Abstract
Post-transcriptional regulation of mRNA by the RNA binding protein HuR (Elavl1) is required in B cells for the germinal centre reaction and for the production of class-switched antibodies in response to T-independent antigens. Transcriptome-wide examination of RNA isoforms, abundance and translation in HuR-deficient B cells, together with direct measurements of HuR-RNA interaction, revealed that HuR-dependent mRNA splicing affects hundreds of transcripts including the dihydrolipoamide S-succinyltransferase (Dlst), a subunit of the 2-oxoglutarate dehydrogenase complex (αKGDH). In the absence of HuR, defective mitochondrial metabolism results in high amounts of reactive oxygen species and B cell death. Our study shows how post-transcriptional processes control the balance of energy metabolism required for B cell proliferation and differentiation.
The generation of antibodies is a hallmark of the fight against infection and is a principal goal in the design of vaccines. B lymphocytes produce distinct antibody isotypes by the process of class switch recombination (CSR). Antibodies arising from extrafollicular differentiation of B cells into antibody-secreting cells (ASC) can prevent pathogen spread at early stages of infection, while switched antibodies quickly recruit effector cells during memory responses. Somatic hypermutation (SHM) of the immunoglobulin genes increases antibody affinity. This requires the positive selection of B cells, bearing high-affinity membrane immunoglobulins and by T cells within germinal centers (GC), the specific anatomical structures formed in peripheral lymphoid organs during many immune responses.
Resting lymphocytes require glucose and glycolysis to survive, but undergo metabolic reprogramming following antigen encounter to meet the energetic and biosynthetic demands of proliferation1. Unlike T cells, which disproportionately increase glycolysis when activated, B cell activation through membrane immunoglobulin is accompanied by a balanced increase in both glycolysis and oxidative phosphorylation2, 3. The molecular mechanisms which control this metabolic reprogramming in lymphocytes have come under intense scrutiny in recent years and have highlighted a link between metabolic flux and cell fate decisions1.
The network of transcription factors required for B cell activation and subsequent differentiation has been elucidated in some detail4, 5. By contrast, despite the established role of splicing and polyadenylation in the control of immunoglobulin gene expression, relatively little is known about how post-transcriptional regulation of gene expression influences the outcome of antigen encounter in B cells. Post-transcriptional fate of mRNA is mediated by its associated RNA-binding proteins (RBP) and non-coding RNAs, which determine the timing and magnitude of protein expression6, 7. B cell specific deletion of the RNA-processing enzyme Dicer inhibits the GC reaction8. Moreover, individual microRNAs, like miR-155, are regulators of ASC and GC formation9-11. Less is known about the importance of RBPs. Roquin is required for the restraint of helper T cell function during the GC reaction, but plays a minor role in B cells12. The RBP HNRPD has been implicated in B cell survival13, but essential functions for sequence specific RBPs have not yet been found in B cells.
HuR (gene name Elavl1) is an RBP implicated in both nuclear and cytoplasmic regulation of RNA fate. HuR binds to uridine (U)-tracts present mostly within introns and 3′UTRs, which correlates with its function as regulator of mRNA splicing and stability14, 15. HuR null mice are embryonic lethal16. Conditional HuR knockout (HuR cKO) mice have proven a useful tool to understand the role of HuR in myeloid cells where it limits proinflammatory cytokine production during chronic inflammation and cancer17, 18. HuR is required for maintenance of haematopoietic stem cells19, and the selection and chemotaxis of T cells20.
Here, we show that regulation of the B cell transcriptome by HuR is essential for normal antibody responses. B cell-specific deletion of HuR reveals an obligate role for HuR in the GC reaction. Moreover, the production of class-switched antibodies induced by T-independent antigens is dependent upon HuR. To identify the direct targets of HuR action, we mapped HuR:RNA interactions at single nucleotide level resolution in primary B cells21, 22. Integration of gene expression profiling using RNAseq and identification of translationally regulated mRNA by ribosome profiling established a direct correlation between changes in gene expression and translation of specific mRNAs bound by HuR. Our analysis indicates that HuR regulates the expression of genes related to B cell energy metabolism and prevents the accumulation of fatal levels of reactive oxygen species upon B cell activation.
