Osx-Cre expresses Cre in several mesenchymal cell types in the bone marrow of post-natal mice, including osteoblasts, osteocytes, chondrocytes, adipocytes, and stromal cell [30]. Thus, it is not entirely clear if Osx-Cre targets a common MSC progenitor of these cell types or if it targets discrete progenitor cell types. We analyzed sections of tibias from 12 month-old control, Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice by hematoxylin and eosin (H&E) staining to determine the consequences of homozygous deletion of Pten with, or without, heterozygous deletion of Rb1. We observed a striking increase in the number of adipocytes in the proximal tibial sections of Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl compared with control mice (Fig 2A). We used histomorphometry to quantify the differences in marrow adipocytes between the three strains. Total marrow fat increased from 3% in control mice to over 50% in Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice (Fig 2B). This difference was due to significant increases in adipocyte density (Fig 2C) and average adipocyte size (Fig 2D). These differences were due to Pten deletion because additional heterozygous loss of Rb1 did not augment the phenotype.
Fig 2. Pten disruption in osteo-progenitor cells increases marrow adipocyte density and size.
(A) Tibias from 12-month old Osx-Cre (control), Osx-Cre; Pten fl/fl, and Osx-Cre; Rb1 fl/+; Pten fl/fl, were cut for H&E staining and proximal end displayed. Scale bars, 500 μM. Tibial marrow contents from 12-month old mice were compared using NIH ImageJ. (B) Percentage of marrow fat; (C) adipocyte number (#/mm2); (D) adipocyte size (μm2) were measured using histomorphometry. Data are the mean ± SD from six mice. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
Pten deletion in mouse osteoblast precursors results in increased calvaria thickness and bone volume
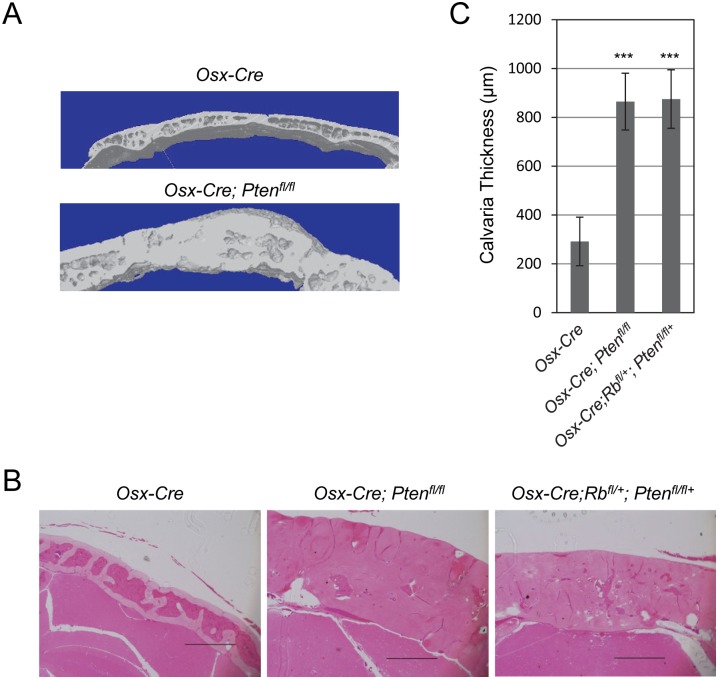
We evaluated the effects of Pten deletion on skeletal anatomy using three-dimensional micro-computed tomography (μCT) analysis and histology. μCT analysis revealed a significant thickening of the calvaria of 12-month-old Osx-Cre; Pten fl/fl mice compared to those from control mice (Fig 3A). We also analyzed calvaria by histology from control, Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice (Fig 3B). Calvaria were on average 3-times thicker in both the Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl compared to control mice (Fig 3C).
Fig 3. Disruption of the Pten gene in osteoblast precursors leads to increased calvaria thickness.
(A) μCT analysis was performed on 12-month-old Osx-Cre (control) and Osx-Cre; Pten fl/fl calvaria. (B) Representative coronal calvarial sections from 12-month old mice. Scale bar, 500 μM. (C) Bar graphs showing the calvarial thickness (mean ± SD; n = 6). ***, p < 0.005.
