Abstract
Viral homologs of the anti-apoptotic Bcl-2 proteins are highly diverged from their mammalian counterparts, yet they perform overlapping functions by binding and inhibiting BH3 motif-containing proteins. We investigated the BH3 binding properties of the herpesvirus Bcl-2 homologs KSBcl-2, BHRF1, and M11, as they relate to those of the human Bcl-2 homologs Mcl-1, Bfl-1, Bcl-w, Bcl-xL, and Bcl-2. Analysis of the sequence and structure of the BH3 binding grooves showed that, despite low sequence identity, M11 has structural similarities to Bcl-xL, Bcl-2, and Bcl-w. BHRF1 and KSBcl-2 are more structurally similar to Mcl-1 than to the other human proteins. Binding to human BH3-like peptides showed that KSBcl-2 has similar specificity to Mcl-1, and BHRF1 has a restricted binding profile; M11 binding preferences are distinct from those of Bcl-xL, Bcl-2 and Bcl-w. Because KSBcl-2 and BHRF1 are from human herpesviruses associated with malignancies, we screened computationally designed BH3 peptide libraries using bacterial surface display to identify selective binders of KSBcl-2 or BHRF1. The resulting peptides bound to KSBcl-2 and BHRF1 in preference to Bfl-1, Bcl-w, Bcl-xL, and Bcl-2, but showed only modest specificity over Mcl-1. Rational mutagenesis increased specificity against Mcl-1, resulting in a peptide with a dissociation constant of 2.9 nM for binding to KSBcl-2 and >1000-fold specificity over human Bcl-2 proteins, and a peptide with >70-fold specificity for BHRF1. In addition to providing new insights into viral Bcl-2 binding specificity, this study will inform future work analyzing the interaction properties of homologous binding domains and designing specific protein interaction partners.
Keywords: KSBcl-2, BHRF1, BH3 peptides, peptide design, bacterial surface display
Introduction
Many proteins function by binding selectively to other proteins. Within homologous families of protein interaction domains, members can have overlapping yet distinct functional specificities that are determined by factors such as expression pattern, subcellular localization, turnover, and intrinsic biochemical properties. The Bcl-2 family of proteins regulates apoptosis using selective interactions between its pro- and anti-apoptotic members. In these regulatory complexes, a helix formed by the BH3 (Bcl-2 homology 3) motif of a pro-apoptotic family member binds into a groove on the surface of a globular anti-apoptotic receptor. In humans, there are five main anti-apoptotic Bcl-2 family receptor proteins: Bcl-xL, Bcl-2, Bcl-w, Mcl-1, and Bfl-1. Three classes of pro-apoptotic Bcl-2 family proteins can engage these receptors via BH3 docking.1,2 First, multi-Bcl-2 homology motif effector proteins Bax and Bak oligomerize in the outer mitochondrial membrane to promote apoptosis; binding of anti-apoptotic proteins to the helical BH3 motifs in Bak or Bax inhibits this process and can block cell death. Second, pro-apoptotic activator BH3-only proteins such as Bid and Bim trigger the oligomerization of Bak and Bax, and this activity is suppressed by anti-apoptotic receptors binding to activator BH3 motifs. Finally, pro-apoptotic BH3-only sensitizer proteins contain BH3 motifs that selectively bind and inhibit subsets of anti-apoptotic receptors.3 Thus, sensitizer proteins promote apoptosis in a manner that depends on the complement of anti-apoptotic proteins expressed in a particular cell. Competitive binding of the three classes of BH3-containing proteins to anti-apoptotic proteins is a key mechanism for regulating apoptosis.4,5
More than 15 viral homologs of anti-apoptotic Bcl-2 proteins have been identified in large double-stranded DNA viruses including adenoviruses, herpesviruses, and poxviruses.6,7 Kaposi’s sarcoma-associated herpesvirus (KSHV or human herpesvirus 8) and Epstein-Barr herpesvirus (EBV or human herpesvirus 4) both express viral Bcl-2 homologs. Current understanding of the function of viral Bcl-2 proteins in infection and virulence is still evolving, and the roles of these proteins likely vary from virus to virus. Both EBV and KSHV latently infect humans, and most carriers present no symptoms. However, both viruses can give rise to malignancies in immunocompromised hosts, such as patients on immunosuppression therapy following organ transplant or AIDS patients. A mutant EBV found in about 15% of Burkitt lymphomas engenders greatly enhanced apoptosis resistance to infected B cells, which was attributed to increased latent cycle expression of the EBV Bcl-2 protein BHRF1.8 This led to the discovery that BHRF1 is expressed at low levels in wild-type EBV-infected, latent Burkitt lymphoma cells and is important in preventing apoptosis triggered by aberrant cell proliferation signals from constitutive c-myc expression.8,9 The function of KSBcl-2, the KSHV viral Bcl-2, in oncogenesis or infection is less clear, but this protein may also act to counteract apoptosis driven by cell proliferation signals, in this case by a viral cyclin.10,11 KSBcl-2 has been shown to be important for the initial stages of lytic reactivation from latent infection.12 The murine gamma herpesvirus γHV68 expresses Bcl-2 homolog M11. Although γHV68 is not associated with cancer in mice, it has been proposed as a useful model system for studying KSHV and EBV, which do not infect mice, and for which cell lines have limited utility as models of viral infection.13 M11 has been shown to be important for persistent replication and virulence during chronic infection.14
Despite playing a similar anti-apoptotic role, the three herpesvirus Bcl-2 proteins are not equivalent. KSHV and γHV68 are gamma-2-herpesviruses, and the synteny of the Bcl-2 genes in these two viruses is similar, suggesting that their viral Bcl-2 proteins KSBcl-2 and M11 may share evolutionary origins.15 However, EBV is a gamma-1-herpesvirus, and its Bcl-2 homolog, BHRF1, is located in a different region in the genome. This suggests that ancestral BHRF1 may have been acquired in a separate horizontal gene transfer event, possibly originating from a different mammalian Bcl-2 homolog than KSBcl-2 and M11.3,15 A key functional difference between the viral Bcl-2 homologs is that although KSBcl-2 and M11 are anti-autophagic, this is not true of BHRF1; EBV upregulates autophagy for non-cell death purposes.6,7,16
Viral Bcl-2 proteins could conceivably function by mimicking a specific mammalian homolog. Alternatively, they could share roles played by several mammalian homologs, or even have distinct functions. The in vivo function of viral Bcl-2 homologs, and how it compares to that of their human counterparts, has not been extensively characterized. But some clues can be gleaned by looking at viral effects on the cell. Herpesvirus gene products can negatively regulate human Bcl-2 and Bcl-xL, suggesting that the viral Bcl-2 homologs may need to compensate for the decreased activity of these human homologs. For example, EBV transcription factor BZLF1 downregulates the cellular protein CD74, resulting in T-cell evasion and decreased expression of Bcl-2 and Bcl-xL in B lymphoblastoid cell lines.8,17,18 An EBV-infected cell line was nevertheless recently shown to be dependent upon Bcl-xL for resistance to apoptosis, but as BHRF1 expression was not detected in this cell line, its role relative to human Bcl-2 homologs remains unclear.8,9,19 In the KSHV-infected cell line Bcbl-1, KSBcl-2 is expressed at low levels and Mcl-1 at high levels. Bcbl-1 cells exhibited a response to a panel of BH3 peptides indicative of a dependence upon both Mcl-1 and KSBcl-2 for protection from apoptosis.10,11,19 KSHV also downregulates Bcl-2 activity by expression of a viral cyclin that directs cellular CDK6 to phosphorylate and inactivate Bcl-2. This may be advantageous for the virus because human Bcl-2 can impair cell cycle progression and be converted into a pro-apoptotic form by caspase cleavage.11,12,20 KSBcl-2 and M11 can also fulfill the anti-autophagic roles of Bcl-2 and Bcl-xL by binding Beclin-1.13,21,22 These findings illustrate that in addition to filling the anti-apoptotic niche, it may be advantageous for herpesviruses to use their Bcl-2 homologs to fulfill additional human Bcl-2 roles (e.g., in autophagy), but not others (e.g. pro-apoptotic and cell cycle regulatory roles). The functional analogies between human and viral Bcl-2 homologs, and how any similarities or differences relate to BH3 binding profiles, remain to be elucidated.
The mechanistic details of protection from apoptosis rely on which pro-apoptotic Bcl-2 family members each anti-apoptotic Bcl-2 homolog binds. The BH3 interaction preferences of the human anti-apoptotic Bcl-2 proteins have been extensively studied, with particular attention focused on the large differences between Bcl-xL and Mcl-1.14,23-28 BH3 motif binding is often tested using peptides ~20 residues in length, here referred to as BH3 peptides. Bim, Bid, and Puma BH3 peptides all bind to the five main anti-apoptotic Bcl-2 proteins, but sensitizer BH3 peptides such as Bad and Noxa are selective for different sets of anti-apoptotic receptors. Notably, Bad binds tightly to Bcl-xL, Bcl-2, and Bcl-w, but not Mcl-1, whereas Noxa preferentially binds Mcl-1.15,29,30 This distinction has long been used to group Bcl-xL, Bcl-2, and Bcl-w into a common specificity class and Mcl-1 into its own class. Bfl-1 is sometimes grouped into a class with Mcl-1, based on not binding to Bad and binding weakly to Noxa, Bik, and Hrk. However, human Bfl-1 does not bind two murine Noxa variants, distinguishing it from Mcl-1, which does bind these proteins.29-31 Viral protein BHRF1 has been shown to have a limited BH3 binding profile, binding only Bim, Bid, and Puma out of a set of 10 mammalian BH3 peptides tested.32 KSBcl-2 and M11 have more permissive binding and exhibit BH3 binding profiles more similar to that of Mcl-1 in that they show moderate binding to Noxa, but only very weak binding to Bad.22,32-34 Further comparison of the binding specificities of viral and human Bcl-2 proteins may shed light on how viral Bcl-2 functions compare to human Bcl-2 functions, as BH3 binding specificity is a crucial determinant of anti-apoptotic Bcl-2 activity.
Studying the binding preferences of viral and human Bcl-2 homologs can also illuminate differences that could be exploited to design specific protein interaction inhibitors. Specific inhibitors of the viral Bcl-2 homologs would be useful for basic research and potentially for therapy. Early peptidic and small molecule inhibitors targeted at human Bcl-xL achieved inter-class specificity in that they showed no binding to Mcl-1, but did not distinguish between members of the Bcl-xL, Bcl-2, and Bcl-w class.23,35 Targeting Bcl-2 and Bcl-xL in cancers with Navitoclax, the clinical form of the small-molecule inhibitor ABT-737, led to thrombocytopenia due to the fact that platelets are dependent upon Bcl-xL for protection from apoptosis.36,37 A Bcl-2-specific small molecule, ABT-199, is achieving better success in clinical trials due to reduced off-target effects.38 This highlights the importance of specific targeting of Bcl-2 homologs for cancer therapy. Significant progress has been made identifying peptide inhibitors of individual human Bcl-2 family members including Mcl-1, Bcl-xL and Bfl-1.25,39,40 For viral Bcl-2 homologs, a computationally designed protein, BINDI, binds BHRF1 with picomolar affinity and >180-fold specificity over the human Bcl-2 homologs.41 BINDI is a 14 kDa protein that incorporates a BH3 helix but gains much of its specificity from contacts outside of the BH3 binding groove. When attached to an antibody-targeted intracellular delivery carrier, BINDI reduced tumor growth in xenograft mouse models of EBV-positive human lymphoma, supporting BHRF1 as a candidate therapeutic target. A peptide variant of the BH3 motif of Beclin-1 was recently engineered that binds selectively to M11 over Bcl-xL, but the peptide had weak affinity for M11 (KD = 6.4 μM).42.
In this paper we report comparisons of viral and human Bcl-2 homologs at the levels of sequence, structure, and binding similarity in order to identify which homologs are most similar. Drawing on experimental data and structural models, we then interrogated the boundaries of this similarity by designing BH3 peptide libraries and screening them for selective binders of KSBcl-2 and BHRF1. Analysis of the peptides identified in the screen shed light on sequence and structural determinants of the BH3 binding preferences of viral and human homologs. Further mutation of library peptides provided molecules with >1000-fold specificity for binding KSBcl-2 and >70-fold specificity for binding BHRF1 over all of the human Bcl-2 homologs. These peptides could serve as reagents to probe and inhibit the function of viral Bcl-2 homologs in viral pathogenesis.
