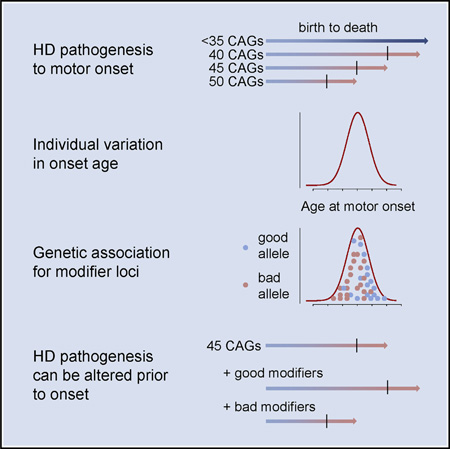
SUMMARY
As a Mendelian neurodegenerative disorder, the genetic risk of Huntington’s disease (HD) is conferred entirely by an HTT CAG repeat expansion whose length is the primary determinant of the rate of pathogenesis leading to disease onset. To investigate the pathogenic process that precedes disease, we used genome-wide association (GWA) analysis to identify loci harboring genetic variations that alter the age at neurological onset of HD. A chromosome 15 locus displays two independent effects that accelerate or delay onset by 6.1 years and 1.4 years, respectively, whereas a chromosome 8 locus hastens onset by 1.6 years. Association at MLH1 and pathway analysis of the full GWA results support a role for DNA handling and repair mechanisms in altering the course of HD. Our findings demonstrate that HD disease modification in humans occurs in nature and offer a genetic route to identifying in-human validated therapeutic targets in this and other Mendelian disorders.
Graphical abstract
INTRODUCTION
For the past three decades, a major goal of genetic analysis in humans has been to understand drivers of disease pathogenesis with the hope that these would implicate targets for developing therapeutic interventions. Initially, the primary approaches were linkage analysis and local association, often using multiallelic simple sequence repeat markers, which enabled the identification of a wide range of causative Mendelian mutations, including that underlying Huntington’s disease (HD) (The Huntington’s Disease Collaborative Research Group, 1993). For the past decade, genome-wide association (GWA) analysis with SNPs has extended the power of human genetic studies to complex diseases by identifying a multitude of contributing risk factors, usually of modest or weak effect (Manolio et al., 2009). In this report, aided by visionary HD community efforts to collect phenotypes and biosamples from large numbers of subjects with this disorder (Dorsey, 2012; Li et al., 2003; Orth et al., 2010; Paulsen et al., 2008), we apply GWA not to identify risk factors for disease but to discover genome-wide significant quantitative modifiers of a Mendelian disorder.
In HD, a CAG trinucleotide expansion mutation in HTT, the gene encoding the large huntingtin protein, causes a progressive movement disorder with dementia and behavioral abnormalities (Ross et al., 2014). Unequivocal clinical signs of HD typically emerge in mid-life, but juvenile onset and elderly onset cases are seen. Diagnosis is based upon the presence of characteristic motor signs, but the disorder includes intellectual decline and psychiatric disturbances, with death ensuing a median of 18 years after onset. The expanded CAG tract is the trigger for HD pathogenesis, and its length is the primary determinant of the rate of the pathogenic process that leads to the onset of diagnostic motor signs (referred to here for simplicity as “motor onset,” although more subtle signs may be present before clinical diagnosis) (Lee et al., 2012b). For individuals with 40 or more CAG repeats, the expansion is necessary and sufficient to cause HD, accounting for essentially all of the life-time risk of developing the disorder. Although symptomatic treatment can improve quality of life, there is no disease-modifying intervention to prevent the onset or delay the progression of HD. Because targets validated to impact on the disease process in humans have been lacking for traditional small-molecule approaches to drug development, a current focus for developing an effective disease-modifying therapy is the exploration of cutting- edge nucleic acid manipulation to prevent expression of mutant huntingtin (Aronin and DiFiglia, 2014). However, such strategies remain unproven in humans and pose numerous hurdles, including achieving efficient delivery and avoiding the potential negative effects of reducing normal huntingtin function. Thus, an effective route for identification of in-human validated therapeutic targets for traditional drug development is also needed.
Here, we postulated that the presence of the expanded CAG in an individual provides a genetically sensitized background on which to search not for risk alleles but for genetic modifiers of the disease process, which may be common in the population but not have detectable effects in the absence of the HD mutation. Consequently, we applied a GWA strategy to quantitative variations in the disease phenotype in an unbiased search for naturally occurring genetic variations associated with modification of HD pathogenesis prior to emergence of clinical disease. Our findings localize several genome-wide significant genetic modifiers of HD age at onset and already suggest at least one biochemical process that alters HD pathogenesis in humans and can be specifically targeted for traditional drug development. The approach of separating disease risk and disease modification in a Mendelian disorder, merged with the power of modern genetic techniques, is widely applicable and has the potential to implicate new therapeutic pathways in human diseases that are individually not common but that taken together constitute a substantial disease burden.
Definition of the Phenotype for Association Analysis
Within a year after the identification of the HD mutation, the inverse relationship between CAG length and age at diagnostic motor onset, which lays the foundation for a genetics-driven approach to understanding HD, was recognized (Gusella and MacDonald, 2006). This relationship establishes that the size of the mutant repeat is the primary determinant of the rate of pathogenesis leading to the emergence of clinical signs of disease (Lee et al., 2012b). The length of the expanded CAG repeat explains much, but not all, of the variation in age at motor onset. We and others have reported that the remaining variance has a large heritable component, implicating the actions of other genetic variations in modifying HD pathogenesis and suggesting that the difference between predicted and observed age of onset could be used in genetic studies to identify modifiers (Djoussé et al., 2003; Wexler et al., 2004). Haplotype analysis of HTT in HD subjects indicates that common genetic variation at the locus is not a major source of disease modification (Lee et al., 2012a), and the length of the normal CAG repeat in heterozygotes shows no statistically significant modifier influence, either alone or in interaction with the expanded allele (Lee et al., 2012b). Indeed, there is also no effect of a second expanded CAG allele on age at onset, indicating that HD pathogenesis is not HTT dosage dependent but rather reflects the completely dominant effects of a single mutant allele. These stringent analyses generated a robust statistical phenotype model, based upon subjects with 40–53 CAG repeats (Figure S1), which was used to calculate the influence of the CAG repeat on log-transformed age at onset of motor signs of HD subjects in the GWA study (GWAS), thereby generating a residual value for each subject. Residual values from the regression model were transformed back into natural scale values as a phenotype for quantitative association analysis to search the genome for genetic variation that influences age at motor onset. The distribution of residuals was similar to a theoretical normal distribution. This “residual age at motor onset” used as the GWA phenotype thus represents the difference in years between observed age at onset and that expected based upon the individual’s CAG repeat size. We analyzed individuals with 40–55 repeats; however, restricting analysis to individuals with 40–53 repeats did not materially alter our results.
