Abstract
Lymphocytes face major metabolic challenges upon activation. They must meet the bioenergetic and biosynthetic demands of increased cell proliferation and also adapt to changing environmental conditions, in which nutrients and oxygen may be limiting. An emerging theme in immunology is that metabolic reprogramming and lymphocyte activation are intricately linked. However, why T cells adopt specific metabolic programs and the impact that these programs have on T cell function and, ultimately, immunological outcome remain unclear. Research on tumor cell metabolism has provided valuable insight into metabolic pathways important for cell proliferation and the influence of metabolites themselves on signal transduction and epigenetic programming. In this Review, we highlight emerging concepts regarding metabolic reprogramming in proliferating cells and discuss their potential impact on T cell fate and function.
The immune system is comprised of a series of specialized cells conditioned to respond rapidly to “danger” signals such as foreign pathogens or inflammatory stimuli. T lymphocytes, or T cells, are sentinels of the adaptive immune system that respond to antigen-specific signals by blasting, proliferating, and differentiating into effector subsets tailored to identify and eliminate threats to the host. Integrated into this program of activation is the regulation of cellular metabolism. Upon activation, T cells dramatically alter their metabolic activity to meet the increased metabolic demands of cell growth, proliferation, and effector function. Metabolism fundamentally underpins T cell function; thus, there is great interest in understanding how metabolic pathways influence immune responses and ultimately affect disease progression. It should be noted that “metabolism” refers to a complex network of biochemical reactions involved in energy production and macromolecular biosynthesis, and comprehensive coverage of such a broad topic is difficult. Several recent reviews have highlighted the molecular mechanisms that govern metabolic reprogramming in the immune system (1–3). This Review will focus on emerging areas in intermediary metabolism in lymphocytes and will discuss their potential impact on T cell fate, plasticity, and effector function.
Differential Regulation of T Cell Metabolism
Lymphocyte Metabolism Is Dynamically Regulated
Maintenance of cellular bioenergetics is an essential function of all living cells, and lymphocytes are no exception. In T lymphocytes, glucose is a critical substrate for adenosine triphosphate (ATP) production (4). During glycolysis, glucose is broken down into two molecules of pyruvate. This process, which does not require oxygen, yields two reduced nicotinamide adenine dinucleotide (NADH) molecules and two net ATP molecules per molecule of glucose. Pyruvate has two alternate fates. Most terminally differentiated, nonproliferating cells can fully oxidize pyruvate in the tricarboxylic acid (TCA) cycle. This process generates NADH and reduced flavin adenine dinucleotide (FADH2), which the cell can use to fuel OXPHOS, an oxygen-dependent process that produces up to 36 molecules of ATP per glucose molecule. Alternatively, pyruvate can be transformed (or fermented) into lactate, regenerating NAD+ for subsequent use in glycolysis (5). From a bioenergetic perspective, engaging OXPHOS maximizes the amount of ATP that can be derived from glucose.
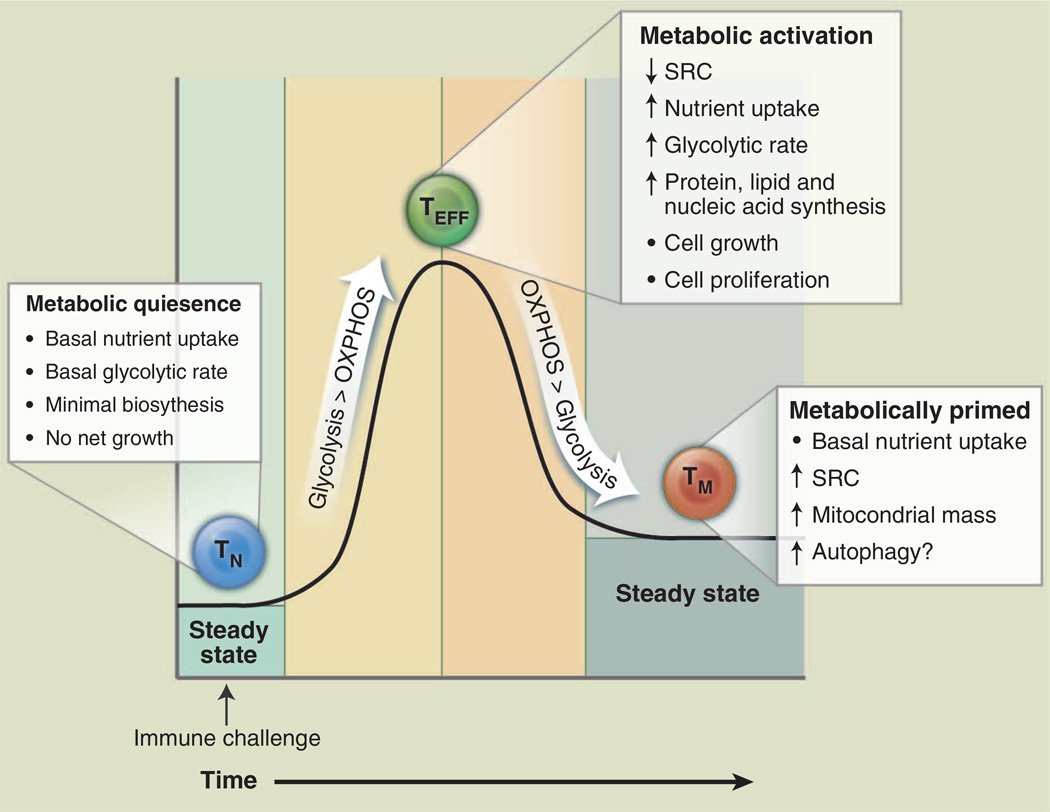
Bioenergetic profiling of T cells has revealed that T cell metabolism changes dynamically with activation state (Fig. 1). Upon antigen encounter, T cells become activated, undergo extensive proliferation, and differentiate into effector T cells (TEFF); upon pathogen clearance, most TEFF cells die, leaving behind a small population of long-lived antigen-specific memory T cells (TM). Consistent with the metabolism of other nonproliferating cells, resting naïve T cells (T cells that have not yet encountered antigen) maintain low rates of glycolysis and predominantly oxidize glucose-derived pyruvate via OXPHOS or engage fatty acid oxidation (FAO) to make ATP. Upon activation, T cells switch to a program of anabolic growth and biomass accumulation to generate daughter cells, which by definition dictates increased demand for ATP and metabolic resources. In this state, T cells are considered to be metabolically activated (Fig. 1). T cell receptor (TCR) signaling directs the metabolic reprogramming of naïve T cells. TCR ligation promotes the coordinated up-regulation of glucose and amino acid transporters (6–8), facilitating nutrient uptake and T cell blastogenesis. TCR-mediated up-regulation of the transcription factors c-Myc (9) and estrogen-related receptor α (ERRα) (10) enhances the expression of genes involved in intermediary metabolism. In addition, catabolic pathways of ATP generation such as fatty acid β-oxidation are actively suppressed (9). The predominant metabolic phenotype of activated T cells is a shift to aerobic glycolysis [reviewed in (11)]. Both CD4+ and CD8+ TEFF cells engage aerobic glycolysis, which is marked by the conversion of glucose-derived pyruvate to lactate despite the availability of oxygen for complete glucose oxidation. This process, also known as the Warburg effect from earlier work in cancer biology, is a common trait of actively proliferating cells (5). It is important to note that OXPHOS is still engaged in TEFF cells (9); however, the production of lactate from pyruvate by aerobic glycolysis is the dominant pathway of glucose metabolism in TEFF cells. Regulating energy metabolism may provide a way for T cells to reversibly switch between quiescent and highly proliferative states (12).
Fig. 1. T cell metabolism changes over the course of an immune response.
T cells display distinct metabolic profiles depending on their state of activation. Naïve T cells (TN, blue) are metabolically quiescent; they adopt a basal level of nutrient uptake and use OXPHOS as their primary pathway of ATP production. Upon immune challenge, TEFF (green) cells shift to a state of metabolic activation characterized by increased nutrient uptake, elevated glycolytic and glutaminolytic metabolism, biomass accumulation, and reduced mitochondrial SRC. TEFF cells preferentially use glycolysis over OXPHOS for ATP production. Transition to the TM (orange) stage is characterized by a quiescent metabolism, with increased reliance on FAO to fuel OXPHOS. Mitochondrial mass and SRC are elevated in TM cells, suggesting that these cells are metabolically primed to respond upon reinfection.
As a quiescent T cell population, TM cells adopt a metabolic profile similar to that of naïve T cells—a catabolic metabolism characterized by increased reliance on OXPHOS and lower rates of nutrient uptake and biosynthesis relative to TEFF cells (Fig. 1). However, TM cells also display a characteristic increase in mitochondrial mass, which translates into greater mitochondrial spare respiratory capacity (SRC) relative to naïve or TEFF populations (13). SRC can be viewed as the maximal respiratory capacity available to a cell, much like the maximum speed that can be achieved by a car engine. Under increased workload, stress, or nutrient limitation, cells engage this reserve capacity to generate more energy and promote cell viability (14, 15). We have recently shown that increased mitochondrial mass and SRC of TM cells allows for rapid mitochondrial ATP production upon TCR engagement, conferring a bioenergetic advantage to TM cells upon secondary exposure to antigen (16). From this vantage, TM cells may be viewed as being metabolically primed, with mitochondrial metabolism fueling the rapid recall response to reinfection. The memory T cell–promoting cytokine interleukin (IL)–15 plays a key role in this catabolic switch by promoting mitochondrial biogenesis (13).