We next analyzed femurs by μCT from control, Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice. Whereas the cortical thickness of the Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl femurs was unchanged compared to control (Fig 4a), both the trabecular volume density and trabecular number were significantly increased in Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice compared to control animals (Fig 4a and 4b). Bone volume over total volume (BV/TV %) significantly increased from 5% in control mice to 10% in Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl 12-month-old mice. Likewise, trabecular number increased from 0.7 (1/mm) to 1.5 (1/mm) at one year in the Pten-deleted and Osx-Cre; Rb1 fl/+; Pten fl/fl mice (Fig 4b). Neither trabecular thickness nor trabecular separation showed any significant differences between control and experimental animals. These results indicate that Pten deletion in OSX+ osteoprogenitor cells results in increased bone mass in vivo. Additional heterozygous loss of the Rb1 tumor suppressor gene (Osx-Cre; Rb1 fl/+; Pten fl/fl) did not increase any of these parameters compared to Pten deletion alone.
Fig 4. Pten deletion in osteoblast precursors increases trabecular bone volume.
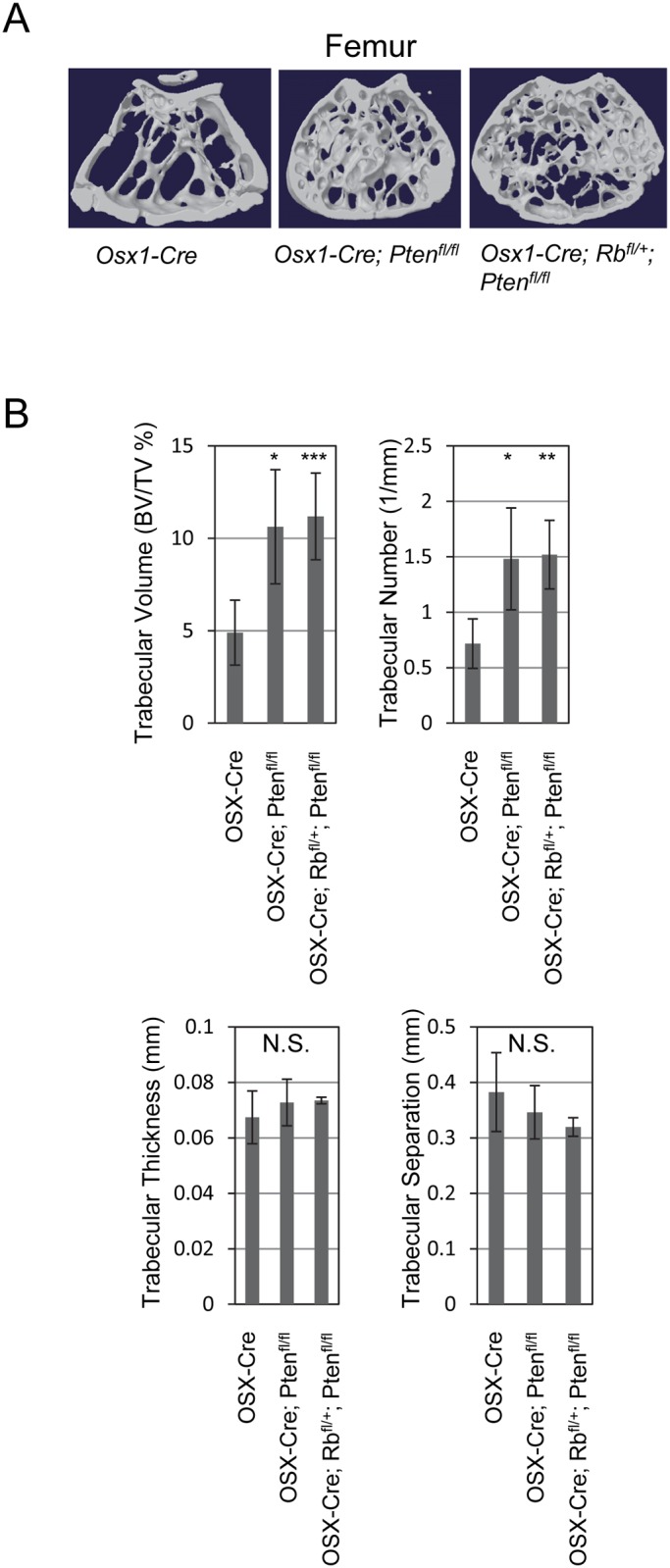
(A) μCT analysis was performed on the distal femur of control, Osx-Cre; Pten fl/fl and Osx-Cre; Rb1 fl/+; Pten fl/fl mice. (B) Bar graphs showing the trabecular volume (BV/TV%), trabecular number (1/mm), trabecular thickness (mm), trabecular separation (mm) (mean ± SD; n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.005; N.S., not significant.
