Abstract
Leukocyte adhesion deficiency (LAD) is an immunodeficiency caused by defects in the adhesion of leukocytes (especially neutrophils) to the blood vessel wall. As a result, patients with LAD suffer from severe bacterial infections and impaired wound healing, accompanied by neutrophilia. In LAD-I, mutations are found in ITGB2, the gene that encodes the β subunit of the β2 integrins. This syndrome is characterized directly after birth by delayed separation of the umbilical cord. In the rare LAD-II disease, the fucosylation of selectin ligands is disturbed, caused by mutations in SLC35C1, the gene that encodes a GDP-fucose transporter of the Golgi system. LAD-II patients lack the H and Lewis Lea and Leb blood group antigens. Finally, in LAD-III (also called LAD-I/variant) the conformational activation of the hematopoietically expressed β integrins is disturbed, leading to leukocyte and platelet dysfunction. This last syndrome is caused by mutations in FERMT3, encoding the kindlin-3 protein in all blood cells that is involved in the regulation of β integrin conformation.
Keywords: LAD-I, LAD-II, LAD-III, ITGB2, SLC35C1, FERMT3, β integrins, GDP-fucose transporter, kindlin-3
Introduction
Leukocyte adhesion deficiency (LAD) is an autosomal recessive disorder caused by decreased expression or functioning of CD18, the β2 subunit of the leukocyte β2 integrins [1]. This deficiency leads to severe impairment of leukocyte adhesion to the vascular wall and leukocyte migration to sites of infection and inflammation. The patients suffer from recurrent, life-threatening bacterial and fungal infections and from impaired wound healing. Characteristic features are delayed separation of the umbilical cord and strong leukocytosis, especially neutrophilia, during periods of infection. Many LAD patients die at young age despite intensive antibiotic therapy. Bone-marrow transplantation is the treatment of choice. LAD is a rare immunodeficiency, but the exact incidence is not known.
The integrins are transmembrane receptors composed of α and β subunits that mediate cellular adhesive interactions throughout the body. At present 18 α and 8 β subunits have been identified that are loosely organized into integrin families. The β2 integrins form a family of four heterodimeric proteins, only expressed on leukocytes, with one of four α subunits coupled to a common β2 subunit: αLβ2 (LFA-1, CD11a/CD18), αMβ2 (Mac-1 or CR3, CD11b/CD18), αXβ2 (p150,95, CD11c/CD18) and αDβ2 (CR4, CD11d/CD18), the latter only being expressed on macrophages. Decreased expression of the common β2 subunit leads to a similar decrease in the expression of all four α subunits on the leukocyte surface. The four β2 integrins act as adhesion proteins, mediating adhesion of leukocytes to other cells and to extracellular matrix proteins. The α subunits and the β2 subunit are transmembrane proteins, intracellularly connected to the leukocyte cytoskeleton. Binding to extracellular ligands leads to a conformational change of the β2 integrins, increased binding of intracellular target proteins and downstream signal transduction to cell spreading and altered gene expression, cell proliferation, differentiation and apoptosis (“outside-in” signaling). Leukocyte activation, e.g. as a result of chemokine binding to chemokine receptors, antigen binding to the T-cell receptor or ligand binding to selectins, induces conformational changes in the extracellular regions of the β2 integrins, leading to a higher affinity for their ligands (“inside-out” signaling) [2].
Classical LAD-I
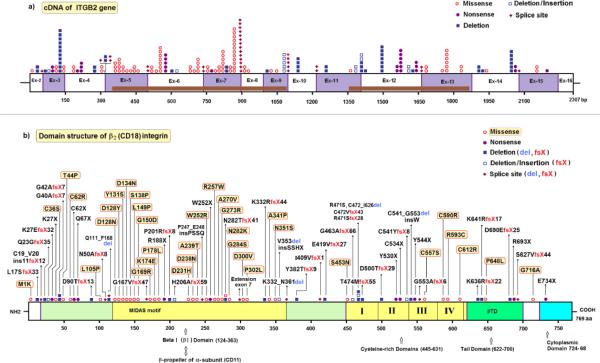
In the most common form of LAD, called LAD-I (OMIM #116920), mutations are found in ITGB2 (integrin beta-2), the gene located at 21q22.3 (OMIM *600065) that encodes the β2 integrin protein. Usually, this leads to the absence or decreased expression of the β2 integrins on the leukocyte surface, but sometimes a normal expression of nonfunctional β2 integrins is found. In a previous publication we listed 34 mutations found in ITGB2 of LAD-I patients [3]. In the present publication, 53 newly identified mutations have been added to Table 1 (marked with * in the last column). Mutations that have not been previously published elsewhere are marked as “Unpubl.”. Most of the point mutations are found in a ~240-residue domain that is highly conserved in all β integrin subunits and coded for by exons 5 – 9 of ITGB2 (figure 1). This “βI domain”, together with its αI counterpart, constitutes the major ligand-binding site of the β2 integrins. Both I domains also contain a metal ion-dependent adhesion site (MIDAS motif) consisting of an Asp-X-Ser-X-Ser sequence.
TABLE I.
Mutations in the LAD-I gene ITGB2
| Nucleotide change | Mutation | Amino-acid or mRNA change | LAD-I type | Families (alleles) | Reference | Accession Number | |
|---|---|---|---|---|---|---|---|
| Not determined | Deletion | Entire gene | LAD-Io | 1(1) | [3, 19] | A0032 | |
| Not determined | Deletion | Entire gene | LAD-Io | 1(1) | [20, 21] | A0051 | * |
| Not determined | Deletion | Entire gene | LAD-I? | 1(1) | Unpubl. | A0085 | * |
| c.-721_58+51del833 | Deletion | Deletion exon 2, including start codon |
LAD-Io | 1(2) | [3, 22] | A0063 A0064 | |
| c.2T>A | Missense | p.Met1Lys | LAD-I– | 1(1) | [3, 23] | A0016 | |
| c.49delC | Deletion | p.Leu17SerfsX33 | LAD-Io | 1(2) | Unpubl. | A0086 | * |