The mechanisms governing the transition of T cells from effector to memory states are still poorly understood, but recent work hints that changes in metabolism may influence this process. We previously demonstrated that mitochondrial FAO stimulated downstream of TNF (tumor necrosis factor) receptor–associated factor 6 (TRAF6) is required for memory CD8+ T cell development (17). Oxidation of free fatty acids (FFAs) generates acetyl–coenzyme A (CoA), which can be metabolized further in the TCA cycle, as well as FADH2 and NADH, which can be used directly by the electron transport chain (ETC) to make ATP. FFAs are energy-dense molecules, and FAO may be a preferred fuel source for TM cells as they rely on OXPHOS-dependent metabolic program. Administration of metformin, a metabolic stressor that activates the energy sensor adenosine monophosphate–activated protein kinase (AMPK), enhances the generation of CD8+ T cell memory (17). One consequence of AMPK activation is the suppression of mammalian target of rapamycin complex 1 (mTORC1) activity in response to energetic stress (18). Consistent with this, the drug rapamycin, which also inhibits mTORC1, enhances the generation of CD8 TM cells (17, 19, 20). These observations suggest that manipulating the metabolism of antigen-specific cells during contraction can influence the development of TM cells. Given these observations, TM formation may be influenced by a number of enzymes and transporters involved in fatty acid synthesis, desaturation, and oxidation, as well as the availability of FFAs to memory precursor cells. Some important players to consider in this regard include acetyl-CoA carboxylase (ACC2) (21), the mitochondrial lipid transporter CPT1A (13, 22), and metabolites such as acetyl-CoA and malonyl-CoA (23). AMPK activation and mTOR inhibition are also both potent activators of autophagy, a catabolic process induced during starvation that promotes the degradation and recycling of cellular components [reviewed in detail in (24)]. Proper induction of autophagy has been shown to be important for the maintenance of cellular bioenergetics and sustained T cell viability after activation (25, 26). It will be interesting to determine whether autophagy, by coupling catabolic fuel supply to mitochondrial metabolism, is important for TM formation after infection.
Mitochondrial OXPHOS and T Cell Activation
Although much focus has been placed on the shift toward glycolysis that accompanies T cell activation, evidence suggests that mitochondrial OXPHOS is also important for T cell activation. Oligomycin, a specific inhibitor of mitochondrial ATP synthase, can block the expression of early activation markers after TCR ligation and blunts subsequent T cell proliferation (27), suggesting that the naïve-to-effector transition requires either de novo production of ATP by mitochondria or specific signals generated during mitochondrial ATP production. Mitochondrial-derived reactive oxygen species (ROS) may function as such a “bioenergetic” second messenger. There has long been evidence that ROS can play critical roles in shaping T cell responses (28–30). However, recent work suggests that mitochondrial ROS produced during OXPHOS is essential for T cell activation. T cells deficient for ubiquinol-cytochrome c reductase (Uqcrfs1), a component of complex III of the ETC, display impaired TCR-dependent ROS production and defects in antigen-specific proliferation (31). Intracellular calcium (Ca2+) flux, an early event in TCR signal transduction, may provide the functional link between TCR ligation, mitochondrial OXPHOS, and cell proliferation. Uptake of Ca2+ by mitochondria stimulates Ca2+-dependent dehydrogenases of the TCA cycle, driving mitochondrial NADH production and ATP production by OXPHOS during early T cell activation (32). T cells lacking the apoptosis regulators Bax and Bak, which display defects in intracellular Ca2+ homeostasis, exhibit reduced Ca2+-dependent mitochondrial ROS production and T cell proliferation after TCR stimulation (33). Restoring Ca2+ signals in Bax/Bak-null T cells restores mitochondrial ROS production and T cell proliferation (33). Thus, although toxic in many biological settings, mitochondrial-dependent ROS may prime T cells and license full T cell activation.
Metabolic Signatures Vary with Differentiation State
Although the paradigm of T cell metabolism as summarized in Fig. 1 holds true with respect to activated versus quiescent states, the metabolic signature of T cells can also vary depending on differentiation state. This was first demonstrated by Michalek et al. (34), who determined that proinflammatory CD4+ T helper (TH) cells (TH1, TH2, and TH17 lineages) displayed a strong bias toward glycolysis over mitochondrial metabolism, whereas induced CD4+ T regulatory (Treg) lineage cells displayed a mixed metabolism involving glycolysis, lipid oxidation, and OXPHOS. In particular, TH17 cells display increased reliance on glycolysis for their development and maintenance. TH17 cell development is promoted by hypoxia inducible factor–1α (HIF-1α) (35, 36), an oxygen-sensitive transcription factor that regulates glycolytic gene expression in TH17 cells. Blocking glycolysis during TH17 cell differentiation reduced the development of TH17 cells and favored the formation of Tregs (35). Added to this are recent results indicating that extracellular salt (NaCl) (37, 38) and short-chain fatty acids (39) can influence TH17 and Treg homeostasis, respectively. This raises the intriguing possibility that the metabolic microenvironment (i.e., nutrient and oxygen availability) can influence T cell polarization (to be discussed later). Determining whether the metabolic signature of differentiated T cells is simply a consequence of lineage-specific cytokine signaling or is instructive for T cell function (i.e., essential for regulating T cell plasticity and/or effector function) remains a question for the field. Examining the influence of key metabolic regulators such as HIF-1α, mTOR, and AMPK on T cell differentiation and plasticity will help in resolving these issues.
Metabolism of Proliferating Cells: Lessons from Tumor Metabolism
Research in cancer metabolism over the past 10 years has increased our understanding of the metabolic requirements of proliferating cells, as well as the metabolic alterations that promote tumor growth. One of the great innovations we have learned from the cancer metabolism field is that signaling pathways, or more specifically the oncogenes and tumor suppressors that comprise and regulate signal transduction pathways, can influence cellular metabolism as part of their program of action. The previous paradigm of metabolic regulation argued that metabolic pathways were exclusively controlled through allosteric regulation of metabolic enzymes, either by ATP levels or metabolites themselves (the reactive model) (40). One example of this is the glycolytic enzyme phosphofructokinase (PFK), which is inhibited allosterically by ATP and citrate (indicating a high energy state in the cell) and stimulated by AMP (indicative of low energy). Although allosteric regulation is important for regulating local flux through metabolic pathways, we now understand that activation of signal transduction pathways (such as phosphatidylinositol 3-kinase or Akt) by growth factor receptors stimulates global changes in metabolic flux independent of ATP levels. This allows a cell that receives a proliferative signal, such as a T cell activated through its TCR, to drive cellular metabolism above the capacity normally maintained in the quiescent state. Overall, metabolism in T cells is likely regulated at several levels: (i) TCR-mediated changes in the expression of metabolic genes facilitate the reprogramming required to match metabolic pathways to biological need; (ii) signal transduction downstream of cell surface receptors (i.e., costimulatory molecules and cytokine receptors) serve to fine-tune flux through these metabolic pathways; and (iii) feedback inhibition and other forms of allosteric regulation can regulate metabolic flux through local nodes in the network. By directly influencing metabolic reprogramming, oncogenes and tumor suppressors gain control over the metabolic currency of the cell, namely, energetic intermediates (ATP, NAD+/NADH, FAD+/FADH2, and NADP+/NADPH) and metabolites involved in bioenergetic and biosynthetic reactions that influence cell growth and survival.
The implication of these findings for immunologists is that metabolic pathways are indirectly connected to cell surface receptors of the immune system via signal transduction pathways. TCR/CD28 stimulation of T cells (6), the stimulation of surface immunoglobulin on B cells (41), and TLR stimulation of macrophages and dendritic cells (DCs) (42, 43) all promote changes in aerobic glycolysis characteristic of the Warburg effect. These results likely just scratch the surface of the complex metabolic networks at work in proliferating cells. The challenge going forward will be to identify key pathways of metabolic flux integral for lymphocyte function. In this regard, research into tumor cell metabolism has provided valuable insight into the metabolic pathways important for cell proliferation. Many of the metabolic pathways abnormally activated in cancer, such as aerobic glycolysis, have been shown to play similar roles in normal lymphocyte physiology. Here, we highlight recent advances in intermediate metabolism observed in cancer that are likely to be relevant to T cell biology.