Rb1 and Pten control adipocyte and osteoblast differentiation
Since Pten deletion in osteoprogenitor cells caused an increase in adipocytes in vivo, it is possible that it directly regulates adipocytic differentiation. We tested this using an established in vitro osteoblast differentiation assay. Primary cells were isolated from the calvaria of newborn mice carrying conditional Pten fl/fl or Rb fl/fl /Pten fl/fl and grown to confluence. Cells were then infected with control or Cre-recombinase expressing adenovirus, and gene deletion was confirmed by PCR (data not shown). Primary plated cells were treated with standard adipogenic or osteogenic differentiation media for 25 days. Using this approach, cells that differentiate into adipocytes contain lipid droplets that can be detected by staining with Oil red O. Likewise, differentiated osteoblasts secrete calcium deposits which can be detected by staining with alizarin red. Oil red O staining on calvarial cells lacking Pten showed increased adipocyte differentiation compared with control cells (Fig 5a). Adipocyte differentiation was more pronounced by co-deleting Rb1 and Pten. Deletion of Pten also increased osteoblast differentiation as detected by alizarin red staining (Fig 5b). The Rb fl/fl /Pten fl/fl cells differentiated to a greater extent than Pten fl/fl cells by alizarin red staining. These data show that loss of Pten acts in an intrinsic manner to increase calvarial cell differentiation into adipocytes and osteoblasts, and that co-deleting Rb1 with Pten augments differentiation into both cell types.
Fig 5. Loss of Pten promotes primary calvaria cell differentaion into adipocytes and osteoblasts and is further accentuated by co-deletion of Rb1.
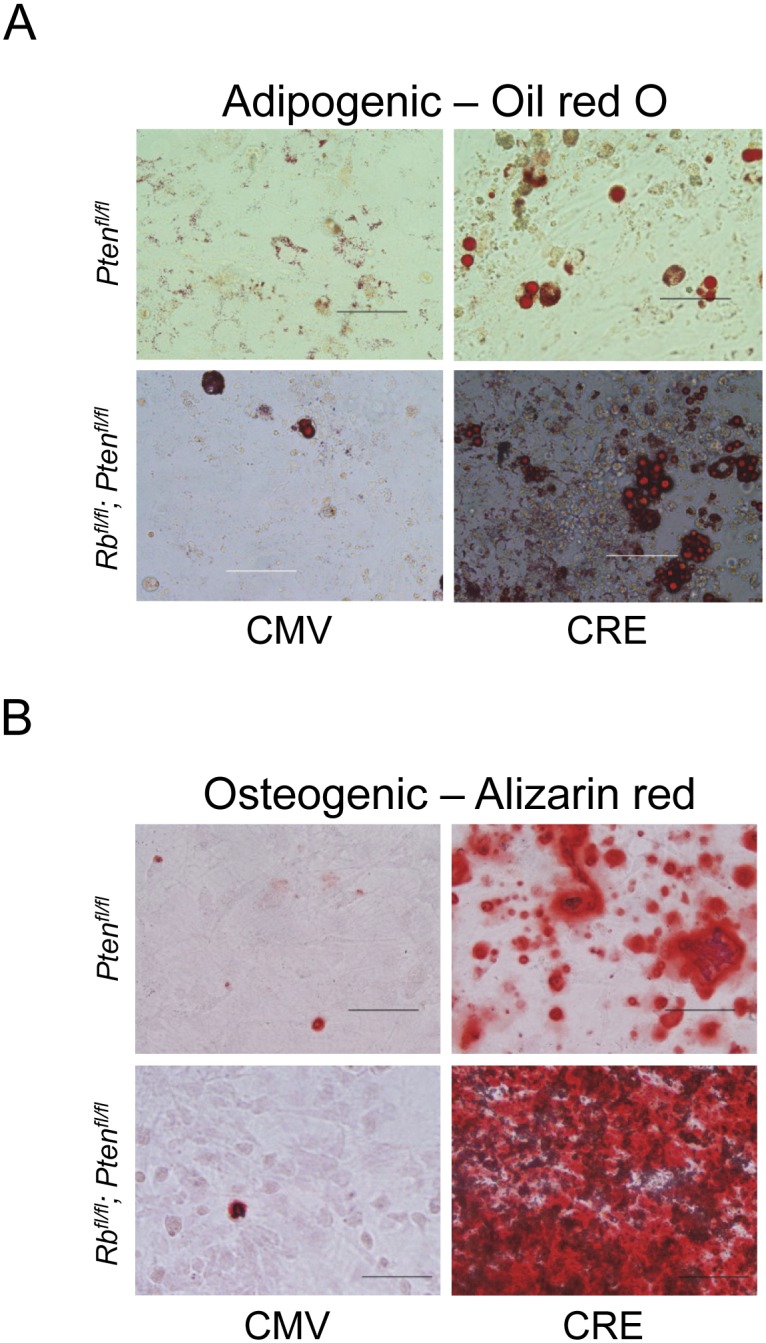