Results
Comparison of eight Bcl-2 homologs based on sequence identity, structure, and binding preferences
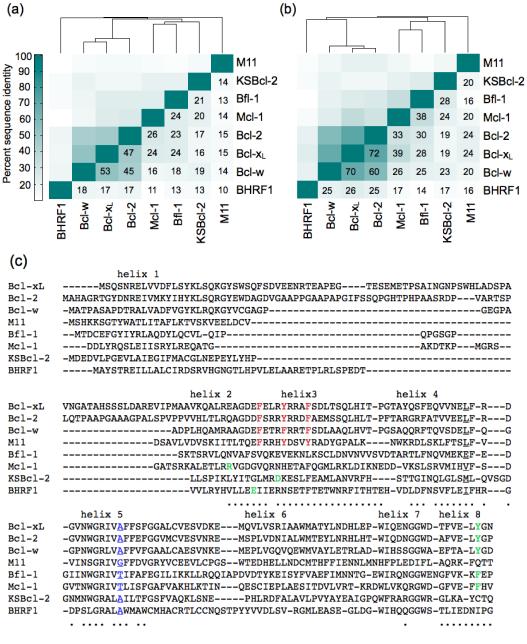
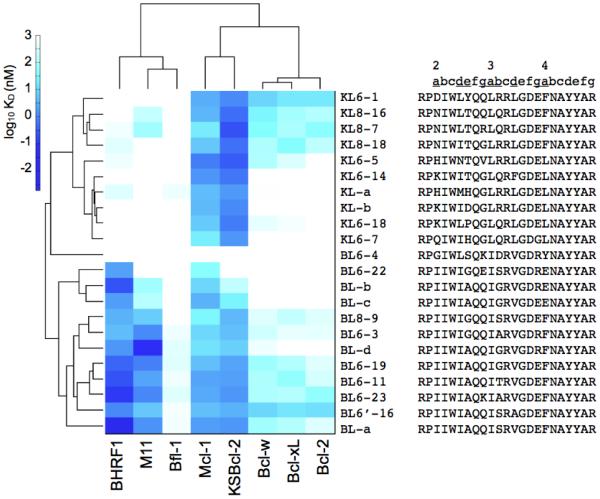
Protein sequence identity is often used to infer functional similarity. We compared the sequences of 8 anti-apoptotic Bcl-2 family homologs across the entire Bcl-2 domain (without the C-terminal trans-membrane helix or the N-terminal PEST domain of Mcl-1) (Fig. 1). The viral Bcl-2 homologs KSBcl-2, BHRF1, and M11 have very low sequence identity to the human homologs and to each other (10-21%, Fig. 1a). To assess the similarity of receptors in the regions that most directly influence BH3 helix binding, we computed the sequence identity over residues that line the BH3 binding groove (Fig. 1b). Residues that were included are within 7 Å of a peptide residue in at least one BH3-bound structure, as described in the Materials and Methods. Sequence identity in the binding groove is higher than in the overall Bcl-2 domain, and Bcl-2, Bcl-xL and Bcl-w form a subgroup sharing high sequence identity in this region (≥ 60%). Human homologs Bfl-1 and Mcl-1 share at most 30% and 39% binding-groove identity with the Bcl-xL/2/w set, respectively, and share 38% identity with each other. The human Bcl-2 homologs with highest sequence identity to the viral Bcl-2 proteins, in the binding groove, are Bfl-1 and Mcl-1 for KSBcl-2 (28%, and 24% identity, respectively), Bcl-xL/2/w for BHRF1 (25-26% identity), and Bcl-xL and Bcl-2 for M11 (24% identity). Thus, the similarities between Mcl-1, Bfl-1, and the viral homologs are much less significant than those between Bcl-xL, Bcl-2, and Bcl-w, although Mcl-1, Bfl-1 and KSBcl-2 do cluster together at a lower similarity threshold.
Figure 1.
Sequence comparison of 5 human and 3 viral Bcl-2 homologs. (a) Percent sequence identity over the entire Bcl-2 domain (without the C-terminal trans-membrane helices or the N-terminal PEST domain of Mcl-1). (b) Percent sequence identity for residues in the BH3 binding groove (see Materials and Methods). (c) Sequence alignment used for the sequence identity calculations. Dots under residues denote residues in the BH3 binding groove. Motifs discussed in the Results section are colored and/or underlined.
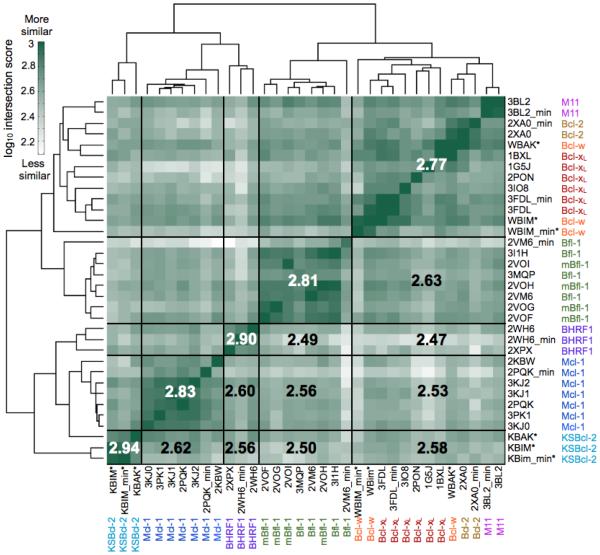
Abundant structural data exist for complexes of anti-apoptotic Bcl-2 proteins bound to short BH3 motif peptides; of the proteins discussed here, only KSBcl-2 lacks a structure bound to a BH3 peptide. The overall helical architecture is conserved between all of the anti-apoptotic Bcl-2 homologs, as illustrated in the structure alignment shown in Supplementary Fig. 1. The Cα RMSD varies from ~0.8-3.2 Å between structures of different Bcl-2 homologs. To examine the chemical similarity of the different binding grooves, we performed analyses using SiteMAP on receptor-peptide complex structures or homology models for all eight Bcl-2 homologs. SiteMAP, originally designed for identifying small molecule binding sites, creates maps of hydrophobic, hydrogen-bond donor, and hydrogen-bond acceptor binding potential proximal to the protein surface.43,44 Our process for defining a similarity score based on the intersection of SiteMAP maps from different receptors is summarized in Supplementary Fig. 1 and explained in greater detail in the Materials and Methods. A summary of the similarities between all pairs of maps, computed as the sum of the intersections of physicochemical property maps for pairs of proteins, is shown in Fig. 2, where protein structures are clustered according to their intersection score profiles. Where available, we ran the SiteMAP analysis on multiple structures for each Bcl-2 homolog, including structures of the receptor with different BH3 peptides bound. Notably, different structures of the same receptor bound to different peptides clustered together, showing that this analysis is robust to small changes in conformation. To further test dependence on small changes in Bcl-2 conformation, we performed a restrained minimization without the peptide present for one structure of each homolog before running SiteMAP. The minimized structure grouped with the other structures of that homolog, though in some cases more distantly.
Figure 2.
Comparison of the physicochemical characteristics of human and viral Bcl-2 homolog BH3 binding groove structures using SiteMAP. The intersection score is a measure of similarity that takes into account the hydrophobic and hydrogen-bond donor/acceptor characteristics of regions of the binding site (see Materials and Methods for details of the metric). The structures of eight Bcl-2 homologs were clustered according to the similarity of their intersection score profiles. Boxed regions partition receptors or groups of receptors; numbers are the average log10(intersection score) over the indicated box and its symmetry-related box across the diagonal. PDB IDs are given, with the receptor name next to them. All mammalian receptors are the human homologs except for several murine Bfl-1 structures (“mBfl-1”). The suffix “min” denotes a structure relaxed without the peptide bound. Asterisks denote homology models.
Consistent with the sequence-based analysis, SiteMAP results show that Bcl-xL, Bcl-2, and Bcl-w form a tight cluster. M11 also joins this group, despite having much lower sequence identity with these proteins than they share with each other. Viral proteins KSBcl-2 and BHRF1 clearly cluster with Mcl-1. But surprisingly, given its higher binding-groove sequence identity to Mcl-1 than to Bcl-xL/2/w, Bfl-1 clusters more closely to the Bcl-xL/2/w/M11 group. Averaged similarity scores (shown in bold for boxed sets of structures in Fig. 2) show that Mcl-1 and KSBcl-2 are most alike in binding-groove chemical structure, despite the fact that Mcl-1 is more similar to Bcl-xL and Bfl-1 than to KSBcl-2 by sequence identity. The KSBcl-2 and Bcl-w maps are based on homology models, which are heavily influenced by the homologs with which they share the greatest sequence similarity, which are Bfl-1 and Mcl-1 for KSBcl-2 and Bcl-xL for Bcl-w. Therefore, the high similarity between these structures may be somewhat artificial, but it is consistent with other trends reported in this paper. While this manuscript was in preparation, a structure of a mutant Bcl-w bound to a peptide corresponding to its own (mutated) BH3 domain was published.45 The Cα RMSD over helices 2-8 between our Bcl-w:Bak BH3 homology model and this Bcl-w:Bcl-w BH3 structure is 1.2 Å, validating our Bcl-w model.
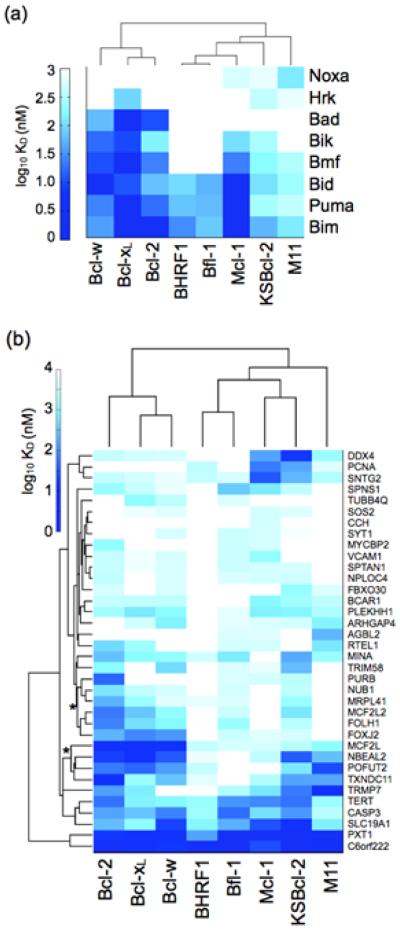
Binding of the viral and human Bcl-2 homologs to a small set of functionally validated human BH3 peptides has been previously reported and is summarized in Fig. 3a.22,30,32 Recently, DeBartolo et al. reported dissociation constants for the five human Bcl-2 homologs binding to 36 new, computationally identified candidate BH3 peptides from the human proteome.46 In Fig. 3b, we compare the binding patterns of the three viral Bcl-2 homologs to those of the human homologs for these 36 BH3-like peptides. The dissociation constants were determined by fluorescence anisotropy binding experiments. Hierarchical clustering of KD values by Bcl-2 receptor shows similar trends for binding of the previously reported and functionally validated vs. new BH3 peptides. Bcl-xL, Bcl-2, and Bcl-w, which have high sequence identity and structural similarity with each other, cluster together as expected. BHRF1 and Bfl-1 are grouped together based on binding tightly to only a small number of peptides. KSBcl-2 and Mcl-1 cluster together in Fig. 3b and are in the same group in Fig. 3a, although there are a few notable differences in the binding profiles of these two proteins. In Fig. 3b, peptides in the clusters marked with asterisks exhibit moderate-to-tight binding to KSBcl-2 and also to Bcl-xL/2/w, but weaker binding to Mcl-1. We return to this point below.
Figure 3.
Comparison of BH3 peptide binding profiles for eight Bcl-2 homologs. (a) Interactions with functionally validated BH3 peptides, using data from the literature. (b) Interactions with 36 BH3-like peptides identified from the human proteome. Receptors are clustered based on correlation of their binding profiles, and peptides are clustered by Euclidean distance. Dissociation constants for the human proteins in panel B were taken from DeBartolo et al. Asterisks denote peptide clusters with tighter binding to KSBcl-2 than to Mcl-1. The heat map indicates affinity measured by fluorescence anisotropy as log10 (KD in nM), with white indicating no detectable binding up to 3000 nM. KD values with 95% confidence intervals are given in Supplementary Table 1 for the 36 BH3-like peptides.
Natural BH3 sequences have high variability, which makes it difficult to parse the determinants that underlie the observed binding patterns. To compare binding preferences in a more interpretable sequence space, we performed Bim BH3 substitution SPOT array analysis for KSBcl-2 and BHRF1. The SPOT array assay is used to test binding to hundreds of membrane-immobilized peptides in parallel, and has proven useful in published analyses of Bcl-2 family binding.47-49 Our viral Bcl-2 SPOT arrays included peptides with 18 individual point mutations (excluding cysteine and methionine) at a set of 10 positions in Bim BH3 that were previously analyzed for human Bcl-2 protein binding (Fig. 4). We refer to BH3 peptide positions using a repeating-heptad nomenclature, with positions labeled a-g, which is shown for the full Bim BH3 sequence in Fig. 5c. The viral Bcl-2 arrays additionally included peptides with mutations in 3 positions in the N-terminal region of Bim, and 2 positions in the C-terminal region. However, varying these terminal positions gave only small changes in binding signal in this assay, not readily distinguished from noise; we do not interpret observed differences at these positions here.
Figure 4.

Bim BH3 and point-mutant peptides on SPOT arrays binding to 100 nM BHRF1 or KSBcl-2. Each row is labeled with the wild-type Bim residue and the position that was varied, and columns are labeled with the substitution. The column labeled “X” included peptides with the following BH3 sequences, from top to bottom: Bad, Bid, Bmf, Hrk, Noxa, Bik, Bak, Bax, Bcl-xL, Bcl-w, Bcl-2, Mcl-1, Bfl-1, BHRF1, KSBcl-2.
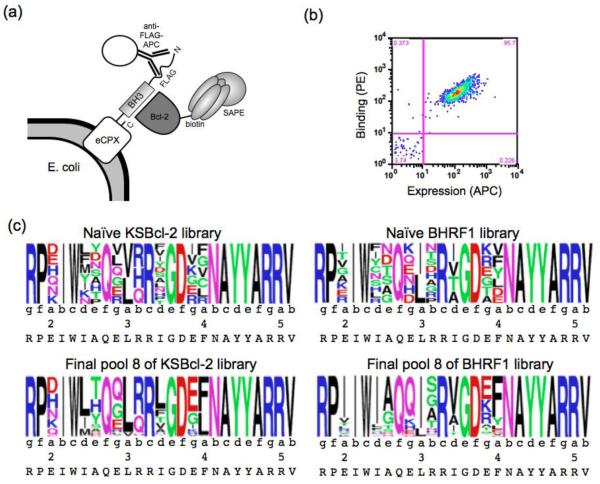
Figure 5.
Bacterial surface display screen for selective binders of KSBcl-2 and BHRF1. (a) The libraries were displayed on the N-terminus of eCPX. A FLAG tag was displayed N-terminal to the peptide, and peptide expression was detected by an anti-FLAG antibody conjugated to APC. Streptavidin-phycoerythrin (SAPE) was used to detect binding of biotinylated Bcl-2 proteins. (b) Representative FACS plot of wild-type Bim BH3 displayed on the surface of E. coli binding to 5 nM biotinylated KSBcl-2. Binding, as reported by PE fluorescence, is plotted as a function of expression reported by APC fluorescence. The lower-left quadrant includes non-expressing cells. (c) equence logo built from deep sequencing of the naïve and eighth (final) library pools, using unique sequences. Heptad positions are indicated, and the wild-type Bim residue is given at each position below the logo. Pool 8 sequences included in these logos were filtered to include only those sequences also present in pools 6 and 7.