The Warburg Effect: More than ATP Synthesis
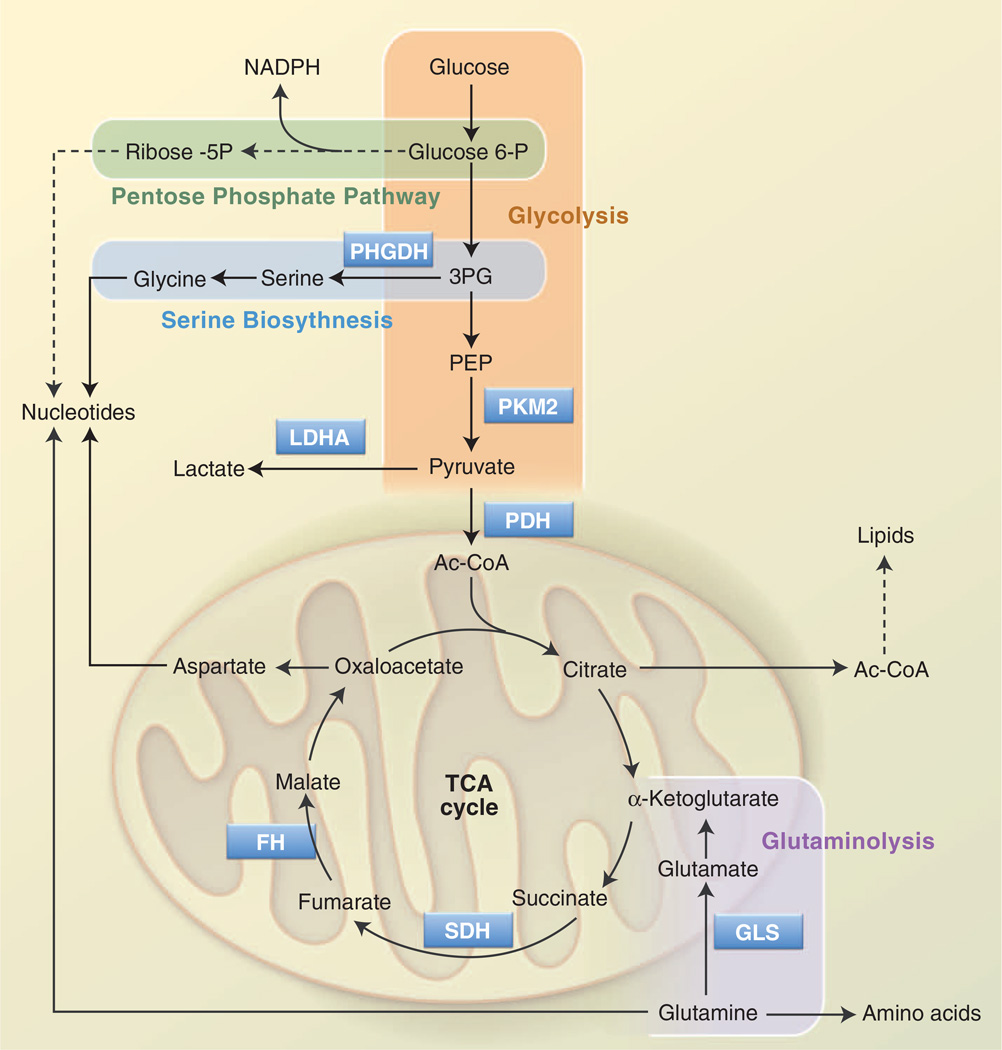
Rapid glucose processing promoted by the Warburg effect allows proliferating T cells to generate ATP quickly; glycolysis also generates metabolic intermediates important for cell growth and proliferation (Fig. 2). Metabolism of glucose through the oxidative or nonoxidative arms of the pentose phosphate pathway (PPP) generates ribose-5-phosphate (Rib-5P), a key intermediate in nucleotide biosynthesis. The oxidative arm of the PPP also produces NADPH, the key metabolic currency for nucleotide and fatty acid biosynthesis. T cell activation promotes a rapid increase in glucose flux through the oxidative PPP (9). Dihydroxyacetone-phosphate (DHAP) is used to generate the glycerol backbone for glycerophos-pholipids, and 3-phosphoglycerate (3PG) is a key intermediate in both amino acid and nucleotide biosynthesis (discussed below). Pyruvate that is not converted to lactate can enter into the mitochondria and be converted into acetyl-CoA by the pyruvate dehydrogenase (PDH) complex. In proliferating cells, mitochondria adopt an additional role as a biosynthetic hub, converting pyruvate and other metabolites into metabolic intermediates involved in protein and fatty acid biosynthesis (44).
Fig. 2. Metabolic pathways that support cell growth and proliferation.
Glycolysis and the TCA cycle are two separate yet connected biochemical pathways that function to generate ATP as well as metabolic precursors for biosynthesis. Glucose is broken down to pyruvate by glycolysis (orange); pyruvate can be further oxidized by the TCA cycle in the mitochondrion. Glycolytic intermediates can be used to generate other metabolites required for growth and proliferation. Glucose 6-phosphate and 3PG produced from glycolysis are metabolized in the PPP (green) and the SBP (blue), respectively, providing important precursors for nucleotide biosynthesis. Similarly, acetyl-CoA, generated from glucose-derived citrate in the TCA cycle, can be used for lipid biosynthesis. OAA, produced as part of the TCA cycle, can be used to generate aspartate, another precursor for nucleotide synthesis. An alternate source of carbon for the TCA cycle occurs via glutaminolysis (purple); in this pathway, glutamine is converted to glutamate and then to α-KG, which joins the TCA cycle. Glutamine is also a precursor for amino acid and nucleotide biosynthesis. Key enzymes in these pathways are PHGDH; PKM2; LDHA, lactate dehydrogenase; PDH; GLS, glutaminase; SDH, succinate dehydrogenase; and FH.
One of the key metabolic intermediates for biosynthesis is acetyl-CoA. Acetyl-CoA has a central role in membrane biogenesis because it provides two-carbon units for fatty acid and isoprenoid biosynthesis, as well as in other diverse processes such as protein prenylation and N-glycosylation (45). The flow of glucose to the cytosolic acetyl-CoA pool is regulated by using TCA cycle intermediates and a truncated TCA cycle (46). In this model, mitochondrial citrate is formed from condensation of oxaloacetate and acetyl-CoA, after which citrate is exported from the mitochondrion to the cytosol and converted back to acetyl-CoA by ATP citrate lyase (ACL) (47). Despite the availability of extracellular lipids for membrane biosynthesis, FFAs are generated de novo from glucose in proliferating cells using this pathway (46). CD8+ T cells unable to engage acetyl-CoA–dependent lipid biosynthetic pathways display defects in antigen-driven blastogenesis and clonal expansion in response to pathogens (48). Acetyl-CoA can also influence metabolic flux through acetylation of metabolic enzymes (49, 50), reinforcing metabolic pathways such as glycolysis when carbon availability is high. Glucose availability and acetyl-CoA production can also influence epigenetics by regulating the cytosolic acetyl-CoA pools available for histone acetylation reactions (51), raising the prospect that glucose-dependent metabolic flux may help drive or reinforce T cell differentiation programs.
One consequence of using glucose-derived mitochondrial citrate for lipid biosynthesis is the potential depletion of TCA cycle intermediates. Oxaloacetate (OAA) is a rate-limiting substrate for acetyl-CoA entry into the TCA cycle. OAA generated from ACL-mediated cleavage of cytosolic citrate can potentially cycle back into the mitochondria to maintain the TCA cycle. Inefficient cycling of this pathway would lead to cumulative depletion of mitochondrial OAA, leading to collapse of the TCA cycle and disruption of mitochondrial function. One way tumor cells counter this is by engaging glutaminolysis, a metabolic shunt that converts glutamine into α-ketoglutarate (α-KG, also known as 2-oxoglutarate) for use in the TCA cycle (Fig. 2). Glutamine has long been known as a key metabolite for supporting T cell function (52). Recent evidence suggests that glutamine metabolism is as dynamically regulated in T cells as glucose metabolism. Glutamine transporters (SNAT1 and SNAT2) as well as key glutaminolysis enzymes (GLS, GPT, GOT, and GLUD) are up-regulated early after T cell activation similar to glycolysis genes (7, 9); several groups have correlated these changes in gene expression to enhanced glutaminolytic flux in lymphocytes (9, 53). Recently, it was found that engagement of the TCR leads to the expression of Slc7a5, an amino acid transporter that mediates the import of large neutral amino acids, such as leucine (8). Amino acid influx via this transporter is required for the activation of mTOR and expression of c-Myc and as such coordinates activation-induced metabolic reprogramming and differentiation of T cells. Thus, amino acids such as glutamine and leucine appear to play additional roles in T cell function beyond protein biosynthesis and may directly influence T cell activation by regulating metabolic reprogramming.
Control of Glycolytic Flux by Pyruvate Kinase M2 (PKM2)
Pyruvate kinase (PK) is a key enzyme of the glycolytic pathway. It catalyzes the terminal reaction of glycolysis by promoting the conversion of phosphoenolpyruvate (PEP) to pyruvate (Fig. 2), and is one of two ATP-generating steps of glycolysis (the second is mediated by phosphoglycerate kinase). The muscle version of PK exists as one of two isoforms, M1 or M2, generated from differential splicing of the PKM primary mRNA transcript, with the PKM2 splice variant expressed in embryonic tissues, proliferating cells, and tumor cells (54). Naïve T cells express both M1 and M2 isoforms at rest; mitogen-dependent activation promotes the rapid accumulation of the M2 isoform, which becomes the dominant isoform expressed in TEFF cells (55). PKM2 exists as either an inactive dimer or an active tetramer, and oscillation between these two states influences the ability of cells to maintain anabolic metabolism (56).
Multiple lines of evidence point to a role for PKM2 in coordinating glycolytic flux and cell proliferation. Tumor cells engineered to exclusively express the M2 isoform display increased lactate production characteristic of the Warburg effect and gain a growth advantage in vivo over M1-expressing tumors (57). Interestingly, the M2 isoform is actually less efficient at converting PEP to pyruvate than the M1 isoform. Moreover, growth-factor–stimulated tyrosine phosphorylation of PKM2 further decreases its activity (58, 59). This prompts the question: Why would proliferating cells, including T cells, promote the expression of a pyruvate kinase isoform that is less efficient at generating ATP? The answer to this question may lie with the second function of Warburg metabolism, namely, supporting anabolic growth. Buildup of PEP because of reduced PKM2 activity promotes the accumulation of glycolytic intermediates, which can then be shunted into upstream biosynthetic pathways to support amino acid, triglyceride, and nucleotide biosynthesis. Activating PKM2 by using small-molecule agonists increases PEP-to-pyruvate conversion, reducing the flux of glycolytic intermediates toward anabolic pathways and slowing tumor cell growth (56, 60). Thus, the ability of PKM2 to support cell proliferation may have less to do with ATP production and more with supporting biosynthetic pathways required for tumor cell growth.
