Abstract
Transposons are ubiquitous genetic elements that drive genome rearrangements, evolution, and the spread of infectious disease and drug-resistance. Many transposons, such as Mu, Tn7 and IS21, require regulatory AAA+ ATPases for function. We use x-ray crystallography and cryo-electron microscopy to show that the ATPase subunit of IS21, IstB, assembles into a clamshell-shaped decamer that sandwiches DNA between two helical pentamers of ATP-associated AAA+ domains, sharply bending the duplex into a 180° U-turn. Biochemical studies corroborate key features of the structure, and further show that the IS21 transposase, IstA, recognizes the IstB•DNA complex and promotes its disassembly by stimulating ATP hydrolysis. Collectively, these studies reveal a distinct manner of higher-order assembly and client engagement by a AAA+ ATPase and suggest a mechanistic model where IstB binding and subsequent DNA bending primes a selected insertion site for efficient transposition.
INTRODUCTION
Transposable elements are ubiquitous, mobile DNAs that rely on dynamic nucleoprotein complexes, termed transposomes, to mediate transposition reactions (Craig et al., 2002). Transposases are among the most prevalent genes in nature, and their DNA reshuffling activity can modify gene expression, promote organismal evolution, and spread antibiotic resistance and virulence factors (Arakawa et al., 1995; Aziz et al., 2010; Chain et al., 2004; Kazazian, 2004; Peters and Craig, 2001; Speek, 2001; Wolff et al., 2010). In humans, where they represent ~45% percent of the genome, transposable elements have been associated with neural development, as well as with multiple diseases, including such as hemophilia, schizophrenia, ataxia telangiectasia and cancer (Ade et al., 2013; Baillie et al., 2011; Bundo et al., 2014; Burns and Boeke, 2012; Coufal et al., 2009, 2011; Lander et al., 2001; Tubio et al., 2014). Moreover, numerous DNA transposases share remarkable structural and functional similarities with retroviral integrases such as the HIV-1 integrase (Cherepanov et al., 2011; Montaño and Rice, 2011), while others have shown a significant potential for use in genetic engineering and gene therapy applications (Ivics et al., 2009).
How transposable elements are regulated has been a long-standing question. Frequently, DNA transposition is tightly controlled both spatially and temporally to prevent chromosome breaks and lethal genome rearrangements. Transposase control often relies on host proteins to aid in complex formation and regulation; for instance, the Tn10 and Mu transposases both require a DNA bending protein (integration host factor, IHF) to form functional synaptic complexes (Sakai et al., 1995; Surette and Chaconas, 1989). In addition, a variety of mobile elements rely on nucleotide cofactors to regulate transposase activity or to choose appropriate target DNAs. For example, GTP is known to stimulate assembly of the Drosophila P element synaptic complex by binding to a nucleotide-dependent regulatory domain appended in cis to a catalytic transposase fold (Kaufman and Rio, 1992; Tang et al., 2005). By comparison, the Mu and Tn7 transposable elements employ dedicated, ATP-dependent molecular matchmaker subunits, which play a key role in selecting suitable insertion sites and in preventing self-insertion, a process whereby a transposase hops back into its own sequence (Gamas and Craig, 1992; Miller et al., 1984; Mizuuchi, 1992; Peters and Craig, 2001).
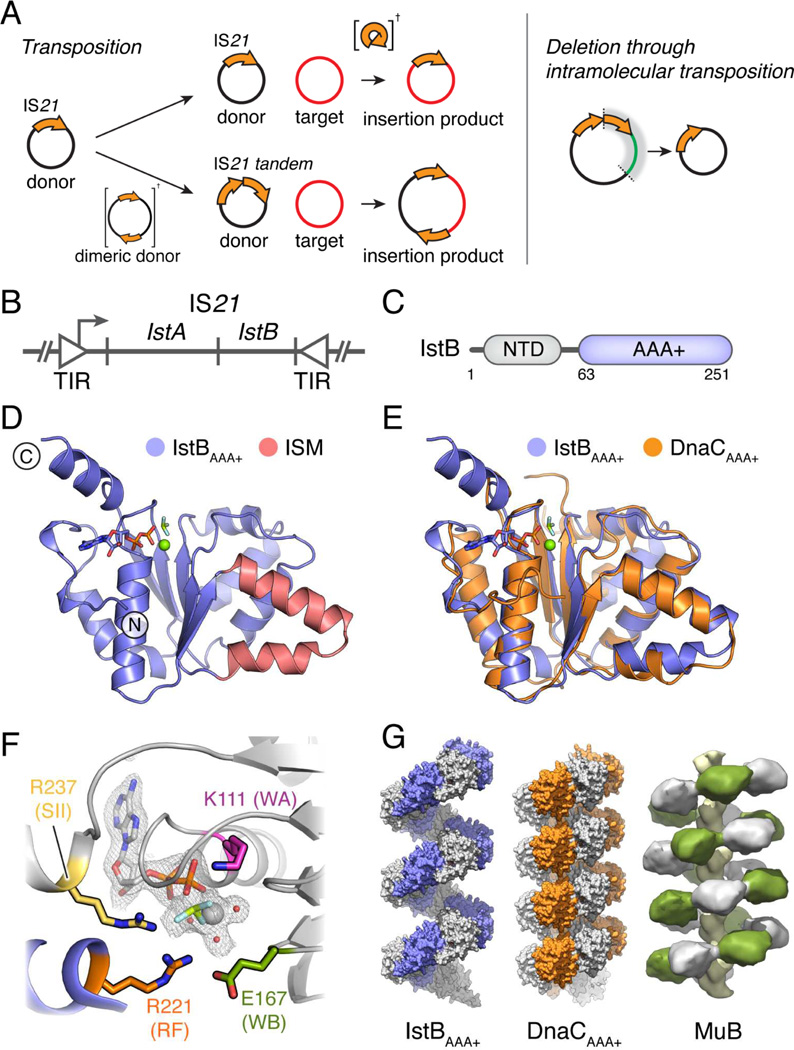
Many DNA transposases and retroviral integrases belong to the large DDE superfamily of polynucleotide transferases. The smallest and most numerous types of DDE transposons only code for proteins involved in the transposition activity, and in bacteria they are known as Insertion Sequences (ISs) (Chain et al., 2004; Parkhill et al., 2001, 2003; Siguier et al., 2014). Of all insertion sequences, the IS21 family is one of the most widespread, distributed broadly throughout bacterial and even archaeal kingdoms (Fig. S1). IS21 has been identified in clinical isolates of pathogenic Escherichia coli and Shigella sonnei strains, and has also been found to flank a pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis (Allué-Guardia et al., 2013; Buchrieser et al., 1998; Burland et al., 1998; Filippov et al., 1995; Hu et al., 1998; Perry et al., 1998; Podladchikova et al., 1994). Is21 has been found to catalyze transposition reactions that can result in different insertion products, and it can generate deletions through intramolecular transposition (Fig. 1A).
Fig. 1.
IS21 organization and function, and IstBAAA+ structure. See also Fig. S1, S2 and Table S1.
(A) DNA transposition events mediated by IS21.
(B) IS21 genetic organization. TIR – Terminal Inverted Repeats.
(C) IstB domain organization.
(D) Crystal structure of IstBAAA+ bound to ADP•BeF3. The Initator/loader Specific Motif (ISM) is shown in red.
(E) Superposition of IstBAAA+ and the AAA+ domain of DnaC (PDB ID 3ECC).
(F) Close-up view of the IstBAAA+ active site. Chains from two protomers are shown in grey and blue. Residues selected for mutational studies are highlighted. The color code is maintained throughout the text. WA – Walker-A; WB – Walker-B; RF – arginine finger; SII – Sensor II.
(G) IstBAAA+ forms a right-handed filament (blue and grey), similar to that of DnaCAAA+ (orange and grey; PDB ID 3ECC) and full-length MuB (green and grey; EMDB-2395).
The ~2 kb IS21 sequence consists of a single operon with two terminal inverted repeats of variable length (11–50 bp) that bracket two open reading frames (IstA and IstB) (Fig. 1B) (Berger and Haas, 2001; Reimmann et al., 1989; Xu et al., 1993). The IstA transposase bears the catalytic DDE motif characteristic of retroviral integrases and numerous bacterial and eukaryotic transposases. By contrast, IstB is homologous to proteins of the AAA+ (ATPases Associated with various cellular Activities) superfamily of nucleotide hydrolases (Koonin, 1992), a group that includes not only the MuB and TnsC helper proteins of the Mu and Tn7 transponsable elements, but also DnaC/DnaI bacterial helicase loaders and DnaA replication initiators (Fig. 1C and S1C). As with MuB and TnsC (Baker et al., 1991; Craigie et al., 1985; Maxwell et al., 1987), IstB activity is crucial for the capture of target DNA segments and accurate donor insertion by its cognate transposase (Reimmann and Haas, 1990; Schmid et al., 1998, 1999). How these factors utilize nucleotide binding and hydrolysis to interface with target DNA segments and regulate transposase activity remains poorly understood, however.