| c.59-10C>A a | Splice site | Ins 43 nts from intron 2 r.59-43_59-1ins43;59- 10C>A p.Cys19_Val20ins11X12 |
LAD-Io | 1(1) | [24] | A0126 | * |
| c.66_67delTC | Deletion | p.Gln23GlyfsX35 | LAD-Io | 1(1) | [3, 22] | A0028 A0029 | |
| c.77dupC | Insertion | p.Lys27GlufsX32 | LAD-Io | 1(1) | Unpubl. | A0079 | * |
| c.79A>T | Nonsense | p.Lys27X | LAD-Io | 2(2) | [20, 21] Unpubl. |
A0051 A0114 | * |
| c.106T>A | Missense | p.Cys36Ser | LAD-Io | 1(2) | Unpubl. | A0088 | * |
| c.119_128delGGCCCGGCTG | Deletion | p.Gly40AlafsX7 | LAD-Io | 5(10)b | [3, 20, 25, 26] Unpubl. |
A0018 A0068 A0083 A0089 A0133 |
|
| c.120delG | Deletion | p.Gly42AlafsX7 | LAD-Io | 1(1) | Unpubl. | A0127 | * |
| c.130A>C | Missense | p.Thr44Pro | LAD-Io | 2(2) | Unpubl. | A0090 A0091 | * |
| Not identified | Splice site? |
Deletion exon 4 p. Asn50AlafsX8 |
LAD-Io | 1(2) | [3, 26] | A0081 | |
| c.184T>C | Missense | p.Cys62Arg | LAD-I? | 1(2) | Unpubl. | A0092 | * |
| c.186C>A | Nonsense | p.Cys62X | LAD-I? | 1(1) | Unpubl. | A0093 | * |
| c.199C>T | Nonsense | p.Gln67X | LAD-Io | 2(3) | [3, 22] Unpubl. | A0005 A0065 A0114 |
|
| c.268delG | Deletion | p.Asp90ThrfsX13 | LAD-I– | 1(2) | Unpubl. | A0094 | * |
| c.314T>C | Missense | p.Leu105Pro | LAD-I– | 1(2) b | [27] | A0095 | * |
| c.328+1G>A a | Splice site | Deletion exon 4 p.Asn50AlafsX8 |
LAD-Io | 1(1) | [28] | A0050 | * |
| c.329-37_461del169 a | Deletion | Deletion exon 5 p.Gln111_Phe168del |
LAD-Io | 1(2) | [3, 29] | A0038 | |
| c.329-6C>A a | Splice site | Deletion exon 5? p.Gln111_Phe168del? |
LAD-Io | 1(2) | [30] | A0053 | * |
| c.382G>A | Missense | p.Asp128Asn | LAD-Io | 2(3) | [3, 31] Unpubl. | A0002 A0117 | |
| c.382G>T | Missense | p.Asp128Tyr | LAD-Io/- | 2(4) | [30] | A0056 A0059 | * |
| c.392A>C c | Missense | p.Tyr131Ser | LAD-Io | 1(2) | [32] | A0046 | * |
| c.400G>A | Missense | p.Asp134Asn | LAD-I? | 1(1) | [33] Unpubl. |
A0076 | * |
| c.412T>C | Missense | p.Ser138Pro | LAD-I+ | 1(1) | [3,34] | A0003 | |
| c.446T>C | Missense | p.Leu149Pro | LAD-I– | 2(2) | [3, 5, 22, 35] | A0017 A0078 |
|
| c.449G>A | Missense | p.Gly150Asp | LAD-I+ | 1(1) | [36] | A0099 | * |
| c.500-12T>G a | Splice site | Deletion part of exon 6 p.Gly167ValfsX47 |
LAD-Io | 1(2) | [3, 22] Unpubl. |
A0033 | |
| c.505G>A | Missense | p.Gly169Arg | LAD-Io | 2(2) | [3, 35, 37] | A0010 A0027 | |
| c.520A>G | Missense | p.Lys174Glu | LAD-Io | 1(1) | Unpubl. | A0125 | * |
| c.533C>T | Missense | p.Pro178Leu | LAD-Io | 5(7) b | [3, 36, 38, 39, 40] Unpubl. |
A0020 A0031 A0073 A0074 A0080 A0099 |
|
| c.562C>T | Nonsense | p.Arg188X | LAD-I? | 3(5) | [32, 41] Unpubl. |
A0076 A0097 A0098 A0122 |
* |
| c.602delC | Deletion | p.Pro201ArgfsX8 | LAD-Io | 1(1) | [28] | A0050 | * |
| c.614_615insA | Insertion | p.His206AlafsX59 | LAD-Io | 1(1) | [3, 42] | A0021 | |
| c.691G>C | Missense | p.Asp231His | LAD-I+ | 2(2) | [3, 43] Unpubl. |
A0013 A0075 |
|
| c.712G>A | Missense | p.Asp238Asn | LAD-I? | 1(1) | Unpubl. | A0113 | * |
| c.715G>A | Missense | p.Ala239Thr | LAD-Io | 2(4) b | [30] Unpubl. | A0055 A0130 | * |
| c.742-14C>A a,c,d | Splice site | p.Pro247_Glu248 insProSerSerGln |
LAD-Io | 2(2) | [3, 4, 5, 22, 32] | A0028 A0029 A0030 |
|
| c.754T>C | Missense | p.Trp252Arg | LAD-Io | 1(2) | [3, 22] | A0004 | |
| c.755G>A | Nonsense | p.Trp252X | LAD-Io | 1(1) | Unpubl. | A0075 | * |
| c.769C>T | Missense | p.Arg257Trp | LAD-Io/- | 1(1) | Unpubl. | A0131 | * |
| c.809C>T | Missense | p.Ala270Val | LAD-Io/- | 2(2) | [3, 44] Unpubl. | A0007 A0111 | |
| c.817G>A | Missense | p.Gly273Arg | LAD-Io | 5(6) b | [3, 34] Unpubl. | A0003 A0093 A0113 A0118 A0123 |
|
| c.843delC | Deletion | p.Asn282ThrfsX41 | LAD-Io | 1(2) | [30] | A0060 | * |
| c.846C>A | Missense | p.Asn282Lys | LAD-I+ | 1(2) b | Unpubl. | A0124 | * |
| c.850G>A c | Missense | p.Gly284Ser | LAD-Io | 10(12) | [3, 4, 5, 32, 43, 44, 46] Unpubl. |
A0011 A0012 A0013 A0014 A0023 A0043 A0044 A0047 A0072 A0091 A0116 |
|
| c.897+1G>A a | Splice site | Extension exon 7 with 64 or 298 nts from intron 7, or with whole intron 7. fsX26 or fsX44 |
LAD-Io | 8(14) | [3, 30, 31, 47] Unpubl. |
A0039 A0049 A0057 A0058 A0085 A0100 A0105 A0121 |
|
| c.899A>T | Missense | p.Asp300Val | LAD-Io | 1(2) | [48] | A0084 | * |
| c.905C>T | Missense | p.Pro302Leu | LAD-Io | 1(1) | Unpubl. | A0079 | * |
| c.995_1004del10 | Deletion | p.Lys332ArgfsX44 | LAD-I? | 1(1) | Unpubl. | A0117 | * |
| c.1021G>C | Missense | p.Ala341Pro | LAD-Io/- | 1(1) | [3, 22, 44] | A0006 | |
| c.1052A>G c | Missense | p.Asn351Ser | Activated LFA-I | 1(1) | [3, 4, 32, 49] | A0030 | |
| c.1057_1059delins35 | Deletion/insertion | p.Val353del insSerSerHisX |
LAD-Io | 3(5) | Unpubl. | A0104 A0106 A0107 |
* |
| c.1083+3G>C a | Splice site | Deletion exon 9 p.Lys332_Asn361del |
LAD-Io | 2(4) b | [3, 46] Unpubl. | A0034 A0035 A0036 A0037 A0119 |
|
| c.1083+5G>C a | Splice site | Deletion exon 9? p.Lys332_Asn361del? |
LAD-Io | 1(1) | Unpubl. | A0125 | * |
| c.1143delC | Deletion | p.Tyr382ThrfsX9 | LAD-I– | 2(4) b | [30] Unpubl. | A0052 A0128 | * |
| c.1225-?_1412+?del | Deletion or splice site |
p.Ile409ValfsX1 | LAD-Io | 1(2) b | Unpubl. | A0132 | * |
| c.1256_1257delAG d | Deletion | p.Glu419ValfsX27 | LAD-I– | 1(2) | [3, 5] | A0019 | |
| c.1358G>A | Missense | p.Ser453Asn | LAD-I? | 1(1) e | Unpubl. | A0112 | * |
| c.1388_1390delGCTinsC A | Deletion/ insertion |
p.Gly463AlafsX66 | LAD-I? | 1(2) | Unpubl. | A0109 | * |
| c.1413-?_1877+?del2114 | Deletion | Deletion exons 12 + 13 p.[Arg471Ser, Cys472_Ile626del] |