(A) Primary calvarial osteoblasts were grown to confluence, infected with control or Adeno-Cre, and grown for 25 days with adipogenic differentiation media and measured for adipocyte differentiation by Oil red O staining. Scale bar: 50 μM. (B) Cells were cultured as in (A) but were treated with osteoblastic differentiation media. Terminal osteoblastic differentiation was measured by alizarin red staining. Results are representative of 6 experiments.
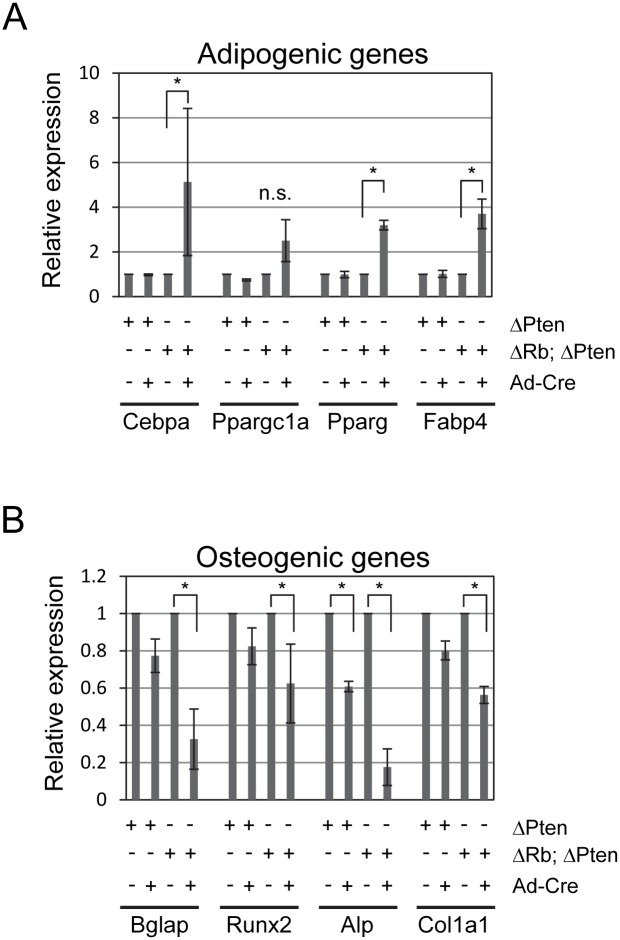
We then measured changes in expression of several adipogenic and osteogenic genes by qPCR to further examine the differentiation characteristics of these cells. Gene expression was assessed 48 hours after adding adipogenic or osteogenic media to detect early effects on differentiation. Deletion of Pten had no effect on any of the pro-adipogenic Cebpa, Ppargc1a, Pparg, or Fabp4 genes tested at this time point (Fig 6a). Co-deleting Rb1 with Pten, however, significantly induced expression of Cebpa, Pparg, and Fabp4, but not Ppargc1a, at this timepoint. Pten deletion caused slight, but non-significant decreased expression of the Bglap, Runx2 and Col1a1 but significantly decreased Alp pro-osteoblastic gene expression. Rb1 and Pten co-deletion, however, significantly reduced expression of each gene. These findings suggest that Pten deletion had only modest effects on adipogenic and osteogenic gene expression at early timepoints but was capable of promoting both adipogenic and osteogenic differentiation in longer assays. Second, co-deleting both Rb1 and Pten significantly increased adipogenic gene expression, concomitant with increased adipogenesis. It is unclear why Rb1 and Pten co-deletion reduced osteogenic gene expression at early time points but robustly induced osteogenesis in this in vitro model.