RESULTS
HuR expression is increased upon B cell activation
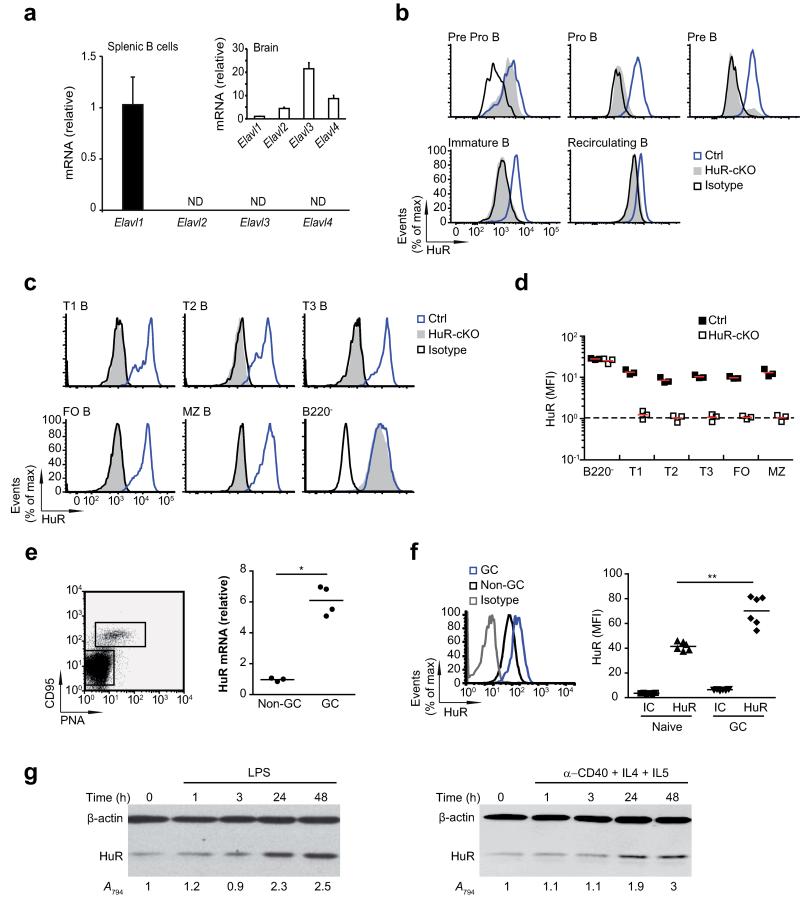
HuR is the only member of the Elavl family expressed in splenic B cells (Fig. 1a). In order to understand the role of HuR during B cell development and activation we generated B cell-specific conditional KO mice (Elavl1fl/fl Mb1-Cre, hereafter HuR-cKO mice). Analysis of HuR protein expression by flow cytometry showed that it was deleted from the pro-B cell stage in the bone marrow (BM) and was undetectable in transitional and mature B lymphocytes (Fig. 1b-d). HuR was similarly expressed in all B cell populations present in the spleen. RT-qPCR analysis of GC B cells, formed in the spleen after immunisation, showed a 6-fold increase in HuR mRNA in GC B cells compared to non-GC B cells (Fig. 1e). Intracellular flow cytometry indicated that HuR protein was increased by 2-fold in GC B cells (Fig. 1f). B cell activation with LPS or αCD40 + IL-4 + IL-5 increased HuR protein expression by up to 3-fold (Fig. 1g). Taken together, these data suggest that HuR expression is increased during B cell activation in vitro and in vivo.
Figure 1. HuR expression is increased during B cell activation.