LAD-Io | 1(2) | [32, 36] | A0070 A0071 | * |
| c.1413-149_2080+839 del2518insAAAA |
Deletion/insertion | Deletion exon 12_14 p.Cys472ValfsX43 |
LAD-Io | 1(2) b | Unpubl. | A0134 | * |
| c.1413-396_?del27703 | Deletion | Deletion exon 12 end p.Arg471SerfsX28 |
LAD-Io | 1(1) | [24] | A0126 | * |
| c.1421delC | Deletion | p.Thr474MetfsX55 | LAD-I? | 1(1) | Unpubl. | A0115 | * |
| c.1498delG | Deletion | p.Asp500ThrfsX29 | LAD-Io | 5(7) b | [50, 51] | A0041 A0042 A0043 A0044 A0045 A0108 |
* |
| c.1590C>G | Nonsense | p.Tyr530X | LAD-Io | 1(2) b | [3] | A0025 | |
| c.1602C>A | Nonsense | p.Cys534X | LAD-Io | 4(6) b | [3, 22, 25, 44] Unpubl. |
A0001 A0006 A0103 A0118 |
|
| c.1622delGins23 | Deletion/ insertion |
p.Cys541TyrfsX8 | LAD-Io | 1(2) | Unpubl. | A0101 | * |
| c.1622_1657del36 (cDNA) |
Deletion? Splice site? |
p.Cys541_Gly553del insTrp |
LAD-I– | 1(1) | [52] | A0040 | * |
| c.1632C>G | Nonsense | p.Tyr544X | LAD-Io | 2(3) | Unpubl. | A0096 A0120 | * |
| c.1658-2A>G a | Splice site | Deletion exon 13? p.Gly553AlafsX6 |
LAD-I? | 1(1) | Unpubl. | A0111 | * |
| Not identified | Splice site | Deletion exon 13 p.Gly553AlafsX6 |
LAD-Io | 1(1) | [3, 38] | ||
| c.1670G>C | Missense | p.Cys557Ser | LAD-Io | 1(2) b | Unpubl. | A0129 | * |
| c.1768T>C | Missense | p.Cys590Arg | Activated LFA-1, no CR3 |
1(1) | [3, 22, 44] | A0007 | |
| c.1777C>T | Missense | p.Arg593Cys | Activated LFA-1, no CR3 |
5(6) b | [3,5,22, 44, 50, 53] |
A0008 A0009 A0012 A0045 A0066 A0067 |
|
| c.1834T>C | Missense | p.Cys612Arg | LAD-I– | 1(2) | [3, 26, 54] | A0082 | |
| c.1907delA | Deletion | p.Lys636ArgfsX22 | LAD-Io | 1(2) | [30] | A0061 A0062 | * |
| c.1920delG | Deletion | p.Lys641ArgfsX17 | LAD-Io | 1(1) | [50] | A0042 | |
| c.1943C>T | Missense | p.Pro648Leu | LAD-I? | 1(1) e | Unpubl. | A0110 | * |
| c.2070delT c | Deletion | p.Asp690GlufsX25 | LAD-Io | 6(6) | [3, 22, 23, 32, 35] Unpubl. |
A0015 A0016 A0017 A0116 A0127 |
|
| c.2077C>T | Nonsense | p.Arg693X | LAD-I? | 1(2) b | Unpubl. | A0077 | * |
| c.2080+1delG a | Splice site | Deletion exon 14 ? p.Ser627ValfsX44? |
LAD-Io | 1(1) | Unpubl. | A0090 | * |
| c.2147G>C f | Missense | p.Gly716Ala | LAD-Io | 1(2) | [30] | A0054 | * |
| c.2200G>T | Nonsense | p.Glu734X | LAD-I– | 1(1) | [52] | A0040 | * |
| Number of different alleles | Total number of alleles | |||
|---|---|---|---|---|
| Deletions | 23 alleles | (26.7%) | 52 alleles | (25.9%) |
| Insertions | 2 alleles | (2.3%) | 2 alleles | (1.0%) |
| Deletion/insertions | 4 alleles | (4.7%) | 11 alleles | (5.5%) |
| Nonsense mutations | 10 alleles | (11.6%) | 23 alleles | (11.4%) |
| Splice site mutations | 12 alleles | (14.0%) | 31 alleles | (15.4%) |
| Missense mutations | 35 alleles | (40.7%) | 82 alleles | (40.8%) |
| Total 86 different allelic mutations | Total 109 families with 201 identified alleles in the 123 patients |
|||
Additional mutations mentioned in refs. 37 and 55-57 can be disregarded as being nonspecific for LAD-I, unconfirmed or printing errors.
indicates that the mutant CD18 molecule supports differentially the expression of the different CD11/CD18 integrins.
Positions of introns in ITGB2: intron 1 c.-4_-3; intron 2 c.58_59; intron 3 c.147_148; intron 4 c.328_329; intron 5 c.499_500; intron 6 c.741_742; intron 7 c.897_898; intron 8 c.993_994; intron 9 c.1083_1084; intron 10 c.1224_1225; intron 11 c.1412_1413; intron 12 c.1657_1658; intron 13 c.1877_1878; intron 14 c.2080_2081; intron 15 c.2247_2248.
One or more patients presumed to be homozygous for this mutation.
And reverse mutations in a small subset of lymphocytes [32].
Corrected after consultation of the authors.
No other mutation found in this patient; variation of unknown significance.
Not certain of clinical significance since transfection of this mutant into a CD18-deficient B cell line induced near normal expression. Moreover, transfection into HEK293 cells together with CD11a induced normal expression and constitutive binding to ICAM-1, and transfection together with CD11b induced expression and constitutive binding to denatured BSA.
Figure 1.
Schematic overview of mutations in ITGB2. a. The number of mutated alleles, the type of mutation and their position along the exons of ITGB2, with active domains depicted in brown (see underneath fig. 1b). Symbols, explained on the right, represent mutated alleles. Numbers refer to nucleotides in cDNA.
b. Domain structure of the β2 integrin (CD18) and the location of each mutation, with active domains depicted in yellow, green and blue. Protein interaction sites are shown for some domains. Symbols, explained on the right, represent separate mutations. Numbers refer to amino acids in β2 integrin.
Figure drawn by Dr. M. Yavuz Köker.
Table 2 contains information on apparently benign polymorphisms that have been identified in ITGB2. The mutation c.1756C>T (p.Arg586Trp) in ITGB2 was found in two non-related patients on the same allele with the mutation c.742-14C>A at the 3’ end of intron 6 (leading to the incorporation of four additional amino acids in exon 7) [4,5]. By itself, the 586Trp CD18 molecule, expressed in COS cells, supports about 75% of normal β2 integrin expression [5], indicating that this mutation may be regarded as a polymorphism. Therefore, the c.1756G/T alternatives are shown as SNPs in Table 2. Other polymorphisms shown in Table 2 have not yet been defined at the functional level.
Table 2.