Fig 6. Effects of Pten deletion and Rb1/Pten co-deletion on adipogenic and osteoblastic gene expression.
Calvarial osteoblasts were isolated, cultured, and infected with control or Adeno-Cre and differentiated with adipogenic (A) or osteogenic (B) differentiation media for 48 hours. Quantitative PCR was performed on isolated RNA and displayed as fold-change in relative expression. Data are the mean ± SD from three independent experiments. *, p < 0.05.
Discussion
In this study we deleted Rb1 and Pten from osteoprogenitor cells to determine the consequences for bone development and tumor formation. We show that Pten loss significantly increased bone accumulation over time, but also caused a greatly increased adipocyte population and size in the bone marrow. Pten deletion also caused lipoma formation outside the marrow, which was robustly accelerated by co-deleting Rb1. A variety of other cre-deleter mouse strains have also been used to demonstrate that Pten is an important contributor to normal skeletal growth. Pten deletion in mature osteoblasts using an Osteocalcin-cre driver increased bone cell proliferation and skeletal mass over the lifespan of the animal [31]. Several groups conditionally deleted Pten in osteochondroprogenitor cells using Col2a1-cre transgenic mice, leading to Pten deletion in cartilage and perichondrium. These mice too exhibited skeletal overgrowth, and also chondrocyte dysplasia, and one displayed increased lipoma formation [32–34]. Deletion of Pten in mesenchymal cells using Dermo-cre caused early lethality in the majority of mutant mice, but surviving mice exhibited increased osteoblast differentiation and bone mass [35, 36]. Thus, Pten deletion across all stages of osteoblast formation, from early mesenchyme, partially differentiated progenitors, and mature osteoblasts, consistently increases bone growth in vivo. Mechanistically, Pten loss may promote bone growth by inactivating the FOXO transcription factors. FOXOs are phosphorylated and inactivated by the AKT serine/threonine kinase, which is constitutively activated following Pten loss. Foxo1, -3, and -4 co-deletion using Osx1-Cre leads to increased osteoblast numbers and high bone mass by increasing β–catenin/TCF mediated transcription, indicating a critical role for FOXOs in suppressing bone formation and osteoblast proliferation [37].
Despite Pten deletion strongly promoting osteoblast proliferation and significantly increasing bone mass, and the genomic region containing the Pten locus being commonly lost in human and canine osteosarcomas [38, 39], Pten deletion in mature osteoblasts or osteoblast precursor cells only rarely leads to osteosarcoma formation. For example, just 2/12 Col2a1-Cre; Pten fl/fl mice developed OS [32], and likewise, we observed only one osteosarcoma formation from 17 Osx1-Cre; Pten fl/fl mice aged to 18 months. Intriguingly, the Pten gene was recovered as one of the key drivers of OS in a murine transposon-based forward genetic screen [8]. Thus, it seems likely that although Pten loss is associated with OS progression in various species, Pten loss is insufficient to initiate OS without other cooperating mutations. Additionally, complete deletion of Pten may favor differentiation towards other lineages, whereas heterozygous deletion of Pten may instead favor OS [8].
We observed a striking increase in marrow adipogenesis in the Pten deleted mice. Pten deletion in the cartilage and perichondrium mesenchymal lineages of Col2a1-cre transgenic mice also displayed increased marrow adipocytes and lipoma formation [33]. In contrast, Pten deletion in mature osteoblasts using Osteocalcin-cre was not reported to affect adipogenesis [31]. Foxo1, -3, and -4 co-deletion using the Osx1-Cre line leads to decreased marrow adiposity, in contrast with our findings that ΔPten increases marrow adiposity, indicating that Pten deletion regulates adipose levels in the marrow through FOXO independent targets [37]. Pten deletion also promotes adipogenesis in other tissues and cell types. For example, hepatocyte-specific deletion of Pten in mice promotes accumulation of adipocytes [40]. Likewise, Pten deletion in the endometrium by Amhr2-Cre results in accumulation of adipocytes in the myometrium [41]. This pro-adipogenic effect is tissue specific, and is not observed when Pten is deleted from skeletal muscles or cardiomyocytes [42, 43]. Deletion of Pten in adipocytes leads to striking lipoma formation when targeting MYF5 expressing cells, but surprisingly did not affect lipid mass when deleted from mature aP2-cre targeted fat cells [44, 45]. The effects of Pten deletion on adipogenesis and fat production appear to depend on the timing of Pten deletion and potentially also of the capacity of the targeted cells to trans- or de-differentiate into adipocytes.