(a) qPCR analysis of Elavl1 (HuR), Elavl2, Elavl3 and Elavl4 mRNA expression in ex vivo splenic B cells (n=6) and brain (n=2). Data shown relative to Elavl1 mRNA expression (ND = not detected). (b) Intracellular HuR staining in B cell subsets in the bone marrow (BM). (c) HuR protein expression in splenic B cell subsets. B cell populations in BM and spleen in Mb1-Cre (unfloxed controls, Ctrl.) and Elavl1fl/flMb1-Cre (HuR-cKO) mice were analysed by flow cytometry as described in online methods. (d) Quantification of HuR expression. Median Fluorescence Intensity (MFI) of HuR staining relative to an isotype control (IC) antibody is shown (n=3 biological replicates). Dashed line indicates the detection limit of the assay. (e) Normalised HuR mRNA expression in GC B cells (B220+CD95+PNA+) relative to the expression of HuR mRNA in non-GC B cells (B220+PNA−CD95−). A representative dot plot is shown to indicate cell sorting strategy. Cells from individual mice were isolated independently for mRNA expression analysis. Data from each individual sample and the mean value are shown (n=3-4 per group, unpaired t-test). (f) HuR protein expression in non-GC and GC B cells from individual immunised mice measured by flow cytometry. Data from each individual sample and the mean value are shown. (n=6, unpaired t-test). Histograms and dot plots shown in b, c, e and f are representative of more than 3 independent stainings using individual mice. (g) Time course analysis of HuR expression after in vitro B cell activation with LPS or αCD40+IL-4+IL-5. Immunoblots are representative of four (activation with LPS) or two (activation with αCD40+IL-4+IL-5) independent experiments respectively. In each experiment, HuR (Abs794) expression was normalised by β-actin levels and quantified relative to non-activated B cells. Data is shown as mean value.
B cell development following HuR deletion in pro-B cells
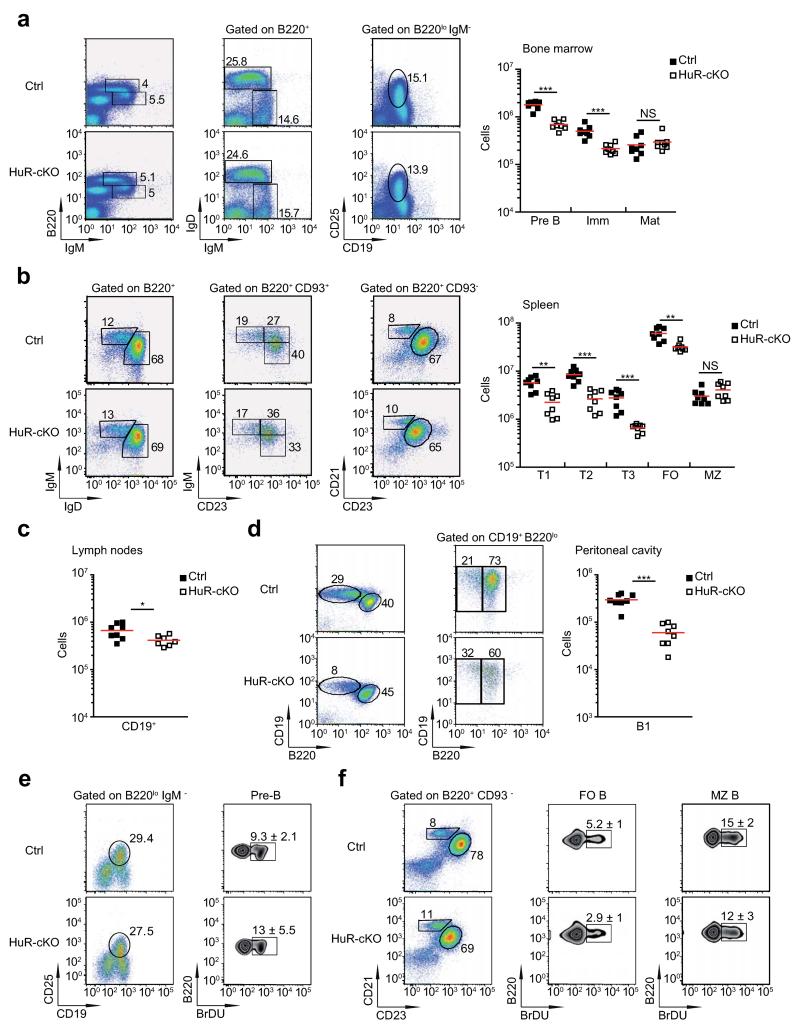