To better understand the means by which nucleotide-dependent helper proteins modulate transposition reactions, we used a combination of structural and biochemical methods to define the higher-order, ATP-dependent organization of IstB, and how this protein interfaces with both DNA and IstA. The structure demonstrates that IstB AAA+ ATPase domain self-associates into a helical assembly, which the full-length protein co-opts to form an unprecedented clamshell-shaped decamer that binds and bends >50bp of duplex DNA. Biochemical analyses support the functional relevance of the observed structural states, and further show that IstA specifically recognizes and disassembles the IstB•DNA supercomplex. Together with comparative studies of distantly-related ATPases, our findings highlight a remarkable plasticity in the ability of AAA+ proteins to bind and remodel client nucleic acid substrates, and help explain how a transpose helper protein can use these interactions to aid DNA transposition.
RESULTS
The IstB AAA+ domain is a close structural paralog of bacterial helicase loaders that can form right-handed helical oligomers
IstB consists of a small N-terminal domain of unknown function and a C-terminal AAA+ domain (Fig. 1C and Fig. S2A). To begin to dissect IstB structure and mechanism, we purified and crystallized the isolated ATPase domain (residues 63–251, termed IstBAAA+) of the Geobacillus stearothermophilus IS21 family member, IS5376, with the non-hydrolyzable ATP-analog, ADP·BeF3. Single-wavelength anomalous dispersion (SAD) phasing from selenomethionine-labeled protein was used to determine the structure; during model building, clear electron density was evident for bound nucleotide (Fig. S2B). The final IstBAAA+ model was refined to a resolution of 2.1 Å, with an Rwork and Rfree of 18.2/20.3% (Table S1).
IstBAAA+ adopts a typical AAA+ ATPase fold, in which five parallel β strands are sandwiched on both sides by α helices (Fig. 1D). Examination of the structure reveals that IstB possesses one of the characteristic signatures of the DNA replication initiator and helicase loader group of AAA+ proteins, namely the presence of an extra α-helix in its ATPase fold, denominated the Initator/loader Specific Motif (ISM) (Fig. 1D) (Dueber et al., 2007; Iyer et al., 2004). The existence of this helix defines IstB as a member of the initiator clade of AAA+ proteins, which includes the bacterial helicase loader DnaC/DnaI, and the bacterial DnaA and archaeal/eukaryotic Cdc6/Orc1 replication initiator proteins (Iyer et al., 2004). Despite relatively limited sequence identity (~15–22%), computational searches of the PDB (using DALI, (Holm and Sander, 1995)) show that the AAA+ domain of IstB is most closely related to that of DnaC/I, followed by DnaA (Fig. 1E).
Phylogenetic studies had indicated previously that IstB was likely to possess AAA+ family ATPase motifs involved in nucleotide binding and hydrolysis (Koonin, 1992). The structure reveals that the active site indeed assumes a canonical AAA+ configuration (Fig. 1F). The Walker-A residues of IstBAAA+ form a phosphate-binding loop (P-loop) in which Lys111 directly contacts the β-phosphate of ADP, while the Walker-B amino acids Asp166 and Glu167 interact with the water coordination shell of an associated Mg2+ ion. The “Sensor II” arginine (Arg237) of the protein lies within the most C-terminal α-helix, which forms a truncated variant of the “lid” subdomain found in a majority of AAA+ proteins (Fig. S2C).
AAA+ proteins frequently assemble into large ring-shaped or helical homo-oligomers (Erzberger and Berger, 2006). Inspection of intermolecular contacts seen in the crystal structure reveals that IstBAAA+ protomers self-associate around a crystallographic 61 axis, creating a right-handed filament reminiscent of those adopted by the ATPase domains of DnaC and the MuB transposase regulator (Fig. 1G) (Mizuno et al., 2013; Mott et al., 2008). Formation of this spiral allows a conserved arginine from one promoter (Arg221) to interact with the active site of a neighboring AAA+ domain (Fig. 1F), implicating this residue as a candidate “arginine finger” motif that typically participates in both ATP hydrolysis and inter-subunit communication. Interestingly, analysis of the electrostatic potential of the helical IstBAAA+ assembly shows that numerous residues, located mainly in the ISM region, create a highly positively charged surface on one side of the ATPase domain oligomer (Fig. S2D).
Full-length IstB forms discrete oligomers
The observation that the IstB AAA+ region could adopt a structural state mirroring that of bacterial initiator/helicase loading proteins and ATP-bound MuB was intriguing. However, in some instances the crystallization of protein fragments can result in the formation of oligomeric states that only partially capture the quaternary arrangement of full-length subunits. For instance, although the ATPase domain of DnaC can crystallize as a continuous 61 helix on its own (Mott et al., 2008), it is restricted to forming a six-subunit helix in the presence of its target helicase, DnaB (Arias-Palomo et al., 2013).
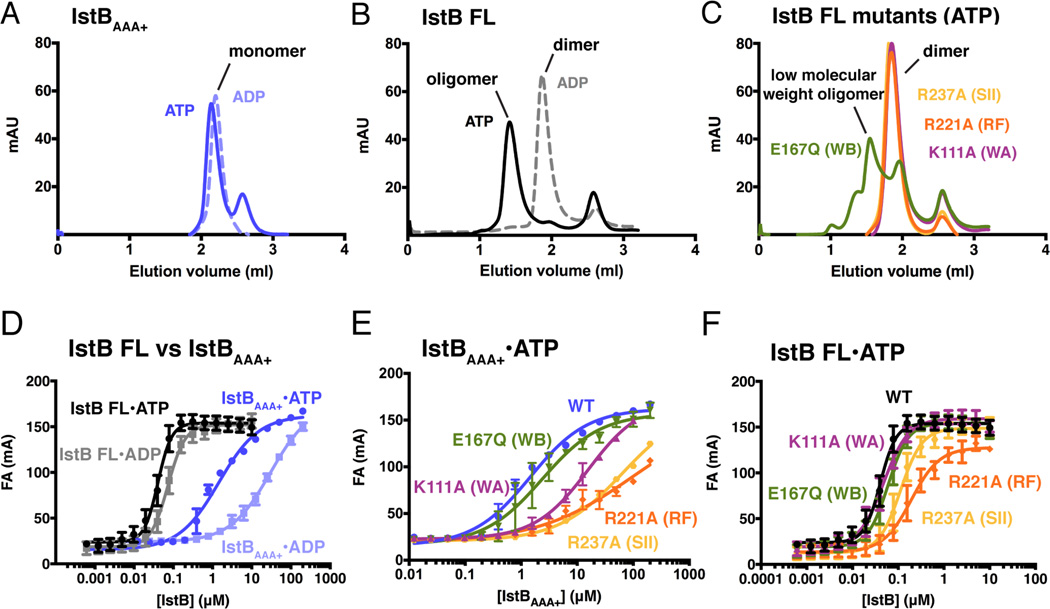
To test whether an IstBAAA+ filament was captured because its N-terminal domain was removed for crystallization, we used analytical size-exclusion chromatography to study the nucleotide-dependent oligomerization of the IstB ATPase domain, full-length IstB, and several IstB active-site mutants. The IstBAAA+ protein elutes as a monomeric species, both in the presence of ADP and ATP, establishing that the domain on its own is unable to oligomerize efficiently under more dilute conditions than those used during crystallization (Fig. 2A). By comparison, full-length IstB formed an apparent dimer in the presence of ADP, suggesting that its N-terminal domain might support protomer-protomer interactions (Fig. 2B). Interestingly, when mixed with ATP, full-length IstB further shifted to a much larger size, eluting as a well-defined high-molecular weight peak with an apparent mass of ~350 kDa (Fig. 2B). To test whether this higher-order assembly depended on the AAA+ domain of IstB, we analyzed a number of mutants predicted to have defects in ATP binding and hydrolysis (e.g., Walker-A (K111A), Walker-B (E167Q), arginine finger (R221A), and sensor-II (R237A)). Following purification, each of the proteins formed dimers in the presence of ADP as per wild-type IstB (not shown). By contrast, none the mutants were able to shift to a higher molecular-weight species in the presence of ATP, with the exception of the Walker-B mutant, which formed a mix of higher and lower (~150 kDa) molecular mass complexes (Fig. 2C). Overall, these results indicate that full-length IstB does not form continuous filaments as has been seen for MuB and DnaC, and that both ATP and an intact active site are required for IstB to self-assemble into a discrete macromolecular complex.