Initial Genome-wide Association Studies
Over almost three decades of investigating HD, the Massachusetts HD Center Without Walls (MaHDC) accumulated a large collection of DNA samples from HD subjects, including collaborations with the HSG PHAROS (Huntington Study Group PHAROS Investigators, 2006), COHORT (Dorsey, 2012), TREND-HD (Huntington Study Group TREND-HD Investigators, 2008), and PREDICT-HD (Paulsen et al., 2008) studies, and from families for linkage and other genetic studies, including a sib-pair linkage scan for modifiers of HD onset with the HDMAPS collaboration (Li et al., 2003). This collection formed the basis for a collaborative effort that led to generation of two sequential GWA datasets. For the initial dataset (GWA1), 1,089 HD subjects were genotyped with the Affymetrix 6.0 array at the Broad Institute of MIT and Harvard. Data cleaning was carried out using standard quality-control criteria (e.g., SNP call rate > 95%, minor allele frequency [MAF] > 1%, Hardy-Weinberg equilibrium p value > 1 × 10−6, sample call rate > 95%). After quality-control analysis, multidimensional scaling analysis revealed 977 unique subjects of European ancestry with CAG repeat lengths in the range 40–55. Analysis of GWA1 (after QC: ~700,000 typed and ~8 million 1000 Genomes-imputed SNPs with MAF > 1%) did not reveal any genome-wide significant signals for association with the phenotype of “residual of age at motor onset” in a linear mixed model with covariates including ancestry characteristics and gender (see Experimental Procedures for details). GWA1 provided the basis for building SNP haplotypes of HTT that revealed that ~50% of European HD subjects share a haplotype indicative of a common ancestor, but the rest are consistent with mutation on multiple other chromosome backbones contributing to HD (Lee et al., 2012a). No effect of HTT haplotype on the age at onset phenotype was detected, permitting all HD subjects to be grouped for our GWA analyses.
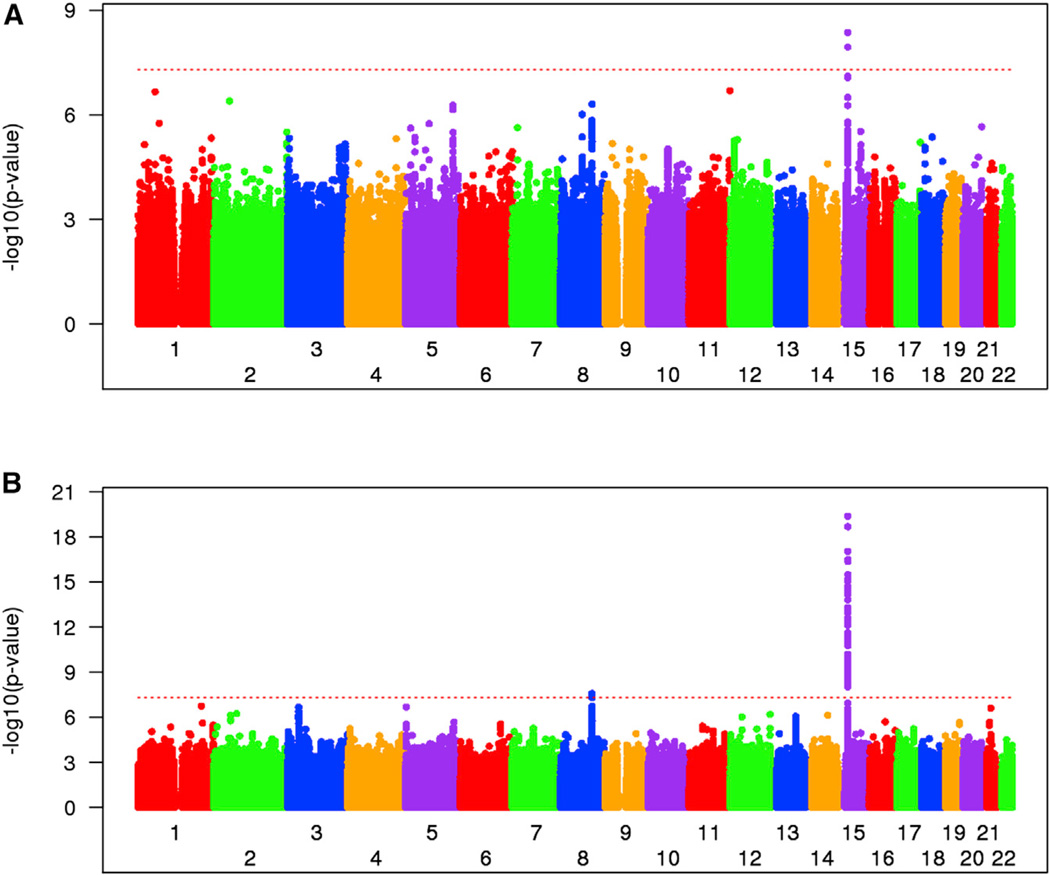
In a second phase of genotyping (GWA2), additional subjects from the MaHDC collection were combined with subjects from the PREDICT-HD natural history study. GWA2 involved 2,874 HD subjects genotyped using the Illumina Omni2.5 array at the Center for Inherited Disease Research (CIDR). Age at onset was known for only a subset of these subjects, mainly those from the MaHDC, as PREDICT-HD enrolled subjects prior to manifestations of diagnostic clinical signs. Data quality-control analysis was similar to GWA1 and yielded 974 unique individuals of European ancestry suitable for our association analysis. Full genotyping data and associated phenotypes for GWA2 have already been deposited into dbGAP, including an extensive and detailed description of the data cleaning steps. Like GWA1, GWA2 (after QC: ~1.5 million genotyped and ~8.6 million 1000 Genomes-imputed SNPs with MAF > 1%) failed to identify a genome-wide significant signal for association to residual age at motor onset. However, a combined analysis of GWA1+GWA2, which increased the effective sample size to 1,951 unique European HD subjects, revealed a genome-wide significant signal, represented by two SNPs on chromosome 15 (chr15) (best SNP, p = 4.36 × 10−9; rs146353869) (Figure 1A).
Figure 1. Genome-wide Association Analysis of Residual Age at Motor Onset.
(A) Manhattan plot of combined GWA1+GWA2 analysis yielding a locus with genome-wide significance on chr15. GWA1 and GWA2 data were combined and tested for association with residual age at onset. Significance of SNPs (−log10[p value], y axis) is plotted against genomic location (x axis). The QQ plot (Figure S1C) did not reveal significant statistical inflation evidenced by an inflation factor of 1.014.
(B) Manhattan plot of meta-analysis of GWA1+2 and 3 showing genome-wide significant peaks at chr15 and chr8 and near-significant on chr3, along with other trails. Association analysis was initially performed independently on GWA3 data (not shown), and then a meta-analysis was performed to summarize the overall association findings of the GWA1+GWA2 and GWA3 analyses. The overall inflation factor of 1.009 suggests the absence of statistical inflation in this analysis (Figure S1D). The red dotted lines in (A) and (B) indicate the genome-wide significance level (p value, 5 × 10−8). The GeM-HD Group has developed a web portal through which interested investigators can access the genome-wide SNP association data by SNP, gene, or genomic location of interest. This can be accessed through the HDinHD portal (https://www.hdinhd.org/). Original data will be made available on request. Please direct inquiries to info@chdifoundation.org with the words “GWAS data” in the subject line.
Follow-up Genome-wide Association Study
To confirm the genome-wide significant signal on chr15 in an independent dataset and to implicate new loci through the increased power of a larger combined analysis, we formed the Genetic Modifiers of HD (GeM-HD) Group to carry out GWA3. Genotyping was performed at the Broad Institute using the Illumina Omni2.5 array for 3,447 HD subject samples from the MaHDC collection and from the European Huntington’s Disease Network (EHDN) Registry study (Orth et al., 2010). After QC, GWA3 comprised 2,131 unique individuals of European ancestry suitable for our analysis of association to the residual age at motor onset. GWA3 alone independently confirmed the chr15 locus at a genome-wide significance level (best SNP, p = 1.35 × 10−12; rs146353869).
Meta-analysis of the two linear mixed-effect model results (combined GWA1+GWA2 and GWA3; n = 4,082) improved the significance of the best SNP on chr15 to p = 4.3 × 10−20 and also yielded a second genome-wide significant locus on chr8 (p = 2.7 × 10−8, rs1037699) with suggestive trails at other locations, such as chr3, chr5, and chr21 (Figure 1B). The most significant variants at all locations that achieved a peak p < 1 × 10−6 are given in Table 1, whereas a more extensive list of all SNPs yielding p < 1 × 10−5 is presented in Table S1.