Normal proportions of B cell subpopulations were present in BM, spleen and lymph nodes of HuR-cKO mice (Fig. 2a-c). We found a small decrease in the number of pre-B cells in BM and in the number of transitional and FO B cells in spleens of HuR-cKO mice. HuR-cKO mice had reduced numbers of B1 B cells in the peritoneal cavity (Fig. 2d). Analysis of the incorporation of 2-bromodeoxyuridine (BrdU) into the DNA of rapidly proliferating pre-B cells was not different in HuR-cKO mice (Fig. 2e and Supplementary Fig. 1a and 1b). Furthermore, B cell turnover in the spleen, inferred from BrdU incorporation over a 7-day period, was not different from control mice (Fig. 2f). Thus, with the exception of B1 B cells, there is little requirement for HuR for either B cell development or homeostasis.
Figure 2. HuR-cKO mice have no major defects in B cell development.
(a) Analysis of B cell populations in the bone marrow of Mb1-Cre (Ctrl) and Elavl1fl/flMb1-Cre (HuR-cKO) mice. Representative dot plots summarise the cell gating strategy and cell frequency. The number of Pre-, immature-, and mature-B cells was calculated based on the expression of surface cell markers. (b) Quantitation of the number of different B cell subsets in the spleen of Ctrl. and HuR-cKO mice. T1, T2, T3, FO and MZ B cells were defined based on cell surface markers as described in the online methods. (c) Analysis of the number of CD19+ cells in peripheral lymph nodes. (d) Analysis of B1 B cells in the peritoneal cavity. Representative dot plots summarise the cell gating strategy and cell frequency. Analysed data shown in a, b, c and d are from individual mice of each genotype and is representative of more than 3 independent experiments (n=8 per group; Mann-Whitney analysis was performed; *p<0.05, **p<0.005, ***p<0.0005). (e) Representative flow cytometry plots of BrdU incorporation in Pre-B cells after 2.5 hours labelling. The mean percentage ± SD of BrdU+ cells is shown (n=8 per group). (f) Analysis of BrdU+ mature B cells following 7 days labelling. Representative dot plots and contour plots are shown. The mean percentage ± SD of BrdU+ cells is shown (n=5 individual mice per group).