Fig. 2.
IstB oligomerization and DNA binding activity. See also Table S2.
(A) Analytical gel filtration chromatogram of IstBAAA+ shows that the isolated domain elutes as a monomer in the presence of 1 mM ATP or ADP.
(B) Analytical gel filtration chromatography shows that full-length IstB protein is as a dimer in the presence of ADP. IstB elutes as a high-order oligomer (~350 kDa) in the presence of 1 mM ATP.
(C) Examination of the active site mutants by analytical gel filtration reveals that most mutations in the ATPase binding site block full-length IstB oligomeriztion. The Walker-B mutant (E167Q) shows limited support for the formation of higher molecular weight assemblies.
(D) Nucleotide effects on dsDNA binding by IstBAAA+ and full-length IstB as determined by fluorescence anisotropy. Experiments were performed in triplicate in the presence of 2 mM nucleotide. Error bars represent the standard deviation between measurements.
(E) Active site mutations disrupt dsDNA binding by IstBAAA+. Experiments were performed in the presence of 2 mM ATP.
(F) ATPase site mutations have a less severe effect on dsDNA binding by full-length IstB. Experiments were performed in the presence of 2 mM ATP.
IstB is a DNA binding protein
Although the exact function of IstB has been enigmatic, its relationship to known DNA binding proteins such as DnaA, DnaC, MuB, and TnsC suggested that it might associate with nucleic-acid substrates. To test this idea and analyze the role that nucleotide coordination and self-oligomerization might have on this activity, we used fluorescence anisotropy to assess the ability of IstB, IstBAAA+, and our panel of IstB active-site mutants to engage DNA substrates. We first examined DNA binding using two 60 bp duplexes, one containing a random sequence, and another having the characteristic repeat sequence that flanks the transposon (the size of the DNA was chosen after screening different lengths of oligonucleotides for optimal affinity and binding behavior). As expected based on the behavior of MuB and TnsC, IstB did not show any significant sequence specificity (not shown); nevertheless, we continued using the transposon-related DNA for all subsequent analyses.
Treatment with nucleotide was found to stimulate duplex DNA binding by both IstBAAA+ and the full-length protein (Fig. 2D). However, the apparent Kd of the full-length protein for DNA in the presence of ATP was ~30-fold higher compared to that of the AAA+ domain alone (Kd,app=40 nM vs. 1.5 µM, Table S2). Interestingly, the binding curves manifest by full-length IstB also displayed evidence of positive cooperativity, whereas the curves seen for the AAA+ region did not (Hill coefficient 2.5 vs. 0.85); this result, coupled with the relatively smaller dependency of the full-length protein for ATP for binding, indicates that the N-terminal domain contributes directly to interactions between IstB and DNA (Fig. 2D). Consistent with this idea, ATPase active-site mutations that significantly compromised DNA binding in the context of the isolated AAA+ domain had a much less pronounced effect when introduced into the full-length protein (Fig. 2E and 2F).
IstB homo-oligomerizes into a pentamer of dimers to remodel dsDNA
Intrigued by the ability of IstB to both bind DNA and form a discrete higher-order oligomer, we turned to single-particle cryo-electron microscopy (cryo-EM) to determine the architecture of the ATP-bound IstB complex with and without a client dsDNA substrate. To ensure complete assembly, we first dialyzed full-length IstB into a buffer containing ATP and subsequently ran the sample over an analytical S200 pre-equilibrated in dialysis buffer. Fractions containing the high-molecular weight IstB oligomer were then collected, diluted into a low salt buffer, and incubated with or without a dsDNA 60mer prior to EM analysis.
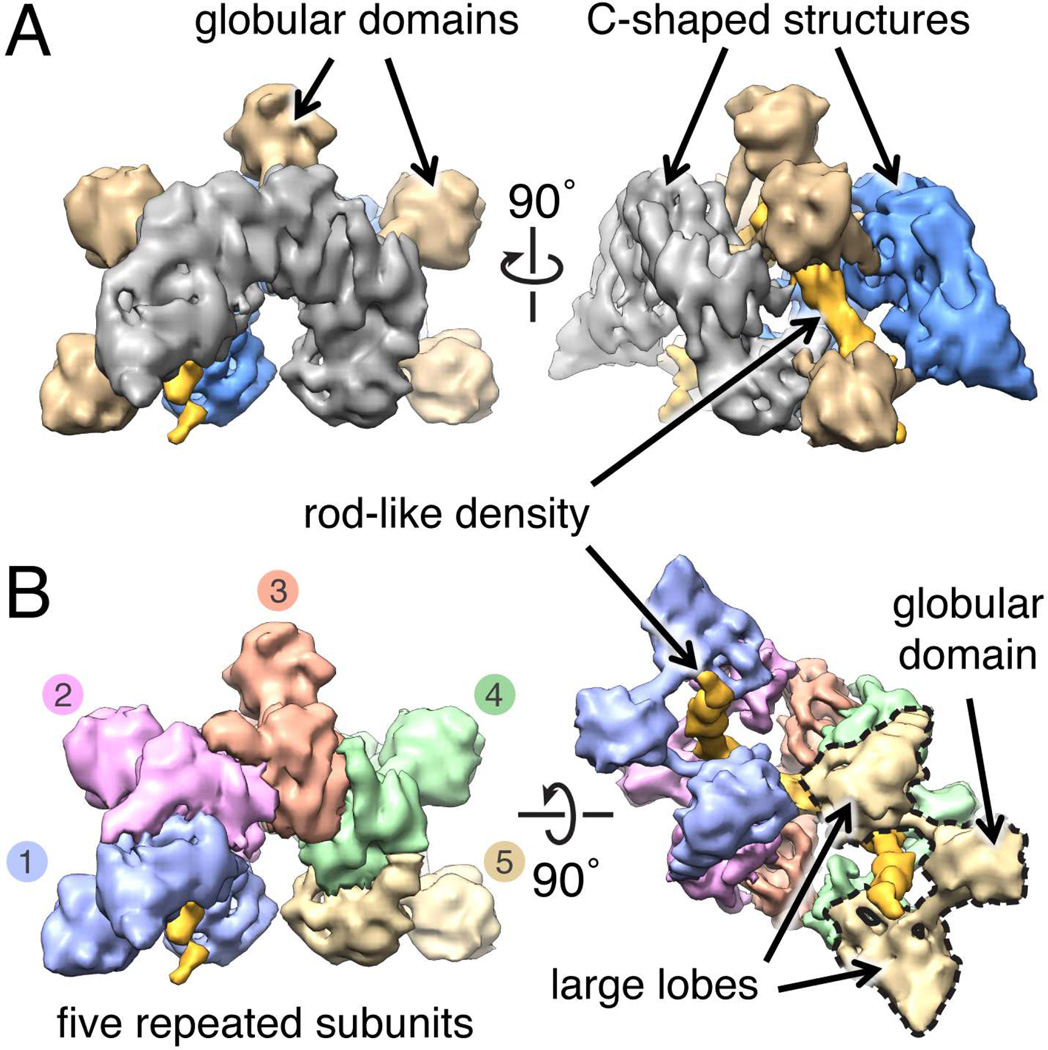
The cryo-EM structure of the ~290 kDa ATP-IstB complex bound to DNA was determined to subnanometer resolution (~9 Å), using an initial model derived from a random conical tilt reconstruction generated from a negatively-stained sample (Methods). The consistency of the structure is supported by the agreement between reference-free 2D class averages and forward reprojections of the cryo-EM model (Fig. S3). In contrast to other known AAA+ systems, IstB neither assembled into a closed/cracked ring nor an elongated filament, but rather formed a distinctive crescent-shaped particle composed of two C-shaped structures connected by five protruding “spikes” of globular density (Fig. 3A). Close inspection of the model reveals that the IstB oligomer is formed by five repeating elements (Fig. 3B), each consisting of two large lobes that connect to one another by two small rods which emanate from a single, small globular domain (Fig. 3B). A semicircular channel traverses the interior of the IstB oligomer, within which continuous, rod-like density is evident.
Fig. 3.
Cryo-EM reconstruction of full-length IstB bound to dsDNA in the presence of ATP. See also Fig. S3.
(A) The IstB•ATP•dsDNA complex comprises two C-shaped densities, connected by five globular domains that sequester a rod-shaped region of density.
(B) The IstB•ATP•dsDNA complex can be divided into five similarly-shaped elements that wrap around the rod-shaped region. Each element is formed by two large lobes that connect through short stalks to a central globular domain.