Table 1.
Most Significant Variants Associated with Residual Age at HD Motor Onset
| SNP | Chr | BP (hg19) | Minor Allele | Major Allele | MAF in Europeans (%)b |
MAF in European HD (%) |
Effect Size (Years/ Minor Allele) |
p Value in meta Analysisa |
|---|---|---|---|---|---|---|---|---|
| rs147804330 | 2 | 56391203 | A | G | 8.0 | 6.3 | −1.6 | 7.6 × 10−7 |
| rs72810940 | 2 | 75555265 | A | G | 3.4 | 2.9 | 2.4 | 5.9 × 10−7 |
| rs144287831 | 3 | 37068079 | C | T | 32.5 | 31.2 | 0.9 | 2.2 × 10−7 |
| rs11133929 | 5 | 2155168 | C | T | 9.4 | 9.3 | 1.5 | 2.1 × 10−7 |
| rs1037699 | 8 | 103250930 | T | C | 8.3 | 9.6 | −1.6 | 2.7 × 10−8 |
| rs11061229 | 12 | 131389783 | C | G | 6.9 | 6.6 | −1.7 | 6.7 × 10−7 |
| rs261453 | 13 | 82324504 | A | C | 9.9 | 11.4 | −1.3 | 9.0 × 10−7 |
| rs148491145 | 14 | 72360176–72360182 | – | GACTCTA | 2.0 | 1.5 | −3.2 | 7.5 × 10−7 |
| rs146353869 | 15 | 31126401 | A | C | 1.1 | 1.7 | −6.1 | 4.3 × 10−20 |
| rs2140734 | 15 | 31243792 | G | T | 30.2 | 30.4 | 1.4 | 7.1 × 10−14 |
| rs143367341 | 21 | 28348433 | G | A | 14.6 | 13.5 | 1.3 | 2.5 × 10−7 |
See also Table S1.
The most significant variant is shown for each independent signal with p < 1 × 10−6. Genome-wide significant signals are shown in bold. See also Table S1.
MAF (%) in Europeans represents the minor allele frequency in 1000 Genomes project data phase 3, except rs143367341 (1000 Genomes Project data, phase 1, release3).
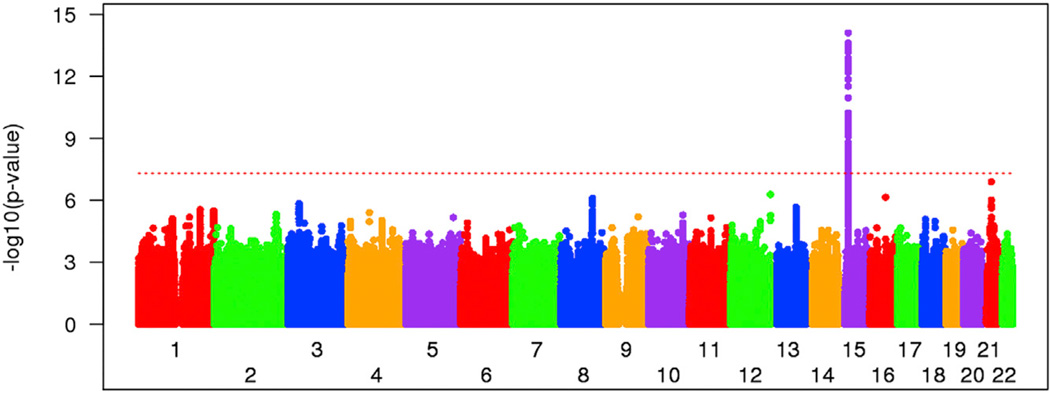
To test the robustness of the quantitative association analysis to outliers with large residuals of age at onset, we also performed a dichotomous analysis. Individuals whose phenotypes fell into the 20% extremes of either earlier or later than expected age at onset were compared for marker allele frequency in a standard “case:control” GWAS design. Logistic regression analysis with ancestry characteristics and gender covariates was implemented using the combined datasets. Results from the genome-wide dichotomous analysis are shown in Figure 2, and details of allele frequency are also provided (in Table S2) for the top SNPs from quantitative analysis. Even though the dichotomous comparison comprised only 40% of the samples, the chr15 region again showed genome-wide significance (best SNP, p = 7.9 × 10−15; rs2140734), and the same secondary peaks on other chromosomes were readily discernible. Thus, detection of these loci does not depend critically on the precise magnitude of the residual of age at onset in the quantitative analysis, as the shift of individuals toward one or the other tail of the distribution creates contrasting allele frequencies between these extremes.
Figure 2. Dichotomous Association Analysis in Extremes of Age at Motor Onset.
Association analysis was carried out to compare SNP allele frequencies between the 20% extremes of residual age at motor onset, showing that the modifier effect on chr15 is captured by the allele frequency distribution in addition to quantitative analysis.
See also Table S2.
Conditional Analysis and Effect Size
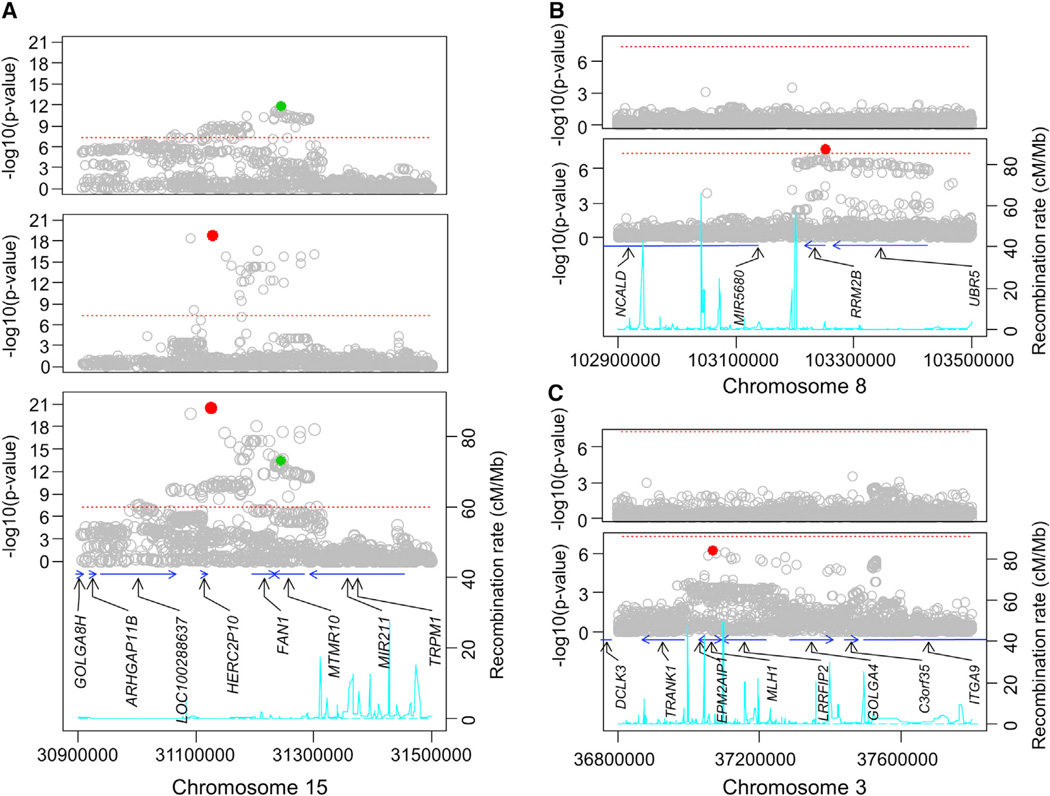
To determine whether any of the top loci show evidence of more than one functional modifier allele, we carried out conditional association analysis using a fixed-effect model of the combined data as shown in Figure 3A for the chr15 region. The bottom panel shows an expanded view of the chr15 locus association plot. When a fixed-effect model was conditioned by the most significant SNP (rs146353869, red circle), many of the most significant signals disappeared (Figure 3A, top panel), but a large number of SNPs remained above the genome-wide significant level (red dotted line, best SNP = rs2140734, green circle), indicating a second modifier effect independent of that captured by rs146353869. This was confirmed by conditioning the analysis on rs2140734 (Figure 3A, middle panel), whose characteristics are also listed in Table 1. Interestingly, the minor alleles for SNPs detecting these two independent signals are associated with opposing effects. The SNPs with the most significant p values all show a relatively low MAF (1.3%–3.0%), and each minor allele corresponds with up to 6.1 years earlier age at onset than expected based upon CAG length (range −2.9 to −6.1 years/minor allele for 34 SNPs). To avoid any contribution of a “winner’s curse,” we also estimated the effect size in only the GWA3 dataset accumulated after genome-wide significance had already been achieved. In this independent confirmation dataset, the effect size for rs146353869 was −6.2 years/minor allele (in a mixed-effect model). The genome-wide significant SNPs at the chr15 locus that detect the second, independent association signal all display a much higher MAF (27.0%–39.1%) and are associated with a delay in motor onset of up to 1.4 years (rs2140734; range +1.1 to 1.4 for 91 SNPs; +1.4 years/minor allele for rs2140734 in GWA3 alone by mixed-effect model analysis). These two independent modifier effects reflect the presence in the population, on different versions of chr15, of two separate functional variants that likely have opposing impacts on the same gene. Unlike the chr15 region, the other loci listed in Table 1 each suggest only a single modifier allele. Examples are shown in Figures 3B and 3C for chr8 and chr3, where conditioning the analysis on the respective top SNPs (rs1037699 and rs144287831) dramatically reduced other association signals in the corresponding region. We did not detect any significant interaction between the four independent SNPs representing chr15 (rs146353869 and rs2140734), chr8 (rs1037699), and chr3 (rs144287831) in pairwise tests. Furthermore, models directly testing interaction between each SNP and HTT CAG repeat length did not support the significance of CAG:SNP interaction term, suggesting independent effects acting equally across the range of expanded repeats.
