Abstract
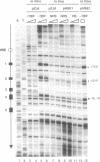
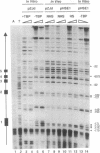
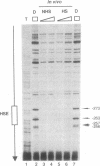
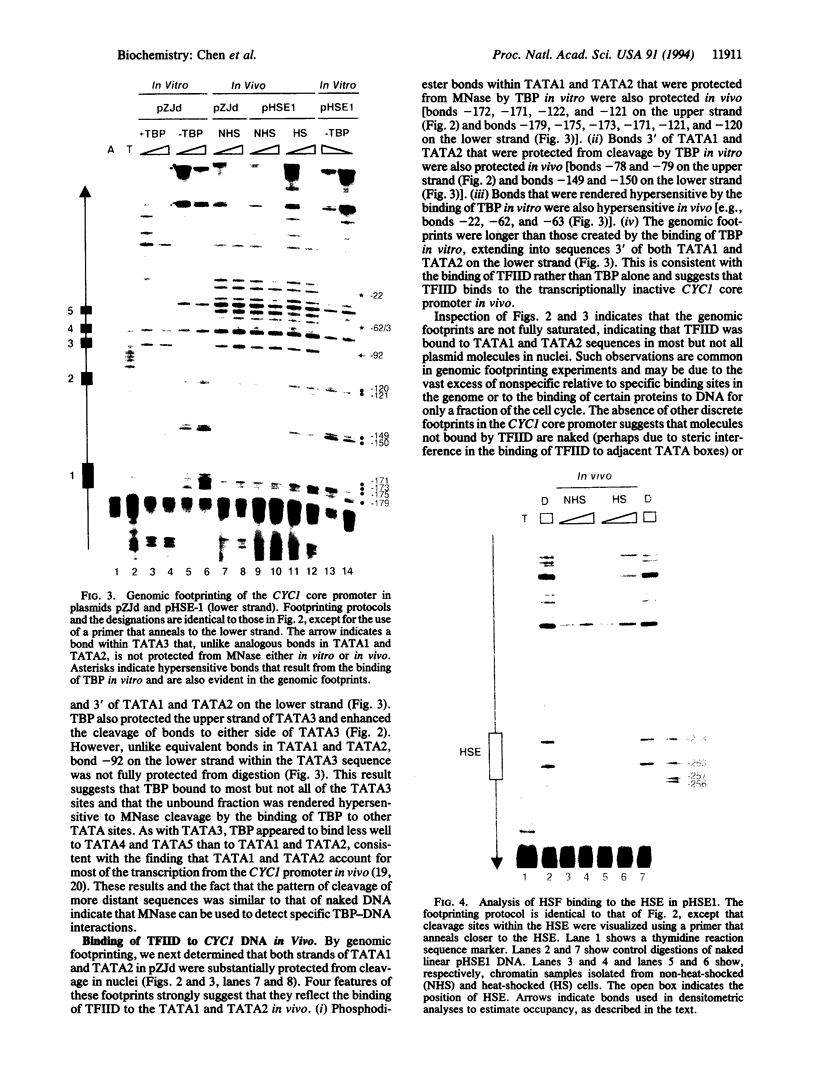
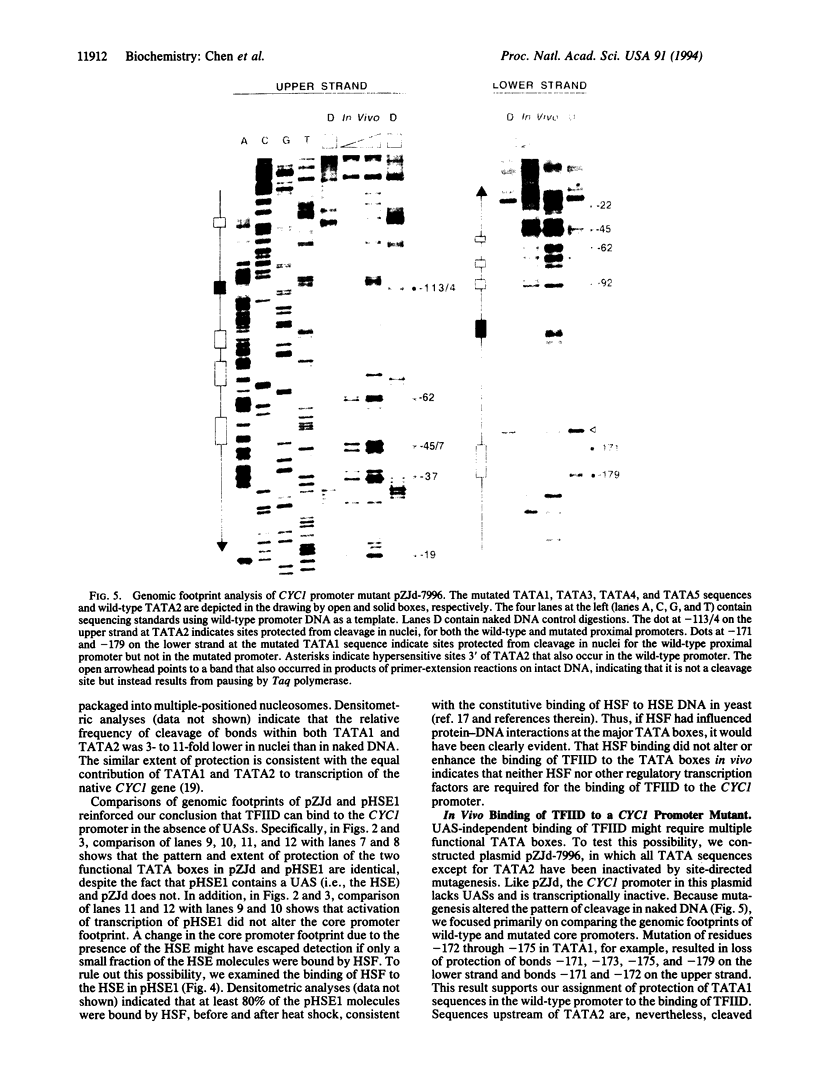
Functional transcription initiation complexes can be assembled in vitro without the aid of regulatory factors that bind to upstream activating sequences. However, promoters that lack upstream activating sequences are transcribed poorly if at all in vivo, suggesting that regulatory factors are necessary for the assembly of transcription initiation complexes in cells. To test this possibility, we asked whether the general transcription factor TFIID can bind to a promoter in yeast that lacks upstream activating sequences and is transcriptionally inactive. Analysis of an inactive CYC1 core promoter by high-resolution genomic footprinting revealed efficient binding of TFIID to either of two TATA box elements. Addition of a heat shock element rendered this promoter highly responsive to induction of transcription by heat shock but did not alter the TATA box footprints in the core promoter. Inactivation of all but one TATA box by site-directed mutagenesis did not prevent TFIID from binding to the remaining wild-type TATA box independently of regulatory sequences. These results indicate that upstream regulatory factors are not required for the in vivo binding of TFIID to the CYC1 promoter and that binding of TFIID to DNA is not necessarily a rate-limiting step in the activation of transcription in cells. Differences in chromatin structure may account for why regulatory transcription factors are required for the binding of TFIID to some promoters but not to others.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod J. D., Reagan M. S., Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993 May;7(5):857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- Carey M. Transcription. Simplifying the complex. Nature. 1994 Mar 31;368(6470):402–403. doi: 10.1038/368402a0. [DOI] [PubMed] [Google Scholar]
- Cavalli G., Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993 Dec;12(12):4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy B., Green M. R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993 Dec 9;366(6455):531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5:153–180. doi: 10.1146/annurev.cb.05.110189.001101. [DOI] [PubMed] [Google Scholar]