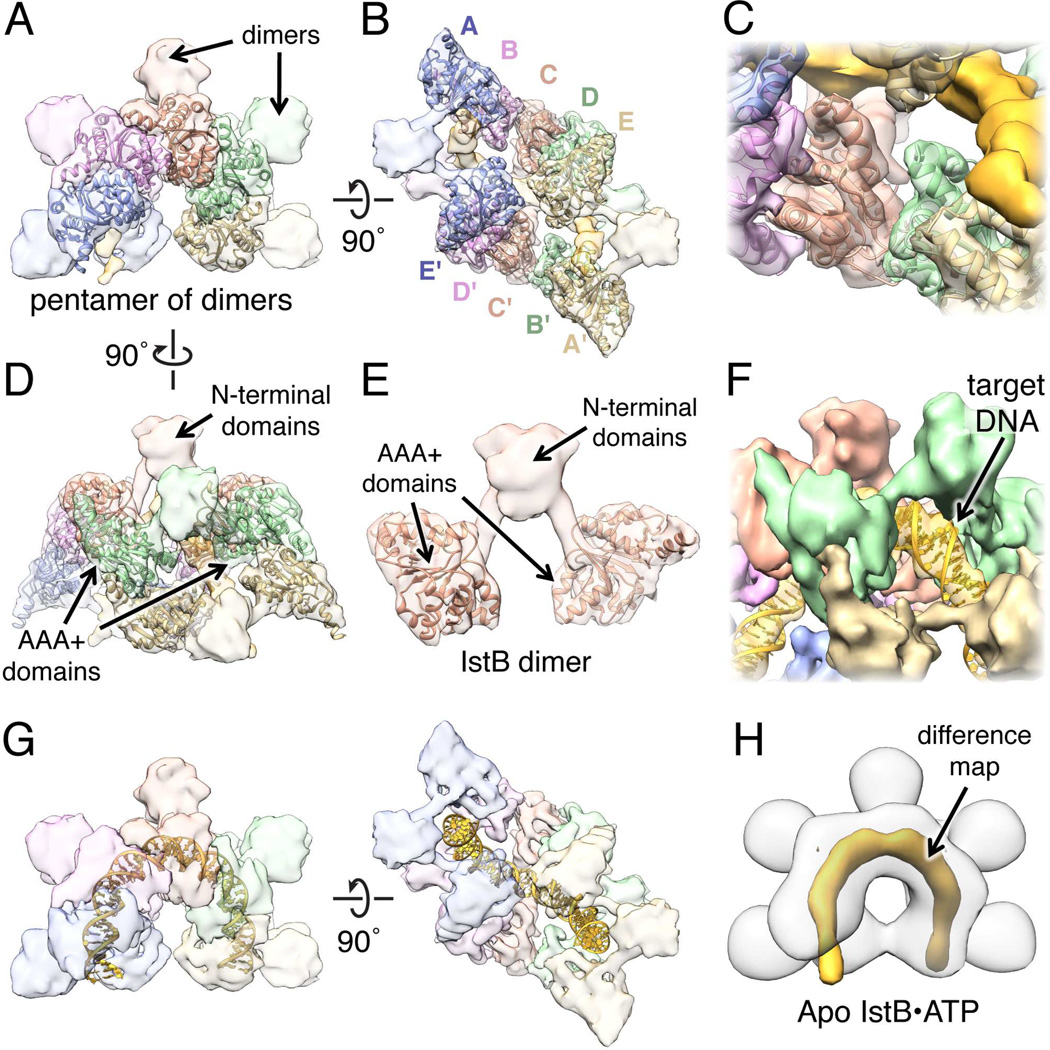
To determine which regions of the DNA-bound ATP-IstB complex correspond to specific domains of the protein and the nucleic acid, we fit available crystal structures into the cryo-EM reconstruction. To date, there is no atomic information of the ~7 kDa IstB N-terminal domain, although PSIPRED predicts it to be α-helical. We therefore attempted to localize the AAA+ fold using our IstBAAA+ crystal structure. Notably, five copies of the ATPase domain, assembled in the same general helical conformation as found in the crystal structure, could be unambiguously fitted into each C-shaped region of density (Fig. 4A and 4B). Only a slight (<7°) rotation of each protomer was necessary to optimize the fit, suggesting that the oligomerization state seen crystallographically closely approximates that used by the full-length assembly.
Fig. 4.
Molecular architecture of the IstB•ATP•dsDNA complex. See also Fig. S4 and Movie S1.
(A) Docking of five IstBAAA+ domains into one of the C-shaped densities.
(B) Two pentamers of ATP-associated IstBAAA+ domains (A-E and A’-E’) can be fitted in the IstB•ATP•dsDNA complex, indicating that full-length IstB assembles into a pentamer of dimers.
(C) Close-up view showing that the secondary structure elements resolved in the cryo-EM reconstruction agree well with the crystal structure.
(D) Side view showing the C-shaped structures fitted with AAA+-domain pentamers.
(E) IstB N-terminal domains mediate dimerization. An IstB dimer (segmented from the complex) is composed of two AAA+ domains connected with an α-helix to the N-terminal region.
(F) A ~50 bp dsDNA molecule can be fitted into the rod-like density. Major and minor grove features can be seen in the reconstruction.
(G) Orthogonal views of the IstB•ATP•dsDNA complex fitted with a 50 bp DNA fitted. IstB introduces a 180° bend in the DNA.
(H) Difference map represented at 7 σ (orange) between the IstB cryo-EM reconstructions determined in the presence and absence (white transparent surface) of DNA.
The global fitting of tw0 pentamers of AAA+ ATPase domains, together with the close agreement between the crystal structure and the secondary structure elements resolved in the EM density, provide strong validation for the cryo-EM reconstruction (Fig. 4C). Given the location of the ATPase elements, the small lobes that protrude from the complex likely correspond to the N-terminal domain of IstB (Fig. 4D and 4E). Each of the five lobes are connected to the AAA+ domains through two short rods of density of the correct dimensions expected for α-helices; together with the two-fold symmetric organization seen in the model, this arrangement indicates that one role of the N-terminal region is to dimerize IstB protomers. This observation is in agreement with the gel-filtration data showing that full-length IstB forms dimers in the presence of ADP, and that the N-terminal region is required for this dimerization (Fig. 2A and 2B).
With the domains of IstB assigned, the only part of the reconstruction left unmodeled was the rod-like density located within the central channel. Notably, this region exhibits clear helical features, with alternating sets of wide and narrow grooves that identify it as duplex DNA (Fig. 4F). Interestingly, to fit within the central groove of the IstB decamer, the DNA must assume a highly bent 180° U-turn structure (Fig. 4G and S4A). To validate the placement of DNA, we determined a cryo-EM reconstruction of ATP-IstB in the absence of the duplex substrate; the difference map between the two structures confirms our assignment (Fig. 4H). Interestingly, in the context of the decameric complex, the highly positively-charged surface of the AAA+ pentamers is oriented towards the internal channel, forming a portion of the nucleic acid binding area (Fig. S4B). Moreover, inspection of the protein-DNA interface revealed that, in agreement with the fluorescence anisotropy data, some residues in the N-terminal region appear capable of participating in DNA binding (Fig. S4C).
Overall, the cryo-EM reconstruction establishes that, in response to ATP binding, IstB assemblies into a pentamer of dimers that engages and markedly reshapes target DNAs. This architectural arrangement is quite distinct compared to other known AAA+ ATPase systems characterized at this time, and was surprising given the close relationship of IstB to proteins such as DnaA and MuB, which form helical filaments. Inspection of the structure reveals why subunit assembly is limited to five IstB dimers: the inverted arrangement of two AAA+ domain pentamers, whose DNA binding surfaces face one another due to the molecular two-fold axes of their associated N-terminal domains, sterically prevents the binding of a sixth ATPase subunit to the complex (Fig. S4D). Thus, the N-terminal region is not only responsible for dimerizing IstB protomers, but also for preventing the formation of contiguous IstB oligomers.