PKM2 may also exert some of its effects on cell proliferation through nonmetabolic functions, including transcriptional and epigenetic regulation (61, 62). Of particular interest to immunologists is that PKM2 can phosphorylate signal transducer and activator of transcription 3 (STAT3) at Tyr705, promoting increased STAT3-dependent transcription (63). The phosphorylation of STAT3 by PKM2 demonstrates that PKM2 possesses both protein and pyruvate kinase activities, the former using PEP as a phosphate donor rather than ATP. The protein kinase activity of PKM2 (and any subsequent effects on STAT3 activity) would be predicted to be sensitive to metabolic flux, favored under high-glycolysis PEP conditions and antagonized by low-energy high-ADP concentrations. The development of mouse models to study the impact of PKM2 activity in vivo will be important for elucidating the role(s) of PKM2 in immune function.
The Serine Biosynthesis Pathway
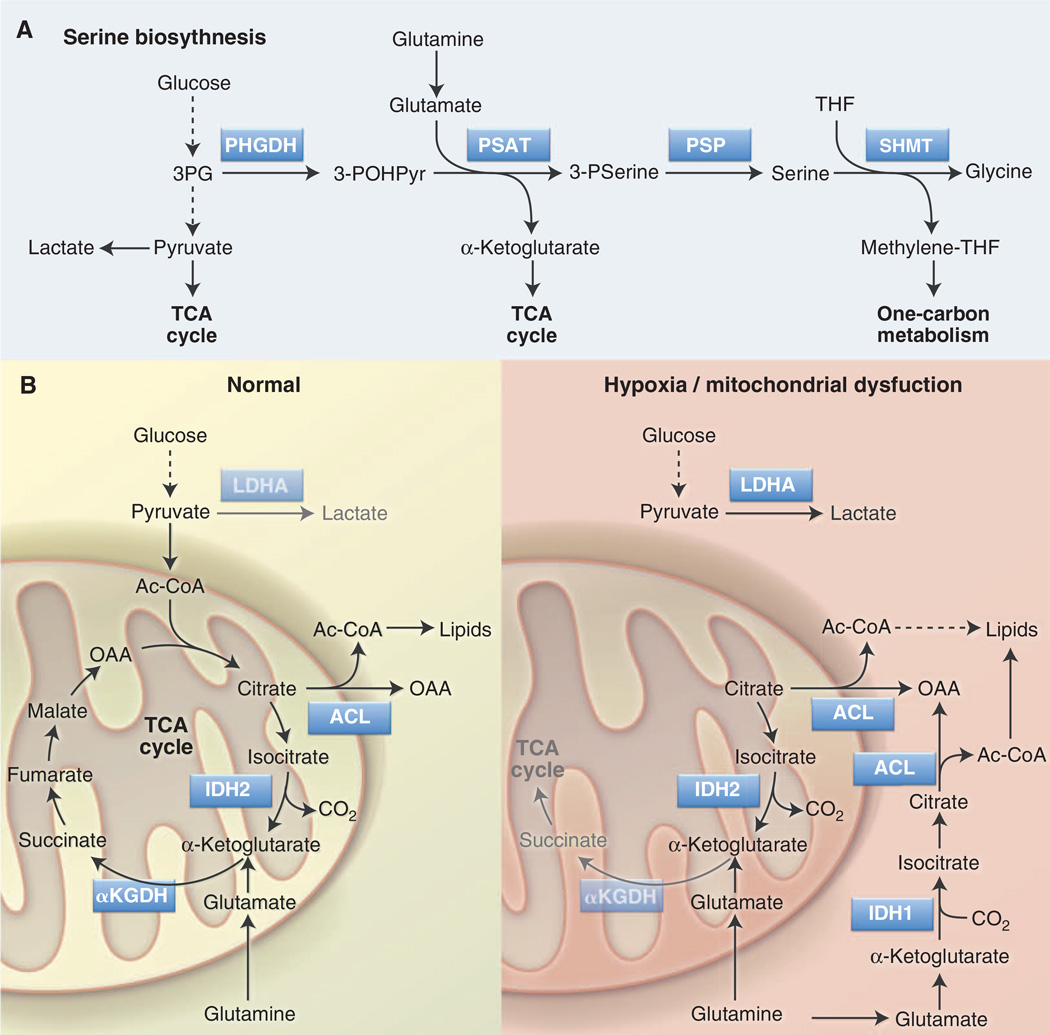
Another glycolytic intermediate that can double as an anabolic precursor is 3PG. 3PG is the starting point for the glucose-dependent biosynthesis of serine and glycine via the serine biosynthesis pathway (SBP) (Fig. 3A). Key enzymes of the SBP are phosphoglycerate dehydrogenase (PHGDH), the rate-limiting step for serine biosynthesis, and serine hydroxymethyltransferase (SHMT), which uses serine as a methyl donor to convert tetrahydrofolate (THF) to methylene-THF, generating glycine in the process. Methylene-THF is a key intermediate in folate-mediated one-carbon metabolism that fuels nucleotide biosynthesis and methylation reactions. Serine is also an allosteric activator of PKM2 (64) and thus provides feedback to the glycolytic pathway to regulate 3PG levels and serine biosynthesis.
Fig. 3. Serine biosynthesis and reductive carboxylation are anabolic pathways that support cell proliferation.
(A) The SBP converts glucose-derived 3PG into serine and glycine, which are precursors for lipid and nucleotide biosynthesis. Serine is also involved in folate-mediated one carbon metabolism by acting as a methyl group donor for THF to methylene-THF conversion. Key enzymes in this pathway are PHGDH, PSAT, and SHMT. (B) Reductive carboxylation is an alternate pathway of glutamine metabolism in which glutamine-derived α-KG is converted to citrate through reverse TCA cycle flux. Under conditions of hypoxia or mitochondrial dysfunction (right), isocitrate dehydrogenase (IDH1 in cytosol, IDH2 in mitochondria) uses CO2 and NADPH to convert α-KG into isocitrate. Citrate produced downstream of this reaction is converted into cytosolic acetyl-CoA without passing through the conventional clockwise steps of the TCA cycle. Acetyl-CoA generated by this pathway can function as a precursor for fatty acid synthesis. α-KGDH, α–KG dehydrogenase.
In mammals, serine and glycine are non-essential amino acids and are widely abundant in serum (and tissue culture medium). However, glucose-dependent serine biosynthesis is actively engaged in some tumors regardless of serine abundance. Amplification of PHGDH has been observed in breast cancer and melanoma (65), and increased PHGDH expression can promote both enhanced serine biosynthesis and the proliferation of cancer cells (65, 66). Enhanced flux through the SBP may confer a growth advantage to tumor cells beyond providing increased serine and glycine for biosynthetic reactions. First, PHGDH produces NADH, which can be used to maintain cytosolic redox balance or fuel mitochondrial OXPHOS to make ATP. The second step of the pathway, the conversion of 3-phosphohydroxypyruvate to 3-phosphoserine by phosphoserine aminotransferase (PSAT), requires glutamate and produces α-KG (Fig. 3A) (66). Thus, the SBP may promote an alternate pathway of α-KG production for mitochondrial metabolism or promote the activity of α-KG– dependent enzymes (to be discussed later). Last, serine and glycine are both intermediates in the production of reduced glutathione (GSH), a key cellular antioxidant. Recent evidence indicates that cancer cells actively produce GSH from glucose via the SBP as a buffer against oxidative damage (67). The expression of SBP enzymes is up-regulated in Myc-driven lymphomas (68); thus, we hypothesize that TEFF cells may actively engage the SBP upon activation, even in the presence of exogenous serine. Future investigation will be needed to determine how the SBP influences T cell biology.
Reductive Carboxylation of α-KG
As mentioned previously, it is now appreciated that in proliferating cells the TCA cycle functions as a source of biosynthetic precursors in addition to its role in ATP production (46). Carbon enters the TCA cycle primarily at one of two entry points: (i) the condensation of glucose-derived acetyl-CoA with OAA to generate citrate and (ii) conversion of glutamine to α-KG via glutaminolysis (Fig. 3B). Carbon-tracing experiments using proliferating glioblastoma cells have established that both glucose and glutamine contribute to mitochondrial citrate pools and subsequent lipid synthesis (69). The oxidative decarboxylation of isocitrate to α-KG by isocitrate dehydrogenase (IDH) is highly favored thermodynamically, such that this reaction is believed to be irreversible and the reason for the “clockwise” flow of metabolic intermediates through the TCA cycle.
Groundbreaking new work suggests that metabolite flow through biochemical pathways does not always conform to conventional dogma. Although oxidation of glutamine-derived α-KG in the TCA cycle serves as a minor source of lipogenic acetyl-CoA under normal growth conditions, α-KG can be converted to citrate through reductive carboxylation under conditions of stress such as hypoxia or mitochondrial dysfunction (70–72). In this reaction, glutamine-derived α-KG is carboxylated, rather than decarboxylated, by IDH1 (in the cytosol) or IDH2 (in mitochondria) to generate citrate (Fig. 3B). This switch to reductive versus oxidative metabolism of α-KG is regulated in part by HIF-1α (71, 72), although the exact mechanism by which HIF-1α does so remains unclear. Engaging reductive carboxylation of α-KG in essence bypasses the conventional TCA cycle by using glutamine to generate the acetyl-CoA required for fatty acid synthesis (Fig. 3B). Under such metabolic reprogramming, cancer cells continue to use glycolysis for ATP production but switch from glucose to glutamine as the major lipogenic precursor.