The peptide binding similarity of different Bcl-2 proteins can be assessed using the correlation of SPOT signals between receptors for a common set of 180 Bim BH3 variants tested for five human and two viral proteins. Previous analyses showed that of the five human proteins tested by SPOT analysis, only the Bcl-xL/w/2 arrays showed a high correlation of binding patterns.48 The correlation of the KSBcl-2 SPOT array signals with SPOT data for most human homologs was moderate (Pearson R = 0.71-0.80), with Bcl-xL having slightly lower similarity (R = 0.66). The BHRF1 SPOT array data were moderately correlated with data for KSBcl-2 and Mcl-1 (R = 0.75 and 0.71, respectively), but correlations with the other homologs were lower. Notably, despite exhibiting a similarly restrictive binding profile for natural BH3 peptides, Bfl-1 and BHRF1 show relatively low correlation between their SPOT array signals (R = 0.66), suggesting they arrive at their restrictive binding profiles through different mechanisms. Differences in positional preferences that can be dissected with the use of the SPOT data are discussed below.
Peptide libraries targeting KSBcl-2 and BHRF1 specificity
Peptide design using library screening has repeatedly led to molecules that discriminate between binding to Bcl-xL/2/w vs. Mcl-1 or Bfl-1 by large margins.23,26,28 It has been more difficult to design specificity between members of more closely homologous groups (e.g., Bcl-xL vs. Bcl-2/w or Bfl-1 vs. Mcl-1).39,40 Above, we showed that KSBcl-2 and BHRF1 have BH3 binding patterns most similar to those of Mcl-1 and Bfl-1, respectively. We interrogated the boundaries of this similarity by designing and screening Bim BH3-based libraries for peptides that could bind KSBcl-2 or BHRF1 selectively (Fig. 5). To increase our chances of success, we used experimental binding data and structure-based models of BH3 peptide binding preferences to design libraries enriched in mutations predicted to provide specificity for KSBcl-2 or BHRF1. The libraries were based on mutating the Bim BH3 motif, for which we have rich binding data from prior work. Specifically, we designed the libraries using mutational data from SPOT arrays and predictions made using STATIUM, a statistical potential that can evaluate mutations based on analysis of human or viral Bcl-2 crystal structures or homology models.46,48 We also used Illumina sequencing data from a yeast surface display library of Bim BH3 variants previously screened for KSBcl-2 binding.25 The computationally assisted library design process (described in further detail in the Materials and Methods) focused on including residues that were tolerated by the viral Bcl-2 proteins and that weakened binding to Mcl-1 and, for the BHRF1 library, Bfl-1. Despite moderate sequence identity between KSBcl-2 and Bfl-1, we did not consider Bfl-1 to be a competitor difficult to discriminate against, because we have previously discovered mutations that favor KSBcl-2 over Bfl-1 binding, and we included these in the library.25 We also prioritized inclusion of a few mutations known to disfavor Bcl-xL, Bcl-2, and Bcl-w binding (e.g. non-aromatic residues at position 4a). 23,24
Nine positions in Bim were mutated, and the theoretical libraries designed using our optimization protocol are shown in Fig. 5c. The heptad register of Bim is given below each position. We chose to vary the four conserved hydrophobic positions 2d, 3a, 3d, and 4a, because the Bcl-2 homologs show strong, differential preferences for mutations at these positions.23,50 We also included variation at position 2a, because the structural environments around this position in the viral receptors exhibit differences from the environment in Mcl-1. Specifically, Mcl-1 has an arginine that forms a salt bridge with glutamate at 2a in Bim that is lacking in the viral receptors.24 The four remaining positions chosen for design, 2e, 2g, 3b, and 3g, are generally occupied by small or polar/charged residues in native BH3 sequences, as they are on the sides of the BH3 helix and are more solvent exposed. The library designs are presented, along with an indication of which residues were included for affinity or specificity, in Supplementary Tables 2 and 3.
The eCPX E. coli surface display system has been used previously to display and sort BH3 peptide libraries.26,51 We optimized it for affinity-based sorting by adding a FLAG-tag N-terminal to the BH3 peptide that can be used to detect the expression level of each library member (Fig. 5a). A representative fluorescence activated cell sorting (FACS) plot illustrating the correlation between peptide cell-surface expression and binding to KSBcl-2 for a single clone is shown in Fig. 5b. We sorted the designed libraries by FACS in a series of positive, negative, and competition sorts to identify high affinity and selective binders of the viral Bcl-2 proteins, as illustrated in Supplementary Fig. 2 and detailed in the Materials and Methods. Human homologs Mcl-1, Bfl-1, and Bcl-w were used as competitors in the KSBcl-2 library sorting, and Mcl-1 and Bfl-1 were used for the BHRF1 library. Pools of clones isolated from rounds of sorting are designated KL1-KL8 for KSBcl-2 and BL1-BL8 or BL5’-BL8’ for BHRF1 (see Methods and Supplementary Figure 2). FACS analysis was performed on the naïve and final pools for both libraries (Supplementary Fig. 3). This analysis showed that although the naïve libraries contained more Mcl-1 binders than viral Bcl-2 binders, the final sorted library pools consisted of tight viral Bcl-2 binders with modest margins of specificity against Mcl-1, and larger margins of specificity against Bfl-1, Bcl-w, and Bcl-2.
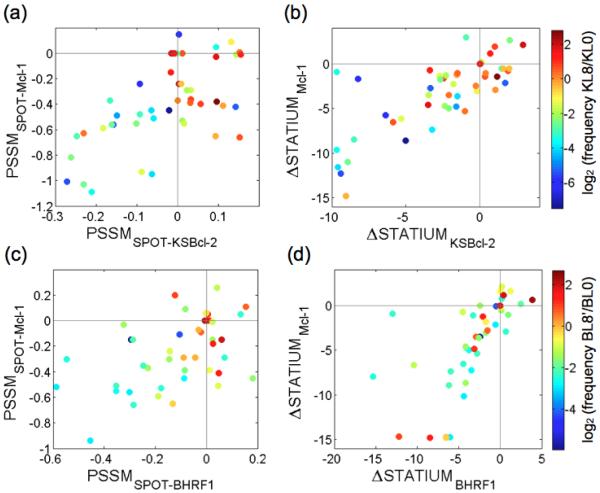
To follow the progress of sorting and to isolate clones for further analysis, we sequenced clones from later library pools. From the KL6 and KL8 library pools, we sequenced 24 and 20 clones, respectively, and found a total of 16 unique sequences. Among four BHRF1 library pools from two different sorting schemes (see Materials and Methods), we obtained 32 unique sequences. Peptide sequences and the frequencies with which they were found in the library pools are given in Supplementary Table 4. To provide more sequences for analysis of library design success and mutation enrichment trends, the naïve libraries (KL0 and BL0) and pools 1, 3, 5, 6, 7, and 8 were sequenced using Illumina technology for each library. The sequence filtering process and number of reads for each pool are described in the Materials and Methods and in Supplementary Table 5. Sequence data show that the library screens resulted in distinct residue preferences at select positions, as shown by logos constructed from unique, filtered sequences in the eighth (final) pools (Fig. 5c). Many of the trends observed support the predictions of the SPOT arrays (PSSMSPOT models) and the STATIUM models. For example, at position 2a, the wild-type glutamate residue was selectively lost from the KSBcl-2 library, while the other five possible mutations were maintained. And in the BHRF1 library, isoleucine was enriched at 2a. Position 2a was not mutated on the SPOT arrays, but the STATIUM models predicted that most mutations from glutamate at 2a – including isoleucine – would provide specificity for binding either of the viral Bcl-2 receptors over Mcl-1, and our library results validate the STATIUM predictions. In the BHRF1 library screen, there was a strong preference for isoleucine over leucine at 2d and 3a. The SPOT arrays indicated that BHRF1 strongly prefers the wild-type isoleucine at 2d, and can tolerate both isoleucine and leucine at 3a. The preference for isoleucine over leucine at 3a in the BHRF1 library results (Fig. 5c) may arise in part because this substitution provides specificity over some of the human receptors, as described in a later section. At peptide positions along the sides of the BH3 binding groove, such as 2e and 3b, the SPOT arrays also accurately reflected preferences. For example, the SPOT arrays indicated that KSBcl-2 is more tolerant of a variety of residues at 2e than are Bcl-xL, Bcl-2, and Bfl-1, whereas BHRF1 has greater tolerance at 3b than this same group of receptors, and this was reflected in the library results.
To evaluate the overall agreement between predictions made by the models used in our library design protocol and the library enrichment results, we compared the model scores for all point mutations with the enrichment of individual mutations during screening (Fig. 6). The SPOT arrays were converted to position-specific scoring matrices (PSSMs) by taking the logarithm of the mutant intensity divided by the average wild-type intensity (giving PSSMSPOT models).23,48 Enrichment was measured as log2(frequency of mutation in unique sequences in final pools KL8 or BL8’/frequency in the naïve pools KL0 or BL0). Enrichment is indicated by color in Fig. 6, with enriched mutations in shades of orange to red. Scores for the wild-type Bim residues are indicated by gray lines on the plots in Fig. 6, such that points that fall in the lower right quadrant of each plot correspond to point mutations predicted by the models to provide enhanced affinity for the target viral Bcl-2 and specificity against Mcl-1. Many of the most enriched mutations in each library fall into this quadrant for the PSSMSPOT models (Fig. 6a and 6c). For example, 83% of the enriched mutations for KSBcl-2 (log2(frequency in final pool/frequency in naïve pool) > 0) fall in this quadrant, as do 58% of the enriched mutations for BHRF1. The STATIUM model predictions did not agree as well with the screening results, but enriched residues had better viral protein binding scores than non-enriched residues: 86% of the mutations with log2(frequency KL8/KL0) > 0 had STATIUMKSBcl-2 scores > −3, whereas only 59% of un-enriched residues had scores this high. For BHRF1, 73% of mutations with log2(frequency BL8’/BL0) > 0 had STATIUMBHRF1 scores > −3, compared to 56% of un-enriched residues. The high correlation of STATIUM scores for KSBcl-2 or BHRF1 binding with scores for Mcl-1 binding reflects the predicted difficulty of distinguishing interactions between the viral proteins and Mcl-1. This is also seen, to a lesser degree, in the SPOT results and is consistent with our experimental observations.
Figure 6.
Enrichment of residues in library sequences versus viral Bcl-2 and Mcl-1 model scores. Each point represents a residue and is colored according to enrichment (log2 (frequency in final pool/frequency in naïve pool)). The frequencies were calculated from unique sequences filtered as described in the Materials and Methods. Residues in the KSBcl-2 library (a, b) and BHRF1 library (c, d) are plotted by (a, c) PSSMSPOT scores and (b, d) ΔSTATIUM scores. For all models, a score of zero is given for the wild-type residue and scores greater than zero indicate tighter binding than wild type.
Binding of library-derived peptides to 8 Bcl-2 homologs
We selected 22 peptides that sample a variety of sequence features to test in direct fluorescence anisotropy binding experiments. Peptide names designate the library (‘KL’ or ‘BL’) followed by the sorting pool from which they were sequenced, followed by an arbitrary numerical designation. References to KL and BL peptides refer specifically to the peptides tested in solution binding experiments. The peptide names with alphabetical designations were chosen from the deep sequencing results, either because they represented a consensus sequence or because they contained enriched substitutions not sampled in the conventionally sequenced clones. The 22 peptides were tested for binding to all 5 human and 3 viral receptors.
A heat map of the KD values is shown in Fig. 7, clustered by receptor binding profile similarity (KD values with 95% confidence intervals are given in Supplementary Table 6). The binding patterns of peptides identified from the two libraries have a clear distinction. Most of the peptides identified by screening for binding to BHRF1 also bound to KSBcl-2, M11 and Mcl-1. In contrast, peptides identified on the basis of binding to KSBcl-2 only bound strongly to KSBcl-2 and Mcl-1. Despite our use of competitive screening against Mcl-1, it is notable that all of the peptides that bound to KSBcl-2 or BHRF1 also bound tightly to Mcl-1. Binding to Bcl-2, Bcl-xL, and Bcl-w was substantially weaker for all of the peptides, consistent with the distinct sequence and structural properties of these receptors. Binding to Bfl-1 was universally very weak for peptides from both libraries, although Bfl-1 bound tightly to positive control peptides. This trend is especially interesting given the similarity between the native BH3 binding profiles of BHRF1 and Bfl-1.
Figure 7.
Binding of human and viral Bcl-2 homologs to peptides identified from library screening. KD values, measured by fluorescence anisotropy, here plotted as log10 (KD in nM), are given with 95% confidence intervals in Supplementary Table 6. White indicates no binding up to 1000 nM. Library peptide sequences are shown to the right, with the heptad register indicated above and with varied positions underlined. Bcl-2 protein receptors (in columns) were clustered by the correlation of their binding profiles and peptides (in rows) were clustered by Euclidean distance.
Specificity mechanisms underlying receptor binding similarity patterns
We analyzed the binding patterns observed for different Bcl-2 proteins by integrating data from the SPOT arrays, the SiteMAP structural analysis and solution binding studies of peptides and selected point mutants. Our analyses suggested mechanisms by which the library peptides achieve specificity. These mechanisms will be discussed on three levels. First, we discuss peptide sequence features that distinguish binding to the three viral homologs KSBcl-2, BHRF1, and M11. Second, we address sequence and structural features that differentiate peptides that bind to the KSBcl-2/Mcl-1/BHRF1 vs. Bcl-xL/2/w/Bfl-1 groups. Finally, we discuss mutations that contribute to the modest level of specificity achieved by some peptides for binding to the viral Bcl-2 proteins over Mcl-1.