IstB introduces positive supercoils in a nucleotide-dependent manner
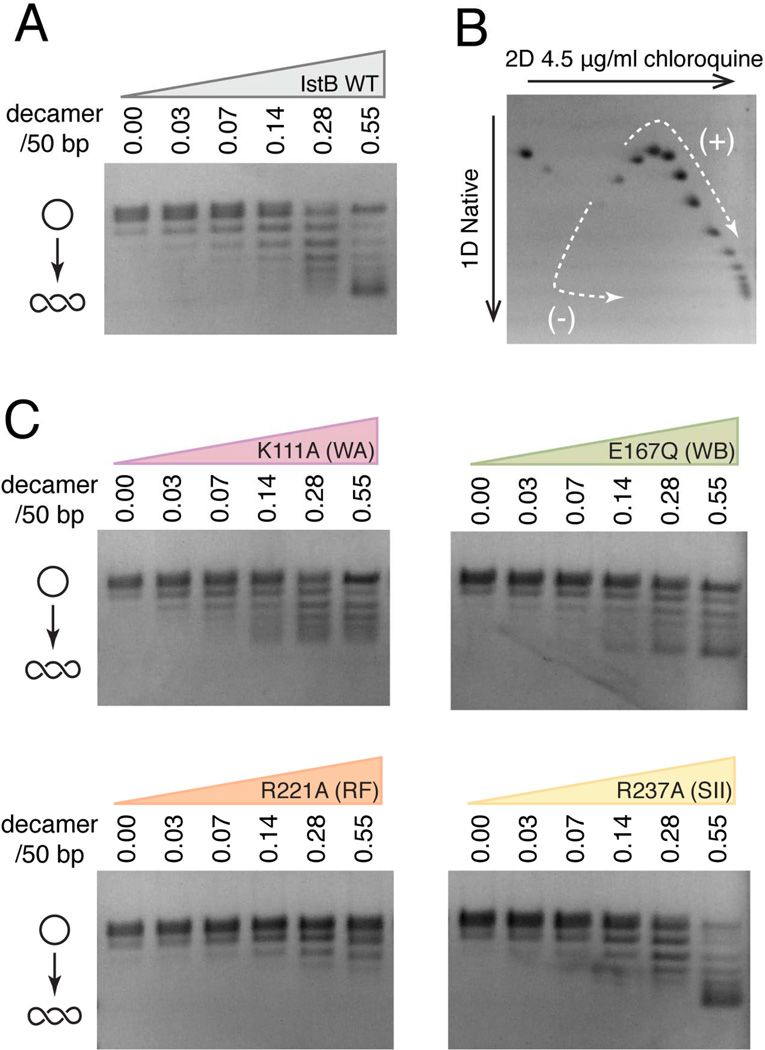
Multiple biochemical studies have indicated that the supercoiling state of nucleic acid substrates can have important effects on the action of different transposition systems, including both IS21 and bacteriophage Mu (Harshey and Jayaram, 2006; Reimmann and Haas, 1990). Given extensive DNA deformation imparted by IstB in the cryo-EM reconstruction, we set out to investigate whether the protein might also alter DNA superstructure in solution. Using a topology-footprint assay and native agarose-gel electrophoresis (Methods), we found that IstB introduces DNA supercoiling in a dose-dependent manner and that ATP stimulates this activity (Fig. 5A and S5).
Fig. 5.
IstB stabilizes positive DNA supercoils in an ATP-dependent manner. See also Fig. S5.
(A) Wild-type IstB topology footprint assay performed in the presence of 2 mM ATP. Relaxed pSG483 (10 nM) was incubated with increasing amounts of IstB (indicated as the ratio of protein decamers to 50 bp DNA segments). The positions of relaxed and supercoiled DNA species are labeled with graphical representations on the left.
(B) 2D electrophoretic analysis of IstB generated topoisomers. The topoisomer distribution indicates that IstB introduces positive supercoils into the DNA.
(C) Mutations in the ATPase site residues negatively impact IstB-mediated topological changes of DNA. Topology footprint experiments were performed in the presence of 2 mM ATP.
To determine whether IstB introduces positive or negative supercoils into DNA, we analyzed the reaction products by two-dimensional gel electrophoresis. Consistent with the right-handed solenoidal wrap seen in the EM reconstruction, the resultant topoisomer distribution revealed that wild-type IstB introduces positive supercoils in the DNA (Fig. 5B). Moreover, ATPase site mutants that lead to assembly defects of the IstB oligomer are highly compromised for supercoil stabilization, with the arginine-finger mutation displaying the most severe defect (Fig. 5C). Of the mutants tested, the sensor II alteration is the lone exception to this trend; however, we note unlike the arginine finger, which plays a key role in stabilizing trans interactions between subunits, the sensor II arginine acts in cis on its own active site. This configuration may impact inter-subunit interactions to a lesser degree than the arginine-finger, such that DNA binding can overcome sensor II-dependent oligomerization defects, allowing the IstB supercoiling signal to be detected. Overall, these experiments support a role for ATP-dependent self-association of IstB in creating an oligomer that not only bends but also overtwists target DNA duplexes.
ATP is required for IstB-IstA interactions
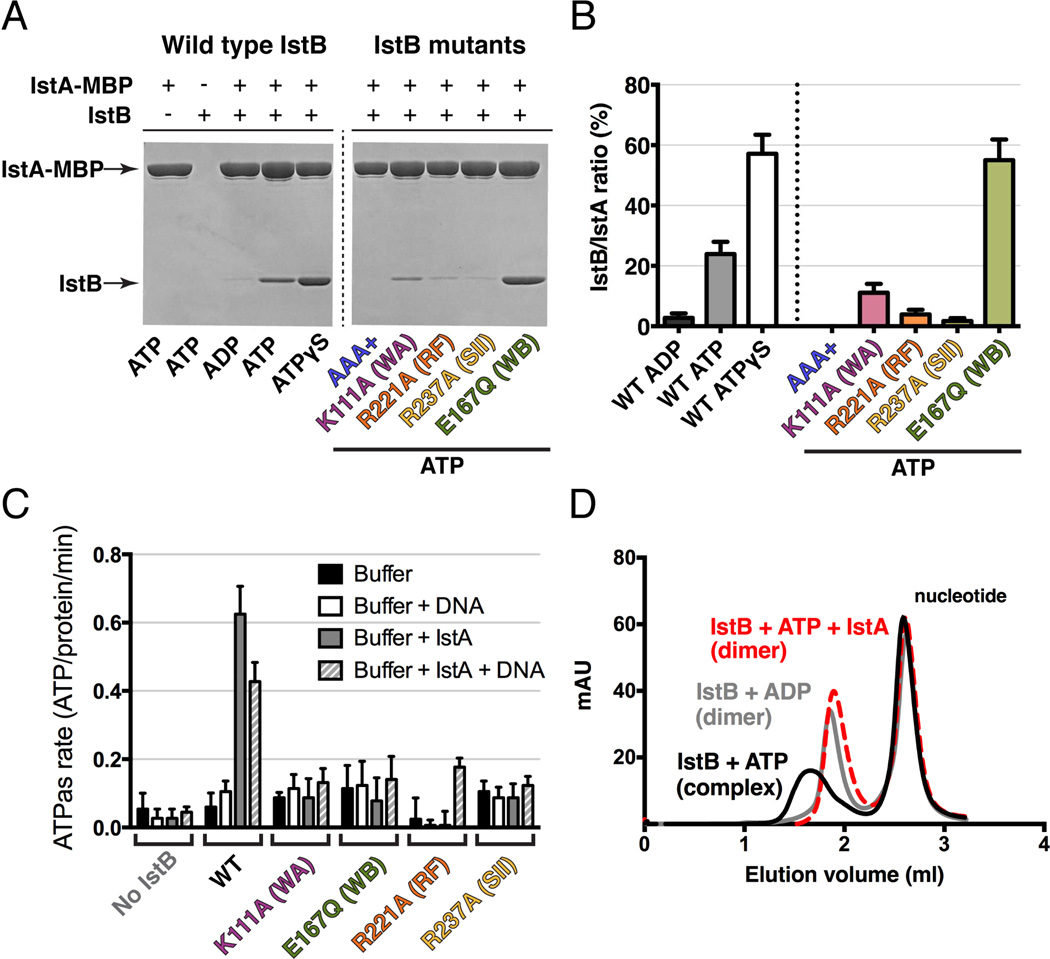
Several studies have demonstrated that helper proteins like TnsC and MuB not only interact with client DNA segments, but that they also directly engage their partner transposases to promote efficient strand cutting and pasting (Craig et al., 2002). To test whether IstB might interact with IstA in an analogous manner, we performed amylose resin pull-down assays using a transposase construct bearing a C-terminal MBP tag as bait and untagged IstB as prey. Experiments were first conducted with different nucleotides to test whether any interaction might be dependent on the identity of the nucleotide present in the reaction. Analysis of the pull-downs by SDS-PAGE revealed that when incubated with ADP, IstB did not associate with IstA-MBP (Fig. 6A). By contrast, IstB and IstA displayed a clear interaction when ATP was present, with the robustness of the association increasing when the slowly-hydrolyzable nucleotide analog, ATPγS, was added.
Fig. 6.
IstA interacts with oligomerized IstB, stimulates its ATPase activity, and promotes decamer dissociation. See also Fig. S6.
(A) Co-precipitation of IstB by IstA fused to maltose binding protein (MBP). WT IstB (left) was examined in the presence of different nucleotides. AAA+ domain and ATPase active site mutants (right) were assessed in the presence of 1 mM ATP.
(B) Quantification of gels shown in (A) shows that IstA preferentially interacts directly with ATP-oligomerized IstB species. Error bars represent the standard deviation between three independent experiments.