One implication of this work is that tumor cells display a high degree of metabolic plasticity and can adapt their metabolism to support proliferation and viability under fluctuating environmental conditions. Whether T cells display similar metabolic flexibility in response to environmental cues requires further investigation. The work from Metallo and colleagues (71) suggests that T cells can use reductive glutamine metabolism for fatty acid biosynthesis under hypoxic conditions. Physiologic oxygen tension varies significantly between tissues, and hypoxic regions can be detected in the bone marrow, thymus, and spleen (73). Moreover, T cells are likely to experience hypoxia at sites of tissue inflammation (74). Stabilization of HIF-1α and metabolic reprogramming to favor reductive carboxylation may help T cells maintain proliferation and/or effector function at hypoxic inflammatory sites. HIF-1α protein expression has also been observed early after T cell activation (75), so it is unclear whether reductive carboxylation may also be engaged as part of the normal program of T cell expansion. As mentioned, HIF-1α has been implicated in the differentiation of both proinflammatory TH17 cells (35, 36) and CD4+FoxP3+ Treg cells (76). It is tempting to speculate that this alternate pathway of glutamine metabolism may influence the expansion or phenotypic stability of these T cell subsets.
Metabolites As Signaling Molecules
Cancer genome sequencing efforts yielded an unexpected discovery in 2008 with the identification of somatic mutations in a metabolic enzyme, the TCA cycle enzyme IDH1, in glioblastoma multiforme (GBM) (77). Mutations in IDH1 presented at significant frequency (∼12% of GBM patients) with high frequency of missense mutations targeting an arginine residue in the active enzyme site (Arg132). This mutation alters the enzymatic ability of IDH1, allowing it to convert α-KG to 2-hydroxyglutarate (2-HG) rather than promote normal isocitrate–to–α-KG inter-conversion (78). In the cancer setting, 2-HG has been shown to influence a number of biological processes, including cell differentiation, DNA methylation, and histone methylation (79–82), leading to its classification as an oncometabolite. These results have shifted thinking in cancer biology to consider that specific metabolites may also act as signaling molecules to influence cell physiology. Given certain similarities between metabolic programming in tumor cells and proliferating T cells, it stands to reason that metabolites will participate in T cell signaling as well. Some examples of how metabolites shape T cell function and fate through metabolic pathways are discussed in detail here.
Regulation of LKB1-AMPK Signaling by Adenylates
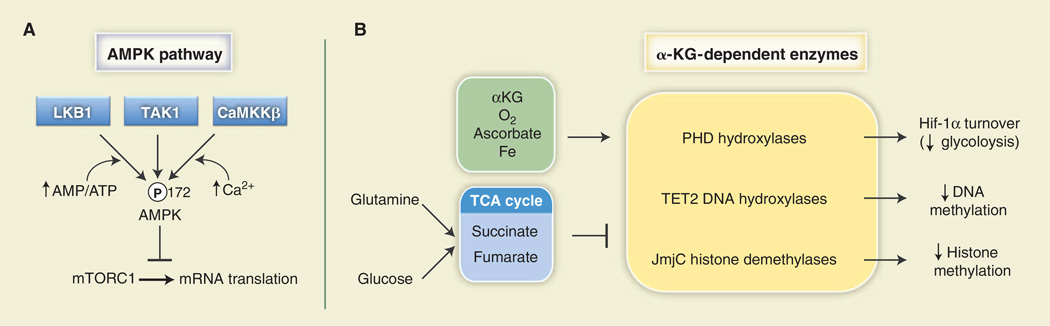
ATP is the primary carrier of chemical energy in the cell and essential for life. Thus, adenylates [ATP, adeonsine diphosphate (ADP), and AMP] are important units of cellular metabolic currency. In mammalian cells, fluctuations in cellular energy are monitored by the heterotrimeric AMPK complex [reviewed in (83)]. ATP, ADP, and AMP compete for nucleotide-binding sites of the γ regulatory subunit of AMPK, with AMP (low energy) promoting and ATP (high energy) antagonizing AMPK activation. As such, AMPK functions as a sensor of the cellular adenylate energy charge (84, 85). Elevation of the cellular AMP:ATP ratio leads to increased phosphorylation of AMPK at Thr172 of its activation loop by the kinase LKB1 (Fig. 4A). AMPK can also be activated by Ca2+ (via CamKKβ) and the cytokine transforming growth factor–β (TGF-β) (via TAK1), although LKB1 appears to be the sole kinase that couples adenylate binding to AMPK activation.
Fig. 4. Metabolites can influence signal transduction.
(A) The AMPK pathway is influenced by adenylate concentration. AMPKα is activated by phosphorylation on Thr172 of its activation loop by the kinases LKB1, TAK1, or CaMKKβ. LKB1 promotes enhanced AMPK phosphorylation under a high AMP:ATP ratio. One biological output of AMPK activity is the inhibition of mRNA translation under low-energy conditions through inhibition of mTORC1 activity. (B) α-KG– dependent enzymes (in yellow) are a class of enzymes regulated by TCA cycle intermediates that require molecular oxygen (O2) and α-KG for their enzymatic activity. Oxygen, α-KG, ascorbate, and iron (green) are positive regulators of these enzymes, whereas the TCA cycle intermediates fumarate and succinate (blue) antagonize their reactions. PHDs destabilize HIF-1α protein, resulting in decreased expression of HIF-1α targets and a reduction in glycolysis. TET2 hydroxylates 5-methylcytosine residues to promote DNA demethylation, whereas JmjC promotes demethylation of trimethylated histones in chromatin.
Together LKB1 and AMPK function as part of an evolutionarily conserved energy-sensing pathway that maintains cellular energy balance by promoting catabolic pathways of ATP production and limiting processes that consume ATP. Protein synthesis is one of the most energy-consuming processes in the cell, accounting for ∼20% of cellular ATP consumption (86). As mentioned, AMPK antagonizes mRNA translation through negative regulation of mTORC1 (Fig. 4A) (18). By regulating AMPK activity, adenylates directly influence pathways of energy usage in the cell. As ATP levels drop, AMP binds to AMPK, and AMPK is switched on to promote ATP production and block its consumption; once ATP homeostasis has been reestablished, increased binding of ATP to AMPK shuts off the kinase.
Recent work indicates that LKB1-AMPK signaling can influence T cell metabolism and function. Lymphocytes exclusively express the a1 catalytic subunit of AMPK (87), and TCR stimulation promotes LKB1-dependent AMPK activation in lymphocytes (87, 88). Glycolysis is enhanced in resting AMPKα-deficient T cells (88), consistent with observations that silencing AMPK in tumor cells promotes the Warburg effect (89). Loss of LKB1-AMPK signaling promotes increased mTORC1 activity in both naïve and TEFF cells (88, 90), which in turn facilitates production of the TH1 cytokine interferon-γ (IFN-γ) by TEFF cells (88). Thus, LKB1 and AMPK act in concert to limit the anabolic growth of T cells by suppressing glycolysis and mTOR-dependent biosynthesis. Perhaps not surprisingly, deletion of either LKB1 or AMPKα1 disrupts normal lymphocyte homeostasis, resulting in an accumulation of activated (CD44hi CD62Llo) CD8+ T cells in animals (88). These results suggest that cellular adenylate levels and AMPK may help regulate lymphocyte pools in the whole organism.
Why would a signaling pathway that normally monitors cellular energy levels regulate T cell function? Similar to tumor cells (91), AMPK may regulate a metabolic checkpoint in T cells, acting as a brake on lymphocyte expansion when energy conditions are poor. TEFF cells with defective AMPK signaling would be freed from these metabolic checkpoints and continue to proliferate and produce cytokines as if cellular bioenergetics were suitable to support T cell function. Additionally, AMPK may regulate the metabolic plasticity of lymphocytes, coordinating metabolic changes in response to nutrient fluctuation and allowing Tcells to survive changes in their environment. As evidence for this, the AMPK agonist metformin, which promotes FAO in activated T cells, enhances the production of CD8+ memory T cells in vivo (17). Furthermore, it was recently shown that AMPK-deficient T cells are defective in their ability to generate CD8+ memory T cells during infection (92). There is also a growing body of evidence implicating AMPK in the control of inflammation (3). Future work will focus on the role of AMPK in regulating the metabolic fitness of lymphocytes, dissecting LKB1-and AMPK-specific effects on immune function and investigating the role(s) of LKB1 and AMPK in regulating inflammation in vivo.
Regulation of α-Ketoglutarate-Dependent Enzymes by TCA Cycle Metabolites
Although TCA cycle metabolites play central roles in energy metabolism, many function as chemical intermediates in other biological reactions. For example, fumarate can be used to chemically modify cysteine residues of proteins, resulting in the formation of S-(2-succinyl)cysteine or succinylation of these residues (93). Cancer cells harboring mutations in fumarate hydratase (FH), the TCA cycle enzyme that converts fumarate to malate, accumulate intracellular fumarate and display increased amounts of succinylated proteins (94). Increased protein succinylation has been associated with renal carcinoma and mechanistically can influence signal transduction pathways similar to protein phosphorylation or acetylation (95).
α-KG is another key metabolite involved in non-TCA cycle biochemical reactions. α-KG is essential for the activity of a class of enzymes known as α-KG–dependent dioxygenases (Fig. 4B). These enzymes use α-KG along with molecular O2, iron, and ascorbate to modify target proteins (96). The most extensively characterized enzymes of this family are the HIF prolyl hydroxylases (PHDs). PHDs use O2 to hydroxylate HIF-1α on two conserved proline residues, facilitating their recognition by the E3 ubiquitin ligase VHL and promoting HIF-1α degradation by the proteosome. When O2 availability is low, HIF-1α protein is stabilized because of reduced PHD activity, resulting in increased HIF-1α-dependent transcription (97). This regulatory circuit allows HIF-1α to promote glycolytic ATP production when O2 cannot support mitochondrial OXPHOS, an example of metabolic adaptation in response to environmental conditions. Other α-KG–dependent enzymes using this chemistry are the TET family of DNA hydroxylases, which hydroxylate 5-methylcytosine residues to promote DNA demethylation, and the Jumonji C (JmjC) class of histone demethylases (Fig. 4B). A detailed summary of these processes has recently been reviewed (98).