B cells require HuR for normal antibody responses
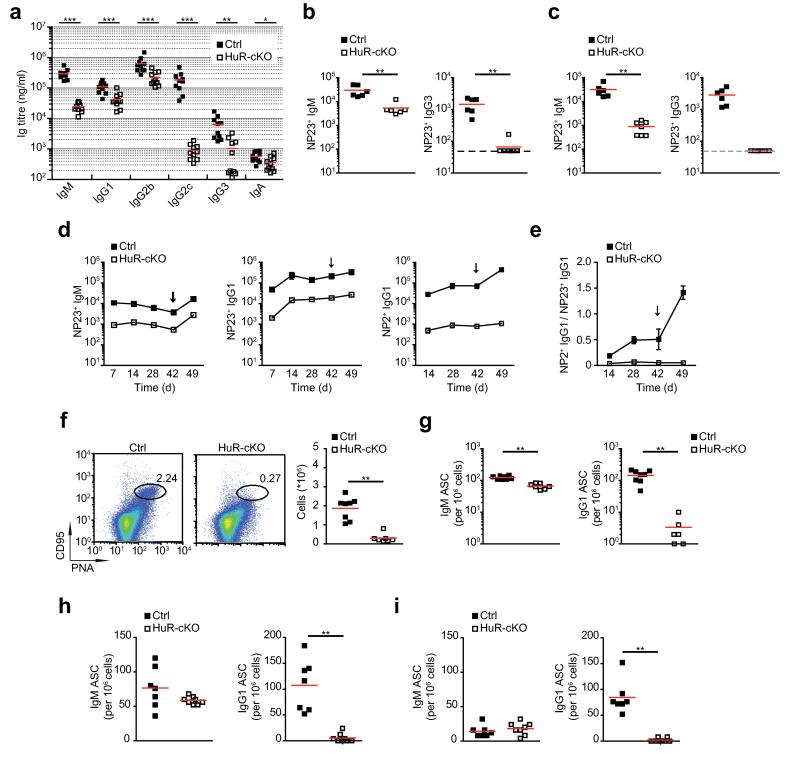
HuR-cKO mice showed a significant reduction in all immunoglobulin (Ig) isotypes present in serum, apart from IgA (Fig. 3a). IgG1 and IgG2b titers were reduced by 2.5-fold in HuR-cKO mice, whereas IgG3 and IgM titers were reduced by 6- and 12-fold, respectively. Decreased IgM titers could result from the reduction of B1 B cells, as these contribute substantially to serum IgM. The biggest difference was found in IgG2c titers that were reduced by 200-fold in HuR-cKO mice. The decrease in almost all antibody isotypes likely reflects a requirement for HuR in B cells for a functional response following antigen encounter.
Figure 3. B cells require HuR for responses to different classes of antigens.
(a) Quantitation of serum immunoglobulin in non-immunised mice. Samples from individual Mb1-Cre (Ctrl) and Elavl1fl/flMb1-Cre (HuR-cKO) mice were collected in two independent experiments (n=10; Mann-Whitney test). (b, c) Analysis of NP-reactive IgM and IgG3 antibodies present in serum samples from individual mice of each genotype collected 7 days after immunization with NP-LPS (b) or NP-Ficoll (c). Dot line indicates the limit of detection in this assay (n=6 per group in b; n=6-7 per group in c; Mann-Whitney test). (d) Time course analysis of NP23 reactive IgM and IgG1 in serum of mice immunised with NP-KLH at day 0 and day 42. (e) Ratio of NP2- bound (high affinity) and NP23-bound (low + high affinity) IgG1 antibodies. Data is shown in d and e as mean ± SEM, n=6/group. Statistical analysis of the data at each time point was performed using a Mann-Whitney test (p<0.005 in all cases). (f) The number of GC B cells present in the spleen of Mb1-Cre (Ctrl) and HuR-cKO mice seven days after immunization with NP-KLH. Representative dot plots show the gating strategy and frequency of GC B cells (B220+ PNA+ CD95+ cells). The number of GC cells in individual mice of each genotype (n= 7-8 mice) from one of the two independent experiments performed is shown (Mann-Whitney test). (g) ELISPOT of the number of IgM and IgG1 ASC present in the spleens of the mice analysed in f (n=7-8, Mann-Whitney test). (h, i) Quantitation of IgM-ASC and IgG1-ASC present in spleen (h) and BM (i) of Mb1-Cre (Ctrl.) and HuR-cKOmice seven days after secondary immunization. Data from each individual mice of each group is shown (n=7-8; Mann-Whitney test) (*p<0.05, **p<0.005, ***p<0.0005).