(C) IstB ATPase activity in the presence of absence of IstA and/or dsDNA. Radiolabeled experiments were performed in triplicate in the presence of 1 mM ATP. Error bars represent the standard deviation between measurements. A representative TLC plate image can be found in Fig. S6.
(D) IstA promotes IstB decamer disassembly as analyzed by size exclusion chromatography.
Having established that IstA binds to IstB in an ATP-dependent manner, we next sought to determine if this interaction required the formation of higher-order IstB oligomers. Using the isolated IstB AAA+ domain, we found no evidence of binding to IstA-MBP even in the presence of ATP, indicating that the ATPase region alone is insufficient for stable transposase engagement (Fig. 6A and 6B). Next, we assessed our panel of ATPase-site mutants in the context of full-length IstB. Neither the Walker A (K111A), arginine finger (R221A), nor sensor II (R237A) mutants showed an ability to interact with IstA; however, the Walker B (E167Q) mutant showed a strong interaction with the transposase when ATP was present, similar to that seen for the wild type protein in the presence of non-hydrolysable nucleotide. These results demonstrate that IstA directly engages IstB in an ATP-dependent manner, likely by recognizing the nucelotide-assembled form of the regulator.
IstA stimulates IstB ATPase activity and triggers decamer disassembly
Previous reports have established that the direct interaction of either TnsC or MuB with their cognate transposases stimulates the ATPase activity of the helper proteins. This stimulation, which is essential for strand transfer and target immunity, triggers DNA release and – in the case of MuB – filament dissociation (Craig et al., 2002). To analyze IstB ATPase activity and determine what effects IstA and DNA might have on nucleotide hydrolysis, we incubated full-length IstB with [γ-32P]ATP and unlabeled ATP, and examined phosphate release using thin layer chromatography. IstB alone, as well as the panel of active-site mutants, showed relatively little ATPase activity overall either in the presence or absence of DNA (Fig. 6C and S6). However, the inclusion of IstA appreciably enhanced nucleotide turnover. Interestingly, the combined presence of DNA and the transposase also resulted in increased Pi release, although to a lesser extent than with IstA alone. All catalytic mutants of IstB were relatively resistant to stimulation by IstA, indicating that the observed hydrolysis activity came from the IS21 helper protein, and not from a potential contaminating ATPase.
Given that IstB oligomerizes when bound to ATP, and that IstA stimulates ATP turnover by IstB, we reasoned that transposase binding might trigger the dissociation of IstB assemblies. To test this idea, we first incubated IstB with ATP, with or without IstA, and then used an analytical gel filtration column, equilibrated in a buffer without nucleotide, to analyze complex formation. When IstB was mixed with ATP, the protein eluted as a single peak consistent with the formation of a decamer species (Fig. 6D). However, when the transposase was added to the reaction, the peak shifted significantly, overlaying with dimeric, ADP-bound IstB. Overall, these findings demonstrate that the IstA transposase stimulates IstB ATPase activity both in the presence and absence of DNA, and that this stimulation triggers decamer dissociation.
DISCUSSION
Since their discovery more than half a century ago (McClintock, 1950), it has become clear that transposable elements and their mechanisms of action are both diverse and highly regulated (Curcio and Derbyshire, 2003). However, while significant strides have been made in understanding the molecular and physical bases of individual DNA transposition reactions (Dyda et al., 2012), the means by which many classes of transposases are directed toward appropriate target sites and activated to catalyze transposition have remained more enigmatic. Insight into this level of control is important, as transposable elements not only constitute a significant proportion of many genomes, but they also serve as major drivers of genomic alterations and the spread of infectious disease (Biémont, 2010; Chain et al., 2004; Kazazian, 2004; Parkhill et al., 2001). In the present study, we used the widespread IS21 element as a model system to better understand how dedicated, accessory NTPase factors – AAA+ ATPases in particular – can participate in the control of transposase target site selection.
IstB utilizes a distinctive arrangement of ATPase domain/substrate interactions in binding compared to other AAA+ proteins
Our investigations into IS21 began with the determination of the crystal structure of the ATPase domain of the transposase chaperone, IstB, bound to a non-hydrolyzable analog (Fig. 1D). In accord with previous sequence predictions (Koonin, 1992; Mott et al., 2008), the structure demonstrates that IstB is a AAA+ ATPase, one that likely emerged from a common ancestor shared by DnaC/DnaI replicative helicase loading proteins and DnaA-family replication initiators (Fig. S1C). Using the IstBAAA+ structure as a guide, mutagenesis-guided biochemical and biophysical experiments show that conserved ATP-coordinating residues of IstB are important for oligomerization, DNA binding/remodeling, and interactions with the IstA transposase (Figs. 2, 4–6). Inspection of inter-subunit contacts shows that – as with DnaC, DnaA and the ATP-dependent phage Mu transposase protein, MuB – the AAA+ domains of IstB possess an ability to self-assembly into right-handed helical oligomers upon ATP binding (Duderstadt et al., 2011; Erzberger et al., 2006; Mizuno et al., 2013; Mott et al., 2008). Strikingly, however, full-length IstB does not polymerize into elongated filaments, but rather uses two pentameric half-spirals to form a decameric, clamshell-shaped structure (Fig. 3 and 4). This organization is unique among AAA+ ATPases that have been characterized to date.
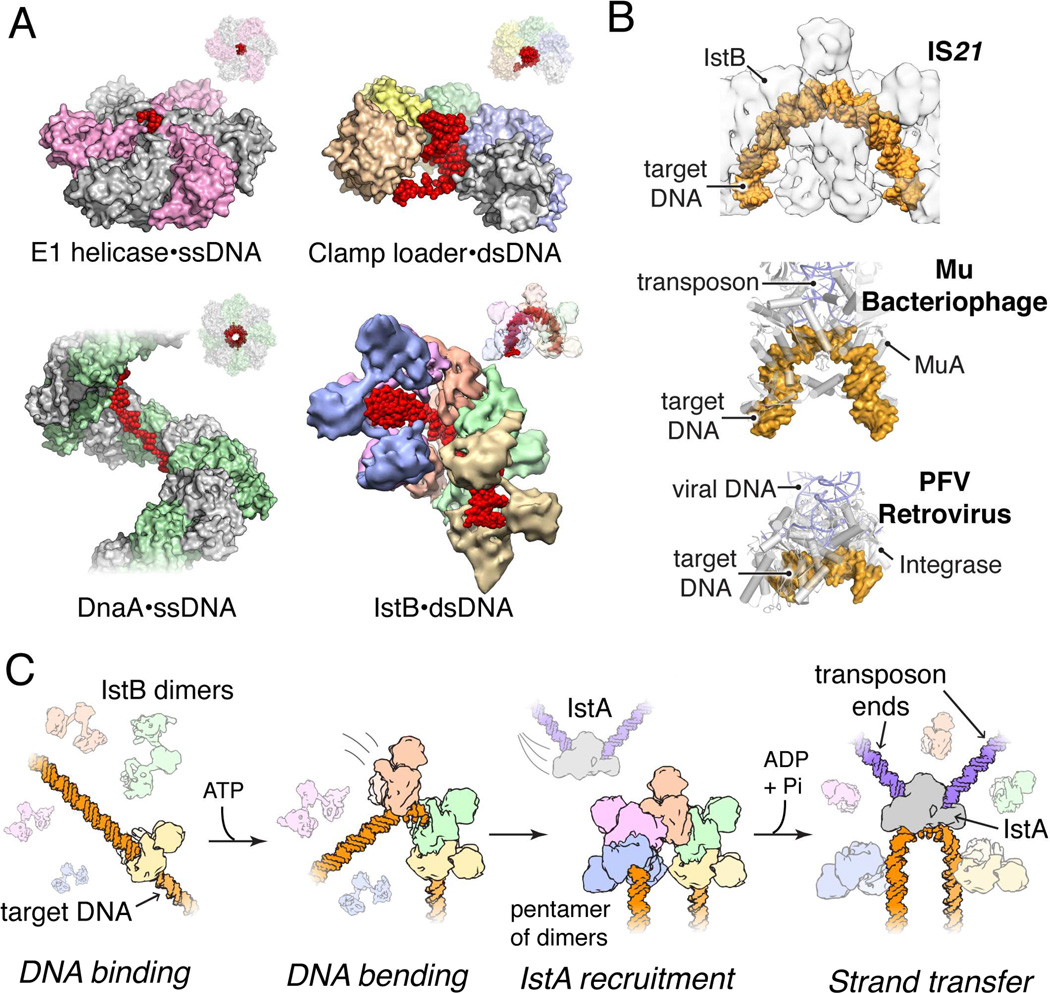
Numerous oligomeric AAA+ enzymes form rings or spirals that accommodate client factors such as DNA within a central pore to promote the detection, translocation, or remodeling of the associated substrate (Duderstadt et al., 2011; Enemark and Joshua-Tor, 2006; Kelch et al., 2011). Interestingly, despite using a common ATP-binding fold and relative positions of secondary structural elements to bind substrate, different AAA+ systems show a remarkable degree of plasticity in how they engage and reshape nucleic-acid segments (Fig. 7A). Surprisingly, we find that IstB does not bind the DNA duplex through the central pore of the AAA+ filament: instead, although the protein interacts with nucleic acid segments using its structurally conserved ISM signature motif, it does so with a different surface as compared to proteins that also possess such an element, such as DnaA or archaeal Orc1 (Fig. S7). Together with its distinctive assembly mechanism, this work reveals that even closely related AAA+ ATPases can undergo an extensive level of higher-order structural rearrangements and divergent adaptations throughout evolution, and that these modifications can lead to significant differences in overall mechanism.