Polymorphisms in the LAD-I gene ITGB2
| Polymorphic nucleotide | Amino acid change | Reference |
|---|---|---|
| c.-403C/T (promoter) | NA | [3, 25] |
| c.-135A/C (5’ UTR) | NA | [Ensembl] |
| c.-128A/T 5’ UTR) | NA | [Ensembl] |
| c.-111T/C (5’ UTR) | NA | [3, 22, 58, 59] |
| c.-110A/G (5’ UTR) | ||
| c.-10_-7AGTT/del | NA | [Ensembl] |
| c.13C/T | p.Arg5Cys | [Ensembl] |
| c.24G/T | p.Leu8Leu (silent) | [3, 16, 22, 24, 30, 58] |
| c.31C/T | p.Leu11Leu (silent) | [Ensembl] |
| c.58G/A | p.Val20Ile | [Ensembl] |
| c.117G/A | p.Ser39Ser (silent) | [Ensembl] |
| c.147+16A/G | NA | Unpubl. |
| c.162G/A | p.Pro54Pro (silent) | [3, 5, 22] |
| c.205_206CT/TC | p.Leu69Ser | [Ensembl] |
| c.222G/A | p.Ala74Ala (silent) | [Ensembl] |
| c.229G/A | p.Asp77Asn | [58] |
| c.294G/A a | p.Leu98Leu (silent) | [58] |
| c.328+15G/A | NA | [3, 22] |
| c.499+7C/T | NA | [3, 22] |
| c.500-103T/G | NA | Unpubl. |
| c.500-29T/C | NA | [24] |
| c.500-11G/T | NA | [3, 22, 24] |
| c.587A/C b | p.Lys196Thr | Unpubl.b |
| c.598C/G | p.Pro200Ala | [Ensembl] |
| c.741+36T/C | NA | Unpubl. |
| c.742-13G/A | NA | [3, 22] |
| c.810G/A | p.Ala270Ala (silent) | [Ensembl] |
| c.819A/G | p.Gly273Gly (silent) | [3, 5, 22, 25, 31, 38, 45, Ensembl] |
| c.849C/T | p.Asp283Asp (silent) | [Ensembl] |
| c.892G/A | p.Glu298Lys | [58] |
| c.906A/G | p.Pro302Pro (silent) | [Ensembl] |
| c.993+41T/C | NA | Unpubl. |
| c.994-47G/A a | NA | [3, 22, 24] |
| c.1002C/T | p.Thr334Thr (silent) | [Ensembl] |
| c.1062T/A | p.His354Gln | [30, 48] |
| c.1101C/A | p.Val367Val (silent) | [3, 22, 24, 25, 30, 37, 58] |
| c.1146C/T | p.Tyr382Tyr (silent) | [Ensembl] |
| c.1186C/T | p.Pro396Ser | [Ensembl] |
| c.1224+48G/T | NA | [3, 22] |
| c.1247C/T | p.Thr416Met | [Ensembl] |
| C.1323C/T | p.Val441Val (silent) | [3, 5, 22, 24, 25, 30, 37, 58] |
| C.1497G ?A | p.Lys499Lys (silent) | [Ensembl] |
| C.1542C/T | p.Cys514Cys (silent) | [Ensembl] |
| C.1635C/T | p.Asn545Asn (silent) | [Ensembl] |
| C.1700G/C | p.Gly567Ala | [58] |
| C.1714G/T | p.Ala572Ser | [58] |
| C.1724G/C | p.Cys575Ser | [58] |
| C.1756C/T | p.Arg586Trp | [3, 4, 5, 58, 59] |
| C.1793G/T | p.Cys598Phe | [Ensembl] |
| C.1888G/A | p.Glu630Lys | [Ensembl] |
| C.1893C/T | p.Cys631Cys (silent) | [Ensembl] |
| C.2058C/G | p.Leu686Leu (silent) | [Ensembl] |
| C.2147G/C c | p.Gly716Ala | N. Parvaneh, unp.c |
| C.2349T/C (3’ UTR) a | NA | [58] |
| C.2433A/G (3’ UTR) a | NA | [3, 22, 58] |
| C.2456C/A (3’ UTr) a | NA | [3, 22, 58] |
| C.2482G/C (3’ UTR) a | NA | [58] |
| C.2484G/A (3’ UTR) | NA | [Ensembl] |
| C.2579A/C (3’ UTR) a | NA | [58] |
| C.2681G/T (3’ UTR) a | NA | [58] |
| c.2721_2722delTG (3’ UTR) | NA | [Ensembl] |
Corrected after Consultation of the authors.
Originally published as an LAD-I– mutation [53].
LAD-II
Two other, extremely rare forms of LAD exist. Patients with LAD-II (OMIM #266265) have a defect in the fucosylation of various cell surface glycoproteins, some of which function as ligands for L-selectin [6]. As a result, the initial “rolling” of leukocytes over the endothelial vessel wall in areas of inflammation, which is mediated by reversible contact between L-selectins on the leukocytes and E- or P-selectins on the endothelial cells with their respective sialated fucosyl ligands on the opposite cells, is disturbed. Both via intracellular signaling and by slowing down the leukocytes, this rolling allows integrins to bind their ligand on endothelial cells, which is needed for stable adhesion. Thus, in LAD-II, one mechanism of β2-integrin activation is lacking, leading to decreased leukocyte adhesion to the vessel wall and decreased transendothelial migration into the tissues. However, the chemokine and T-cell receptor pathways of β2-integrin activation are still operative, and the infectious episodes in LAD-II patients are therefore in general less severe than those seen in LAD-I patients. On the other hand, the fucosylation defect affects not only selectin ligands but also other essential glycoproteins, leading to severe mental and growth retardation. The molecular defect in LAD-II has been identified as a deficiency in a GDP-fucose transport protein in the Golgi system [7,8]. This protein is encoded by SLC35C1 (Solute carrier family 35 member C1) at 11p11.2 (OMIM *605881). Table 3 lists the mutations found in this gene in six families with LAD-II patients. Two of these mutations concern Arg147 and Thr308, both highly conserved amino acids in the family of nucleotide-sugar transporters group 2 and suggested to be involved in substrate recognition [9]. Supplementation of fucose led to a substantial clinical improvement and correction of hypofucosylation in the patient homozygous for the Arg147Cys mutation, whereas it was of no benefit to the patients homozygous for the Thr308Arg mutation [7,8]. Possible polymorphisms in SLC35C1 are also listed in Table 3.
TABLE 3.