The activity of α-KG–dependent enzymes is directly affected by the abundance of TCA cycle intermediates. For example, 2-HG acts as a competitive inhibitor of α-KG, and its production by mutant IDH1 consumes α-KG (81, 99), leading to reduced TET2 and JmjC activity in tumor cells (79, 80). High levels of succinate and fumarate can inhibit PHD2 activity through product-mediated inhibition of PHD function, leading to HIF-1α protein stabilization under normoxic (20% O2) conditions (100, 101). Additionally, total abundance of α-KG is low in most cell types. The implication of these findings is that metabolic flux through the TCA cycle can affect gene transcription and/or epigenetic programs. It was recently shown that succinate plays a central role in production of the cytokine IL-1 β by lipopolysaccharide (LPS)–stimulated macrophages (102). LPS induces the de novo production of succinate from glutamine, leading to PHD inhibition, stabilization of HIF-1α, and increased HIF-1α-dependent IL-1β production. Mutant IDH2 [Arg140→Gln140 (R140Q)], which promotes α-KG depletion/2-HG production, promotes DNA hypermethylation in hematopoietic cells and can inhibit myeloid differentiation when overexpressed in bone marrow stem cells (79). Differentiation of T cells to specific TH lineages is driven by specific transcription factors but reinforced by epigenetic modifications on histone tails (H3K4 and H3K27 trimethylation) and DNA methylation of CpG dinucleotides (103). Differential TCA cycle flux or buildup of intermediates such as succinate or fumarate may influence the activity of α-KG-dependent enzymes that regulate T cell epigenetics. This may be one way in which the environment (O2, nutrient levels) can influence the plasticity of TH lineages at sites of infection or inflammation in vivo. Thus, much like the role of oncometabolites in tumorigenesis, studying the metabolism of T cell responses may reveal the existence of “immunometabolites” that influence T cell responses and inflammation.
Connecting Metabolism and Gene Regulation
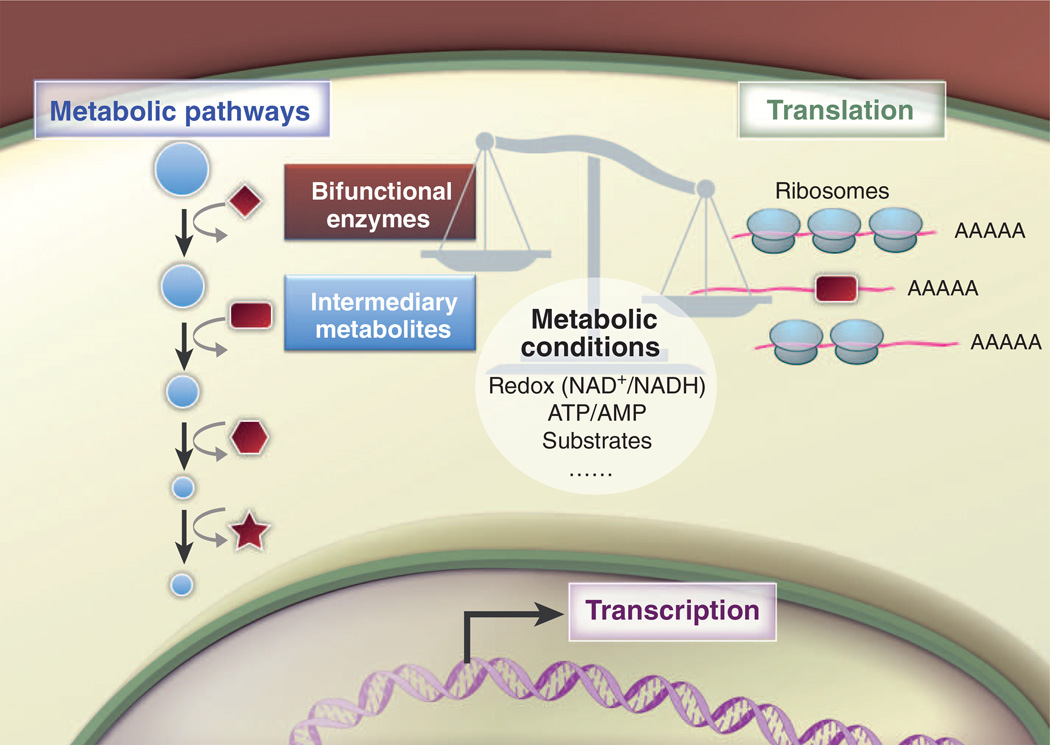
A transformative conceptual change in the way we consider metabolism within cells is that changes in metabolism can be linked to changes in gene expression through posttranscriptional regulatory networks involving RNA, metabolites, and metabolic enzymes “moonlighting” as RNA binding proteins and regulating specific target mRNAs (104) (Fig. 5). Many enzymes, including those connected with the TCA cycle, glycolysis, PPP, fatty acid metabolism, and other pathways, have been shown to bind RNA in vitro and in cultured cells (105, 106). In addition, the RNA binding function of enzymes can be influenced by interactions with their metabolites or cofactors, illustrating how the metabolic state of the cell can control an enzyme’s RNA binding function.
Fig. 5. Bifunctional metabolic enzymes connect metabolism and gene regulation.
Metabolic enzymes can moonlight as RNA binding proteins and regulate the translation of specific target mRNAs. The RNA binding function of enzymes can be influenced by interactions with intermediary metabolites and cofactors, leading to posttranscriptional regulation of protein expression. Posttranslational modification of metabolic enzymes could influence their RNA binding function directly or by altering the enzyme’s subcellular location. Changes in metabolic conditions, such as bioenergetic demand, hypoxia, stress, and substrate availability, may affect the consequences of the REM interactions. The overall balance of the network between RNA, enzymes, and metabolites can potentially influence T cell fate and function.
The metabolic enzyme perhaps best characterized for its physiological role as an RNA binding protein is cytosolic aconitase (107, 108), a key regulator of cellular iron metabolism (109). In the early 1990s, it was shown that cytosolic aconitase and the RNA binding protein IRP-1 represent the same polypeptide and that the availability of iron triggers insertion or removal of an iron sulfur cluster—switching the protein’s function between RNA binding activity (iron low, IRP-1) and metabolic enzyme activity (iron high, aconitase) (110). Remarkably, IRP-1 binds to and regulates mRNAs encoding proteins that function in iron homeostasis. Work from this group and others has led to the idea that bifunctional enzymes and RNA binding proteins may represent a general mechanism of how metabolism and gene expression are coordinated through RNA/enzyme/metabolite (REM) networks [proposed by Hentze and Preiss in (104)].
Lending weight to the REM network hypothesis is a recent study showing that the enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH), by engaging or disengaging the glycolysis pathway and through fluctuations in its expression, regulates the posttranscriptional production of IFN-γ by T cells (27). GAPDH is a multifunctional enzyme that can bind a range of RNAs, including AU-rich regions in the 3’ untranslated region (UTR) of cytokine mRNAs, including IFN-γ and IL-2 (111). Activated T cells can use either OXPHOS or glycolysis to generate energy to support proliferation and survival. When T cells switch between these ATP-generating programs, as can occur with changes in nutrient availability, or costimulatory or growth factor signals, GAPDH switches from its function as a metabolic enzyme (glycolysis) to its function as an RNA binding protein controlling expression of immunomodulatory factors. When GAPDH is not engaged in glycolysis and OXPHOS is used for ATP production, it binds the 3′UTR of cytokine mRNAs, and translation of these mRNAs is dampened. Thus although OXPHOS can support T cell survival and proliferation, only aerobic glycolysis can facilitate full effector status. This regulatory mechanism provides a checkpoint to allow for uncoupling of T cell proliferation and survival from cytokine production. This is a desirable control mechanism over effector function, because T cells are required to undergo both homeostatic proliferation, when IFN-γ production is neither required nor desirable, and antigen-driven proliferation during an immune response, when effector cytokine production is essential.
The activity of GAPDH is not only controlled by pathway engagement as dictated by substrate availability but is also heavily influenced by redox balance within the cell. For example, GAPDH requires NAD+ for its enzymatic function, but NAD+ also interferes with mRNA binding, at least in vitro (112). Thus, NAD+ controls both enzyme activity and RNA binding in a mutually exclusive fashion. Metabolic enzymes that regulate NAD+/NADH balance, including LDHA and PHGDH, may also influence this process. Redox changes may also affect posttranslational modifications of GAPDH, altering its binding to mRNA, metabolites, as well as its localization within the cell (113, 114). This level of regulation between redox balance and RNA binding would be expected to occur with other RNA binding enzymes that share similar dinucleotide binding motifs with GAPDH. Although much work needs to be done to fully understand the biological importance of the interactions between enzymes, RNA, and metabolites, these observations clearly demonstrate how cofactors and substrates generally considered for their direct effects on metabolism may also coordinate metabolism with gene expression.
Future Challenges and Other Considerations
Technical Challenges in Studying Lymphocyte Metabolism
Although interest in studying lymphocyte metabolism and technological advances in metabolomics have increased over the past several years, the field faces many challenges going forward. For instance, although equipment for studying basic metabolism (i.e., oxygen consumption and proton production) is becoming more commonplace, specialized equipment for metabolite measurements [i.e., mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy] and analytical expertise is not routinely accessible to investigators. Another limitation is that, unlike microarray or sequencing technologies, metabolomic analyses do not adhere to a single global platform. For example, gas chromatography coupled to MS (GC-MS) is effective at quantifying organic acids such as TCA cycle intermediates but not most glycolytic intermediates. Thus, multiple platforms must be used to generate complete metabolite data sets. Current extraction methods do not allow for the measurement of subcellular metabolite pools, so information on metabolite localization or channeling between organelles is also lost. One caution of measuring steady-state metabolite levels is that these data provide no measurement of metabolite flux, that is, the speed or direction of metabolite flow through a given pathway (115). Metabolic flux analysis using isotopically labeled metabolites such as 13C-glucose or 13C-glutamine will be essential for delineating pathways of metabolite flow in T cells.
