Abstract
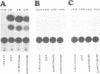
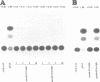
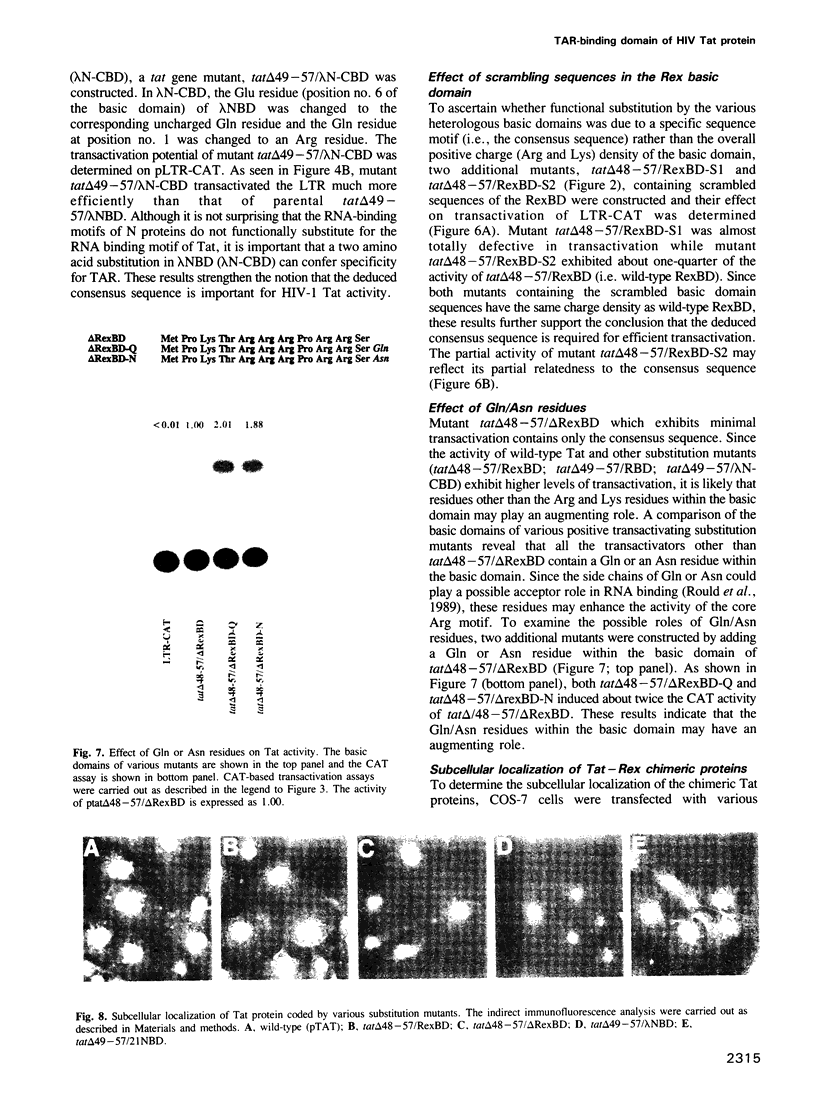
The Tat protein coded by HIV-1 is a unique eukaryotic transactivator. It activates gene expression from the viral LTR by its interaction with a nascent RNA element (TAR) located at the 5' end of all HIV-1 transcripts. Tat appears to bind to its target RNA structure in a highly sequence-specific manner. The TAR-binding activity of Tat has been localized in an Arg-rich basic domain located between residues 49 and 57 of the Tat protein. We have carried out domain substitution studies with heterologous basic domains which are also implicated in RNA binding. Here, we report that a 19 or a 12 amino acid region from the N-terminus of HTLV-I Rex can functionally substitute for the Tat basic domain. In contrast, the Arg-rich domains of the N gene products of bacteriophages lambda and 21 do not functionally substitute for the Tat basic domain. The positive and negative effects of various domain substitution mutants have facilitated identification of a consensus sequence (Arg/Lys-X-X-Arg-Arg-X-Arg-Arg) in the basic domain required for Tat activity. Conversion of the functionally inactive basic domain of the lambda N protein to the consensus motif restored the transactivation function of the Tat-N chimeric protein. Similarly, the Rex basic domain containing scrambled sequences unrelated or partially related to the consensus motif were either totally defective in transactivation or exhibited reduced activity. Our results further suggest that the activity of the core Arg motif may be enhanced by the presence of Gln or Asn within the basic domain.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Braddock M., Chambers A., Wilson W., Esnouf M. P., Adams S. E., Kingsman A. J., Kingsman S. M. HIV-1 TAT "activates" presynthesized RNA in the nucleus. Cell. 1989 Jul 28;58(2):269–279. doi: 10.1016/0092-8674(89)90841-6. [DOI] [PubMed] [Google Scholar]
- Cochrane A. W., Perkins A., Rosen C. A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990 Feb;64(2):881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990 Nov 16;63(4):655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Kumar A., Rabson A., Jeang K. T. Identification of cellular proteins that bind to the human immunodeficiency virus type 1 trans-activation-responsive TAR element RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7828–7832. doi: 10.1073/pnas.86.20.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R., Soultanakis E., Kuwabara M., Garcia J., Sigman D. S. Specific binding of a HeLa cell nuclear protein to RNA sequences in the human immunodeficiency virus transactivating region. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4858–4862. doi: 10.1073/pnas.86.13.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Malim M. H., Cullen B. R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol. 1989 Mar;63(3):1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Perkins A., Heimer E. P., Cullen B. R. Trans-activation of human immunodeficiency virus gene expression is mediated by nuclear events. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6364–6368. doi: 10.1073/pnas.84.18.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M., Subramanian T., Srinivasan A., Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989 May 11;17(9):3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Calnan B. J., Frankel A. D., Sharp P. A. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990 Nov 16;63(4):791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Garcia-Blanco M. A., Sharp P. A. Identification and characterization of a HeLa nuclear protein that specifically binds to the trans-activation-response (TAR) element of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1990 May;87(9):3624–3628. doi: 10.1073/pnas.87.9.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
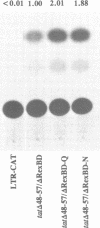
- Nosaka T., Siomi H., Adachi Y., Ishibashi M., Kubota S., Maki M., Hatanaka M. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9798–9802. doi: 10.1073/pnas.86.24.9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B. M., Luciw P. A., Barr P. J., Walker M. D. Elevated levels of mRNA can account for the trans-activation of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9734–9738. doi: 10.1073/pnas.83.24.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Ruben S., Perkins A., Purcell R., Joung K., Sia R., Burghoff R., Haseltine W. A., Rosen C. A. Structural and functional characterization of human immunodeficiency virus tat protein. J Virol. 1989 Jan;63(1):1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Peterlin B. M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990 Aug 24;62(4):769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Marciniak R. A. HIV TAR: an RNA enhancer? Cell. 1989 Oct 20;59(2):229–230. doi: 10.1016/0092-8674(89)90279-1. [DOI] [PubMed] [Google Scholar]
- Siomi H., Shida H., Maki M., Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990 Apr;64(4):1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Shida H., Nam S. H., Nosaka T., Maki M., Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988 Oct 21;55(2):197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- Sodroski J., Patarca R., Rosen C., Wong-Staal F., Haseltine W. Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science. 1985 Jul 5;229(4708):74–77. doi: 10.1126/science.2990041. [DOI] [PubMed] [Google Scholar]
- Southgate C., Zapp M. L., Green M. R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990 Jun 14;345(6276):640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989 Apr;14(4):137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- Subramanian T., Kuppuswamy M., Venkatesh L., Srinivasan A., Chinnadurai G. Functional substitution of the basic domain of the HIV-1 trans-activator, Tat, with the basic domain of the functionally heterologous Rev. Virology. 1990 May;176(1):178–183. doi: 10.1016/0042-6822(90)90242-j. [DOI] [PubMed] [Google Scholar]
- Venkatesh L. K., Mohammed S., Chinnadurai G. Functional domains of the HIV-1 rev gene required for trans-regulation and subcellular localization. Virology. 1990 May;176(1):39–47. doi: 10.1016/0042-6822(90)90228-j. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]