Mutations and polymorphisms in the LAD-II gene SLC35C1
| Nucleotide change | Mutation | Amino-acid or mRNA change | LAD type | Families (alleles) | Reference | Accession Number | |
|---|---|---|---|---|---|---|---|
| c.439C>T | Missense | p.Arg147Cys | LAD-II | 1(2) | [3, 7, 8] | A0001 | |
| c.588delG | Deletion | p.Trp196CysfsX35 | LAD-II | 1(2) | [60] | A0004 | * |
| c.923C>G | Missense | p.Thr308Arg | LAD-II | 2(4) | [3, 8, 61] | A0002 A0003 A0005 |
|
| c.969G>A | Nonsense | p.Trp323X | LAD-II | 1(2) | [62] | A0008 | * |
| c.1010A>G | Missense | p.Tyr337Cys | LAD-II | 1(2) | [63] | A0009 | * |
| c.718A/G | SNP a | p.Ile240Val b | [61] | ||||
| c.772T/C | SNP a | p.Phe258Leu b | [61] |
LAD-III (LAD-I/variant)
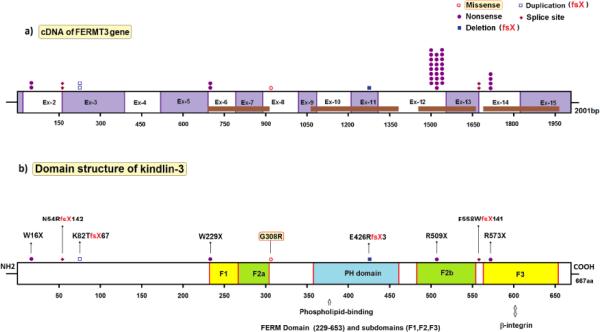
Finally, patients have been described with a defect in the “inside-out” signaling of leukocytes required for activation of β2 integrins into structures that bind their ligands with high affinity [10]. These patients, in addition to infections, also present with a bleeding disorder, indicating that the signaling defect also affects the β3 integrin fibrinogen receptor αIIbβ3 on blood platelets. The molecular defect of this variant form of LAD (LAD-III or LAD-I/variant, OMIM #612840) has recently been assigned to mutations in FERMT3 (fermitin family homolog 3) at 11q13.1 (OMIM *607901), the gene encoding kindlin-3, a protein involved in inside-out signaling to all blood cell-expressed β integrins (β1, β2 and β3) [11-14]. Kindlin-3 is apparently expressed not only in hematopoietic cells, but also in endothelial cells [15]; the biological significance of its deficiency in endothelial cells in LAD-III is not yet clear. A discussion has raged in the literature about the importance of a genetic variation in the gene encoding CalDAG-GEF1 (a guanine nucleotide exchange factor for Rap1, involved in integrin activation) in some patients with LAD-III, in addition to mutations in FERMT3 found in these patients [11,12,16]. However, since the functional defect in such patients can be corrected by reconstitution with kindlin-3 but not by reconstitution with CalDAG-GEF1 [14], this variation in CalDAG-GEF1 is of no importance for the functional defect in LAD-III patients. Table 4 and figure 1 list the mutations found in FERMT3 in 21 families with LAD-III patients. A hotspot of p.Arg509X mutations in kindlin-3 points to a founder effect, since these mutations are all found in Turkish families originating from Anatolia.
TABLE 4.
Mutations in the LAD-III gene FERMT3
| Nucleotide change | Mutation | Amino-acid or mRNA change | LAD type | Families (alleles) | Reference | Accession Number | |
|---|---|---|---|---|---|---|---|
| c.48G>A | Nonsense | p.Trp16X | LAD-III | 1(2) | [64] | F0019 F0020 | * |
| c.161-2A>C a | Splice site | p.Asn54ArgfsX142 | LAD-III | 1(2) | [65] | F0025 | * |
| c.238_244dup7 | Insertion | p.Lys82ThrfsX67 | LAD-III | 1(2) | Unpubl. | F0024 | * |
| c.687G>A | Nonsense | p.Trp229X | LAD-III | 1(2) | [12, 66] | F0014 F0015 | * |
| c.922G>A | Missense | p.Gly308Arg | LAD-III | 1(1) | [67] | F0021 | * |
| c.1275delT | Deletion | p.Glu426ArgfsX3 | LAD-III | 1(1) | [67] | F0021 | * |
| c.1525C>T | Nonsense | p.Arg509X | LAD-III | 12(24) | [11, 12, 14, 64] | F0001 F0002 F0003 F0004 F0005 F0006 F0007 F0008 F0009 F0010 F0011 F0012 F0017 F0018 |
* |
| c.1671-2A>G a | Splice site | Deletion exon 14 p.Phe558TrpfsX141 |
LAD-III | 1(2) | [14] | F0016 | * |
| c.1717C>T | Nonsense | p.Arg573X | LAD-III | 2(4) | [12, 68] | F0013 F0022 F0023 |
* |
Position of introns in FERMT3: intron 1 c.-15_-14; intron 2 c.160_161; intron 3 c.394_395; intron 4 c.514_515; intron 5 c.683_684; intron 6 c.786_787; intron 7 c.894_895; intron 8 c.1029_1030; intron 9 c.1079_1080; intron 10 c.1204_1205; intron 11 c.1311_1312; intron 12 c.1545_1546; intron 13 c.1670_1671; intron 14 c.1812_1813.
Final remarks
Additional information about the tabulated mutations and about LAD in general can be found in a recent review [16] and in the cited literature. In Table 1 we have used the notation LAD-Io, LAD-I− and LAD-I+ for differentiating the various phenotypes of LAD-I. In this nomenclature the superscript symbol indicates whether the protein is present at < 5% of normal expression (o), diminished in expression (−), i.e. between 5% and 20% of normal expression, or normally present but nonfunctional (+).This information is based on immune reactivity of the patients’ leukocytes with monoclonal antibodies analyzed by flow cytometry and sometimes on similar analyses of COS cells cotransfected with mutant CD18 molecules and wild-type CD11 molecules. In case this information is not known, this is indicated as (?). In a number of cases functionality of the mutant CD18 proteins was tested in cellular adherence assays to β2 ligands.
The nucleotide numbering system we have used is based on the cDNA sequence and follows the convention that +1 is the A of the ATG initiation codon. This differs from the numbering of the GenBank sequences; for ITGB2 (GenBank Accession Number M15395) subtract 72 from the GenBank sequence number to make the initiator A +1. The notation of the mutations follows the recommendations of the Human Genome Variation Society [17] (see also www.hgvs.org/mutnomen). The consequences of the mutations for protein composition have been checked with the Mutalyzer program (www.lovd.nl/mutalyzer) [18]. Where possible we have cross-referenced the mutations indicated in the present article with those in an LAD-I, LAD-II and LAD-III database that lists these patients by accession number. These databases contain additional biochemical, genetic and clinical information and are available at www.uta.fi/imt/bioinfo/ITGB2base, www.uta.fi/imt/bioinfo/SLC35C1base and www.uta.fi/imt/bioinfo/FERMT3base, respectively. Moreover, information can also be found in the HGMD database at www.hgmd.cf.ac.uk/ac/search.php.
Figure 2.
Schematic overview of mutations in FERMT3. a. The number of mutated alleles, the type of mutation and their position along the exons of FERMT3, with active domains depicted in brown (see underneath fig. 2b). Symbols, explained on the right, represent mutated alleles. Numbers refer to nucleotides in cDNA.
b. Domain structure of kindlin-3 and the location of each mutation, with active domains depicted in yellow, green and blue. Protein interaction sites are shown for some domains. Symbols, explained on the right, represent separate mutations. Numbers refer to amino acids in kindlin-3.
Figure drawn by Dr. M. Yavuz Köker.