Figure 3. Conditional Association Analysis at Top Loci.
(A) Chromosome 15 locus.
Bottom panel: The single SNP association analysis of the combined dataset using a fixed-effect model is shown above the recombination rate (cyan line), based upon HapMap samples, and the largest transcript for each annotated gene in the region (blue arrows). The red and green circles represent the most significant independent SNPs that emerged from the conditional analyses shown in the middle and top panels.
Middle panel: Single SNP association analysis conditioned by rs2140734 (green in bottom and top panels) revealing a group of SNPs that remain significant after removing the effect associated with rs2140734.
Top panel: Single SNP association analysis conditioned by rs146353869 (red in bottom and middle panels) revealing a group of SNPs that remain significant after removing the effect associated with rs146353869.
(B) Chromosome 8 locus.
Bottom panel: The chr8 locus single SNP association analysis of the combined dataset using a fixed-effect model is shown above the recombination rate (cyan line), based upon HapMap samples, and the largest transcript for each annotated gene in the region (blue arrows). The red circle represents the most significant SNP that was used in the conditional analysis.
Top panel: Single SNP association analysis conditioned by rs1037699 (red in bottom panel) revealing that all SNPs that showed association in the original association analysis were no longer significant after removing the effect associated with rs1037699.
(C) Chromosome 3 locus.
Bottom panel: The chr3 locus single SNP association analysis of the combined dataset using a fixed-effect model is shown above the recombination rate (cyan line), based upon HapMap samples, and the largest transcript for each annotated gene in the region (blue arrows). The red circle represents the most significant SNP that was used in the conditional analysis.
Top panel: Single SNP association analysis conditioned by rs144287831 (red in bottom panel) revealing that all SNPs that showed association in the original association analysis were no longer significant after removing the effect associated with rs144287831.
Genes near Top Association Signals
As with any GWA analysis, the location of the significant SNPs does not immediately identify which gene mediates the consequences of the as yet unknown functional variant, but several candidates are evident for the genome-wide significant loci. At the chr15 locus, a recombination frequency peak (cyan line in Figure 3A) on the telomeric side coincides with the loss of both independent significant association signals, which extend proximally in a region containing the two highest priority candidate genes, MTMR10 (myotubularin related protein 10) and FAN1 (Fanconi anemia FANC1/FANCD2-associated [endo] nuclease 1), along with the pseudogene HERC2P10 in a segment that also specifies several putative large intergenic non-coding RNAs (lincRNAs). On chr8 (Figure 3B), the significant association signal also extends across two high-priority candidate genes, RRM2B (a subunit of DNA damage p53-inducible ribonucleotide reductase M2 B) and UBR5 (an HECT domain E3 ubiquitin-protein ligase). The region also contains the microRNA gene MIR5680 and the 50 end of NCALD (neurocalcin delta). Among the top loci that did not reach genome-wide significance, the most notable is that on chr3 (Figure 3C), which centers on MLH1 (the human homolog of the E. coli DNA mismatch repair gene mutL), whose mouse homolog, Mlh1, was discovered in a genome-wide genetic screen to modify somatic instability of the CAG repeat and the timing of CAG length-dependent phenotypes in the striatum of genetic HD replica CAG knockin mice (Pinto et al., 2013).
Pathway Analysis
To examine systematically whether variants associated with altered age at onset, extending beyond the most significant hits, cluster in genes with common biological function, we performed pathway analyses using three approaches chosen to have different characteristics: Setscreen, ALIGATOR, and gene-set enrichment analysis (GSEA). Setscreen (Moskvina et al., 2011) combines p values from all SNPs in a pathway, making it advantageous for genes and pathways containing multiple quasi-independent signals of modest size. However, this approach may lose power when the pathways contain a few strong signals with many SNPs showing no association. ALIGATOR (Holmans et al., 2009) defines genes as “significant” based on their most significant SNP and tests whether a pathway contains a higher number of significant genes than would be expected by chance, taking into account gene size and linkage disequilibrium between genes. This gives good power to detect pathways in which there is one strong association signal per gene but has the disadvantage of requiring that a criterion be set for defining significant SNPs and genes. GSEA (Wang et al., 2007) ranks genes in order of a gene-wise significance measure, then tests whether pathway genes have a significantly high rank, weighting by the significance measure. For each analysis, we conservatively assigned SNPs between the start of the first and the end of the last exon of any transcript to the corresponding gene. To avoid making a priori assumptions about the areas of biology involved in the modification of age at motor onset, we deliberately chose to use a large pathway set covering as many areas of biology as possible, comprising 14,706 functional gene sets, many with overlapping members, containing between 3 and 500 genes: 10,741 from GO, 265 from KEGG, 1897 from MGI, 119 from PANTHER, 217 from Biocarta, 1248 from Reactome, and 219 from NCI.