To investigate the origins of binding differences between the viral Bcl-2 homologs, we tested residue substitutions in Bim BH3 that were common in the peptides identified from the libraries. Threonine was enriched at position 2e in the KSBcl-2 library (Fig. 5c), and the specificity of Bim_A2eT for KSBcl-2 and Mcl-1 has been addressed in prior work.25 Our nomenclature for mutants lists the parent peptide (Bim) followed by the wild-type residue that was mutated (A), the heptad position (2e) and the new residue identity at that site (T). The alanine-to-threonine mutation in Bim_A2eT weakens binding to Bfl-1, Bcl-xL, Bcl-2 and Bcl-w.25 Here, we show that this mutation also weakens BHRF1 and M11 binding (Table 1). Another peptide position that can favor KSBcl-2 binding is 2g. In contrast to the BHRF1-binding peptides in Fig. 6, which have glutamine, lysine or glutamate at position 2g, many of the KSBcl-2-binding peptides that we analyzed have a glycine at this site. BHRF1 binding is weakened dramatically by the two mutations in Bim_A2eT_E2gG, and 60% of the KL peptides tested combine the E2gG mutation with a larger residue (proline, threonine, aspartate, or histidine) at 2e. This likely explains why BHRF1 shows limited binding to many of the KSBcl-2 library peptides. As for BHRF1 specificity determinants, serine at position 3b is common in the BHRF1 library peptides. Bim_R3bS binds tightly to BHRF1 but the serine mutation weakens binding to Bfl-1, Bcl-w, Bcl-2, and M11 by ~4-9-fold when introduced into Bim BH3 (Table 1), providing a partial explanation of the BHRF1 library peptide specificity. Notably, many of the BHRF1 library peptides bind more tightly to M11 than does wild-type Bim BH3, suggesting that even though R3bS weakens M11 binding, other mutations present in the BL peptides must compensate for this effect.
Table 1.
Dissociation constants for Bcl-2 homologs binding to mutants of Bim BH3.
| KD (nM) a |
||||
|---|---|---|---|---|
| Bim | Bim_R3bS | Bim_A2eT | Bim_A2eT_E2gG | |
|
|
||||
| BHRF1 | 1.2 ± 0.74 | ~1.2b | 64 ± 37 | >1000c |
| KSBcl-2 | 1.2 ± 0.79 | 1.5 ± 0.57 | 1.8 ± 0.81 | 1.1 ± 0.49 |
| Mcl-1 | <1 c | <1 c | 0.62 ± 0.55 | 1.6 ± 0.57 |
| Bfl-1 | <1 c | 5.2 ± 1.7 | 32 ± 6.9 | 260 ± 64 |
| Bcl-w | 2.6 ± 2.1 | 18 ± 5.3 | 40 ± 9.3 | 98 ± 35 |
| Bcl-xL | 2.6 ± 1.9 | 3.8 ± 1.4 | 18 ± 4.9 | 42 ± 7.1 |
| Bcl-2 | 1.9 ± 1.3 | 7.2 ± 3.1 | 44 ± 3.1 | 150 ± 27 |
| M11 | 23 ± 10 | 210 ± 160 | 78 ± 19 | 15 ± 5.3 |
Values are given ± 95% confidence intervals.
An approximate value is given where the confidence interval was very large, which was a problem for some BHRF1 curves that had a low dynamic range for the anisotropy signal.
Values designated >1000 or <1 nM were too weak or tight to quantify using this assay.
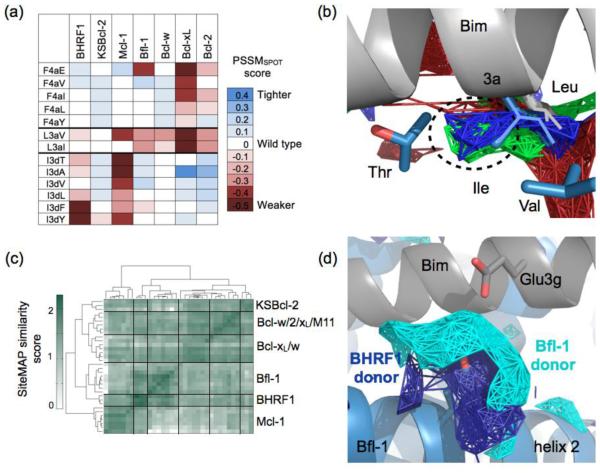
The four BH3 positions that are conserved as hydrophobic (2d, 3a, 3d and 4a) have been demonstrated to make strong contributions to human Bcl-2 specificity patterns.23,24,27,50 Here, we observed that these positions are important for differentiating binding to KSBcl-2/Mcl-1/BHRF1 vs. Bcl-xL/2/w/Bfl-1. The SPOT arrays indicated that viral proteins BHRF1 and KSBcl-2, and human proteins Mcl-1 and Bcl-w, are broadly tolerant of substitutions at position 4a (Fig. 8a). We observed the same trend in library screening. Several of the tested KL peptides had an F4aL substitution, and an F4aE mutation was found in a minority of the BL peptides. Notably, Bcl-xL and Bcl-2 showed no binding to these peptides (Fig. 7), in keeping with the observation made in several past studies that Bcl-xL and Bcl-2 have a more enclosed 4a pocket that favors aromatics of complementary size and shape.23-25,52 Human Mcl-1 and viral KSBcl-2, M11, and BHRF1 appear to have similar preferences at hydrophobic position 3a. The BHRF1 library peptides showed a strong preference for isoleucine at position 3a, where most natural BH3 peptides have a leucine. The Bim substitution SPOT arrays indicate that BHRF1, KSBcl-2 and Mcl-1 exhibit no loss in affinity for the L3aI mutation, whereas Bfl-1, Bcl-xL, Bcl-2, and Bcl-w binding is weakened (Fig. 8a). The shape of the 3a pocket is influenced by two positions in helices 4 and 5 that are underlined in Fig. 1c. KSBcl-2, Mcl-1 and BHRF1 have either a β-branched isoleucine or valine, or a long but flexible methionine on helix 4, where the other receptors have glutamate or leucine, which leave less space for a branched Cβ at peptide position 3a. The extra space for a branched Cβ can be seen in KSBcl-2, BHRF1, and Mcl-1 hydrophobic SiteMAPs (Fig. 8b). M11 also has a leucine on helix 4, but has a unique glycine on helix 5, which may allow more space for a branched Cβ than is provided by the alanine or threonine found in the other receptors. Thus, preferences at two of the conserved hydrophobic positions, 4a and 3a, distinguish the binding properties of the library peptides, with KSBcl-2, BHRF1, Mcl-1, and M11 exhibiting broader tolerance for residue substitution than Bfl-1, Bcl-xL, and Bcl-2.
Figure 8.
Specificity mechanisms employed by library peptides. (a) PSSMSPOT scores for substitutions at Bim peptide positions L3a, I3d, and F4a. (b) SiteMAP hydrophobic density near position 3a shows extra density (circled) for KSBcl-2, BHRF1, and Mcl-1 (red, green, and blue, respectively). Bim with leucine at position 3a (2PQK)24 is in gray, and isoleucine at 3a from an Mcl-1-specific peptide (3KZ0)23 is shown in blue. The threonine on helix 5 and valine on helix 4 from 3KZ0 are also shown in blue. (c) SiteMAP similarity score for the region around position 3d, clustered by structure. Black lines divide clusters, and Mcl-1 structures form a cluster distinct from the other receptors. (d) SiteMAPs for BHRF1 and Bfl-1 have significant donor density near the peptide 3g position.
Our libraries were designed and screened for peptides that would bind to the viral proteins in preference to all human proteins including Mcl-1. Specificity over Bcl-xL, Bcl-2, Bcl-w, and Bfl-1 was readily achieved, but the library peptides showed only modest specificity (2- to 20-fold) over Mcl-1. We traced much of the viral protein vs. Mcl-1 binding specificity to hydrophobic interactions at positions 3d and electrostatic interactions at positions 2g and 3g. Most KSBcl-2 binders that we characterized had leucine at 3d, and most BHRF1 binders had valine. SPOT arrays indicate that Bcl-xL, Bcl-2, Bcl-w, Bfl-1, and KSBcl-2 are highly tolerant of mutations at 3d, whereas BHRF1 prefers valine and tolerates alanine (Fig. 8a). However, Mcl-1 is highly intolerant of any mutation of I3d according to both the SPOT array and a previous BH3 peptide library sorted for Mcl-1 specificity (Fig. 8a).23 Comparing the region surrounding position 3d using SiteMAP, we found that Mcl-1 structures form a cluster separate from the other homologs (Fig. 8c). The structural mechanism behind the position 3d tolerance exhibited by Bcl-xL may be related to the multiple different conformations accessible at the helix 2-3 bend (Supplementary Fig. 4). These conformations are stabilized by different arrangements of a cluster of three aromatic residues highlighted in red in Fig. 1c, and they result in different environments for the peptide side chain at position 3d in different structures. These three aromatic residues form a motif that is also present in Bcl-2, Bcl-w, and M11. In contrast to Bcl-xL, all solved Mcl-1 structures have a similar conformation in the helix 2-3 bend. Until more structures are solved of KSBcl-2 and BHRF1 bound to diverse peptides it will remain unclear how they accommodate residues other than isoleucine at 3d. However, the different degrees of tolerance at 3d between KSBcl-2 (very tolerant), BHRF1 (moderately tolerant), and Mcl-1 (very intolerant) may arise from the different residues at the structural motif between helices 2 and 3 (L/K/F for KSBcl-2, I/N/F for BHRF1 and V/H/F for Mcl-1, in the same positions as the aromatic residues in the aforementioned Bcl-xL motif; see Fig. 1).
Charge patterns at positions 2g and 3g also contribute to selectivity for viral proteins over Mcl-1. For example, KL8-7 and KL8-16 both bind with KD < 1 nM to KSBcl-2, but KL8-7 has a KD of 21 nM for Mcl-1, whereas KL8-16 has a KD of 3.9 nM. The only difference between these peptides is an arginine at position 2g in KL8-7 versus a glutamine in KL8-16 (Supplementary Table 6). Likewise, mutation of E3g (the wild-type residue in Bim) to arginine in BL peptides reduced affinity to Mcl-1 by approximately 3-fold. By comparing BL6-19 (E3g) to BL_d (R3g), or BL_c (E3g) to BL_b (R3g), it can be seen that only BHRF1, Bfl-1, and M11 tolerate R3g without a significant loss of affinity (Supplementary Table 6; the glutamate to arginine mutation is the only difference between these peptide pairs). Consistent with this, BHRF1 and Bfl-1 are the only receptors that show large patches of donor density proximal to position 3g in their SiteMAPs (Fig. 8d).
Designed peptides with improved specificity for KSBcl-2 and BHRF1 vs. Mcl-1
To increase the specificity of our designed peptides against Mcl-1, we considered positions beyond the 9 that were varied in our libraries. We searched for differences between the viral homologs and Mcl-1 in sequence and structure, and mined our binding data and the literature for observations that could guide further optimization. Based on this analysis, we made additional mutations at C-terminal positions 3f and 4e that improved specificity, as described below.
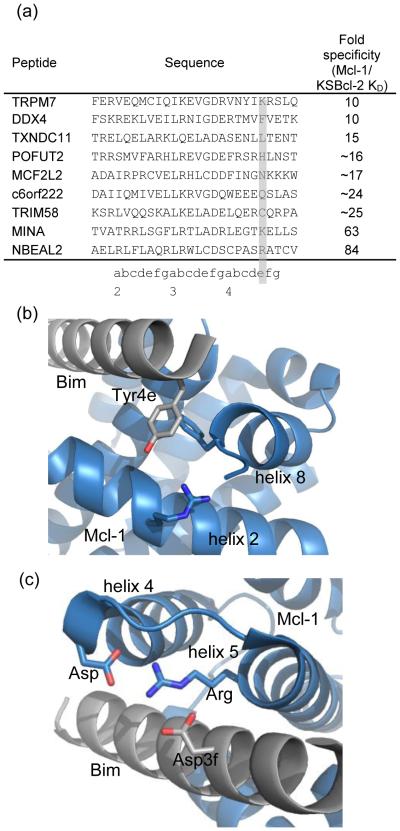
Among the 36 BH3-like peptides in Fig. 3, 9 peptides showed ≥10-fold tighter binding to KSBcl-2 than to Mcl-1 (fold specificity and sequences are in Fig. 9a). Three out of the nine KSBcl-2-specific peptides had arginine or lysine at position 4e. Boersma et al. showed that Mcl-1 binding to Bim BH3 is weakened by a Y4eK mutation, which may arise in part from repulsion from an arginine present on helix 2 and/or from the loss of a favorable aromatic interaction with a phenylalanine on helix 8 (both highlighted in green in Fig. 1c, with the interactions in the Mcl-1:Bim BH3 structure shown in Fig. 9b).50 KSBcl-2 and BHRF1 have negatively charged residues along the peptide interface with helix 2 (green in Fig. 1c), and helix 8 in these proteins lacks the aromatic residue present in the human homologs. Finally, STATIUM predicts that substitution of tyrosine with lysine imparts a preference for KSBcl-2 over Mcl-1 binding, but is also moderately disruptive for KSBcl-2 binding. To test the role of lysine at position 4e, we made a Y4eK substitution in the background of the most specific KL and BL peptides (KL6-7 and BL6-22). KL6-7_Y4eK maintained tight binding to KSBcl-2 (KD of 2.9 nM) but Mcl-1 binding was weakened by 350-fold; this peptide was 195-fold more specific for KSBcl-2 over Mcl-1 than KL6-7 (Table 2). The other Bcl-2 homologs displayed no binding to KL6-7_Y4eK (Supplementary Table 6). Y4eK also improved the specificity of BL6-22 for BHRF1 by 3.6-fold, though it reduced BHRF1 binding affinity from a KD of 2.2 nM to 30 nM. BL6-22_Y4eK had 70-fold specificity for BHRF1 over Mcl-1 and >100-fold specificity over the other human Bcl-2 homologs (no binding detected, Supplementary Table 6). Thus, Y4eK is the single strongest contributor to specificity between KSBcl-2/BHRF1 and Mcl-1 that has been identified.
Figure 9.