Although the Rb1 gene is also frequently mutated in human osteosarcoma [46], previous studies indicated that Rb1 deletion in osteoblast precursors only has modest effects and does not promote OS [27, 28]. In general, Rb1 promotes osteogenesis through interactions with the RUNX2 transcription factor, and inhibits adipogenesis by repression expression of PPARγ[12, 13]. Accordingly, Rb1 deletion in the embryo using Meox1-Cre reduces bone formation and osteogenic gene expression, and simultaneously increases expression of adipogenic markers and adipogenesis [11]. The ability of pRB to suppress adipogenesis has also been observed in the context of mesenchymal p53 deletion. p53 is a major osteosarcoma tumor suppressor, and its homozygous deletion in early mesenchymal precursors (Prx1-cre) or partially differentiated osteoblasts (Osx1-cre) leads to robust osteosarcoma emergence [11, 27, 28]. Consistent with a role for Rb1 in suppressing adipogenesis, homozygous Rb1 co-deletion dramatically shifted the tumor spectrum towards fatty tumors and reduced incidence of OS. Surprisingly, retaining a single copy of Rb1 in p53 deleted cells shifted the tumor spectrum strongly back towards osteosarcomas, consistent with the notion that Rb1 is a positive regulator of osteogenesis.
Because Pten deletion was insufficient to drive robust OS onset, we tested if co-deleting Rb1 and Pten could synergistically promote osteosarcoma formation. Co-deleting Rb1 with Pten greatly accelerated lipoma formation and increased the incidence to 100%, but did not induce osteosarcoma. It is possible that we failed to observe osteosarcoma because the rapid enlargement of lipoma tumors required mice to be euthanized around 3–4 months of age. We also aged Osx1-Cre; Rb fl/+; Pten fl/fl mice to determine of retaining a single copy of Rb1 promoted osteosarcoma formation. However, these mice only induced osteosarcoma in a single mouse (1/7), and developed lipomas at a rate and size intermediate between Osx1-Cre; Pten fl/fl and Osx1-Cre Rb fl/fl; Pten fl/fl mice. These findings suggest that Osx1-Cre expresses cre in certain fat cells, or adipocyte precursors, outside the marrow, and that co-deleting Rb1 and Pten in these cells promotes their sustained proliferation. In contrast, Rb1 heterozygous co-deletion with Pten did not affect marrow adiposity or Pten driven bone outgrowth in the calvaria or femur trabeculation. Thus, mesenchymal cell fate outcomes, in regards to lipoma- vs osteosarcoma-genesis appear to depend heterozygous vs. homozygous gene deletion, the timing of gene loss during mesenchymal cell specification, and the capacity of the target cell to trans- or de-differentiate into other mesenchymal lineages.
Supporting Information
A. Representative presentation of OS on the sternum of 12 month-old (A) Osx-Cre; Pten fl/fl and (B) Osx-Cre; Rb1 fl/+; Pten fl/fl mice. C. Tumor histology of OS from Osx-Cre; Pten fl/fl mouse. Bar = 500 μm. D. Tumor histology of OS from Osx-Cre; Rb1 fl/+; Pten fl/fl mouse. Bar = 500 μm. E & G. Histology of OS from Osx-Cre; Pten fl/fl mouse. Bar = 50 μm. Arrow notes adipocytes within the OS. F. Histology of OS from Osx-Cre; Rb1 fl/+; Pten fl/fl mouse. Bar = 50 μm. Arrow notes adipocytes within the OS. H. Osteoid-rich section of the tumor. Bar = 50 μm.