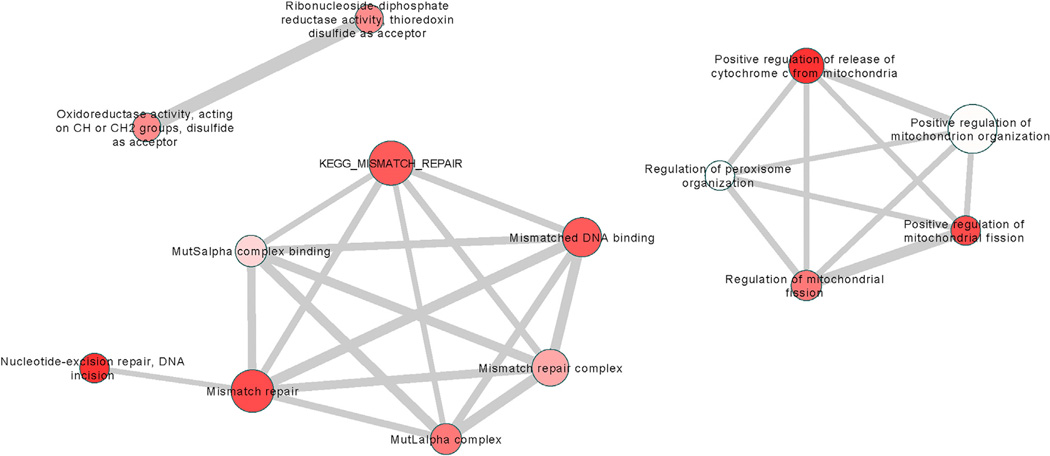
In the primary Setscreen analysis, 14 pathways were significant after correcting for multiple testing (q < 0.05). These are listed in Table 2, together with their enrichment p values under the ALIGATOR and GSEA analyses. Enrichment p values under all analysis methods are given in Table S3 for the 326 pathways with p < 0.05 in the Setscreen analysis. Figure 4 shows that the 14 significant pathways from Table 2 group into three clusters by gene membership: DNA repair, mitochondrial fission, and oxidoreductase activity. Pathways in the DNA repair cluster also show significant enrichment under the ALIGATOR and GSEA analysis (Table 2), increasing confidence that the enrichments are genuine. Best SNP and gene-wide p values for the genes in these clusters are given in Table S4. Gene-wide p values are calculated using the method of Brown (Brown, 1975), similar to that used in Setscreen. Note that FAN1 (together with ERCC3) is a member only of GO:33683 (nucleotide-excision repair, DNA incision), although it is a member of the broader pathway GO:6281 (DNA repair), which has nominally significant (p = 0.012) evidence for enrichment (see Tables S3 and S5). Thus, the significant enrichments observed in the DNA-repair pathways in Table 2 are achieved independently of the association region on chr15. If genome-wide significant (p < 5 × 10−8) SNPs are removed from the Setscreen analysis, GO:33683 is still significant (p = 8.65× 10−5), with the other pathways in Table 2 being unaltered. Genes with gene-wide p < 0.1 from all 326 pathways significantly enriched (p < 0.05) in the Setscreen analysis are shown in Table S5.
Table 2.
Pathways Significant after Multiple-Testing Correction (q < 0.05) in the Primary Setscreen Analysis and Enrichment p Values For ALIGATOR And GSEA
| Pathway | p(Set-screen) | q(Set-screen) | p(ALIGATOR) | p(GSEA) | Description |
|---|---|---|---|---|---|
| GO:0090200 | 8.89 × 10−8 | 0.0007 | NA | 0.1040 | positive regulation of cytochrome c release from mitochondria |
| GO:0033683 | 1.69 × 10−6 | 0.0063 | 0.0087 | 0.0030 | nucleotide-excision repair, DNA incision |
| GO:0090141 | 2.30 × 10−6 | 0.0063 | NA | 0.1314 | positive regulation of mitochondrial fission |
| GO:0006298 | 3.25 × 10−6 | 0.0066 | 0.0086 | 0.0074 | mismatch repair |
| KEGG:3430 | 6.65 × 10−6 | 0.0101 | 0.0732 | 0.0280 | mismatch repair |
| GO:0030983 | 7.43 × 10−6 | 0.0101 | 0.00254 | 0.0062 | mismatched DNA binding |
| GO:0090140 | 1.57 × 10−5 | 0.0169 | NA | 0.1560 | regulation of mitochondrial fission |
| GO:0032389 | 1.66 × 10−5 | 0.0169 | 0.00072 | 0.0382 | MutLalpha complex |
| GO:0004748 | 2.66 × 10−5 | 0.0217 | NA | 0.0380 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor |
| GO:0016728 | 1.65 × 10−5 | 0.0217 | NA | 0.0380 | oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor |
| GO:0032300 | 3.82 × 10−5 | 0.0283 | 0.00088 | 0.0058 | mismatch repair complex |
| GO:0032407 | 5.74 × 10−5 | 0.0390 | 0.00127 | 0.0062 | MutSalpha complex binding |
| GO:0010822 | 7.63 × 10−5 | 0.0478 | NA | 0.0436 | positive regulation of mitochondrion organization |
| GO:1900063 | 8.39 × 10−5 | 0.0488 | NA | 0.0376 | regulation of peroxisome organization |
Figure 4. Fourteen Significant Pathways (q < 0.05) from the Main Setscreen Analysis Clustered by Gene Membership.
Thickness of line connecting two pathways is proportional to the number of genes shared between them. The size of the node is proportional to the number of SNPs. The intensity of shading is inversely proportional to the q value; deep shades of red have low q values, and pale shading is close to the 5% threshold. Pathways were assigned to clusters as follows: For each pair of pathways, an overlap measure K was defined as the number of genes common to both pathways divided by the number of genes in the smaller pathway. A pathway was assigned to a cluster if the average K between it and the pathways already in the cluster was greater than 0.4. If it was not possible to assign a pathway to an existing cluster, a new cluster was started. This procedure was carried out recursively, in descending order of enrichment significance.
Effects on Age at Onset of Cognitive and Psychiatric Signs
In addition to motor signs, HD subjects also show cognitive difficulties and sometimes exhibit prominent psychiatric disturbance that may precede their movement disorder. The age at onset for cognitive and psychiatric signs was noted for limited subsets of our genotyped subjects of European ancestry. Consequently, we tested the hypothesis that the SNPs showing genome-wide significant association with residual age at motor onset also reveal modification of these non-motor phenotypes. The sample sizes of subjects with CAG 40–55 repeats for these analyses comprised 843 subjects with a recorded age at onset of cognitive signs and 1,515 with a known age at onset of psychiatric signs, as scored by an expert rater familiar with HD. Each of these phenotypes shows a negative correlation with CAG repeat length, explaining 49.3% and 39.2% of the variance in age at onset, respectively (Figure S2), as compared with 59.4% for the motor phenotype. Both are very significantly correlated (Pearson correlation p value < 2.2× 10−16) with age at motor onset (Pearson correlation coefficients of 0.891 and 0.804, respectively), but we sought to assess whether differential effects might be revealed by the actions of the modifier loci. Because stringent phenotype models describing the relationship between age at onset of cognitive or psychiatric onsets and CAG repeat length have not yet been developed, we tested the effects of the independent significant SNPs associated with residual age at motor onset by modeling log-transformed age at onset of cognitive signs or psychiatric signs as a function of CAG repeat length, SNP, gender, and four ancestry covariate values in a combined fixed-effect model analysis framework. For comparison, we also applied this analytical approach to age at motor onset, which yielded p values that were only slightly different than the residual-based analysis above.
For both age at cognitive onset and age at psychiatric onset, the independent signals at the chr15 locus both showed a nominally significant association, hastening or delaying cognitive or psychiatric onset in the same direction as the respective effect on motor onset (Table 3). The chr8 modifier was also nominally significant for association with age at cognitive onset and age at psychiatric onset. Interestingly, although the chr3 locus at MLH1 showed near-significance for psychiatric onset, there was no evidence of any effect on cognitive onset, contrasting with the other modifier loci and suggesting that the different modifiers may act on different processes to accomplish their effect on HD pathogenesis. This suggests that not all modifiers of motor onset influence other HD phenotypes and that future analyses of larger HD datasets specifically for cognitive and psychiatric phenotypes may reveal additional modifying genetic variants distinct from those that alter age at motor onset.