Specificity mechanisms that disfavor Mcl-1 binding. (a) Natural BH3-like sequences showing specificity for binding KSBcl-2 over Mcl-1 have diverse residues at position 4e (highlighted), including positively charged residues. (b) Tyrosine at position 4e in an Mcl-1:Bim BH3 complex (2PQK, blue:gray).24 (c) Aspartate at position 3f in the same Mcl-1:Bim BH3 complex.
Table 2.
Dissociation constants for Bcl-2 homologs binding to peptides designed for increased specificity against Mcl-1.
| KD in nM with 95% confidence interval |
||||
|---|---|---|---|---|
| Peptide | Sequence | BHRF1 | KSBcl-2 | Mcl-1 |
| KL6-7 | RPQIWIHQGLQRLGDGLNAYYAR | >1000a | 1.6 ± 0.45 | 22 ± 2.8 |
| KL6-7_Y4eK | RPQIWIHQGLQRLGDGLNAYKAR | >1000 a | 2.9 ± 1.3 | ~7800b |
| KL6-7_D3fA | RPQIWIHQGLQRLGAGLNAYYAR | >1000 a | 180 ± 80 | >5000a |
| KScomp | RPNIWLTQRLQRTGDGLNAYYAR | >1000 a | 13 ± 7.3 | 510 ± 190 |
| BL6-22 | RPIIWIGQEISRVGDRENAYYAR | 2.2 ± 1.3 | 1100 ± 860 | 45 ± 6.1 |
| BL6-22_Y4eK | RPIIWIGQEISRVGDRENAYKAR | 30 ± 17 | 750 ± 300 | ~2200 |
No binding was observed and the value given is the highest concentration of receptor used for that curve.
Approximate values had large 95% confidence intervals due to the absence of an upper baseline.
Our library design strategy favored residues predicted to be non-disruptive for viral Bcl-2 binding. While appropriate for obtaining high affinity binders, this strategy may exclude mutations that provide specificity at the cost of affinity. We identified an example of such a mutation at peptide position 3f. Huang et al. showed that mutation of aspartate at 3f to alanine in a Bak BH3 peptide weakened Bcl-xL binding more than KSBcl-2 binding.53 SPOT arrays also showed that mutation of aspartate at 3f in Bim to any other residue abolished binding to Mcl-1, Bfl-1, Bcl-2, and Bcl-xL, but only weakened KSBcl-2 binding. To test the idea that substitutions at position 3f might provide specificity by decreasing Mcl-1 binding more than KSBcl-2 binding, we introduced D3fA into KL6-7. This mutation reduced binding to KSBcl-2 110-fold (KD 180 nM), but weakened binding to Mcl-1 to the extent that it was undetectable up to 5000 nM (Table 2). Structural analysis suggests a plausible mechanism for this differential effect. Mcl-1 forms a salt bridge between an aspartate at the end of helix 4 and the BH1 motif arginine at the N-terminus of helix 5. This interaction appears to position the arginine optimally for interaction with the BH3 aspartate at 3f (Fig. 9c). Although KSBcl-2 also has an aspartate in the helix 4-helix 5 loop, it has an insertion of 3 residues preceding the aspartate, relative to Mcl-1 (Fig. 1c). In homology models of KSBcl-2 complexes built on three different alignments with Mcl-1, the interactions observed in Mcl-1 were not able to form in an optimal configuration. Thus, the interaction between the 3f aspartate and the BH1 arginine may be weaker in KSBcl-2:BH3 complexes than in Mcl-1 complexes. We tested two other mutations with the potential to improve specificity for KSBcl-2 over Mcl-1 (G3eC and I2dR, see notes in Supplementary Table 6), but these had equally detrimental effects on KSBcl-2 and Mcl-1 binding.
We also synthesized and tested a composite of the most KSBcl-2 specific library peptides, KL6-7 and KL8-7. KScomp has the N-terminal 5 substitutions present in the KL8-7 peptide, including the 2gR specificity mutation, and the 2 C-terminal substitutions of KL6-7 (3gG and 4aL). A 3dT substitution was also introduced, as the SPOT arrays suggested that threonine would provide more specificity against Mcl-1 than leucine. According to deep sequencing data, KScomp was preserved in our library from the naïve pool to pool KL5, but at a very low frequency (<0.0007%). We found that KScomp had reduced affinity for KSBcl-2 (13 nM KD) when compared to library peptides that were tested from pools KL6 and KL8 (all had KD values of ~ 1 nM), but KScomp had 39-fold specificity over Mcl-1, which is greater than the specificity of the other tested KL peptides. Given its low frequency, KScomp may have been lost from the library by chance. However, its 10-fold lower affinity than the peptides that survived the screen suggests that our sorting conditions were too stringent for KSBcl-2 affinity to enrich this peptide.
Discussion
As evolutionarily distant homologs of the mammalian anti-apoptotic Bcl-2 proteins, viral Bcl-2 proteins offer an interesting opportunity to study structure-function relationships in binding. In this paper, we characterized KSBcl-2, BHRF1 and M11 in terms of their sequence, structural, and binding-profile similarity to the human Bcl-2 homologs. The binding specificities of the different proteins provided insights into possible functional analogy, whereas analysis of sequence and structural similarity was useful for testing whether or not these measures are a good proxy for biochemical function similarity. We found that the low sequence identity of the human and viral proteins primarily emphasizes the high degree of divergence and is not particularly useful in identifying similarities between these groups. The SiteMAP results capture the clear similarities between Bcl-xL, Bcl-2, and Bcl-w that are apparent from sequence analysis, but present a more nuanced view of the more dissimilar homologs Mcl-1, Bfl-1, KSBcl-2, and BHRF1. Though by sequence identity Bfl-1 is closest to Mcl-1, the SiteMAP results suggests that Bfl-1 has structural features closer to those of the xL/2/w/M11 group. Conversely, BHRF1 has higher sequence identity to Bcl-xL and Bcl-2, but appears structurally more similar to Mcl-1. KSBcl-2 is similar to Mcl-1 by structure, but to a lesser degree than the highly similar xL/2/w/M11 group. The structural relatedness of both KSBcl-2 and BHRF1 to Mcl-1 was reflected in the library screening experiments, in which it was difficult to achieve large margins of binding specificity over Mcl-1.
By looking at parallels in binding similarity and structure we can make hypotheses about functional similarity between the viral and human Bcl-2 homologs. For example, BHRF1 shows structural similarity to Mcl-1, and appears to have BH3 binding preferences representative of a narrower subset of the binding preferences of Mcl-1. The restrictive binding of BHRF1 to only Bim, Bid, and Puma and the pro-apoptotic effectors Bak and Bax is reminiscent of human Bcl-b and viral Bcl-2 homolog F1L from vaccinia virus. Bcl-b only binds Bim, Bik and Bax, and F1L is reported only to bind Bim, Bak, and Bax, out of a set of pro-apoptotic BH3-only proteins and Bax and Bak.54,55 BHRF1 binds Bak in cells, and its interaction with Bim appears to be especially critical to its anti-apoptotic activity in lymphocytes.56 The restrictive binding profiles of BHRF1 and F1L may be a strategy employed by viruses either to focus on only the most critical activators and effectors of apoptosis, or to prevent off-target effects by avoiding the “moonlighting” functions of human Bcl-2 homologs in autophagy, mitochondrial homeostasis, or other pathways.57
M11 and KSBcl-2 have binding preferences similar in many respects to those of Mcl-1 (Fig. 3). However, structural similarity suggests that M11 also has features similar to the Bcl-xL/2/w class, whereas KSBcl-2 is closest to Mcl-1 by both structure and binding. As previously noted, both KSBcl-2 and M11 bind Beclin-1 and prevent autophagy as an important part of their cellular function.21,22 Bcl-2 and Bcl-xL also bind Beclin-1 to prevent autophagy; Mcl-1 binds Beclin-1 more weakly.58 However, KSBcl-2 and M11 do not bind tightly to the classical Bcl-2/xL/w binder, Bad, but do bind the Mcl-1 binder, Noxa.32,34,53 It will be interesting to investigate functionally why KSBcl-2 and M11 have binding characteristics of both the Bcl-2/xL/w and Mcl-1 classes. Much of the viral Bcl-2 literature only focuses on comparisons between viral Bcl-2 and human Bcl-2 or Bcl-xL function. Our results suggest that an investigation of the relationship between viral Bcl-2 and Mcl-1 function would be informative in discerning the relationship between the roles played by these viral and human Bcl-2 homologs.
Several lessons can be learned from our attempts to design KSBcl-2 and BHRF1 specific BH3 peptides. First, trade-offs between affinity and specificity must be considered when dealing with homologs with very similar binding preferences, such as the viral Bcl-2s and Mcl-1. The importance of such trade-offs has been previously discussed in the context of protein-protein interaction design.59,60 Our library screen imposed criteria on both affinity and specificity, but with an emphasis on tight binding. As a consequence, we did not identify viral Bcl-2 selective peptides that had weakened binding to KSBcl-2 and BHRF1, such as KScomp (Table 2). Only very tight binders (Kd ~1 nM) survived to round 8. Screening conditions could be adjusted to maintain destabilizing but specific mutations by relaxing target affinity requirements in the design stage and reducing the stringency of screening. Peptides identified in this way could then be further optimized for tight binding. Another consideration in achieving specific binders is library diversity. Discriminating homologs with very similar binding preferences such as KSBcl-2 and Mcl-1 may require broader exploration of sequence space than can be achieved from screening just one library of ~107 molecules, or may require more precise selection of the sequences to be screened. For example, we did not vary position 4e in our library, the site of our most specific mutation, because we were limited by library size. Yet modeling using STATIUM, as well as hints from the literature, indicated this as a promising substitution. A recent paper by Dutta et al. also found that the Y4eK mutation imparted specificity for Bcl-xL over Bcl-2, suggesting that positions outside the core BH3 motif can strongly influence specificity.39 Searching sequence space more extensively, with larger libraries or multiple rounds of library design and sorting, could accelerate discovery.
In this work we ultimately identified selective peptide inhibitors of viral Bcl-2 family proteins KSBcl-2, BHRF1 and M11 by various means, including library screening, rational mutagenesis and profiling of BH3-like sequences previously identified in the human proteome. Although we did not sort a library for M11 specificity, one of the 36 BH3-like peptides identified from the proteome, AGBL2 showed moderate binding to M11 (210 nM), and at least 20-fold weaker binding to all of the other Bcl-2 homologs (Supplementary Table 1). For use as a research reagent to specifically inhibit M11, this offers a 30-fold improvement in affinity over the existing M11-specific Beclin-1 mutant.42 Several of our BL peptides also have higher affinity for M11 (~1 nM) than is offered by natural BH3 sequences, although these peptides show only moderate specificity over Mcl-1. The Y4eK mutants of the most specific KL and BL peptides (KL6-7_Y4eK and BL6-22_Y4eK) achieved large margins of specificity over all of the human Bcl-2 homologs. KL6-7_Y4eK is an especially promising peptide for future studies of KSBcl-2, as it binds very tightly and selectively to this protein.
As recently demonstrated by the use of a designed protein inhibitor of BHRF1 to suppress tumor growth in a mouse xenograft model of Epstein-Barr-positive human lymphoma, anti-viral Bcl-2 agents have potential for use in disease therapy.41 Large proteins face obstacles in terms of intracellular delivery, and in this respect the much smaller designed peptides presented here may have advantages. More immediately, designed potent and selective inhibitors of viral Bcl-2 proteins can provide useful reagents for studying the function of viral Bcl-2 homologs in herpesvirus-associated pathologies, and for testing the extent to which these proteins contribute to cancer onset or progression.
Materials and Methods
Sequence identity, homology modeling and SiteMAP
Multiple-sequence alignments of Bcl-2 family proteins were manually constructed based on preliminary Clustal alignments and structural analysis. For calculation of sequence identity in the BH3 binding groove, Pymol was used to select residues of the Bcl-2 homologs within 7 Å of the BH3 peptide. Structures used for this were: 2PQK(Mcl-1:Bim)24, 2WH6 (BHRF1:Bim)64, 2VM6 (Bfl-1:Bim)63, 3FDL (Bcl-xL:Bim)62, 2XA0 (Bcl-2:Bax)65, 3BL2 (M11:Beclin-1)22, and the homology models of KSBcl-2 and Bcl-w bound to Bim. An inclusive set of groove residues was defined to include any residue found within 7 Å of any peptide atom in any structure. See Fig. 1c for alignments.
Homology models of KSBcl-2 and Bcl-w were built using MODELLER (version 9.1).71,72 The KSBcl-2:Bim complex was modeled on 2PQK24, 2WH664, 1O0L73, 2VM663, 3FDL62, and 1G5M74. The Bcl-w:Bim complex was modeled on 3FDL, 1G5M, 2PQK, and 2VM6. The KSBcl-2:Bak complex was modeled on 3PK175 and 2XPX64. The Bcl-w:Bak complex was modeled on 1BXL76 and 2XA065.
