Abstract
We used a transcriptional run-on assay in permeabilized yeast cells to study the distribution of RNA polymerase II (pol II) complexes before and after activation by Gal4. Polymerases were found engaged on the gene at the 5' end before activation, but only appeared at the 3' end after activation. Mutations of the pol II C-terminal domain (CTD), the CTD kinase Kin28 and the holoenzyme subunit Srb2 all inhibited the formation of 3' polymerases in response to activator. However, these mutations did not inhibit the establishment of polymerases at the 5' end. The differences between 3' and 5' ternary complexes suggest that they represent qualitatively distinct 'activated' and 'non-activated' forms of polymerase. The results implicate CTD phosphorylation in a switch from 'non-activated' transcription, which is confined to the 5' end, to an 'activated' mode that traverses the length of the gene.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Ingles C. J. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. M., Ricupero-Hovasse S., Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995 Apr 3;14(7):1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A., Pearlberg J., Simkovich N., Farrell S., Reinagel P., Bamdad C., Sigal G., Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995 May 5;81(3):359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- Bentley D. L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995 Apr;5(2):210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Piña B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992 Jul 24;70(2):251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Blau J., Xiao H., McCracken S., O'Hare P., Greenblatt J., Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996 May;16(5):2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Luse D. S. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J Biol Chem. 1987 Jan 5;262(1):298–304. [PubMed] [Google Scholar]
- Chen J., Ding M., Pederson D. S. Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski M. J., Laff G. M., Solomon M. J., Reed S. I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995 Jun;15(6):2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B. A., Donelson J. E. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol. 1984 Apr;158(1):269–278. doi: 10.1128/jb.158.1.269-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R., Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994 Nov;19(11):504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estojak J., Brent R., Golemis E. A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995 Oct;15(10):5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
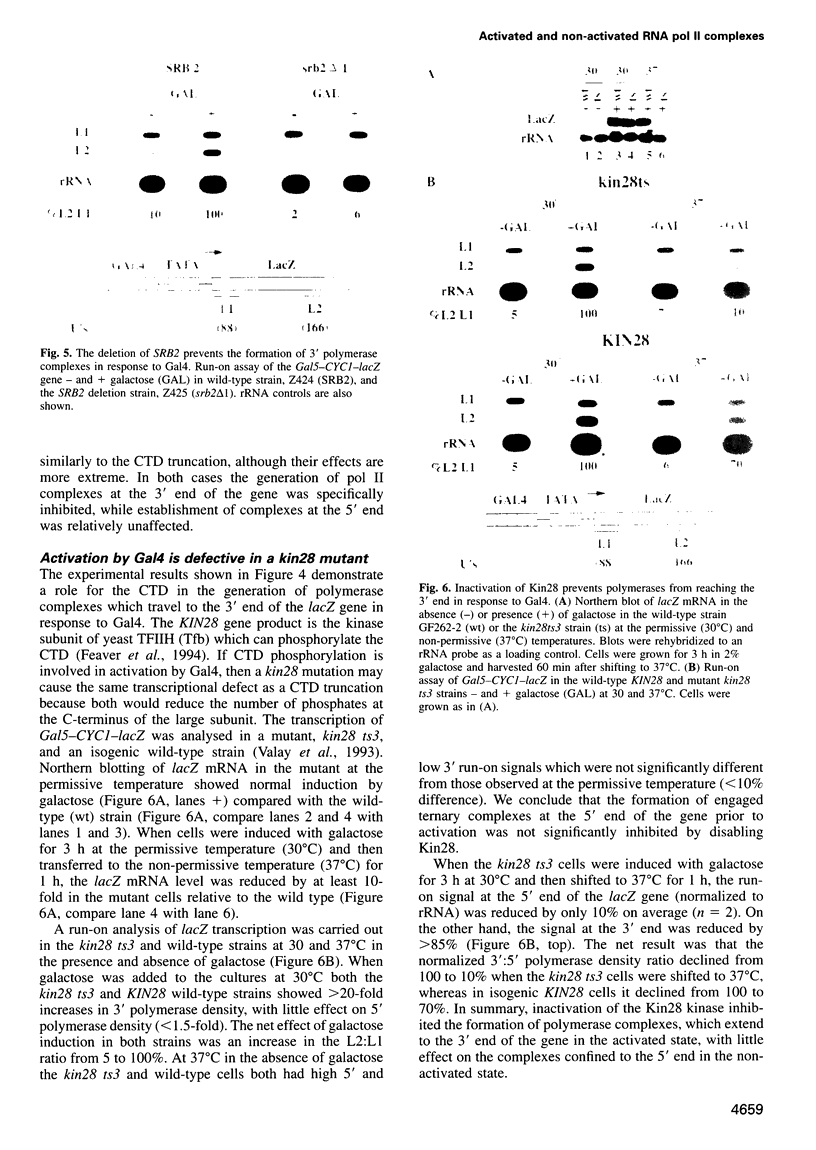
- Feaver W. J., Svejstrup J. Q., Henry N. L., Kornberg R. D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994 Dec 16;79(6):1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Feilotter H. E., Hannon G. J., Ruddell C. J., Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994 Apr 25;22(8):1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gallwitz D., Sures I. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 May;77(5):2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H. P., Hagmann M., Seipel K., Georgiev O., West M. A., Litingtung Y., Schaffner W., Corden J. L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995 Apr 13;374(6523):660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Giardina C., Lis J. T. DNA melting on yeast RNA polymerase II promoters. Science. 1993 Aug 6;261(5122):759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P. A., Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989 Dec;9(12):5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner C. J., Thompson C. M., Zhang J., Chao D. M., Liao S. M., Koleske A. J., Okamura S., Young R. A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995 Apr 15;9(8):897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Eghtedarzadeh M. K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987 Dec;117(4):711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991 Apr;5(4):683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jerome J. F., Jaehning J. A. mRNA transcription in nuclei isolated from Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1633–1639. doi: 10.1128/mcb.6.5.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. W., Stillman D. J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Yan M., Gralla J. D. A three-step pathway of transcription initiation leading to promoter clearance at an activation RNA polymerase II promoter. Mol Cell Biol. 1996 Apr;16(4):1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. E., Dahmus M. E. RNA polymerases IIA and IIO have distinct roles during transcription from the TATA-less murine dihydrofolate reductase promoter. J Biol Chem. 1993 Nov 25;268(33):25033–25040. [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kim T. K., Hashimoto S., Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D., Horikoshi M., Roeder R. G. Effects of activation-defective TBP mutations on transcription initiation in yeast. Nature. 