Acknowledgements
EvdV was supported by the Landsteiner Foundation for Blood Transfusion Research, Amsterdam, The Netherlands (LSBR 0619). The Jeffrey Modell Foundation is acknowledged for their support of Dr Somech.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Anderson DC, Smith CW. Leukocyte adhesion deficiencies. In: Scriver CR, Beaudet AL, Sly S, Volle D, editors. The Metabolic and Molecular Basis of Inherited Disease. McGraw-Hill; New York: 2001. pp. 4829–4856. [Google Scholar]
- 2.Hynes RO. Integrins: versatility, modulation and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 3.Roos D, Law SKA. Hematologically important mutations: Leukocyte Adhesion Deficiency. Blood Cells Mol. Dis. 2001;27:1000–1004. doi: 10.1006/bcmd.2001.0473. [DOI] [PubMed] [Google Scholar]
- 4.Nelson C, Rabb H, Arnaout MA. Genetic cause of leukocyte adhesion deficiency. Abnormal splicing and a missense mutation in a conserved region of CD18 impair cell surface expression of β2 integrins. J. Biol. Chem. 1992;267:3351–3357. [PubMed] [Google Scholar]
- 5.Wright AH, Douglass WA, Taylor GM, Lau Y-L, Higgins, Davies KA, Law SKA. Molecular characterization of leukocyte adhesion deficiency in six patients. Eur. J. Immunol. 1995;25:717–722. doi: 10.1002/eji.1830250313. [DOI] [PubMed] [Google Scholar]
- 6.Etzioni A, Frydman M, Pollack S, Avidor I, Phillips ML, Paulson JC, Gershioni-Baruch R. Recurrent severe infections caused by a novel leukocyte adhesion deficiency. N. Engl. J. Med. 1992;327:1789–1792. doi: 10.1056/NEJM199212173272505. [DOI] [PubMed] [Google Scholar]
- 7.Luhn K, Wild MK, Eckhardt M, Gerardy-Schahn R, Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- 8.Lubke T, Marquardt T, Etzioni A, Hartmann E, von Figura K, Korner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- 9.Handford M, Rodriguez-Furlán C, Orellana A. Nucleotide-sugar transporters: structure, function and roles in vivo. Braz. J. Med. Biol. Res. 2006;39:1149–1158. doi: 10.1590/s0100-879x2006000900002. [DOI] [PubMed] [Google Scholar]
- 10.Kuijpers TW, van Lier RAW, Hamann D, de Boer M, Thung LY, Weening RS, Verhoeven AJ, Roos D. Leukocyte adhesion deficiency type 1 (LAD-1)/variant. A novel immunodeficiency syndrome characterized by dysfunctional β2 integrins. J. Clin. Invest. 1997;100:1725–1733. doi: 10.1172/JCI119697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mory A, Feigelson SW, Yarali N, Kilic SS, Bayhan GI, Gershoni-Baruch R, Etzioni A, Alon R. Kindlin-3: a new gene involved in the pathogenesis of LAD-III. Blood. 2008;112:2591. doi: 10.1182/blood-2008-06-163162. [DOI] [PubMed] [Google Scholar]
- 12.Kuijpers TW, van de Vijver E, Weterman MA, de Boer M, Tool AT, van den Berg TK, Moser M, Jakobs ME, Seeger K, Sanal O, Unal S, Cetin M, Roos D, Verhoeven AJ, Baas F. LAD-1/variant syndrome is caused by mutations in FERMT3. Blood. 2009;113:4740–4746. doi: 10.1182/blood-2008-10-182154. [DOI] [PubMed] [Google Scholar]
- 13.Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fässler R. Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat. Med. 2009;15:300–305. doi: 10.1038/nm.1921. [DOI] [PubMed] [Google Scholar]
- 14.Svensson L, Howarth K, McDowall A, Patzak I, Evans R, Ussar S, Moser M, Metin A, Fried M, Tomlinson I, Hogg N. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat. Med. 2009;15:306–312. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bialkowska K, Ma YQ, Bledzka K, Sossey-Alaoui K, Izem L, Zhang X, Malinin N, Qin J, Byzova T, Plow EF. J. Biol. Chem. 2010;285:18640–18649. doi: 10.1074/jbc.M109.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etzioni A. Genetic etiologies of leukocyte adhesion defects. Curr. Opin. Immunol. 2009;21:481–486. doi: 10.1016/j.coi.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum. Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 19.Rivera-Matos IR, Rakita RM, Mariscalco MM, Elder FFB, Dreyer SA, Cleary TG. Leukocyte adhesion deficiency mimicking Hirschsprung disease. J. Pediatr. 1995;127:755–757. doi: 10.1016/s0022-3476(95)70169-9. [DOI] [PubMed] [Google Scholar]
- 20.Castriconi R, Dondero A, Cantoni C, Della Chiesa M, Prato C, Nanni M, Fiorini M, Notarangelo L, Parolini S, Moretta L, Notarangelo L, Moretta A, Bottino C. Blood. 2007;109:4873–4881. doi: 10.1182/blood-2006-08-038760. [DOI] [PubMed] [Google Scholar]
- 21.Fiorini M, Piovani G, Schumacher RF, Magri C, Bertini V, Mazzolari E, Notarangelo L, Notarangelo LD, Barlati S. ITGB2 mutation combined with deleted ring 21 chromosome in a child with leukocyte adhesion deficiency. J. Allergy Clin. Immunol. 2009;124:1356–1358. doi: 10.1016/j.jaci.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 22.Roos D, Meischl C, de Boer M, Simsek S, Weening RS, Sanal O, Tezcan I, Güngör T, Law SKA. Genetic analysis of patients with leukocyte adhesion deficiency. Genomic sequencing reveals otherwise undetectable mutations. Exp. Hematol. 2002;30:252–261. doi: 10.1016/s0301-472x(01)00782-2. [DOI] [PubMed] [Google Scholar]
- 23.Sligh JE, Jr., Hurwitz MY, Zhu C, Anderson DC, Beaudet AL. An initiation codon mutation in CD18 in association with the moderate type of leukocyte adhesion deficiency. J. Biol. Chem. 1992;267:714–718. [PubMed] [Google Scholar]
- 24.Cher THB, Chan HS, Klein GF, Jabkowski J, Schadenböck-Kranzl G, Zach O, Law SKA. A novel 3'splice-site mutation and a novel gross deletion in leukocyte adhesion deficiency (LAD)-1. Biochem. Biophys. Res. Commun. 2011;404:1099–1104. doi: 10.1016/j.bbrc.2010.12.124. [DOI] [PubMed] [Google Scholar]
- 25.Lopez Rodriguez C, Nueda A, Grospierre B, Sanchez-Madrid F, Fischer A, Springer TA, Corbi AL. Characterization of two new CD18 alleles causing severe leukocyte adhesion deficiency. Eur. J. Immunol. 1993;23:2792–2798. doi: 10.1002/eji.1830231111. [DOI] [PubMed] [Google Scholar]
- 26.Fiorini M, Vermi W, Facchetti F, Moratto D, Alessandri G, Notarangelo L, Caruso A, Grigolato P, Ugazio AG, Notarangelo LD, Badolato R. Defective migration of monocyte-derived dendritic cells in LAD-1 immunodeficiency. J. Leukoc. Biol. 2002;72:650–656. [PubMed] [Google Scholar]