For SiteMAP (SiteMap, version 3.0, Schrödinger, LLC, New York, NY, 2014) structure analysis, the following PDB IDs were aligned to 2PQK (Mcl-1:Bim)24: 2VM6 (Bfl-1:Bim)63, 3I1H (Bfl-1:Bak) and 3MQP (Bfl-1:Noxa) (NESG, unpublished), 2VOG (murine Bfl-1: Bmf)77, 2VOF (murine Bfl-1:Puma)77, 2VOH (murine Bfl-1:Bak)77, 2VOI (murine Bfl-1:Bid)77, 2WH6 (BHRF1:Bim)64, 2XPX (BHRF1:Bak)64, 3FDL (Bcl-xL:Bim)62, 1BXL (Bcl-xL:Bak)76, 1G5J (Bcl-xL:Bad)78, 3IO8 (Bcl-xL:Bim L12F)62, 2PON (Bcl-xL:Beclin-1)79, 3PK1 (Mcl-1:Bax)75, 2KBW (Mcl-1:Bid)80, 3KJ0 (Mcl-1:Bim I2dY)24, 3KJ1 (Mcl-1:BimI2dA)24, 3KJ2 (Mcl-1:Bim F4aE) 24, 3BL2 (M11:Beclin-1)22, 2XA0 (Bcl-2:Bax)65, and homology models of Bcl-w and KSBcl-2 bound to Bak and Bim BH3s. The following structures were relaxed using minimization in Maestro in the absence of the peptide (minimization uses the OPLS_2005 force field and proceeds until no movements are >0.05 Å) and were aligned to 2PQK using just the equivalent receptor positions: 2PQK, 2WH6, 2VM6, 3FDL, 3BL2, 2XA0, and the KSBcl-2 and Bcl-w:Bim models. Cealign in PyMOL (version 1.3, Schrödinger, LLC) was used to do the alignments based on the Cα atoms of helix 2, the helix4/5 region, and peptide regions of the structures. The following residues were used for the alignments: 208-220, 243-280, and 6-17 for 2PQK; 32-44, 68-105, and 148-159 for 2VM6; 45-57, 80-117, and 58-69 for 2WH6; 32-103, 127-163, and 59-70 for 2XA0; 36-48, 68-104, and 110-121 for 3BL2; 85-97, 120-156, and 90-101 for 3FDL; 31-43, 63-103, and 153-164 for KSBcl-2; 41-53, 76-112, and 164-175 for Bcl-w. The numbering for KSBcl-2 and Bcl-w refers to the residue number in the natural gene, with the third set of numbers referring to BH3 positions 2d-4a. Equivalent residue numbers to those listed above were used to align all structures of each homolog. Maestro (version 9.7) Prepwizard was then run on the aligned coordinates (complex structures) with the following settings: find disulfides, fill sidechains, propKa_pH 7.0, OPLS force field version 2005, RMSD minimization cutoff of 0.3, cap termini. SiteMAP (version 3.0, analysis, Schrödinger, LLC) was then run on only the receptor coordinates, restricting map finding to a zero Å box around the 2PQK Bim peptide, with the enclosure setting at 0.4 for finding shallow grooves, and with verbose file output.43,44 SiteMAP identified the BH3 binding groove as the largest site for all structures. Scripts were written in Python (version 2.6.1) to process the “.smpot” SiteMAP potential output files (lists of coordinates and potentials). All points that met the SiteMAP default potential values for hydrophilic (-8 kcal/mol) and hydrophobic (-0.5 kcal/mol) maps were extracted. The environment around each peptide position was analyzed by selecting all map points (donor, acceptor, and hydrophobic maps) that were within 6 Å of the Cβ of each position in the Bim peptide in structure 2PQK. The intersection between receptors for each type of map at each position was computed by counting all points within 1 Å of a point in another receptor’s map. A similarity score was computed by summing the intersection counts from the acceptor, donor, and hydrophilic maps at all positions.
Clustering
Clustering of proteins by sequence identity, binding profile, or SiteMAP similarity was performed in Matlab (version R2012b) with the clustergram function in the Bioinformatics toolbox using hierarchical clustering based on correlation. The exception was clustering of the peptides (rows in Fig. 3 and 6), which were clustered by Euclidean distance to give better ordering according to affinity.
Expression and purification of Bcl-2 proteins
Biotin-acceptor peptide (BAP-tagged) and His6-tagged variants of all eight Bcl-2 homologs were made in vector pDW363 (used for analysis of direct binding on the bacterial cell surface, and, in the case of M11, for the fluorescence anisotropy experiments).61 Sequence encoding MBP in the parent vector was replaced by the Bcl-2 homolog, resulting in a product with the BAP-tag and His6-tag at the N-terminus of the Bcl-2. C-myc-tagged versions of all Bcl-2 homologs were in the pSVM vector. Constructs are given in Supplementary Table 7.
pDW363 constructs were expressed in BL21 (DE3) pLysS Rosetta cells. 50 mL overnights were grown at 37 °C with shaking, and 10 mL of the overnight culture was used to inoculate 1 L of LB including 100 μg/mL ampicillin, 25 μg/mL chloramphenicol, and 12-15 mg of D-(+)-biotin. Cells were grown at 37 °C with shaking to an optical density at 600 nm (O.D. 600) of 0.6 and then induced with 1 mM IPTG for ~5 hours before harvesting. pSVM constructs were grown similarly, with 50 μg/mL kanamycin, 25 μg/mL chloramphenicol, and no biotin.
Purification of pSVM constructs was performed as follows. 1 L cell pellets were resuspended in 25 mL of 5 mM imidazole, 500 mM NaCl, 1 mM DTT, 20 mM Tris pH 8.0, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF) protease inhibitor. Cells were sonicated ten times for 30 seconds followed by 30 seconds of rest. The supernatant from the centrifuged lysis product was filtered through 0.2 μm filters before application to 3 mL of Ni-nitrilotriacetic acid agarose resin equilibrated in 20 mM Tris pH 8.0, 500 mM NaCl. After the supernatant was applied, the resin was washed 3 times with 8 mL of 20 mM imidazole, 500 mM NaCl, 20 mM Tris pH 8.0. The His6-MBP-c-myc-Bcl-2 construct was eluted with 8 mL 20 mM Tris pH 8.0, 500 mM NaCl, 300 mM imidazole. At this point, the protein in the eluate was quantitated by absorbance at 280 nm and diluted to 1 mg/mL with 50 mM NaCl, 50 mM Tris pH 8.0, 1 mM DTT, 0.5 mM EDTA, before the addition of TEV protease at a ratio of 50 mg Bcl-2 protein:1 mg TEV. The Bcl-2 protein and TEV were dialyzed overnight at 4 °C against 1 L of 50 mM NaCl, 50 mM Tris pH 8.0. The dialyzed protein was purified by nickel affinity chromatography as before, with the exception that the wash buffer did not contain imidazole, to minimize dissociation of His6-MBP and His6-TEV from the resin. The flow-through (and if not overly contaminated, the wash) was applied to a S75 26/60 size exclusion column equilibrated in 20 mM Tris pH 8.0, 150 mM NaCl, 1% glycerol, 1 mM DTT. Purity was verified by SDS-PAGE, and proteins were frozen at −80 °C in the final buffer. pDW363 constructs were purified similarly, but with no cleavage step, and with just one nickel affinity step followed by the size exclusion column.
Binding affinity measurements by fluorescence anisotropy
Library peptides, the Bim BH3 peptide, and Bim BH3 peptide mutants were 23 residues long with N-terminal 5/6-fluorescein amidite and C-terminal amidation and were synthesized by the MIT Biopolymers Laboratory. The crude synthesis product was verified by mass spectrometry to contain predominantly the desired species and then purified by HPLC on a C18 column with a linear gradient of acetonitrile in water. Proteome-derived BH3 peptides (26-mers, N-fluoresceinated, C-amidated) in Fig. 3 were the same as those used by DeBartolo et al.46 Direct fluorescence anisotropy experiments were performed as in Foight et al., with a titration of twelve receptor concentrations, with a maximum concentration of 1 μM receptor for the library peptide curves and 3 μM for the proteome peptide curves.25 C-myc-tagged receptors were used for all Bcl-2 homologs, with the exception of M11, for which a BAP and His6-tagged construct was used. All KD values are from averaged data from three replicates done over three days. Data were fit as described for direct fluorescence anisotropy experiments in Foight et al., with the upper baseline and KD fit, while the lower baseline and concentration of the fluoresceinated peptide were fixed as described.25
SPOT arrays
SPOT arrays were synthesized on activated nitrocellulose support using Fmoc protection/deprotection chemistry by an Intavis AutoSpot robot in the MIT Koch Biopolymers Laboratory. The peptides were synthesized with PEG3 (three ethylene glycol units) linkers at the carboxy terminus. All peptides were 26 residues long (wild-type Bim sequence MRPEIWIAQELLRIGEDEFNAYYARV). Native BH3 peptides in column “X” in Fig. 4, top to bottom: Bad, Bid, Bmf, Hrk, Noxa, Bik, Bak, Bax, Bcl-xL, Bcl-w, Bcl-2, Mcl-1, Bfl-1, BHRF1, KSBcl-2, with the BH3 residues equivalent to those described for Bim BH3. Arrays were processed by first hydrating the membranes in 100% methanol, followed by water. Arrays were blocked in blocking buffer (50 mM Tris pH 8.0, 100 mM NaCl, 0.01% Triton X-100, 1% BSA) for 20 min. 100 nM c-myc-BHRF1 or KSBcl-2 in blocking buffer was applied to the arrays and incubated 1 hr at ~ 23 °C with rocking. Arrays were washed 3 times with 10 mL blocking buffer before addition of 5 mL blocking buffer + anti-c-myc-Cy3 (Sigma Aldrich) at 1:100 dilution. Arrays were incubated in the dark for 30 minutes at ~23 °C with rocking. Finally, the array was washed 3 times with 10 mL blocking buffer, imaged on a Typhoon 9400, and analyzed with ImageQuant.
Models used for library design
Illumina sequencing data of a yeast display library sorted for selective binding to KSBcl-2 but not Mcl-1 and Bcl-xL was used to inform selection of substitutions that are tolerated by KSBcl-2 binding.25 Enrichment of residues was computed as the percentage of unique sequences with a residue after five rounds of screening minus the percentage of unique sequences with that residue in the naïve library. Sequences surviving the selection were likely to be moderate to high-affinity KSBcl-2 binders, but not all were selective for KSBcl-2. Peptides tested individually in solution bound KSBcl-2 with dissociation constants of ~1 nM but also bound to Mcl-1 with similar affinities.25
Position-specific scoring matrices based on SPOT array intensities for KSBcl-2 and BHRF1 (PSSMSPOT models) were built by computing log10 of each mutant peptide fluorescence intensity divided by the average wild-type intensity, as described previously for the human Bcl-2 proteins.23,48,49
STATIUM is a statistical potential built to evaluate the fit of a sequence on a particular structural template. STATIUM models were built using the STATIUM_sidechain method reported by DeBartolo et al., with the exception of Bcl-w, for which the original STATIUM method was used.46,48 The template structures used were as follows: a KSBcl-2:Bim BH3 homology model described above, 2PQK (Mcl-1:Bim BH3)24, 3IO8 (Bcl-xL:Bim BH3_L12F)62, 2VM6 (Bfl-1:Bim BH3)63, 2WH6 (BHRF1:Bim BH3)64, and 2XA0 (Bcl-2:Bax BH3)65. The Bcl-w model was built using template 1ZY366, an NMR-based docking model with Bid BH3.
Library design
KSBcl-2 library
To guide library design, all possible point mutants of Bim at positions 2a-4e were scored with the three models described above, with some positions not having data from all models. Substitutions were rated as non-disruptive for KSBcl-2 binding if they met at least one of the following criteria:
Frequency increase of 1% in the library screening experiment targeting KSBcl-2, or present in at least 10% of unique sequences after 5 rounds of screening
PSSMKSBcl-2 score greater than the median score for all mutations across all positions
ΔSTATIUM KSBcl-2 (raw Bim score – raw mutant score) greater than the median for all mutations across all positions
Substitutions were counted as specific for KSBcl-2 over an alternative receptor if ΔPSSM = PSSMKSBcl-2 -PSSMalternative receptor or ΔΔSTATIUM = ΔSTATIUMKSBcl-2 -ΔSTATIUMalternative receptor was greater than 0.2. As for ΔSTATIUM KSBcl-2, ΔSTATIUMalternative receptor was defined as the score for Bim on that receptor minus the mutant score on that receptor. The specificity score for each substitution was defined as the number of scores that met the aforementioned cutoff (e.g., a specificity score of 10 would be given if all ΔPSSM and all ΔΔSTATIUM scores for all 5 human Bcl-2 proteins were greater that 0.2). The 0.2 cutoff was chosen because it was close to the average over all receptor comparisons of the median ΔPSSM and ΔΔSTATIUM values for all mutations, and seemed to represent a reasonable minimal margin of specificity when compared to model scores for known specificity mutations.
Libraries were constructed using degenerate codons chosen by a computational optimization protocol.67 To guide the selection of a set of degenerate codons to consider at each position, residue substitutions were divided into two categories, “preferred” or “required”. Substitutions were included in the preferred category if they were non-disruptive by at least one out of the three categories previously mentioned and had a specificity score of at least 6 (or at least 3 for positions with information available from only PSSMSPOT or STATIUM). Additionally, some substitutions were included that did not meet this criteria but had large ΔPSSM or ΔΔSTATIUM scores for Mcl-1. Required residues included the wild-type residues and a subset of the preferred residues that were hand-selected for a high degree of specificity, with emphasis placed on specificity against Mcl-1. Degenerate codons selected for consideration encoded all of the required residues, and the choices were further narrowed by the elimination of any codon that included more trinucleotides than another but encoded fewer preferred residues.
Optimization of degenerate codon combinations was performed by integer linear programming, as previously described, with a limit on the DNA size of the library set at 1 × 107.67 At positions where more than one degenerate codon of equal DNA size encoded the same number of “preferred” residues, one codon was selected to maximize specificity against Mcl-1. Codon options at certain positions were narrowed to encourage the ILP code to compose a library in which the majority of positions had a modest number of variants (more than 3). This strategy was employed with the aim of decreasing the likelihood that the library sequences would be overly reliant on one or two positions to achieve specificity. Several possible library designs arose from different choices of positions to mutate and codon choices to include. The final library design was chosen because it contained a large number of protein sequences (5.23 × 106), and it had the highest number of sequences that scored in a range predicted to have high affinity for KSBcl-2 and weaker binding than Bim to Mcl-1 on the PSSMSPOT and STATIUM models. BHRF1 library Design of the BHRF1-targeted library used the same PSSMSPOT and STATIUM models as described above for the cellular receptors. BHRF1 models used to categorize mutations included a PSSMSPOT model and STATIUM models built on two templates, 2WH6 (BHRF1:Bim BH3) and 2XPX (BHRF1:Bak BH3).64 Preferred residues were defined as all residues that were non-disruptive by at least one out of three models (i.e., a PSSMBHRF1 or ΔSTATIUMBHRF1 score above the median across all mutations at all positions). Required residues were selected to be non-disruptive and highly specific for BHRF1, with preference given to residues that disfavor binding to Mcl-1. A few residues were also included as required because PSSMSPOT indicated high affinity binding to BHRF1. Codon selection was performed as for the KSBcl-2 library. In selecting the final library, a similar emphasis was placed on including 4-8 mutations at as many positions as possible; manual selection of codons and adjustment of required residues achieved this goal. The final BHRF1 library had a large number of protein sequences (6.72 × 106), as well as large numbers of sequences that scored as high for BHRF1 affinity but low for Mcl-1 affinity according to the PSSMSPOT and STATIUM models. The final library designs are presented in Supplementary Tables 2 and 3.