Fig. 7.
DNA recognition strategies of AAA+ proteins and proposed role for IstB in mediating transposition. See also Fig. S7.
(A) Structures of DNA-bound AAA+ assemblies involve in nucleic-acid transactions reveal different DNA binding and reshaping approaches used by a common nucleotide-binding fold. E1 helicase – PDB ID 2GXA (Enemark and Joshua-Tor, 2006); bacterial clamp loader complex – PDB ID 3U60 (Simonetta et al., 2009); bacterial replication initiator DnaA – PDB ID 3R8F (Duderstadt et al., 2011).
(B) IstB bends DNA in a manner reminiscent to that of bacteriophage MuA (PDB ID 4FCY; (Montaño et al., 2012)) and the PFV retroviral integrase (PDB ID 3OS2; (Maertens et al., 2010)).
(C) Proposed stepwise model for how IstB may facilitate DNA transposition by IstA.
A proposed role for IstB in IstA-mediated transposition
ATP-dependent transposase co-chaperones such as IstB, MuB and TnsC use nucleotide binding and hydrolysis to promote appropriate DNA strand cutting and pasting by their cognate transposases. As such, these three factors fall within the category of molecular matchmakers, proteins that enforce the correct assembly of multisubunit complexes to regulate essential biological processes such as DNA replication, transcription, and repair (Sancar and Hearst, 1993). Although a wide variety of nucleotide-coupled transposase systems have been identified and studied to date, how dedicated NTPase factors interact with DNA and help control transposase activity has not been well understood. For example, in Tn7, the TnsC ATPase is critical both for transposome assembly and modulating transposition (Gamas and Craig, 1992; Stellwagen and Craig, 1997), but how this element physically coordinates such tasks is not known. Similarly, while the MuB ATPase can form filaments on duplex DNA (Greene and Mizuuchi, 2002a, 2002b; Mizuno et al., 2013), how this protein engages a substrate duplex and interfaces with MuA to promote or repress transposition has yet to be defined.
The structure/function studies of IstB presented here provide new insights into the role of ATP-dependent transposase helper proteins, and into the action of AAA+-type co-chaperones for IS21 elements in particular. We find that full-length IstB forms dimers, and that in the presence of ATP, these dimers self-assemble into a decamer that sequesters and bends ~50 base pairs of DNA into a 180° U-turn (Fig. 4). Interestingly, alterations in nucleic acid structure, curvature, and supercoiling are known to be critical for many transposition and retroviral systems (Craigie and Mizuuchi, 1986; Davies et al., 2000; Hallet et al., 1994; Harshey and Jayaram, 2006; Hickman et al., 2014; Kuduvalli et al., 2001; Maertens et al., 2010; Montaño et al., 2012; Müller and Varmus, 1994; Pribil and Haniford, 2003; Pruss et al., 1994; Reimmann and Haas, 1990; Schmid et al., 1999). Although it is unclear why IstB should bend and overwind DNA upon binding, effects on local twist might help counteract the innate, negatively-supercoiled state of target DNA to render binding more energetically favorable. Alternatively, IstB’s action may help preferentially direct the protein to certain regions of the genome that have a desirable local topology. DNA bending, in turn, could be used to facilitate access of the active site of the transposase to the scissile bonds, as well as to help control the directionality of strand transfer, an approach that has been exploited by other transposition systems (Davies et al., 2000; Hickman et al., 2014; Maertens et al., 2010; Montaño et al., 2012). Indeed, the DNA configuration adopted in the presence of the IstB decamer is remarkably similar to ones seen for target DNAs bound to both the MuA transposase and the PFV (prototype foamy virus) retroviral integrase (Maertens et al., 2010; Montaño et al., 2012) (Fig. 7B). Consistent with these concepts, the absence of IstB drastically reduces transposition efficiency of IS21 and generates atypical duplications and/or deletions in the target DNA, indicating that IstB action is required for the correct alignment between the donor-transposase complex and the nucleic acid substrate (Schmid et al., 1999).
Given the duplex reshaping activities of IstB, together with the discovery that IstA specifically recognizes ATP-assembled IstB oligomers, we propose that IstB serves a landing pad that both marks and prepares DNA for IstA-mediated transposition of IS21 elements (Fig. 7C). In this model, an IstB decamer would first associate with a target DNA, which would then recruit IstA, along with a pair of pre-cleaved inverted IS21 repeats, to generate a synaptic complex. The inability of DNA to promote ATP turnover by IstB would impart sufficient stability to the complex to allow time for recognition by IstA. A potential problem arising from the direct recognition of a DNA bend by IstA is that IstB masks the target duplex; however, the capacity of IstA to stimulate ATP hydrolysis and IstB dissociation would be expected to promote clearing of the DNA, thereby allowing the transposase to access the target nucleic acid segment. Future studies aimed at imaging and biochemically characterizing higher-order IstA•IstB complexes in conjunction with DNA will be necessary to help establish the details of these interactions and events further.
EXPERIMENTAL PROCEDURES
A detailed description of protocols can be found in Extended Experimental Procedures.
Protein expression and purification
All proteins were overexpressed as N or C-terminal His6-MBP fusion proteins in E. coli, using BL21codon-plus (DE3) RIL (Stratagene) or C41 (Lucigen) cells. Following lysis by sonication, proteins were purified by a combination of affinity chromatography, proteolysis to remove the fusion tags, and size-exclusion chromatography. Purified samples were concentrated and flash-frozen for subsequent studies.
Crystallization, data collection, structure solution and refinement
Se-Met IstBAAA+ was concentrated to 8 mg/mL and spiked with 2 mM ADP·BeF3. IstBAAA+ crystals were grown by mixing 0.65 µL of protein solution with 0.65 µL of well solution containing 100 mM Bis-Tris propane pH 6.5, 300 mM ammonium sulfate, and 18.5% PEG 3350. Following harvesting and cryo-cooling, diffraction data were collected from a single crystal on Beamline 8.3.1 at the Advanced Light Source. Data were phased by SAD, and model building and refinement were performed with COOT and PHENIX (Adams et al., 2010; Emsley and Cowtan, 2004).
Analysis of IstB oligomeric state
IstB, IstBAAA+ and IstB active site mutants were dialyzed over-night at 4°C into 20 mM HEPES pH 7.5, 300 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM β-mercaptoethanol, with either 1 mM ATP or ADP. 40 µL at 1.5 mg/mL of each protein were then run over a Superdex 200 5/150 GL analytical gel filtration column (GE Healthcare) in the same buffer and monitored by A280.
DNA binding assays
Binding of a 5’-FAM-tagged dsDNA 60mer oligonucleotide was monitored at different IstB concentrations by fluorescence anisotropy using a Clariostar (BMG Labtech) microplate reader. Data were plotted and analyzed using PRISM.
EM and image analysis
IstB•ATP samples, both in the presence or absence of DNA, were either stained with 2% (w/v) uranyl acetate or frozen into liquid ethane for cryo-EM analysis. All EM data were collected using LEGINON (Suloway et al., 2005) and preprocessed with APPION (Lander et al., 2009). The initial random conical tilt reconstruction was obtained using XMIPP (Scheres et al., 2008) and refined with EMAN2 and SPARX (Hohn et al., 2007; Tang et al., 2007). Fitting of the atomic structure of the ATPase domain, and segmentation and rendering of the cryo-EM densities was done in CHIMERA (Pettersen et al., 2004).
DNA supercoiling assays
Nicked pSG483 plasmid, a derivative of pUC19, was used as a DNA substrate. Different amounts of IstB were incubated with the DNA for 20 min at 37 °C. Following ligation with T4 ligase, reactions were quenched and run for 18 h on 1% (w/v) TAE agarose gels. To analyze the effect that ATP and ADP might have on supercoiling activity, the plasmid was ligated with the ATP-independent E. coli ligase.