Table 3.
Association of Top Loci with Other Phenotypes
| Age at Onset of Motor Signs |
Age at Onset of Cognitive Signs |
Age at Onset of Psychiatric Signs |
||
|---|---|---|---|---|
| SNP | Chr | p value | p value | p value |
| rs146353869 | 15 | 7.0 × 10−19 | 7.6 × 10−3 | 2.7 × 10−7 |
| rs2140734 | 15 | 4.7 × 10−13 | 1.1 × 10−3 | 4.8 × 10−3 |
| rs1037699 | 8 | 1.2 × 10−7 | 8.7 × 10−4 | 4.1 × 10−2 |
| rs144287831 | 3 | 5.1 × 10−7 | 9.7 × 10−1 | 7.6 × 10−2 |
See also Figure S2.
DISCUSSION
Investigation of humans with an expanded HTT CAG allele and studies of model systems in which full-length mutant huntingtin is expressed, including both human cells and CAG knockin mice, support the view that the HD mutation has effects throughout life due to a completely dominant gain-of-function mechanism that leads after decades to onset of clinical signs (Gusella et al., 2014). The precise biological differences distinguishing individuals who possess expanded CAG alleles and will develop HD from those with normal-length CAG alleles who will not are not well understood. However, the proof-of-principle that HD disease modification is possible is demonstrated not by medical treatment but by observations of a heritable portion of the variance in age at onset that is not explained by either the size of the CAG repeat or other HTT region polymorphisms (Lee et al., 2012a). Instead functional variants exist in the human population that do not themselves confer risk of HD but are capable of modifying the course of the disorder during the long phase that precedes emergence of clinical disease, resulting in earlier or later onset than expected based upon the individual’s expanded CAG repeat length. In essence nature has achieved disease modification, the goal of those seeking therapeutic interventions, and it has remained for investigators to identify the means by which it occurs. Genetic modifiers could lead to a better understanding of the genes and processes that impact on HD pathogenesis and provide in-human validated targets for traditional small-molecule therapies. The significant loci that have emerged from our unbiased genome-wide search for variants associated with altered age at diagnostic motor onset offer a different entrée into influencing pathogenesis in this long-studied but still intractable disorder.
The previous investigation of potential genetic modifiers of HD has largely relied upon biased candidate gene studies, but none has identified a locus of genome-wide significance. The findings have been weak and inconsistent even for the same gene, likely reflecting a lack of power and statistical stringency, variable phenotype definition, and population stratification. Indeed, in our GWAS, none of the previously suggested candidate modifiers achieved p < 1 × 10−5. However, two previous unbiased genetic-linkage modifier searches in HD sib pairs from North America, Europe, and Australia (Li et al., 2003, 2006) or in families limited to Venezuela (Gayá n et al., 2008) yielded genome-wide significant peaks at 6q23-q24 (LOD = 4.05) or at 2p25 (LOD = 4.29), respectively, with trends in the latter at 2q35 (LOD = 3.39), 5p14 (LOD = 3.31), and 5q32 (LOD = 3.14). None of the most significant association signals and none of the trending SNPs (p < 1 × 10−5) from our European GWA analysis correspond to any of these linkage regions. The lack of overlap between our GWA and the Venezuela linkage scan could be explained simply by population differences in the modifier alleles present. The difference with the other linkage study, which included subjects expected to be primarily of European ancestry, more likely represents either a diversity of modifier alleles at the 6q23-q24 locus detectable by linkage but not by association in this sample or inaccuracy in precisely localizing the linkage peak, as there is a nearby association signal at 6q23 (top SNP rs6934819, p = 2.8 × 10−6). A similar discrepancy between GWA and linkage results has been seen for risk factors in some complex disorders (Weiss et al., 2009).
The genome-wide significant loci identified here permit discovery of the specific functional variants responsible for the modifier effects and the genes through which they act. Both the chr15 and chr8 loci offer attractive candidates. On chr15, the presence of two independent genome-wide significant signals in the same region reflects two functional variants, with Occam’s razor arguing that these are likely to affect the same gene. The two strongest locational candidates, FAN1 and MTMR10, are implicated in functions previously suggested in studies of HD pathogenesis: structure-specific DNA handling and inositol-phosphate signaling, respectively. The FAN1 nuclease plays a role in repair of DNA inter-strand cross-links but not of double-strand breaks (Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010) and has recently been identified as essential for restart of paused replication forks in DNA synthesis (Chaudhury et al., 2014). MTMR10, although catalytically inactive, is thought, like other such myotubularin-related family members, to heterodimerize with an active phosphatase subunit to act on phosphatidylinositol phosphates (Hnia et al., 2012). The top SNPs for each of the two independent signals are located upstream of FAN1 and within MTMR10, respectively, although each is backed by a distinct, extensive set of associated SNPs spanning both genes. FAN1 and MTMR10 lie within a larger 2 Mb region of copy-number variation (CNV) due to non-allelic homologous recombination of flanking repeats. Both deletion and duplication of the region have been associated with intellectual disability, epilepsy, and autism, with the former also associated with schizophrenia. Although no genome-wide significant GWA signal has been reported in this segment for autism, schizophrenia, or other psychiatric disorders, it has been suggested that FAN1 may drive neurodevelopmental susceptibility based upon an increased frequency of rare missense variants (Ionita-Laza et al., 2014). Analysis of our GWA data using (1) the number of CNV segments (p = 0.7819), (2) total CNV size (p = 0.853), and (3) average of CNV segment size (p = 0.5201) did not support a contribution of CNV to our HD modifier association signals, indicating that more subtle but relatively common genetic variants are responsible for the HD modifier effect. Very recent data show that the same 2 Mb segment of 15q13.3 is inverted without change in copy number at low but readily measurable frequency in the normal population (Antonacci et al., 2014). It will be interesting to determine whether SNPs tagging either of the functional effects that we have observed are present on inversion chromosomes. However, in either circumstance, the fact that our genome-wide association signals extend over < 250 kb of the internal region suggests that it will be possible to use haplotyping and sequence analysis to home in on the functional variants, which are likely to act via one of these two genes.
The same strategies can apply to the single functional variant implicated on chr8, where the association signals extend over < 250 kb spanning two prime locational candidates, RRM2B and UBR5. These genes are involved in additional functions previously suggested to play a role in HD pathogenesis: mitochondrial energetics and oxidative stress, and proteostasis. As a subunit in quiescent cells of the rate-limiting enzyme in new deoxyribonucleotide triphosphate synthesis (Pontarin et al., 2011), RRM2B has effects on DNA synthesis and repair (Pontarin et al., 2012). It also regulates mitochondrial DNA content (Bourdon et al., 2007) and suppresses activation of the oxidative stress pathway (Kuo et al., 2012). Proteasome-mediated protein degradation due to tagging by E3 ubiquitin ligases like UBR5 is critical to many cellular processes and has been a focus of HD research due to the intracellular accumulation of misfolded polyglutamine fragments of mutant huntingtin (Ortega and Lucas, 2014).
Although we believe it most likely that the modifiers act through intersection with the biology of HD pathogenesis, it is formally possible that they act through an independent effect on motor control in aging and modify HD age at onset additively. None of the top SNPs on chr15, chr8, or chr3 SNPs has been associated with any phenotype in previous GWASs, including analysis of age at onset of Parkinson disease, a movement disorder wherein a similar additive effect might be predicted. The effect sizes of −6 to +1.4 years, determined from CAG repeat lengths typically associated with a mean onset range of ~20 to 60 years, would indicate a substantial effect in mid-life, but it is conceivable that more detailed phenotyping of normal individuals may reveal HD-independent effects of these loci. Investigation of other movement disorders might also reveal an influence of these loci, particularly if the other disorder shares aspects of pathogenesis with HD.