Library construction
The pBAD33-eCPX-FLAG-Bim BH3 construct used for bacterial surface display of the Bim variant libraries and the primers used for library construction are shown in Supplementary Table 7. The protocol for library assembly was similar to that presented in Getz et al.68 The vector into which the library was cloned was pBAD333-eCPX with a FLAG tag included for use as an expression control and linkers optimized for Bim BH3 expression and binding. A vector containing the non-binding mutant BimL3aD was used as a cloning template to prevent background from undigested vector from influencing the screening results. 200 mL of this vector expressed in DH5α E. coli was minipreped over 8 Qiagen miniprep columns and digested with SfiI according to the NEB protocol for 8 hours. Digested vector was PCR purified over an appropriate number of Qiagen PCR purification columns and eluted with water. The digested vector was then dephosphorylated with Antarctic phosphatase (1 μL phosphatase: 1 μg DNA) at 37 °C for 2 hours, followed by 10 min at 65 °C for deactivation of the enzyme. This prevented re-ligation of the original insert back into the vector. This protocol yielded 8-10 μg of DNA, which was the amount of vector needed per library.
To prepare the library inserts, a DNA fragment corresponding to the eCPX gene was generated by performing PCR with the primers eCPX_rev_library and BimC_fwd_library (Supplementary Table 7) on the BimL3aD-eCPX vector. Platinum Taq HiFi polymerase was used according to manufacturers instructions. The product was run on an agarose gel using GelGreen DNA dye and blue light for visualization to minimize DNA damage. The appropriate band was extracted with a Zymo DNA gel recovery kit. Overlap PCR was then performed on this eCPX fragment to add the varied Bim region. For each library, the appropriate library primer (1.5 μL at 10 μM) was combined with 1 μL of the eCPX product in standard Platinum Taq HiFi conditions. Seven PCR cycles were run with an annealing temperature of 54 °C, and then 1 μL of a 12.5 μM mix of the eCPX_rev_library and SfiI_fwd primers were added and a further 28 PCR cycles were completed (Supplementary Table 7). 15, 50 μL reactions were done per library to obtain sufficient insert DNA. The PCR products were purified over Zymo Clean & Concentrate columns and digested with SfiI for 4 hours. Digestion reactions were then purified again. This protocol yielded ~7 μg of insert DNA for each library.
Digested vector and library insert were ligated at a 5:1 molar ratio of insert:vector, using ~8 μg vector. An 800 μL reaction volume with standard T4 DNA ligase conditions was used, and ligations were performed at 14 °C overnight. A 20 μL control ligation of digested vector alone was also performed to allow estimation of vector background. Following ligation, the ligase was deactivated at 70 °C for 10 minutes, and the ligation was concentrated over four Zymo Clean & Concentrate columns, eluting each column in 12.5 μL DNA elution buffer (from the kit). Each ligation aliquot was then desalted for 20 minutes on a Millipore 0.025 μm filter suspended on an Eppendorf tube of sterile MilliQ water. The ligations were then pooled on ice and split between 4, 250 μL aliquots of thawed MC1061 competent E. coli cells. MC1061 competent cells were prepared as in Getz et al.68 Each aliquot was electroporated in a cold 2 mm BioRad cuvette at 2.5 kV, 50 μF, 100 Ω on a BioRad Gene Pulser electroporator, and then immediately rinsed three times with 1 mL warm SOC and combined with 7 mL warm SOC and then allowed to recover at 37 °C for 1 hr on a rotator wheel. The library was then added to 500 mL LB + 25 μg/mL chloramphenicol + 0.2% w/v sterile-filtered glucose in a 2 L flask and grown at 37 °C until an O.D.600 of ~1.5 was reached (~7 hours). 400 mL of the library culture was pelleted at 3000 relative centrifugal force (rcf) for 15 minutes and re-suspended in 10 mL SOB + 15% v/v sterile glycerol and aliquoted and frozen at −80 °C. The number of transformants was 3.9 × 109 for the BHRF1 library and 1.8 × 109 for the KSBcl-2 library, with vector background estimated at < 0.001%.
Library Sorting
The general protocol for preparation of library samples for sorting was as follows. A quantity of glycerol stock sufficient to oversample the estimated library diversity by at least 10-fold was used to inoculate 5 mL LB plus 0.2% glucose and 25 μg/mL chloramphenicol and grown overnight at 37 °C on a rotator wheel. ~100 μL of overnight culture (or enough to maintain library diversity) was pelleted and used to inoculate 5 mL LB plus 25 μg/mL chloramphenicol. The culture was then grown to an O.D. 600 of 0.5-0.6 and induced with 0.04% w/v arabinose for 1 hour. Sufficient cells to oversample the library diversity were pelleted at 3000 rcf for 5 minutes and mixed with 100 μL PBS + 0.1% BSA (PBSA) and 100 μL of an appropriate 2x biotinylated viral Bcl-2 stock. Cells were incubated at room temperature with shaking for 1 hour. Cells were pelleted, washed with 200 μL PBSA, pelleted again, and then 210 μL of a mix of streptavidin-PE and anti-FLAG-APC (both at a 1:100 dilution in PBSA) was added. For three-color competition sorts, a two-step antibody labeling process was used. The first incubation was with streptavidin-PE (Molecular Probes), anti-FLAG-APC (Perkin Elmer), and rabbit-anti-c-myc (Sigma), and the second incubation (after a wash step) was with anti-rabbit-FITC (Sigma). Gates for the three-color competition sorts were set to exclude FITC-positive cells and include PE-positive cells. Cells were incubated with the labeling reagents for 15 minutes on ice in the dark. The pelleting and washing steps were repeated, and cells were resuspended in 1.5 mL PBSA for sorting on a BD FACS Aria. Cells were collected in PBSA, which was diluted in SOC after sorting and cells were allowed to recover at 37 °C overnight, at which point some cells were harvested for glycerol stocks and some were used to inoculate cultures for the next day’s sort.
The sorting scheme for the KSBcl-2 library was as follows (summarized in Supplementary Fig. 2): sort for binding to 1 μM biotinylated-KSBcl-2 (positive sort; 2.7% of cells collected, cells were frozen as a glycerol stock and used to inoculate overnight cultures at a later date), sort for cells that do not bind to 500 nM biotinylated-Mcl-1 (negative sort; 7.1% of cells collected, the next four sorts after this point were done on consecutive days), positive sort at 100 nM biotinylated-KSBcl-2 (2.1% collected), negative sort at 300 nM Bfl-1 and Bcl-w (21.5% collected), sort for binding to 100 nM biotinylated-KSBcl-2 and in the presence of 500 nM c-myc-Mcl-1 (competition sort; 3.4% collected), competition sort at 100 nM biotinylated-KSBcl-2 and 500 nM c-myc-Mcl-1 (9.9% collected, glycerol stocks were made at this point and used to inoculate overnight cultures), competition sort at 100 nM biotinylated-KSBcl-2 and 500 nM c-myc-Bcl-w and Bfl-1 (23.7% collected), and finally, competition sort at 20 nM biotinylated-KSBcl-2 and 1 μM c-myc-Mcl-1 (0.6% collected). Competition sorts were all three-color. As often as possible, sorts were done on consecutive days to minimize growth time for the libraries, thereby minimizing potential for the occurrence of secondary mutations that imparted growth advantages.
The sorting scheme for the BHRF1 library was as follows: positive sort at 1 μM biotinylated-BHRF1 (0.4% of cells collected, cells were frozen as a glycerol stock and used to inoculate overnight cultures at a later date), negative sort at 500 nM biotinylated-Mcl-1 (21.1% of cells collected), positive sort at 100 nM biotinylated-BHRF1 (3.3% collected), negative sort at 500 nM biotinylated-Mcl-1 (39.5% collected), competition sort at 100 nM biotinylated-BHRF1 and 500 nM c-myc-Mcl-1 (19% collected), competition sort at 100 nM biotinylated-BHRF1 and 500 nM c-myc-Mcl-1 (11.3% collected), negative sort at 500 nM biotinyalted-Bfl-1 (33.3% collected), and finally, competition sort at 100 nM biotinylated-BHRF1 and 500 nM c-myc-Mcl-1 and Bfl-1 (5.5% collected). These competition sorts were done with no labeling of the c-myc labeled competitor. Clones were conventionally sequenced from the sixth and eighth pools of this sorting process. The sorts were largely not done on consecutive days, and we noticed that one clone with a growth advantage (clone BL8-11) had taken over the library by the final sort. Therefore, to get better diversity for Illumina sequencing, we went back to the fourth pool and re-cloned the library into fresh vector and fresh MC1061 cells using Gibson assembly, at which point the growth advantage disappeared. The BHRF1 library was re-sorted from this point on consecutive days with the following scheme (in which competition sorts were three-color): positive sort at 100 nM BHRF1 (1.5% collected), competition sort at 100 nM biotinylated-BHRF1 and 400 nM c-myc-Mcl-1 and Bfl-1 (3.4% collected), negative sort at 500 nM biotinylated-Mcl-1 and Bfl-1 (34.4% collected), competition sort at 100 nM biotinylated-BHRF1 and 500 nM c-myc-Mcl-1 and Bfl-1 (7.9% collected). We sequenced clones from the sixth and eight pools from the first sorting attempt (pools BL6 and BL8). From the second sorting attempt, we sequenced 25 clones each from pools BL6’ and BL7’. The accelerated growth clone from the first sorting attempt, BL8-11, was not found in the BL6’ and BL7’ pools.
FACS analysis samples were prepared as were samples for sorting, and were analyzed on a BD FACS Calibur.
Illumina sequencing and data processing
The KSBcl-2 Bim variant library sorted by yeast surface display was described by Foight et al.25 To decrease contamination by peptides that bound the antibody detection reagents, we performed two rounds of negative sorting against antibody binding on the fifth library pool. The resulting pool and the naïve, unsorted library were prepared for Illumina sampling on a Genome Analyzer II, and the data were processed as in DeBartolo et al.48 The information from this screen was used in the design of the KSBcl-2 bacterial display library as described above.
Bacterial display pools were prepared for Illumina sequencing with a scheme similar to that reported by Hietpas et al. for yeast libraries.69 First, 10 mL LB, 0.2% w/v glucose, 25 μg/mL chloramphenicol cultures of each library pool were started from glycerol stocks and grown overnight at 37 °C. The entire culture was mini prepped with a Qiagen mini prep kit and eluted in sterile water. The DNA was diluted to ~50-100 ng/μL, and 1 μL was used for the first PCR. The first PCR added an MmeI restriction enzyme site to the 5’ end and a universal Illumina sequencing region on the 3’ end. Primers MmeI_fwd and 3prime_rev were used in standard Phusion polymerase conditions with no annealing step and 25 cycles with a 30 second extension period in each cycle. All primer and adapter sequences are given in Supplementary Table 7. PCR products were purified with the Qiagen PCR purification kit and eluted in 30 μL sterile water. MmeI digestion was performed with 3.45 pmol DNA:2 μL MmeI (NEB) for 1 hour at 37 °C, followed by 20 minutes at 80 °C for enzyme deactivation. Double stranded DNA fragments containing Illumina adapters and 5-mer barcodes were then ligated onto the 5’ end of the digested DNA. The adapter was double stranded through the end of the barcode, leaving a single strand “TC” overhang to anneal with the digestion product.
Barcodes were all different by at least 2 bases. 3 barcodes were used for each naïve library, 2 barcodes each for pools 1, 3, and 5, and 1 barcode each for pools 6, 7, and 8, for a total of 24 barcodes. Ligations were performed on 30 μL MmeI-digested DNA, with 4 μL 6 μM adapter, 4 μL 10x T4 ligase buffer, and 2 μL T4 DNA ligase for 30 minutes at room temperature, followed by deactivation of the ligase at 65 °C for 10 minutes. Ligation products were run on an agarose gel and ~200 bp bands were extracted with a Zymoclean Gel DNA recovery kit and eluted in 15 μL water. The second PCR amplified the ligation product and extended the 5’ region to encompass the universal Illumina forward read sequencing primer. Standard Phusion polymerase conditions described above were used on 15 μL of gel-purified ligation product with 0.5 μM finalPCR_fwd and rev primers, with 25 cycles. PCR products were run an on agarose gel and extracted with the Zymoclean Gel DNA recovery kit and eluted in 30 μL water. Samples were then multiplexed and run in one lane on an Illumina Hiseq2000 with paired-end reads of 80 bp using the universal Illumina forward sequencing primer and rev_seq primer. A PhiX lane was also run for control.
Illumina sequencing data from the bacterial display library pools were filtered using in-house scripts written in Matlab and Python. First, the constant positions in the library between I2d and R4g were required to match the wild-type DNA sequence exactly, though no quality score filtering was imposed upon the constant positions. Second, the variable position bases were required to have Illumina quality scores greater than 20 (99% base calling accuracy). Finally, sequences were sorted into pools according to their barcodes, which had to exactly match a theoretical barcode, though no quality score filtering was imposed. Sequence logos were made using Weblogo.70
Supplementary Material
Highlights.
KSBcl-2, BHRF1, and M11 are herpesvirus homologs of anti-apoptotic Bcl-2 proteins.
Sequence/structure/binding properties of viral and human Bcl-2 homologs were compared.
SPOT arrays and library screening revealed binding determinants for KSBcl-2 and BHRF1.
Peptides were designed to bind viral Bcl-2 proteins in preference to human homologs.