Perhaps the most relevant challenge for immunologists is the amount of material required for metabolomic analysis. Because of advances in flow cytometry and the identification of new biomarkers to define T cell subsets, immunology has entered an era of cellular subspecialization, where rare cell populations are readily characterized. In comparison, a typical metabolic flux experiment requires millions of cells. This raises the issue of having limited material to analyze the metabolism of cells grown in vitro, let alone in vivo. The development of instrumentation with increased sensitivity will help reduce this gap, but better sample preparation and techniques to amplify metabolite signals are badly needed. The development of genetically encoded fluorescent biosensors for metabolic activity, such as those recently developed for NADH (116) and cellular energy charge (117), will be particularly powerful for studying T cell metabolism at the single-cell level.
Microenvironmental Effects on Metabolism: Are Our Model Systems Correct?
One of the ongoing questions regarding current experimental models in immunology is how closely cell culture models recapitulate immune responses in vivo. From a metabolic perspective, one can conclude that the two systems are worlds apart. Activated T cells cultured in standard medium (Iscove’s modified Dulbecco medium plus 10% serum) experience 25 mM glucose (five times standard blood glucose of 5 mM), 4 mM glutamine (eight times the standard concentration of 0.5 mM in blood), and 20% O2 [two to four times the oxygen tension in blood, which varies depending on tissue type (73)]. Most in vitro assays to assess T cell function are performed at nutrient and O2 levels much higher than seen in normal physiology; these conditions model a hyperglycemic and hyperoxic environment never seen in vivo.
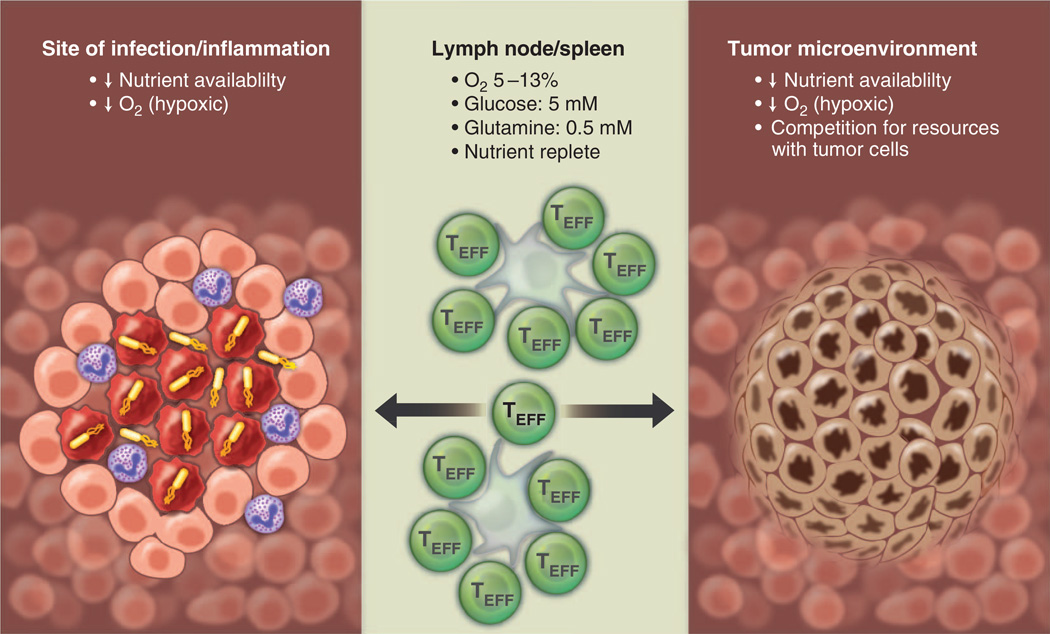
As TEFF cells move from a nutrient-replete environment in the lymph node or spleen to distant sites of infection, they are likely to experience more restrictive metabolic environments (Fig. 6). Some tissue sites, such as the thymus, bone marrow, and the gastrointestinal epithelium, are naturally hypoxic, whereas inflammation can promote local hypoxic regions in tissues (73). Unlike O2, which can freely diffuse into tissues, nutrients such as glucose move through space by Brownian motion and require transport into cells, and thus they are likely to have a much more restricted distribution in tissues. Local metabolic activity of immune cells at the site of infection can rapidly consume available O2 and nutrients, resulting in metabolic stress for infiltrating T cell populations (Fig. 6). There is evidence that TEFF cells in the tumor microenvironment compete with tumor cells for available glucose, and this competition model of nutrient restriction limits the ability of TEFF cells to produce effector cytokines such as IFN-γ (27). Thus, metabolic and environmental influences on T cell function in vivo may elicit very different responses and may account for experimental variance between T cell responses in a petri dish versus what is observed in animal models. Although these points provide sobering food for thought, the development of in vitro methods that control metabolic parameters (e.g., hypoxia incubators, perfusion systems for culture medium) may help to reconcile these differences. Studying metabolite flux of T cells in vivo by using isotopomer labeling techniques will further our understanding of metabolic pathways relevant for T cell function. These techniques are currently being developed in the cancer biology field (118, 119) and should be readily transferable to immunology research.
Fig. 6. T cells must display metabolic plasticity to adapt to changes in nutrient and oxygen availability in vivo.
TEFF (green) cells must adapt to varying oxygen and nutrient levels depending on environmental context. Lymphoid organs (middle) are considered to be nutrient- and oxygen-replete areas, whereas sites of inflammation (left) and the tumor micro-environment (right) contain hypoxic areas with fluctuations in nutrient availability. At sites of inflammation, nutrient and oxygen availability may become limited because of the metabolic activity of cells at the site of inflammation, necrosis of infected cells, and oxygen consumption by neutrophils. Tumor microenvironments can be highly hypoxic resulting from insufficient vascularization. Additionally, T cells must compete with tumor cells for nutrients such as glucose.
If activated T cells frequently transition between nutrient-replete states (lymph nodes) and nutrient-deficient states (sites of infection), then management of metabolic resources is an important consideration for lymphocytes in order to ensure maintenance of proliferation and/or effector function. Metabolic insufficiency may be a fundamental mechanism by which environmental context regulates T cell function, potentially influencing T cell tolerance and anergy. Metabolic interference mechanisms, such as indoleamine 2,3-dioxygenase (IDO)–1–dependent degradation of tryptophan by antigen presenting cells (APCs), may represent key regulatory mechanisms at sites of infection or inflammation (120). Tumors may similarly restrict antitumor immunity by influencing T cell metabolism. Competition between tumor cells and tumor-infiltrating T cells for available glucose can impose nutrient deprivation on TEFF cells that limits their ability to produce effector cytokines (27). Tumor-derived lactate can also suppress CTL function directly by blocking lactate export by T cells, thus disrupting their ability to maintain glycolysis (121).
It remains to be determined whether T cells deal with nutrient restriction by displaying metabolic plasticity similar to tumor cells. In this context, AMPK and HIF-1α, which monitor cellular energy and O2 levels, respectively, are likely to have a profound effect on the adaptive immune response. Loss of HIF-1α impairs TH17 expansion and induction of experimental autoimmune encephalitis (35), consistent with a requirement for HIF-1α in promoting TH17 function in vivo. Overall, we know very little regarding pathways that regulate metabolism and adaptation to metabolic stress in lymphocytes, particularly in vivo; this area of immunology is poised for important discoveries in the years to come.
Concluding Thoughts
Recent findings in metabolism and cancer biology have rapidly closed the gap between cell signaling and biochemical pathways. One can now consider all parameters of these fields as being directly intertwined, comprising an interconnected network from gene expression to metabolite production. T cells provide a unique opportunity to understand how metabolism is used in normal biology to achieve proliferation versus abnormal biology, such as that observed in cancer. Characterizing how these pathways are integrated in T cells, how perturbations in the system (i.e., nutrient availability, O2 tension, and metabolite flux) influence T cell responses, and how metabolic responses are regulated in vivo in the context of infection will be some of the challenges facing scientists in this field. Understanding how environmental cues and cellular metabolism influence the outcome of T cell–mediated immune responses will continue to be an active area of research in the future. Interfering with metabolic pathways by using agents such as metformin and rapamycin has already revealed substantial and unexpected effects on T cell– mediated immunity. Understanding how metabolic reprogramming influences T cell fate and effector function has the potential to uncover new ways of modulating T cell responses.
Acknowledgments
We thank J. Blagih, E. Clambey, M. Hentze, C. Krawczyk, M. Vander Heiden, and members of the Jones and Pearce laboratories for insight and comments on this manuscript and L. Donnelly and M. Maslowska for administrative help. This work was supported by grants to R.G.J. from the Canadian Institute for Health Research (MOP-93799 and a New Investigator Career Award) and the Arthritis Society of Canada and by grants to E.L.P. from National Institute of Allergy and Infectious Diseases (AI091965) and National Cancer Institute (CA158823).