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grants to T.H. from National Institutes of Health (R01CA168622) and the Children’s Cancer Research Fund, Minneapolis, MN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75(7):1241–4. Epub 1993/12/31. 0092-8674(93)90610-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2. Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–56. Epub 1994/12/30. 0092-8674(94)90006-X [pii]. . [DOI] [PubMed] [Google Scholar]
- 3. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–71. Epub 1997/05/30. S0092-8674(00)80259-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 4. Hoshi K, Komori T, Ozawa H. Morphological characterization of skeletal cells in Cbfa1-deficient mice. Bone. 1999;25(6):639–51. Epub 1999/12/11. S8756-3282(99)00223-9 [pii]. . [DOI] [PubMed] [Google Scholar]
- 5. Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW. HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Miner Res. 2006;21(6):921–33. Epub 2006/06/07. . [DOI] [PubMed] [Google Scholar]
- 6. Rodda S, McMahon A. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–44. [DOI] [PubMed] [Google Scholar]
- 7. Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7(1):104–12. Epub 2014/04/08. S2211-1247(14)00165-X [pii] 10.1016/j.celrep.2014.03.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, et al. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–24. Epub 2015/05/12. ng.3293 [pii] 10.1038/ng.3293 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–62. [DOI] [PubMed] [Google Scholar]
- 10. Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1-/- mice. Proc Natl Acad Sci U S A. 2008;105(47):18402–7. Epub 2008/11/21. 10.1073/pnas.0805925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466(7310):1110–4. Epub 2010/08/06. 10.1038/nature09264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8(2):303–16. Epub 2001/09/08. . [DOI] [PubMed] [Google Scholar]
- 13. Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J. E2Fs regulate adipocyte differentiation. Dev Cell. 2002;3(1):39–49. [DOI] [PubMed] [Google Scholar]
- 14. Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–7. [DOI] [PubMed] [Google Scholar]
- 15. Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. Epub 1998/10/20. S0092-8674(00)81780-8 [pii]. . [DOI] [PubMed] [Google Scholar]
- 16. Cantley L. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–7. [DOI] [PubMed] [Google Scholar]
- 17. Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–74. Epub 2007/07/03. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie C, Lu H, Nomura A, Hanse EA, Forster CL, Parker JB, et al. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol Cancer. 2015;14(1):93 Epub 2015/04/25. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–8. [DOI] [PubMed] [Google Scholar]
- 20. Lee M-H, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, et al. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–32. [DOI] [PubMed] [Google Scholar]
- 21. Trotman L, Niki M, Dotan Z, Koutcher J, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biology. 2003;1(3):385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egan KP, Brennan TA, Pignolo RJ. Bone histomorphometry using free and commonly available software. Histopathology. 2012;61(6):1168–73. Epub 2012/08/14. 10.1111/j.1365-2559.2012.04333.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parlee SD, Lentz SI, Mori H, MacDougald OA. Quantifying size and number of adipocytes in adipose tissue. Methods Enzymol. 2014;537:93–122. Epub 2014/02/01. B978-0-12-411619-1.00006–9 [pii] 10.1016/B978-0-12-411619-1.00006-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broege A, Pham L, Jensen ED, Emery A, Huang TH, Stemig M, et al. Bone morphogenetic proteins signal via SMAD and mitogen-activated protein (MAP) kinase pathways at distinct times during osteoclastogenesis. J Biol Chem. 2013;288(52):37230–40. Epub 2013/11/16. 10.1074/jbc.M113.496950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu H, Hallstrom TC. The nuclear protein UHRF2 is a direct target of the transcription factor E2F1 in the induction of apoptosis. J Biol Chem. 2013;288(33):23833–43. Epub 2013/07/09. 10.1074/jbc.M112.447276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. 10.1006/meth.2001.1262 S1046-2023(01)91262-9 [pii]. . [DOI] [PubMed] [Google Scholar]