- Giardina C., Pérez-Riba M., Lis J. T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992 Nov;6(11):2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Adams C. C., Lee S., Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993 Oct;12(10):3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Maguire K., Kralli A., Maldonado E., Reinberg D., Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann J., Smale S. T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994 Apr 1;8(7):821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Lee H. L., Archer T. K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994 Jan;14(1):32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Li W. Z., Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. M., Taylor I. C., Kingston R. E., Young R. A. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991 Dec;5(12B):2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Lohr D., Torchia T., Hopper J. The regulatory protein GAL80 is a determinant of the chromatin structure of the yeast GAL1-10 control region. J Biol Chem. 1987 Nov 15;262(32):15589–15597. [PubMed] [Google Scholar]
- Pederson D. S., Fidrych T. Heat shock factor can activate transcription while bound to nucleosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Jan;14(1):189–199. doi: 10.1128/mcb.14.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. S., Morse R. H. Effect of transcription of yeast chromatin on DNA topology in vivo. EMBO J. 1990 Jun;9(6):1873–1881. doi: 10.1002/j.1460-2075.1990.tb08313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell B. A., Emanuel P. A., Gilmour D. S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994 Apr 1;8(7):830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Roy A. L., Carruthers C., Gutjahr T., Roeder R. G. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature. 1993 Sep 23;365(6444):359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- Schmid A., Fascher K. D., Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992 Nov 27;71(5):853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Dimitrov S. Histone-modulated gene activity: developmental implications. Crit Rev Eukaryot Gene Expr. 1993;3(3):167–191. [PubMed] [Google Scholar]
- Wolffe A. P. New approaches to chromatin function. New Biol. 1990 Mar;2(3):211–218. [PubMed] [Google Scholar]
- Wong J. M., Liu F., Bateman E. Cloning and expression of the Acanthamoeba castellanii gene encoding transcription factor TFIID. Gene. 1992 Aug 1;117(1):91–97. doi: 10.1016/0378-1119(92)90494-a. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Buchman A. R. Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci. 1993 Mar;18(3):90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]