In HD itself, the effect is substantial, representing ~8%–33% of the typical disease duration and as much as 25%–30% of the life span prior to diagnosis. This is most evident in the odds-ratios from extreme dichotomous analysis, which, at 1.8 to 18.1 (Table S2), far exceed those in most disease GWAS where small additive risk effects are the norm. Although larger effect sizes may prove to be more frequent in modifier-based than in risk-based GWAS, they are not a prerequisite for judging a modifier’s value in directing therapeutic development, as pharmaceutical interventions have the potential for stronger effects than naturally occurring human variation on disease-modifying processes.
Although not quite achieving genome-wide significance, prior discovery in CAG repeat knockin mouse genetic modifier screen strongly supports the candidacy of MLH1 at the chr3 locus and implicates DNA mismatch repair as a process that modifies HD pathogenesis (Pinto et al., 2013). This proposal receives further support from our pathway analyses. Genes involved in DNA mismatch repair pathways were enriched for association with HD residual age at onset. In humans, inactivation of MLH1 and other mismatch repair genes, predominantly MSH2, MSH6, and PMS2, is associated with dinucleotide repeat instability in certain cancers (Sehgal et al., 2014). By contrast, inactivation of some mismatch repair genes (Msh2, Msh3, Mlh1, Mlh3) eliminates somatic instability of the CAG repeat in Htt CAG knockin mice, and knock out of Mlh1, and other mismatch repair genes, also slows the pathogenic process in these precise genetic HD replicas (Dragileva et al., 2009; Pinto et al., 2013). Given that Mlh1 and other mismatch repair genes influence CAG instability in knockin mice, and longer HTT CAG repeat expansions in HD post-mortem brain are associated with earlier disease onset (Swami et al., 2009), one would predict that MLH1 modifies disease onset in patients as a consequence of an effect on somatic HTT CAG repeat instability. However, although attractive, this assumption should be tested further. The HD knockin mouse studies do not a priori rule out the possibility that DNA repair genes might modify HD pathogenesis via DNA structure-specific functions unrelated to their activities in enhancing CAG instability.
The identification of genetic modifier loci that alter the course of HD prior to disease onset opens an entirely new direction for development of therapies to prevent the onset of this devastating disorder. To truly understand the modifier mechanisms, additional work is needed to identify the functional DNA variant and gene(s) responsible for mediating the modifier effect at each locus. Follow-up to our initial lead is enabled by the ongoing collection of additional well-phenotyped DNA samples from large HD natural history studies and clinical trials. Adding samples will power genome-wide association to detect many more modifiers and to potentially reveal additional pathways by which HD pathogenesis can be altered. Similarly, the analysis of non- European subjects offers the potential for identifying additional alleles at these modifier loci and/or additional loci specific to certain populations. Applying this approach to other HD phenotypes, both before and after disease onset, may, as our findings already suggest, reveal modifiers that distinguish different disease domains. All of these pursuits will accelerate the ability to translate natural disease modification by genetic factors into directed disease modification by human intervention.
The genetic modifiers defined here have already identified one pathway, DNA handling, whose manipulation may provide therapeutic benefit in HD. Investigation of the chr15 and chr8 loci is likely to provide additional targets. Even before these modified loci are fully understood and additional modifiers are found, our findings will have a profound impact in a number of areas. As a proof-of-principle, they will spur pursuit of this genetic modifier strategy in other Mendelian disorders with quantitative or discrete qualitative differences in phenotype. In basic research, they will open paths into understanding the pathogenic process in HD by providing human-relevant tools with which to perturb it. In clinical research, they will provide the basis for genotype-phenotype studies to more fully explore the phenotypic correlates of each mechanism of disease modification. For clinical trials, stratification of the patient population and/or quantitation of the outcomes in a manner that takes into account the effects of patient genotype at these loci will permit smaller, less costly trials with greater power to detect therapeutic benefit. Finally, for HD families, our findings present the hope of novel in-human validated targets to accelerate development of treatments and represent a validation of their active and willing participation in HD studies that have made such large-scale investigations possible.
EXPERIMENTAL PROCEDURES
Subjects and Residual Phenotype
Patient consents and the overall study were reviewed and approved by the Partners HealthCare Institutional Review Board. The sources of subjects are described in Supplemental Experimental Procedures. We used age at onset of motor signs and CAG repeat length to derive residual age at onset of motor signs, which we used as the primary phenotype for the GWAS to identify genetic modifiers of HD. In order to subtract the effects of CAG repeats from the age at onset of motor signs, we used a phenotype model previously developed through stringent data analysis (Lee et al., 2012b). This model relates natural log-transformed age at onset of motor signs to CAG repeat length using only normally distributed data points. In the current study, the previously established phenotype model (intercept, 7.01; slope, −0.073) was interrogated with CAG repeat lengths of study subjects of CAG lengths 40–55 to obtain individual predicted age at onset of motor signs, which we transformed into natural scale for calculating the difference between predicted and actual age at motor onset, i.e., the residual age at motor onset. For example, a residual age at onset of −10 for a HD subject indicates that the individual developed motor signs 10 years earlier than expected from his or her CAG repeat length. The distribution of residual age at onset of motor signs of study subjects was very similar to a theoretical normal distribution.
GWA Analysis
Quantitative GWA analysis used mixed-effect linear regression analysis, modeling residual age at motor onset as a function of minor allele count of a SNP and gender and ancestry covariates. For extreme dichotomous analysis, we directly compared genome-wide SNP allele frequencies of those individuals whose residual age at motor onset was among the20% extremes, respectively representing earlier and later than expected onset. As no prior statistical model was available for other onset phenotypes, for these we modeled log-transformed age at onset of cognitive signs or psychiatric signs as a function of CAG repeat length, minor allele count of a test SNP, and gender and ancestry covariates in a linear regression analysis framework to determine the extent to which the test SNP explains the amount of variance in phenotype.
Pathway Analysis
The primary pathway analysis was performed using Setscreen (Moskvina et al., 2011), with secondary analyses using ALIGATOR (Holmans et al., 2009), and GSEA (Wang et al., 2007) as described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
GWA signals reveal loci that modify the age at onset of Huntington’s disease
Effects at the chr15 locus hasten or delay onset by 6 or 1.4 years, respectively
A single effect at the chr8 locus hastens onset by 1.6 years
MLH1 association & pathway analysis implicate DNA handling in disease modification
ACKNOWLEDGMENTS
This work was supported by the CHDI Foundation, by grants X01HG006074, U01NS082079, R01NS091161, R01HG002449, and P50NS016367 from the National Institutes of Health (USA), and by grants G0801418 and MR/ L010305/1 from the Medical Research Council (UK). Although not directly related to this work, E.R.D. has received grant support from Auspex Pharmaceuticals and Prana Biotechnology Ltd, and as of May 12, 2014, after completion of his term leading the Huntington Study Group, I.S. became a compensated non-executive director for Prana Biotechnology Ltd.