- 27.Hinze CH, Lucky AW, Bove KE, Marsh RA, Bleesing JH, Passo MH. Leukocyte adhesion deficiency type I presenting with recurrent pyoderma gangrenosum and flaccid scarring. Pediatr. Dermatol. 2010;27:500–503. doi: 10.1111/j.1525-1470.2010.01260.x. [DOI] [PubMed] [Google Scholar]
- 28.Tone Y, Wada T, Shibata F, Toma T, Hashida Y, Kasahara Y, Koizumi S, Yachie A. Somatic revertant mosaicism in a patient with leukocyte adhesion deficiency type 1. Blood. 2007;109:1182–1184. doi: 10.1182/blood-2007-08-039057. [DOI] [PubMed] [Google Scholar]
- 29.Allende LM, Hernandez M, Corell A, Garcia-Perez MA, Varela P, Moreno A, Caragol I, Garcia-Martin F, Guillen-Perales J, Olive T, Espanol T, Arnaiz-Villena A. A novel CD18 genomic deletion in a patient with severe leucocyte adhesion deficiency: a possible CD2/lymphocyte function-associated antigen-1 functional association in humans. Immunology. 2000;99:440–450. doi: 10.1046/j.1365-2567.2000.00960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parvaneh N, Mamishi S, Rezaei A, Rezaei N, Tamizifar B, Parvaneh L, Sherkat R, Ghalehbaghi B, Kashef S, Chavoshzadeh Z, Isaeian A, Ashrafi F, Aghamohammadi A. Characterization of 11 new cases of leukocyte adhesion deficiency type 1 with seven novel mutations in the ITGB2 gene. J. Clin Immunol. 2010;30:756–760. doi: 10.1007/s10875-010-9433-2. [DOI] [PubMed] [Google Scholar]
- 31.Matsuura S, Kishi F, Tsukahara M, Nunoi H, Matsuda I, Kobayashi K, Kajii T. Leukocyte adhesion deficiency: identification of novel mutations in two Japanese patients with a severe form. Biochem. Biophys. Res. Commun. 1992;184:1460–1467. doi: 10.1016/s0006-291x(05)80047-6. [DOI] [PubMed] [Google Scholar]
- 32.Uzel G, Tng E, Rosenzweig SD, Hsu AP, Shaw JM, Horwitz ME, Linton GF, Anderson SM, Kirby MR, Oliveira JB, Brown MR, Fleisher TA, Law SKA, Holland SM. Reversion mutations in patients with leukocyte adhesion deficiency type-1 (LAD-1) Blood. 2008;111:209–218. doi: 10.1182/blood-2007-04-082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorusso F, Kong D, Jalil AK, Sylvestre C, Tan SL, Ao A. Preimplantation genetic diagnosis of leukocyte adhesion deficiency type I. Fertil. Steril. 2006;85:494.e15–494.e18. doi: 10.1016/j.fertnstert.2005.03.092. [DOI] [PubMed] [Google Scholar]
- 34.Hogg N, Stewart MP, Scarth SL, Newton R, Shaw JM, Law SKA, Klein N. A novel leukocyte adhesion deficiency caused by expressed but nonfunctional β2 integrins Mac-1 and LFA-1. J. Clin. Invest. 1999;103:97–106. doi: 10.1172/JCI3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wardlaw AJ, Hibbs ML, Stacker SA, Springer TA. Distinct mutations in two patients with leukocyte adhesion deficiency and their functional correlates. J. Exp. Med. 1990;172:335–345. doi: 10.1084/jem.172.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uzel G, Kuhns D, Hussey A, Spalding C, Stoddard J, Hsu A, Holland S. The new face of leukocyte adhesion deficiency type 1 (LAD-1). XIVth Meeting of the European Society for Immunodeficiencies; 132: 2010. (Abstr. P256) [Google Scholar]
- 37.Corbi AL, Vara A, Ursa A, Garcia Rodriguez MC, Fontan G, Sanchez-Madrid F. Molecular basis for a severe case of leukocyte adhesion deficiency. Eur. J. Immunol. 1992;22:1877–1881. doi: 10.1002/eji.1830220730. [DOI] [PubMed] [Google Scholar]
- 38.Back AL, Kwok WW, Hickstein DD. Identification of two molecular defects in a child with leukocyte adhesion deficiency. J. Biol. Chem. 1992;267:5482–5487. [PubMed] [Google Scholar]
- 39.Ohashi Y, Yambe T, Tsuchiya S, Kikuchi H, Konno T. Familial genetic defect in a case of leukocyte adhesion deficiency. Hum. Mutat. 1993;2:458–467. doi: 10.1002/humu.1380020606. [DOI] [PubMed] [Google Scholar]
- 40.Malawista SE, de Boisfleury Chevance A, Brown EJ, Boxer LA, Law SKA. Chemotaxis of non-compressed blood polymorphonuclear leukocytes from an adolescent with severe leukocyte adhesion deficiency. Am. J. Hematol. 2003;73:115–120. doi: 10.1002/ajh.10311. [DOI] [PubMed] [Google Scholar]
- 41.Simon AJ, Lev A, Wolach B, Gavrieli R, Amariglio N, Rosenthal E, Gazit E, Eyal E, Rechavi G, Somech R. The effect of gentamycin-induced readthrough on a novel premature termination codon of CD18 leukocyte adhesion deficiency patients. PLoS ONE. 2010;5:e13659. doi: 10.1371/journal.pone.0013659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard AJ, Heale J-P, Tsang A, Massing B, Speert DP. Nonopsonic phagocytosis of Pseudomonas aeruginosa: insights from an infant with leukocyte adhesion deficiency. Pediatr. Infect. Dis. J. 2001;20:452–454. doi: 10.1097/00006454-200104000-00019. [DOI] [PubMed] [Google Scholar]
- 43.Mathew EC, Shaw JM, Bonilla FA, Law SKA, Wright DA. A novel point mutation in CD18 causing the expression of dysfunctional CD11/CD18 leucocyte integrins in a patient with leucocyte adhesion deficiency. Clin. Exp. Immunol. 2000;121:133–138. doi: 10.1046/j.1365-2249.2000.01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw JM, Al-Shamkhani A, Boxer LA, Buckley CD, Dodds AW, Klein N, Roberts I, Roos D, Scarth SL, Simmons DL, Tan S-M, Law SKA. Identification of four CD18 mutations in leukocyte adhesion deficient (LAD) patients with differential abilities to associate with the CD11a, CD11b and CD11c antigens. Clin. Exp. Immunol. 2001;126:311–318. doi: 10.1046/j.1365-2249.2001.01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Back AL, Kerkering M, Baker D, Bauer TR, Embree LJ, Hickstein DD. A point mutation associated with Leukocyte Adhesion Deficiency Type I of moderate severity. Biochem. Biophys. Res. Commun. 1993;193:912–918. doi: 10.1006/bbrc.1993.1712. [DOI] [PubMed] [Google Scholar]
- 46.Kishimoto TK, O'Connor K, Springer TA. Leukocyte adhesion deficiency. Aberrant splicing of a conserved integrin sequence causes a moderate deficiency phenotype. J. Biol. Chem. 1989;264:3588–3595. [PubMed] [Google Scholar]
- 47.Tsai Y-C, Lee W-I, Huang J-L, Hung I-J, Jaing T-H, Yao T-C, Chen M-T, Kuo M-L. Neutrophil function and molecular analysis in severe leukocyte adhesion deficiency type I without separation delay of the umbilical cord. Pediatr. Allergy Immunol. 2008;19:25–32. doi: 10.1111/j.1399-3038.2007.00590.x. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Jin YY, Cao RM, Chen TX. A novel point mutation in CD18 causing leukocyte adhesion deficiency in a Chinese patient. Chin. Med. J. (Engl) 2010;123:1278–1282. [PubMed] [Google Scholar]