1994 May 19;369(6477):252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994 May 20;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Klein C., Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994 Oct 14;266(5183):280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Buratowski S., Nonet M., Young R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992 May 29;69(5):883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994 Mar 31;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Kruger W., Peterson C. L., Sil A., Coburn C., Arents G., Moudrianakis E. N., Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995 Nov 15;9(22):2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- Krumm A., Hickey L. B., Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995 Mar 1;9(5):559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- Krumm A., Meulia T., Brunvand M., Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992 Nov;6(11):2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. Synergy between HIV-1 Tat and adenovirus E1A is principally due to stabilization of transcriptional elongation. Genes Dev. 1990 Dec;4(12B):2397–2408. doi: 10.1101/gad.4.12b.2397. [DOI] [PubMed] [Google Scholar]
- Liao S. M., Zhang J., Jeffery D. A., Koleske A. J., Thompson C. M., Chao D. M., Viljoen M., van Vuuren H. J., Young R. A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995 Mar 9;374(6518):193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Lu H., Flores O., Weinmann R., Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Welsh T. M., Peterlin B. M. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J Virol. 1993 Apr;67(4):1752–1760. doi: 10.1128/jvi.67.4.1752-1760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä T. P., Parvin J. D., Kim J., Huber L. J., Sharp P. A., Weinberg R. A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci U S A. 1995 May 23;92(11):5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Young R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989 Dec;123(4):715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- O'Brien T., Hardin S., Greenleaf A., Lis J. T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994 Jul 7;370(6484):75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Payne J. M., Laybourn P. J., Dahmus M. E. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989 Nov 25;264(33):19621–19629. [PubMed] [Google Scholar]
- Peterson C. L., Kruger W., Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991 Mar 22;64(6):1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- Roberts S., Bentley D. L. Distinct modes of transcription read through or terminate at the c-myc attenuator. EMBO J. 1992 Mar;11(3):1085–1093. doi: 10.1002/j.1460-2075.1992.tb05147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Scafe C., Chao D., Lopes J., Hirsch J. P., Henry S., Young R. A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990 Oct 4;347(6292):491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Simon M., Seraphin B., Faye G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986 Oct;5(10):2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell L. A., Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995 Jul 7;269(5220):75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- Strobl L. J., Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992 Sep;11(9):3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. M., Koleske A. J., Chao D. M., Young R. A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993 Jul 2;73(7):1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Usheva A., Maldonado E., Goldring A., Lu H., Houbavi C., Reinberg D., Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992 May 29;69(5):871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- Valay J. G., Simon M., Dubois M. F., Bensaude O., Facca C., Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995 Jun 9;249(3):535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Valay J. G., Simon M., Faye G. The kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993 Nov 20;234(2):307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- Weeks J. R., Hardin S. E., Shen J., Lee J. M., Greenleaf A. L. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993 Dec;7(12A):2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- West M. L., Corden J. L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995 Aug;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Friesen J. D., Lis J. T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995 Oct;15(10):5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K., Blau J., Purton T., Roberts S., Bentley D. L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994 Jun 3;77(5):749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Yankulov K., Yamashita K., Roy R., Egly J. M., Bentley D. L. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995 Oct 13;270(41):23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]