CONSORTIA
The Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium was organized into the following groups: GeM Group 1: Jong-Min Lee, Vanessa C. Wheeler, Michael J. Chao, Jean Paul G. Vonsattel, Ricardo Mouro Pinto, Diane Lucente, Kawther Abu-Elneel, Eliana Marisa Ramos, Jayalakshmi Srinidhi Mysore, Tammy Gillis, Marcy E. MacDonald, and James F. Gusella; GeM Group 2: Denise Harold, Timothy C. Stone, Valentina Escott-Price, Jun Han, Alexey Vedernikov, Peter Holmans, and Lesley Jones; GeM Group 3: Seung Kwak and Mithra Mahmoudi; GeM Group 4: Michael Orth and G. Bernhard Landwehrmeyer; Registry Investigators: Jane S. Paulsen; PREDICT-HD Investigators: E. Ray Dorsey and Ira Shoulson; COHORT, PHAROS, and TREND-HD Investigators; Richard H. Myers; and HD-MAPS Investigators.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, two figures, five tables, and a full list of GeM-HD Consortium Members and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.07.003.
AUTHOR CONTRIBUTIONS
Conceptualization, J.-M.L., M.E.M., J.F.G., L.J., and S.K.; resources, R.H.M., J.P.G.V., D.L., M.O., M.M., G.B.L., J.S.P., E.R.D., I.S., and the Registry, PREDICT-HD, COHORT, PHAROS, TREND-HD, and HD-MAPS Investigators; investigation, K.A.-E., E.M.R., J.S.M., and T.G.; data curation, J.-M.L., M.J.C., K.A.-E., D.H., M.O., T.C.S., J.H., A.V., L.J., and P.H.; formal analysis, J.-M.L., P.H., D.H., J.H., M.J.C., T.C.S., and V.E.-P.; writing: J.F.G., J.-M.L., M.E.M., V.C.W., R.M.P., D.H., P.H., L.J., S.K., M.O., and R.H.M.; supervision: J.-M.L., M.E.M., J.F.G., G.B.L., M.O., L.J., P.H., and S.K.
REFERENCES
- Antonacci F, Dennis MY, Huddleston J, Sudmant PH, Steinberg KM, Rosenfeld JA, Miroballo M, Graves TA, Vives L, Malig M, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronin N, DiFiglia M. Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov. Disord. 2014;29:1455–1461. doi: 10.1002/mds.26020. [DOI] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chrétien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Brown MB. A method for combining non-independent, one-sided tests of significance. Biometrics. 1975;31:978–992. [Google Scholar]
- Chaudhury I, Stroik DR, Sobeck A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 2014;34:3939–3954. doi: 10.1128/MCB.00457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoussé L, Knowlton B, Hayden M, Almqvist EW, Brinkman R, Ross C, Margolis R, Rosenblatt A, Durr A, Dode C, et al. Interaction of normal and expanded CAG repeat sizes influences age at onset of Huntington disease. Am. J. Med. Genet. A. 2003;119A:279–282. doi: 10.1002/ajmg.a.20190. [DOI] [PubMed] [Google Scholar]
- Dorsey E Huntington Study Group COHORT Investigators. Characterization of a large group of individuals with huntington disease and their relatives enrolled in the COHORT study. PLoS ONE. 2012;7:e29522. doi: 10.1371/journal.pone.0029522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, Wheeler VC. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayán J, Brocklebank D, Andresen JM, Alkorta-Aranburu G, Zameel Cader M, Roberts SA, Cherny SS, Wexler NS, Cardon LR, Housman DE US-Venezuela Collaborative Research Group. Genomewide linkage scan reveals novel loci modifying age of onset of Huntington’s disease in the Venezuelan HD kindreds. Genet. Epidemiol. 2008;32:445–453. doi: 10.1002/gepi.20317. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME, Lee JM. Genetic modifiers of Huntington’s disease. Mov. Disord. 2014;29:1359–1365. doi: 10.1002/mds.26001. [DOI] [PubMed] [Google Scholar]
- Hnia K, Vaccari I, Bolino A, Laporte J. Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol. Med. 2012;18:317–327. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N Wellcome Trust Case-Control Consortium. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington Study Group PHAROS Investigators. At risk for Huntington disease: The PHAROS (Prospective Huntington At Risk Observational Study) cohort enrolled. Arch. Neurol. 2006;63:991–996. doi: 10.1001/archneur.63.7.991. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group TREND-HD Investigators. Randomized controlled trial of ethyl-eicosapentaenoic acid in Huntington disease: the TREND-HD study. Arch. Neurol. 2008;65:1582–1589. doi: 10.1001/archneur.65.12.1582. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I, Xu B, Makarov V, Buxbaum JD, Roos JL, Gogos JA, Karayiorgou M. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc. Natl. Acad. Sci. USA. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Kuo ML, Sy AJ, Xue L, Chi M, Lee MT, Yen T, Chiang MI, Chang L, Chu P, Yen Y. RRM2B suppresses activation of the oxidative stress pathway and is up-regulated by p53 during senescence. Sci Rep. 2012;2:822. doi: 10.1038/srep00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Gillis T, Mysore JS, Ramos EM, Myers RH, Hayden MR, Morrison PJ, Nance M, Ross CA, Margolis RL, et al. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am. J. Hum. Genet. 2012a;90:434–444. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, Warby SC, Morrison P, Nance M, Ross CA, et al. PREDICT-HD study of the Huntington Study Group (HSG); REGISTRY study of the European Huntington’s Disease Network; HD-MAPS Study Group; COHORT study of the HSG. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012b;78:690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Hayden MR, Almqvist EW, Brinkman RR, Durr A, Dodé C, Morrison PJ, Suchowersky O, Ross CA, Margolis RL, et al. A genome scan for modifiers of age at onset in Huntington disease: The HD MAPS study. Am. J. Hum. Genet. 2003;73:682–687. doi: 10.1086/378133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Hayden MR, Warby SC, Durr A, Morrison PJ, Nance M, Ross CA, Margolis RL, Rosenblatt A, Squitieri F, et al. Genome-wide significance for a modifier of age at neurological onset in Huntington’s disease at 6q23–24: the HD MAPS study. BMC Med. Genet. 2006;7:71. doi: 10.1186/1471-2350-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- MacKay C, Déclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V, O’Dushlaine C, Purcell S, Craddock N, Holmans P, O’Donovan MC. Evaluation of an approximation method for assessment of overall significance of multiple-dependent tests in a genomewide association study. Genet. Epidemiol. 2011;35:861–866. doi: 10.1002/gepi.20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z, Lucas JJ. Ubiquitin-proteasome system involvement in Huntington’s disease. Front. Mol. Neurosci. 2014;7:77. doi: 10.3389/fnmol.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth M, Handley OJ, Schwenke C, Dunnett SB, Craufurd D, Ho AK, Wild E, Tabrizi SJ, Landwehrmeyer GB Investigators of the European Huntington’s Disease Network. Observing Huntington’s Disease: the European Huntington’s Disease Network’s REGISTRY. PLoS Curr. 2010;2:RRN1184. doi: 10.1371/currents.RRN1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, et al. Predict-HD Investigators and Coordinators of the Huntington Study Group. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, Panigrahi GB, Hou C, Holloway K, Gillis T, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 2013;9:e1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarin G, Ferraro P, Rampazzo C, Kollberg G, Holme E, Reichard P, Bianchi V. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J. Biol. Chem. 2011;286:11132–11140. doi: 10.1074/jbc.M110.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarin G, Ferraro P, Bee L, Reichard P, Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. USA. 2012;109:13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- Sehgal R, Sheahan K, O’Connell PR, Hanly AM, Martin ST, Winter DC. Lynch syndrome: an updated review. Genes (Basel) 2014;5:497–507. doi: 10.3390/genes5030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swami M, Hendricks AE, Gillis T, Massood T, Mysore J, Myers RH, Wheeler VC. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A Gene Discovery Project of Johns Hopkins & the Autism Consortium. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayán J, et al. US-Venezuela Collaborative Research Project. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. USA. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.