- 49.Cheng M, Foo SY, Shi ML, Tang RH, Kong LS, Law SKA, Tan SM. Mutation of a conserved asparagine in the I-like domain promotes constitutively active integrins alphaLbeta2 and alphaIIbbeta3. J. Biol. Chem. 2007;282:18225–18232. doi: 10.1074/jbc.M701386200. [DOI] [PubMed] [Google Scholar]
- 50.Fathallah DM, Jamal T, Barbouche MR, Bejaoui M, Hariz MB, Dellagi K. Two novel frame shift, recurrent and de novo mutations in the ITGB2 (CD18) gene causing leukocyte adhesion deficiency in a highly inbred North African population. J. Biomed. Biotechnol. 2001;1:114–121. doi: 10.1155/S1110724301000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halioui-Louhaichi S, Ben Hariz M, Fathallah Dahmani M, Barbouch MR, Mahmoudi J, Bejaoui M, Fischer A, Dellagi K, Ben Ammar B, Maherzi A. Le défaut d'expression des protéines d'adhésion leucocytaires. A propos d'une nouvelle observation Tunesienne [Deficient expression of leukocyte adhesion proteins. A new Tunisian case] Tunis Med. 2006;84:464–466. [PubMed] [Google Scholar]
- 52.Hixson P, Smith CW, Shurin SB, Tosi MF. Unique CD18 mutations involving a deletion in the extracellular stalk region and a major truncation of the cytoplasmic domain in a patient with leukocyte adhesion deficiency type 1. Blood. 2004;103:1105–1113. doi: 10.1182/blood-2003-08-2780. [DOI] [PubMed] [Google Scholar]
- 53.Arnaout MA, Dana N, Gupta SK, Tenen DG, Fathallah DM. Point mutations impairing cell surface expression of the common β subunit (CD18) in a patient with leukocyte adhesion molecule (Leu-CAM) deficiency. J. Clin. Invest. 1990;85:977–981. doi: 10.1172/JCI114529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eyerich K, Cifaldi L, Notarangelo LD, Porta F, Notarangelo L, Mazzolari E, Fiorini M, Paradisi A, Cavani A. Chronic eczema in a patient with Leukocyte Adhesion Deficiency (LAD) type I. Eur. J. Dermatol. 2009;19:78–79. doi: 10.1684/ejd.2008.0552. [DOI] [PubMed] [Google Scholar]
- 55.Hogg N, Bates PA. Genetic analysis of integrin function in man: LAD-I and other syndromes. Matrix Biol. 2000;19:211–222. doi: 10.1016/s0945-053x(00)00066-4. [DOI] [PubMed] [Google Scholar]
- 56.Arnaout AM, Michishita M. Genetic abnormalities in Leukocyte Adhesion Molecule deficiency. In: Gupta S, Griscelli C, editors. Immunodeficiency Diseases. John Wiley; Chichester: 1993. pp. 191–202. [Google Scholar]
- 57.Vihinen M, et al. Primary immunodeficiency mutation database. In: Hall JC, Dunlap JC, Friedmann T, Gianelli F, editors. Advances in Genetics. Academic Press; San Diego: 2001. pp. 103–188. [DOI] [PubMed] [Google Scholar]
- 58.Moore SW, Sidler D, Zaahl MG. The ITGB2 immunomodulatory gene (CD18), enterocolitis, and Hirschsprung's disease. J. Pediatr. Surg. 2008;43:1439–1444. doi: 10.1016/j.jpedsurg.2007.12.057. [DOI] [PubMed] [Google Scholar]
- 59.Meller S, Jagiello P, Borgmann S, Fricke H, Epplen JT, Gencik M. Novel SNPs in the CD18 gene validate the association with MPO-ANCA+ vasculitis. Genes Immun. 2001;2:269–272. doi: 10.1038/sj.gene.6363781. [DOI] [PubMed] [Google Scholar]
- 60.Hidalgo A, Ma S, Peired AJ, Weiss LA, Cunningham-Rundles C, Frenette PS. Insights into leukocyte adhesion deficiency type 2 from a novel mutation in the GDP-fucose transporter gene. Blood. 2003;101:1705–1712. doi: 10.1182/blood-2002-09-2840. [DOI] [PubMed] [Google Scholar]
- 61.Etzioni A, Sturla L, Antonellis A, Green ED, Gershoni-Baruch R, Berninsone PM, Hirschberg CB, Tonetti M. Leukocyte adhesion deficiency (LAD) type II/carbohydrate deficient glycoprotein (CDG) IIc founder effect and genotype/phenotype correlation. Am. J. Med. Genet. 2002;110:131–135. doi: 10.1002/ajmg.10423. [DOI] [PubMed] [Google Scholar]
- 62.Helmus Y, Denecke J, Yakubenia S, Robinson P, Lühn K, Watson DL, McGrogan PJ, Vestweber D, Marquardt T, Wild MK. Leukocyte adhesion deficiency II patients with a dual defect of the GDP-fucose transporter. Blood. 2006;107:3959–3966. doi: 10.1182/blood-2005-08-3334. [DOI] [PubMed] [Google Scholar]
- 63.Gazit Y, Mory A, Etzioni A, Frydman M, Scheuerman O, Gershoni-Baruch R, Garty BZ. Leukocyte adhesion deficiency type II: long-term follow-up and review of the literature. J. Clin. Immunol. 2010;30:308–313. doi: 10.1007/s10875-009-9354-0. [DOI] [PubMed] [Google Scholar]
- 64.Malinin NL, Zhang L, Choi J, Ciocea A, Razorenova O, Ma YQ, Podrez EA, Tosi M, Lennon DP, Caplan AI, Shurin SB, Plow EF, Byzova TV. A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat. Med. 2009;15:313–318. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robert P, Canault M, Farnarier C, Nurden A, Grosdidier C, Barlogis V, Bongrand P, Pierres A, Chambost H, Alessi MC. A novel Leukocyte Adhesion Deficiency III variant: Kindlin-3 deficiency results in integrin and nonintegrin-related defects in different steps of leukocyte adhesion. J. Immunol. 2011;186:5273–5283. doi: 10.4049/jimmunol.1003141. [DOI] [PubMed] [Google Scholar]
- 66.Manevich-Mendelson E, Feigelson SW, Pasvolsky R, Aker M, Grabovsky V, Shulman Z, Kilic SS, Rosenthal-Allieri MA, Ben-Dor S, Mory A, Bernard A, Moser M, Etzioni A, Alon R. Loss of kindlin-3 in LAD-III eliminates LFA-1 but not VLA-4 adhesiveness developed under shear flow conditions. Blood. 2009;114:2344–2353. doi: 10.1182/blood-2009-04-218636. [DOI] [PubMed] [Google Scholar]
- 67.McDowall A, Svensson L, Stanley P, Patzak I, Chakravarty P, Howarth K, Sabnis H, Briones M, Hogg N. Two mutations in the KINDLIN3 gene of a new leukocyte adhesion deficiency III patient reveal distinct effects on leukocyte function in vitro. Blood. 2010;115:4834–4842. doi: 10.1182/blood-2009-08-238709. [DOI] [PubMed] [Google Scholar]
- 68.Jurk K, Schulz AS, Kehrel BE, Räpple D, Schulze H, Möbest D, Friedrich WW, Omran H, Deak E, Henschler R, Scheele JS, Zieger B. Novel integrin-dependent platelet malfunction in siblings with leukocyte adhesion deficiency-III (LAD-III) caused by a point mutation in FERMT3. Thromb. Haemost. 2010;103:1053–1064. doi: 10.1160/TH09-10-0689. [DOI] [PubMed] [Google Scholar]