Binding data and peptide reagents inform analysis of viral Bcl-2 function.
Acknowledgements
The authors would like to thank C. Kougentakis for making the pDW363-Mcl-1, Bcl-w, Bcl-2, and Bcl-xL constructs, O. Levsh for assistance with the fluorescence anisotropy assays and S.V. Gullá for performing the viral Bcl-2 SPOT arrays. We thank the MIT Koch Flow Cytometry core staff for assistance with FACS sorting and MIT Koch Biopolymers for peptide and SPOT array synthesis. This work was funded by the National Institute of General Medical Sciences through award R01GM110048. Computational resources to support this work were provided by the National Science Foundation under Grant No. 0821391. G.W.F. was supported by a National Science Foundation Graduate Research Fellowship.
Abbreviations
- BH3
Bcl-2 homology 3
- EBV
Epstein Barr virus
- KSHV
Kaposi’s sarcoma herpesvirus
- BL
Burkitt lymphoma
- FACS
fluorescence activated cell sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- (4).Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- (5).Llambi F, Moldoveanu T, Tait SWG, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Polster BM, Pevsner J, Hardwick JM. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim Biophys Acta. 2004;1644:211–227. doi: 10.1016/j.bbamcr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- (7).Galluzzi L, Brenner C, Morselli E, Touat Z. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hwang S, Oh S, Lee J-S, Jeong JH, Gwack Y, Kowalik TF, Sun R, Jung JU, Liang C. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009;5:e1000609. doi: 10.1371/journal.ppat.1000609. E, X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Watanabe A, Maruo S, Ito T, Ito M, Katsumura KR, Takada K. Epstein-Barr virus-encoded Bcl-2 homologue functions as a survival factor in Wp-restricted Burkitt lymphoma cell line P3HR-1. J Virol. 2010;84:2893–2901. doi: 10.1128/JVI.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ojala PM, Tiainen M, Salven P, Veikkola T, Castaños-Vélez E, Sarid R, Biberfeld P, Mäkelä TP. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- (11).Ojala PM, Yamamoto K, Castaños-Vélez E, Biberfeld P, Korsmeyer SJ, Mäkelä TP. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates Bcl-2. Nat Cell Biol. 2000;2:819–825. doi: 10.1038/35041064. [DOI] [PubMed] [Google Scholar]
- (12).Gelgor A, Kalt I, Bergson S, Brulois KF, Jung JU, Sarid R. KS-Bcl-2 Encoded by the Kaposi's Sarcoma-Associated Herpesvirus is Vital for Virus Reactivation. J Virol. 2015 doi: 10.1128/JVI.00098-15. doi:10.1128/JVI.00098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hardwick JM, Bellows DS. Viral versus cellular BCL-2 proteins. Cell Death Differ. 2003;10(Suppl 1):S68–76. doi: 10.1038/sj.cdd.4401133. [DOI] [PubMed] [Google Scholar]
- (14).Gangappa S, van Dyk LF, Jewett TJ, Speck SH, Virgin HW. Identification of the in vivo role of a viral bcl-2. J Exp Med. 2002;195:931–940. doi: 10.1084/jem.20011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cheng EH-Y, Nicholas J, Bellows DS, Hayward GS, Guo H-G, Reitz MS, Hardwick JM. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Taylor GS, Blackbourn DJ. Infectious agents in human cancers: lessons in immunity and immunomodulation from gammaherpesviruses EBV and KSHV. Cancer Lett. 2011;305:263–278. doi: 10.1016/j.canlet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- (17).Zuo J, Thomas WA, Haigh TA, Fitzsimmons L, Long HM, Hislop AD, Taylor GS, Rowe M. Epstein-Barr Virus Evades CD4 T Cell Responses in Lytic Cycle through BZLF1-mediated Downregulation of CD74 and the Cooperation of vBcl-2. PLoS Pathog. 2011;7:e1002455. doi: 10.1371/journal.ppat.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lantner F, Starlets D, Gore Y, Flaishon L, Yamit-Hezi A, Dikstein R, Leng L, Bucala R, Machluf Y, Oren M, Shachar I. CD74 induces TAp63 expression leading to B-cell survival. Blood. 2007;110:4303–4311. doi: 10.1182/blood-2007-04-087486. [DOI] [PubMed] [Google Scholar]
- (19).Cojohari O, Burrer CM, Peppenelli MA, Abulwerdi FA, Nikolovska-Coleska Z, Chan GC. BH3 Profiling Reveals Selectivity by Herpesviruses for Specific Bcl-2 Proteins to Mediate Survival of Latently Infected Cells. J Virol. 2015 doi: 10.1128/JVI.00236-15. doi:10.1128/JVI.00236-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hardwick JM. Cyclin' on the viral path to destruction. Nat Cell Biol. 2000;2:E203–4. doi: 10.1038/35041126. [DOI] [PubMed] [Google Scholar]
- (21).Pattingre S, Tassa A, Qu X, Garuti R, Liang X. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- (22).Ku B, Woo J-S, Liang C, Lee K-H, Hong H-S, Kim K-S, Jung JU, Oh B-H. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. E, X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dutta S, Gullá S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fire E, Gullá S, Grant R, Keating A. Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–519. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Foight GW, Ryan JA, Gullá SV, Letai A, Keating AE. Designed BH3 Peptides with High Affinity and Specificity for Targeting Mcl-1 in Cells. ACS Chem Biol. 2014;9:1962–1968. doi: 10.1021/cb500340w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang S, Long A, Link AJ. A comparison of two strategies for affinity maturation of a BH3 peptide toward pro-survival Bcl-2 proteins. ACS Synth Biol. 2012;1:89–98. doi: 10.1021/sb200002m. [DOI] [PubMed] [Google Scholar]
- (27).Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN, Puthalakath H, Bouillet P, Colman PM, Huang DCS, Fairlie WD. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol. 2008;180:341–355. doi: 10.1083/jcb.200708096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lee E, Fedorova A, Zobel K, Boyle M. Novel Bcl-2 homology-3 domain-like sequences identified from screening randomized peptide libraries for inhibitors of the pro-survival Bcl-2 proteins. J Biol Chem 284(45), 31315-31326. 2009 doi: 10.1074/jbc.M109.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393, 403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- (30).Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- (31).Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ. 2012;19:67–74. doi: 10.1038/cdd.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Flanagan AM, Letai A. BH3 domains define selective inhibitory interactions with BHRF-1 and KSHV BCL-2. Cell Death Differ. 2008;15:580–588. doi: 10.1038/sj.cdd.4402292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Huang Q, Petros A, Virgin H, Fesik S. Solution structure of the BHRF1 protein from Epstein-Barr virus, a homolog of human Bcl-2. J Mol Biol. 2003;332:1123–1130. doi: 10.1016/j.jmb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- (34).Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- (36).Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, Carney DA, He SZ, Huang DCS, Xiong H, Cui Y, Busman TA, McKeegan EM, Krivoshik AP, Enschede SH, Humerickhouse R. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rudin CM, Hann CL, Garon EB, Ribeiro de Oliveira M, Bonomi PD, Camidge DR, Chu Q, Giaccone G, Khaira D, Ramalingam SS, Ranson MR, Dive C, McKeegan EM, Chyla BJ, Dowell BL, Chakravartty A, Nolan CE, Rudersdorf N, Busman TA, Mabry MH, Krivoshik AP, Humerickhouse RA, Shapiro GI, Gandhi L. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res. 2012;18:3163–3169. doi: 10.1158/1078-0432.CCR-11-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DCS, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park C-M, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature Medicine. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- (39).Dutta S, Ryan J, Scott Chen T, Kougentakis C, Letai A, Keating AE. Potent and specific peptide inhibitors of human pro-survival protein Bcl-xL. J Mol Biol. 2014;427:1241–1253. doi: 10.1016/j.jmb.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dutta S, Chen TS, Keating AE. Peptide ligands for pro-survival protein Bfl-1 from computationally guided library screening. ACS Chem Biol. 2013;8:778–788. doi: 10.1021/cb300679a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Procko E, Berguig GY, Shen BW, Song Y, Frayo S, Convertine AJ, Margineantu D, Booth G, Correia BE, Cheng Y, Schief WR, Hockenbery DM, Press OW, Stoddard BL, Stayton PS, Baker D. A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells. Cell. 2014;157:1644–1656. doi: 10.1016/j.cell.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting γ-herpesvirus 68 Bcl-2 mediated down-regulation of autophagy. J Biol Chem. 2014;289(12):8029–8040. doi: 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- (44).Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- (45).Lee EF, Dewson G, Evangelista M, Pettikiriarachchi A, Gold GJ, Zhu H, Colman PM, Fairlie WD. The functional differences between pro-survival and pro-apoptotic B cell lymphoma 2 (Bcl-2) proteins depend on structural differences in their Bcl-2 homology 3 (BH3) domains. J Biol Chem. 2014;289:36001–36017. doi: 10.1074/jbc.M114.610758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).DeBartolo J, Taipale M, Keating AE. Genome-wide prediction and validation of peptides that bind human prosurvival Bcl-2 proteins. PLoS Comput Biol. 2014;10:e1003693. doi: 10.1371/journal.pcbi.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Dutta S, Gullá S, Chen T, Fire E, Grant R. Determinants of BH3 binding specificity for Mcl-1 vs. Bcl-xL. J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).DeBartolo J, Dutta S, Reich L, Keating AE. Predictive Bcl-2 family binding models rooted in experiment or structure. J Mol Biol. 2012;422:124–144. doi: 10.1016/j.jmb.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).London N, Gullá S, Keating AE, Schueler-Furman O. In silico and in vitro elucidation of BH3 binding specificity toward Bcl-2. Biochemistry. 2012;51:5841–5850. doi: 10.1021/bi3003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Boersma M, Sadowsky J, Tomita Y. Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: Application to the Bim BH3 domain. Protein Sci. 2008;17:1232–1240. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhang S, Link AJ. Bcl-2 family interactome analysis using bacterial surface display. Integr Biol (Camb) 2011;3:823–831. doi: 10.1039/c1ib00023c. [DOI] [PubMed] [Google Scholar]
- (52).Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM, Fairlie WD. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14:1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
- (53).Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc Natl Acad Sci USA. 2002;99:3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kvansakul M, Yang H, Fairlie WD, Czabotar PE, Fischer SF, Perugini MA, Huang DCS, Colman PM. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 2008;15:1564–1571. doi: 10.1038/cdd.2008.83. [DOI] [PubMed] [Google Scholar]
- (55).Rautureau GJP, Yabal M, Yang H, Huang DCS, Kvansakul M, Hinds MG. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012;3:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Desbien AL, Kappler JW, Marrack P. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc Natl Acad Sci USA. 2009;106:5663–5668. doi: 10.1073/pnas.0901036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hardwick JM, Chen Y-B, Jonas EA. Multipolar functions of BCL-2 proteins link energetics to apoptosis. Trends Cell Biol. 2012;22:318–328. doi: 10.1016/j.tcb.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, Hirsch JA, Stein R, Pinkas-Kramarski R. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561–568. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- (59).Chen TS, Keating AE. Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods. Protein Sci. 2012;21:949–963. doi: 10.1002/pro.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Tsao K, Debarbieri B, Michel H. A versatile plasmid expression vector for the production of biotinylated proteins by site-specific, enzymatic modification in Escherichia coli. Gene. 1996;169:59–64. doi: 10.1016/0378-1119(95)00762-8. [DOI] [PubMed] [Google Scholar]
- (62).Lee E, Czabotar P, Yang H, Sleebs B. Conformational changes in BCL-2 pro-survival proteins determine their capacity to bind ligands. J Biol Chem. 2009;284(44):30508–30517. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Herman MD, Nyman T, Welin M, Lehtiö L, Flodin S, Trésaugues L, Kotenyova T, Flores A, Nordlund P. Completing the family portrait of the anti-apoptotic Bcl-2 proteins: crystal structure of human Bfl-1 in complex with Bim. FEBS Lett. 2008;582:3590–3594. doi: 10.1016/j.febslet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- (64).Kvansakul M, Wei AH, Fletcher JI, Willis SN, Chen L, Roberts AW, Huang DCS, Colman PM. Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog. 2010;6:e1001236. doi: 10.1371/journal.ppat.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Ku B, Liang C, Jung JU, Oh B-H. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Denisov AY, Chen G, Sprules T, Moldoveanu T, Beauparlant P, Gehring K. Structural model of the BCL-w-BID peptide complex and its interactions with phospholipid micelles. Biochemistry. 2006;45:2250–2256. doi: 10.1021/bi052332s. [DOI] [PubMed] [Google Scholar]
- (67).Chen TS, Palacios H, Keating AE. Structure-based redesign of the binding specificity of anti-apoptotic Bcl-x(L) J Mol Biol. 2013;425:171–185. doi: 10.1016/j.jmb.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Getz JA, Schoep TD, Daugherty PS. Peptide discovery using bacterial display and flow cytometry. Meth Enzymol. 2012;503:75–97. doi: 10.1016/B978-0-12-396962-0.00004-5. [DOI] [PubMed] [Google Scholar]
- (69).Hietpas R, Roscoe B, Jiang L, Bolon DNA. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat Protoc. 2012;7:1382–1396. doi: 10.1038/nprot.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- (72).Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. Chapter 2, Unit 2.9. [DOI] [PubMed] [Google Scholar]
- (73).Hinds M, Lackmann M, Skea G, Harrison P. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 2003;22(7):1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Petros A, Medek A, Nettesheim D. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci USA. 2001;98(6):3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J Biol Chem. 2011;286:7123–7131. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- (77).Smits C, Czabotar PE, Hinds MG, Day CL. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure. 2008;16:818–829. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- (78).Petros A, Nettesheim D, Wang Y, Olejniczak E. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2001;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- (80).Liu Q, Moldoveanu T, Sprules T, Matta-Camacho E, Mansur-Azzam N, Gehring K. Apoptotic regulation by MCL-1 through heterodimerization. J Biol Chem. 2010;285:19615–19624. doi: 10.1074/jbc.M110.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.