References and Notes
- 1.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat. Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. pmid: 22990888. [DOI] [PubMed] [Google Scholar]
- 2.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. pmid: 23601682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. pmid: 23325217. [DOI] [PubMed] [Google Scholar]
- 4.Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. J. Biol. Chem. 1994;269:31484–31490. pmid: 7989314. [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. pmid: 19460998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. pmid: 12121659. [DOI] [PubMed] [Google Scholar]
- 7.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. pmid: 20554958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. pmid: 23525088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. pmid: 22195744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalek RD, et al. Estrogen-related receptor-α is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. pmid: 22042850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. pmid: 23298210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce EL. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. pmid: 20189791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. pmid: 22206904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J. Neurosci. 2007;27:7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. pmid: 17611283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem. Soc. Trans. 2009;37:1385–1388. doi: 10.1042/BST0371385. pmid: 19909281. [DOI] [PubMed] [Google Scholar]
- 16.van der Windt GJ, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. pmid: 23940348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. pmid: 19494812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. pmid: 19629071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. pmid: 20060330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. pmid: 19543266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. pmid: 11283375. [DOI] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J. Biol. Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. pmid: 17030509. [DOI] [PubMed] [Google Scholar]
- 23.McGarry JD, Mannaerts GP, Foster DW. A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J. Clin. Invest. 1977;60:265–270. doi: 10.1172/JCI108764. pmid: 874089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundu M, Thompson CB. Autophagy: Basic principles and relevance to disease. Annu. Rev. Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. pmid: 18039129. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard VM, et al. Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. pmid: 21059894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. pmid: 17190837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. pmid: 23746840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhri G, Clark IA, Hunt NH, Cowden WB, Ceredig R. Effect of antioxidants on primary alloantigen-induced T cell activation and proliferation. J. Immunol. 1986;137:2646–2652. pmid: 2944959. [PubMed] [Google Scholar]
- 29.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: Selective regulation of mitogen-activated protein kinase activation and fas ligand expression. J. Exp. Med. 2002;195:59–70. doi: 10.1084/jem.20010659. pmid: 11781366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson SH, Devadas S, Kwon J, Pinto LA, Williams MS. T cells express a phagocyte-type NADPH oxidase that is activated after T cell receptor stimulation. Nat. Immunol. 2004;5:818–827. doi: 10.1038/ni1096. pmid: 15258578. [DOI] [PubMed] [Google Scholar]
- 31.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. pmid: 23415911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath—New quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. pmid: 11672532. [DOI] [PubMed] [Google Scholar]
- 33.Jones RG, et al. The proapoptotic factors Bax and Bak regulate T Cell proliferation through control of endoplasmic reticulum Ca(2+) homeostasis. Immunity. 2007;27:268–280. doi: 10.1016/j.immuni.2007.05.023. pmid: 17692540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. pmid: 21317389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi LZ, et al. HIF1a–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. pmid: 21708926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. pmid: 21871655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, et al. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. pmid: 23467085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinewietfeld M, et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. pmid: 23467095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. pmid: 23828891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. pmid: 22439925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doughty CA, et al. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: Role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood. 2006;107:4458–4465. doi: 10.1182/blood-2005-12-4788. pmid: 16449529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. pmid: 20351312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haschemi A, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. pmid: 22682222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. pmid: 18387799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellen KE, Thompson CB. A two-way street: Reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. pmid: 22395772. [DOI] [PubMed] [Google Scholar]
- 46.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. pmid: 18177721. [DOI] [PubMed] [Google Scholar]
- 47.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. pmid: 16226706. [DOI] [PubMed] [Google Scholar]
- 48.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. pmid: 23563690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. pmid: 20167787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. pmid: 20167786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. pmid: 19461003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffiths M, Keast D. The effect of glutamine on murine splenic leukocyte responses to T and B cell mitogens. Immunol. Cell Biol. 1990;68:405–408. doi: 10.1038/icb.1990.54. pmid: 2097296. [DOI] [PubMed] [Google Scholar]
- 53.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. pmid: 22225880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. pmid: 20156581. [DOI] [PubMed] [Google Scholar]
- 55.Marjanovic S, Eriksson I, Nelson BD. Expression of a new set of glycolytic isozymes in activated human peripheral lymphocytes. Biochim. Biophys. Acta. 1990;1087:1–6. doi: 10.1016/0167-4781(90)90113-g. pmid: 2169315. [DOI] [PubMed] [Google Scholar]
- 56.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. pmid: 22922757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. pmid: 18337823. [DOI] [PubMed] [Google Scholar]
- 58.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. pmid: 18337815. [DOI] [PubMed] [Google Scholar]
- 59.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. pmid: 19920251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kung C, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem. Biol. 2012;19:1187–1198. doi: 10.1016/j.chembiol.2012.07.021. pmid: 22999886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. pmid: 21620138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang W, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. pmid: 22901803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. pmid: 22306293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. pmid: 23064226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. pmid: 21804546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. pmid: 21760589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maddocks OD, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. pmid: 23242140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilsson LM, et al. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet. 2012;8:e1002573. doi: 10.1371/journal.pgen.1002573. pmid: 22438825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. pmid: 18032601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. pmid: 22101431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. pmid: 22101433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. pmid: 22106302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McNamee EN, Korns Johnson D, Homann D, Clambey ET. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol. Res. 2013;55:58–70. doi: 10.1007/s12026-012-8349-8. pmid: 22961658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N. Engl. J. Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. pmid: 21323543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura H, et al. TCR engagement increases hypoxia-inducible factor-1 alpha protein synthesis via rapamycin-sensitive pathway under hypoxic conditions in human peripheral T cells. J. Immunol. 2005;174:7592–7599. doi: 10.4049/jimmunol.174.12.7592. pmid: 15944259. [DOI] [PubMed] [Google Scholar]
- 76.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2784–E2793. doi: 10.1073/pnas.1202366109. pmid: 22988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. pmid: 18772396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. pmid: 19935646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. pmid: 21130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. pmid: 22343901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. pmid: 21251613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. pmid: 22343889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hardie DG, Ross FA, Hawley SA. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. pmid: 22436748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao B, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. pmid: 21399626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oakhill JS, et al. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. pmid: 21680840. [DOI] [PubMed] [Google Scholar]
- 86.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995;312:163–167. doi: 10.1042/bj3120163. pmid: 7492307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tamás P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J. Exp. Med. 2006;203:1665–1670. doi: 10.1084/jem.20052469. pmid: 16818670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacIver NJ, et al. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J. Immunol. 2011;187:4187–4198. doi: 10.4049/jimmunol.1100367. pmid: 21930968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. pmid: 23274086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tamás P, et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur. J. Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. pmid: 19830737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. pmid: 15866171. [DOI] [PubMed] [Google Scholar]
- 92.Rolf J, et al. AMPKa1: A glucose sensor that controls CD8 T-cell memory. Eur. J. Immunol. 2013;43:889–896. doi: 10.1002/eji.201243008. pmid: 23310952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alderson NL, et al. S-(2-Succinyl)cysteine: A novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 2006;450:1–8. doi: 10.1016/j.abb.2006.03.005. pmid: 16624247. [DOI] [PubMed] [Google Scholar]
- 94.Bardella C, et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 2011;225:4–11. doi: 10.1002/path.2932. pmid: 21630274. [DOI] [PubMed] [Google Scholar]
- 95.Adam J, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. pmid: 22014577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaelin WG., Jr Cancer and altered metabolism: Potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb. Symp. Quant. Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. pmid: 22089927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. pmid: 22304911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. pmid: 23630074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. pmid: 20171147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. pmid: 16098467. [DOI] [PubMed] [Google Scholar]
- 101.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. pmid: 15652751. [DOI] [PubMed] [Google Scholar]
- 102.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. pmid: 23535595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: Molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. pmid: 22224760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hentze MW, Preiss T. The REM phase of gene regulation. Trends Biochem. Sci. 2010;35:423–426. doi: 10.1016/j.tibs.2010.05.009. pmid: 20554447. [DOI] [PubMed] [Google Scholar]
- 105.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. pmid: 22658674. [DOI] [PubMed] [Google Scholar]
- 106.Cieśla J. Metabolic enzymes that bind RNA: Yet another level of cellular regulatory network? Acta Biochim. Pol. 2006;53:11–32. pmid: 16410835. [PubMed] [Google Scholar]
- 107.Hentze MW, Argos P. Homology between IRE-BP a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991;19:1739–1740. doi: 10.1093/nar/19.8.1739. pmid: 1903202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rouault TA, Stout CD, Kaptain S, Harford JB, Klausner RD. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: Functional implications. Cell. 1991;64:881–883. doi: 10.1016/0092-8674(91)90312-m. pmid: 2001588. [DOI] [PubMed] [Google Scholar]
- 109.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: Regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. pmid: 20603012. [DOI] [PubMed] [Google Scholar]
- 110.Constable A, Quick S, Gray NK, Hentze MW. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: Switching between enzymatic and genetic function? Proc. Natl. Acad. Sci. U.S.A. 1992;89:4554–4558. doi: 10.1073/pnas.89.10.4554. pmid: 1584791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nagy E, Rigby WFC. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD (+)-binding region (Rossmann fold) J. Biol. Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. pmid: 7531693. [DOI] [PubMed] [Google Scholar]
- 112.Nagy E, et al. Identification of the NAD(+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem. Biophys. Res. Commun. 2000;275:253–260. doi: 10.1006/bbrc.2000.3246. pmid: 10964654. [DOI] [PubMed] [Google Scholar]
- 113.Colell A, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. pmid: 17540177. [DOI] [PubMed] [Google Scholar]
- 114.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. pmid: 20727968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: What do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. pmid: 22424225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. pmid: 21982714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsou P, Zheng B, Hsu CH, Sasaki AT, Cantley LC. A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab. 2011;13:476–486. doi: 10.1016/j.cmet.2011.03.006. pmid: 21459332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. pmid: 22682223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marin-Valencia I, et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012;25:1177–1186. doi: 10.1002/nbm.2787. pmid: 22383401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mellor AL, Munn DH. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. pmid: 15459668. [DOI] [PubMed] [Google Scholar]
- 121.Fischer K, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. pmid: 17255361. [DOI] [PubMed] [Google Scholar]