For two-dimensional gels, purified DNA fractions were run for 18 h at 4°C on a 1.2% (w/v) TPE agarose gel (36 mM Tris–HCl pH 7.9, 30 mM NaH2PO4, 1 mM EDTA pH 8.0). The gel was then washed three times for 30 min with TPE buffer supplemented with 4.5 µg/ml chloroquine, rotated 90 degrees, and run for 18 h at 4°C in TPE buffer plus 4.5 µg/mL chloroquine.
IstB and IstA pull-down experiments
Coprecipitation studies were performed by incubation of amylose beads (New England Biolabs) with bait and prey proteins in binding buffer. Following washing, samples were analyzed by SDS-PAGE.
ATPase assays
50 µL reactions containing different combinations of 10 µM IstB, 1 µM IstA (0–5 µM for IstA titration assay), and 10 µM dsDNA (random 60mer) were incubated during 1 h at 37 °C in a buffer containing 1 mM cold ATP and 5 nM [γ32P]ATP (1.125 µCi). Quenched reactions were analyzed using thin-layer chromatography.
IstB disassembly
IstB (32 µM) was incubated in the presence or absence of IstA (17 µM) in 40 µL of reaction buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM β-mercaptoethanol), supplemented with either 1 mM ATP or ADP, for 30 min at 37 °C. Samples were subsequently run in over a Superdex 200 5/150 GL analytical gel filtration column (GE Healthcare), pre-equilibrated in reaction buffer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to: Eva Nogales and her research group, and the beamline 8.3.1 staff at the Advanced Light Source, for help with structural data collection and analysis; Fang Wu from the Macrolab (UC, Berkeley) for her assistance with the site-directed mutagenesis; Nancy Craig for critically reading the manuscript; and the Berger laboratory for helpful discussions. Atomic coordinates for IstBAAA+ have been deposited to the PDB (accession number 5BQ5). The 3D cryo-EM models for DNA-bound and DNA-free full-length IstB have been deposited to the EMDB (www.emdatabank.org, accession number EMD-3031 and EMD-3032 respectively). This work has been supported by a post-doctoral fellowship from the “Programa Nacional de Movilidad de Recursos Humanos del Plan Nacional de I+D+i 2008–2011” from the Spanish Ministry of Education (to EAP), and the NIGMS (GM071747, to JMB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade C, Roy-Engel AM, Deininger PL. Alu elements: an intrinsic source of human genome instability. Curr. Opin. Virol. 2013;3:639–645. doi: 10.1016/j.coviro.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allué-Guardia A, Imamovic L, Muniesa M. Evolution of a self-inducible cytolethal distending toxin type V-encoding bacteriophage from Escherichia coli O157:H7 to Shigella sonnei. J. Virol. 2013;87:13665–13675. doi: 10.1128/JVI.02860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Palomo E, O’Shea VL, Hood IV, Berger JM. The bacterial DnaC helicase loader is a DnaB ring breaker. Cell. 2013;153:438–448. doi: 10.1016/j.cell.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards Ra. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Mizuuchi M, Mizuuchi K. MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell. 1991;65:1003–1013. doi: 10.1016/0092-8674(91)90552-a. [DOI] [PubMed] [Google Scholar]
- Berger B, Haas D. Transposase and cointegrase?: specialized transposition proteins of the bacterial insertion sequence IS 21 and related elements. Cell. Mol. Life Sci. 2001;58:403–419. doi: 10.1007/PL00000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, et al. Increased L1 Retrotransposition in the Neuronal Genome in Schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MCN, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- Craigie R, Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Craigie R, Arndt-Jovin DJ, Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Dyda F, Chandler M, Hickman AB. The emerging diversity of transpososome architectures. Q. Rev. Biophys. 2012;45:493–521. doi: 10.1017/S0033583512000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Filippov AA, Oleinikov PV, Motin VL, Protsenko OA, Smirnov GB. Sequencing of two Yersinia pestis IS elements, IS285 and IS100. Contrib. Microbiol. Immunol. 1995;13:306–309. [PubMed] [Google Scholar]
- Gamas P, Craig NL. Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res. 1992;20:2525–2532. doi: 10.1093/nar/20.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC, Mizuuchi K. Dynamics of a protein polymer: the assembly and disassembly pathways of the MuB transposition target complex. EMBO J. 2002a;21:1477–1486. doi: 10.1093/emboj/21.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC, Mizuuchi K. Direct observation of single MuB polymers: evidence for a DNA-dependent conformational change for generating an active target complex. Mol. Cell. 2002b;9:1079–1089. doi: 10.1016/s1097-2765(02)00514-2. [DOI] [PubMed] [Google Scholar]
- Hallet B, Rezsöhazy R, Mahillon J, Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol. 1994;14:131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- Harshey RM, Jayaram M. The mu transpososome through a topological lens. Crit. Rev. Biochem. Mol. Biol. 2006;41:387–405. doi: 10.1080/10409230600946015. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ewis HE, Li X, Knapp JA, Laver T, Doss A-L, Tolun G, Steven AC, Grishaev A, Bax A, et al. Structural Basis of hAT Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca domestica. Cell. 2014;158:353–367. doi: 10.1016/j.cell.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker RR, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A, Izsvák Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Rio DC. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992;69:27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kelch Ba, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuduvalli PN, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A, Craigie R, Mizuuchi K. B protein of bacteriophage mu is an ATPase that preferentially stimulates intermolecular DNA strand transfer. Proc. Natl. Acad. Sci. U. S. A. 1987;84:699–703. doi: 10.1073/pnas.84.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U. S. A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JL, Anderson SK, Fujita DJ, Chaconas G, Baldwin DL, Harshey RM. The nucleotide sequence of the B gene of bacteriophage Mu. Nucleic Acids Res. 1984;12:8627–8638. doi: 10.1093/nar/12.22.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Dramićanin M, Mizuuchi M, Adam J, Wang Y, Han Y-W, Yang W, Steven AC, Mizuuchi K, Ramón-Maiques S. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2441–E2450. doi: 10.1073/pnas.1309499110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. TRANSPOSITIONAL RECOMBINATION: Mechanistic Insights from Studies of Mu and Other Elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Montaño SP, Rice Pa. Moving DNA around: DNA transposition and retroviral integration. Curr. Opin. Struct. Biol. 2011;21:370–378. doi: 10.1016/j.sbi.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaño SP, Pigli YZ, Rice Pa. The µ transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MTG, Churcher CM, Bentley SD, Mungall KL, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Perry RD, Straley SC, Fetherston JD, Rose DJ, Gregor J, Blattner FR. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Podladchikova ON, Dikhanov GG, Rakin AV, Heesemann J. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol. Lett. 1994;121:269–274. doi: 10.1111/j.1574-6968.1994.tb07111.x. [DOI] [PubMed] [Google Scholar]
- Pribil PA, Haniford DB. Target DNA bending is an important specificity determinant in target site selection in Tn10 transposition. J. Mol. Biol. 2003;330:247–259. doi: 10.1016/s0022-2836(03)00588-6. [DOI] [PubMed] [Google Scholar]
- Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C, Haas D. The istA gene of insertion sequence IS21 is essential for cleavage at the inner 3’ ends of tandemly repeated IS21 elements in vitro. EMBO J. 1990;9:4055–4063. doi: 10.1002/j.1460-2075.1990.tb07627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C, Moore R, Little S, Savioz A, Willetts NS, Haas D. Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet. 1989;215:416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- Sakai J, Chalmers RM, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Hearst JE. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Seitz T, Haas D. Cointegrase, a naturally occurring, truncated form of IS21 transposase, catalyzes replicon fusion rather than simple insertion of IS21. J. Mol. Biol. 1998;282:571–583. doi: 10.1006/jmbi.1998.2041. [DOI] [PubMed] [Google Scholar]
- Schmid S, Berger B, Haas D. Target joining of duplicated insertion sequence IS21 is assisted by IstB protein in vitro. J. Bacteriol. 1999;181:2286–2289. doi: 10.1128/jb.181.7.2286-2289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Gourbeyre E, Chandler M. Bacterial Insertion Sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Surette MG, Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J. Biol. Chem. 1989;264:3028–3034. [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tang M, Cecconi C, Kim H, Bustamante C, Rio DC. Guanosine triphosphate acts as a cofactor to promote assembly of initial P-element transposase-DNA synaptic complexes. Genes Dev. 2005;19:1422–1425. doi: 10.1101/gad.1317605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science (80) 2014;345:1251343–1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM, Byun H-M, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, He ZQ, Mao YM, Sheng RQ, Sheng ZJ. On two transposable elements from Bacillus stearothermophilus. Plasmid. 1993;29:1–9. doi: 10.1006/plas.1993.1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.