- 27. Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22(12):1662–76. 10.1101/gad.1656808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105(33):11851–6. Epub 2008/08/14. 10.1073/pnas.0805462105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wirt SE, Sage J. p107 in the public eye: an Rb understudy and more. Cell Div. 2010;5:9 Epub 2010/04/03. 10.1186/1747-1028-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen J, Shi Y, Regan J, Karuppaiah K, Ornitz DM, Long F. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9(1):e85161 Epub 2014/01/24. 10.1371/journal.pone.0085161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X, Bruxvoort KJ, Zylstra CR, Liu J, Cichowski R, Faugere MC, et al. Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc Natl Acad Sci U S A. 2007;104(7):2259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ford-Hutchinson AF, Ali Z, Lines SE, Hallgrimsson B, Boyd SK, Jirik FR. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J Bone Miner Res. 2007;22(8):1245–59. Epub 2007/04/26. . [DOI] [PubMed] [Google Scholar]
- 33. Hsieh SC, Chen NT, Lo SH. Conditional loss of PTEN leads to skeletal abnormalities and lipoma formation. Mol Carcinog. 2009;48(6):545–52. Epub 2008/11/01. 10.1002/mc.20491 [DOI] [PubMed] [Google Scholar]
- 34. Yang G, Sun Q, Teng Y, Li F, Weng T, Yang X. PTEN deficiency causes dyschondroplasia in mice by enhanced hypoxia-inducible factor 1alpha signaling and endoplasmic reticulum stress. Development. 2008;135(21):3587–97. Epub 2008/10/04. 10.1242/dev.028118 [DOI] [PubMed] [Google Scholar]
- 35. Guntur AR, Reinhold MI, Cuellar J Jr, Naski MC. Conditional ablation of Pten in osteoprogenitors stimulates FGF signaling. Development. 2011;138(7):1433–44. Epub 2011/03/10. 10.1242/dev.058016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tiozzo C, Carraro G, Al Alam D, Baptista S, Danopoulos S, Li A, et al. Mesodermal Pten inactivation leads to alveolar capillary dysplasia- like phenotype. J Clin Invest. 2012;122(11):3862–72. Epub 2012/10/02. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iyer S, Ambrogini E, Bartell SM, Han L, Roberson PK, de Cabo R, et al. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest. 2013;123(8):3409–19. Epub 2013/07/23. 10.1172/JCI68049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Freeman S, Allen S, Ganti R, Wu J, Ma J, Su X, et al. Copy number gains in EGFR and copy number losses in PTEN are common events in osteosarcoma tumors. Cancer. 2008;113(6):1453–61. 10.1002/cncr.23782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levine R, Forest T, Smith C. Tumor Suppressor PTEN is mutated in canine osteosarcoma cell lines and tumors. Vet Pathol. 2002;39(3):372–8. [DOI] [PubMed] [Google Scholar]
- 40. Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113(12):1774–83. Epub 2004/06/17. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Daikoku T, Jackson L, Besnard V, Whitsett J, Ellenson LH, Dey SK. Cell-specific conditional deletion of Pten in the uterus results in differential phenotypes. Gynecol Oncol. 2011;122(2):424–9. Epub 2011/05/17. 10.1016/j.ygyno.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110(6):737–49. Epub 2002/09/26. . [DOI] [PubMed] [Google Scholar]
- 43. Wijesekara N, Konrad D, Eweida M, Jefferies C, Liadis N, Giacca A, et al. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol Cell Biol. 2005;25(3):1135–45. Epub 2005/01/20. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanchez-Gurmaches J, Hung CM, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2013;16(3):348–62. Epub 2012/09/04. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurlawalla-Martinez C, Stiles B, Wang Y, Devaskar SU, Kahn BB, Wu H. Insulin hypersensitivity and resistance to streptozotocin-induced diabetes in mice lacking PTEN in adipose tissue. Mol Cell Biol. 2005;25(6):2498–510. Epub 2005/03/04. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wadayama B, Toguchida J, Simizu T, Ishizaki K, Sasaki MS, Kotoura Y, et al. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994;54(11):3042–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Representative presentation of OS on the sternum of 12 month-old (A) Osx-Cre; Pten fl/fl and (B) Osx-Cre; Rb1 fl/+; Pten fl/fl mice. C. Tumor histology of OS from Osx-Cre; Pten fl/fl mouse. Bar = 500 μm. D. Tumor histology of OS from Osx-Cre; Rb1 fl/+; Pten fl/fl mouse. Bar = 500 μm. E & G. Histology of OS from Osx-Cre; Pten fl/fl mouse. Bar = 50 μm. Arrow notes adipocytes within the OS. F. Histology of OS from Osx-Cre; Rb1 fl/+; Pten fl/fl mouse. Bar = 50 μm. Arrow notes adipocytes within the OS. H. Osteoid-rich section of the tumor. Bar = 50 μm.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.