To investigate the requirement for HuR during the response of B cells to antigen, we immunised HuR-cKO mice with the thymus (T)-independent antigens, Ficoll or LPS coupled to the hapten 4-hydroxy-3 nitrophenolic acid (NP). NP-reactive IgM titers in serum showed a 5-fold reduction with both antigens (Fig. 3b and 3c). By contrast, we were unable to detect NP-reactive IgG3 antibodies in serum from HuR-cKO mice. Next, we evaluated the ability of HuR-cKO B cells to participate in a thymus-dependent antibody response by assessing the serum antibody responses following primary and secondary immunisation with NP coupled to keyhole limpet haemocyanin (KLH) (Fig. 3d). Serum NP-reactive IgM titers showed an average 8.5-fold reduction. NP23-reactive IgG1 titers, which reflect the combination of both low and high affinity antibodies, were 15-times lower in HuR-cKO mice than in control mice. Memory responses evaluated seven days after secondary immunisation were clearly detectable in HuR-cKO mice in the form of IgM antibodies. By contrast, NP2-reactive IgG1 titers did not increase, suggesting high affinity IgG1 memory cells did not form, or, if they did, their reactivation failed to generate ASC. As a measure of affinity maturation, we determined the ratio of NP2- and NP23-reactive IgG1 antibodies, which indicated that NP-reactive antibodies in HuR-cKO mice failed to undergo affinity maturation (Fig 3e).
Affinity maturation takes place in GC, where helper T cells select B cells with the highest affinity for antigen. GC formation after seven days of immunisation with NP-KLH was diminished in HuR-cKO mice. The number of GC B cells was reduced by more than 6-fold (Fig. 3f). Similarly, following administration of highly immunogenic sheep red blood cells the GC response was defective over time in HuR-cKO mice (Supplementary Fig. 1c and 1d). Consistent with reduced titers of serum antibodies, the number of NP-reactive IgM ASC in the spleen of HuR-cKO mice immunised with NP-KLH was reduced by 2-fold, whereas the number of NP-reactive IgG1 ASC was reduced by 50-fold (Fig. 3g). A similar reduction was found in the number of NP-reactive IgM and IgG1 ASC in the spleen of HuR-cKO mice seven days after secondary immunisation with NP-KLH (Fig. 3h). Further analysis of ASC in the BM showed that HuR-cKO mice contained a similar number of NP-reactive IgM ASC, but NP-reactive IgG1 ASC were barely detectable (>100-fold reduction) when compared to control mice (Fig. 3i). These data show that HuR is required for proper B cell activation and/or differentiation following antigen encounter.
Regulation of the B cell transcriptome by HuR
To examine whether an early stage of B cell activation was affected by the absence of HuR, we measured changes in intracellular Ca2+ following crosslinking of cell surface IgM. The Ca2+ flux elicited after activation of Mb1-Cre unfloxed controls (defined as “Ctrl.”) and HuR-cKO LN B cells was similar using either a highly crosslinking anti-IgM F(ab’)2 or a monoclonal anti-IgM (mAb B7.6) (Supplementary Fig. 2a). Additionally, cell surface expression of CD25, CD40, CD69 and CD86 was increased to a similar extent in response to a variety of stimuli (Supplementary Fig. 2b and 2c), indicating that, by these criteria at least, B cell activation was normal in the absence of HuR.
To discover the molecular mechanisms regulated by HuR in resting and activated B cells, we performed an integrated analysis of three high throughput measurements of the B cell transcriptome. Firstly, we searched for differential mRNA expression in HuR-cKO B cells by mRNAseq. Secondly, we determined the differences in mRNA translation between HuR-cKO and control B cells using ribosome footprinting (Ribo-Seq). Thirdly, we characterised HuR-RNA interactions at the single nucleotide level in LPS-activated B cells using iCLIP. Taken together, these measurements have the power to discriminate between changes in RNA isoform and abundance as well as translational regulation, and can relate these changes to HuR binding. Thus detected changes can be ascribed to be direct or indirect effects of HuR:RNA interactions. None of the changes were due to compensatory regulatory mechanisms affecting the abundance or ribosome loading of mRNAs encoding other members of the Elavl-family or of other AU-rich element binding proteins (Supplementary Fig. 3).
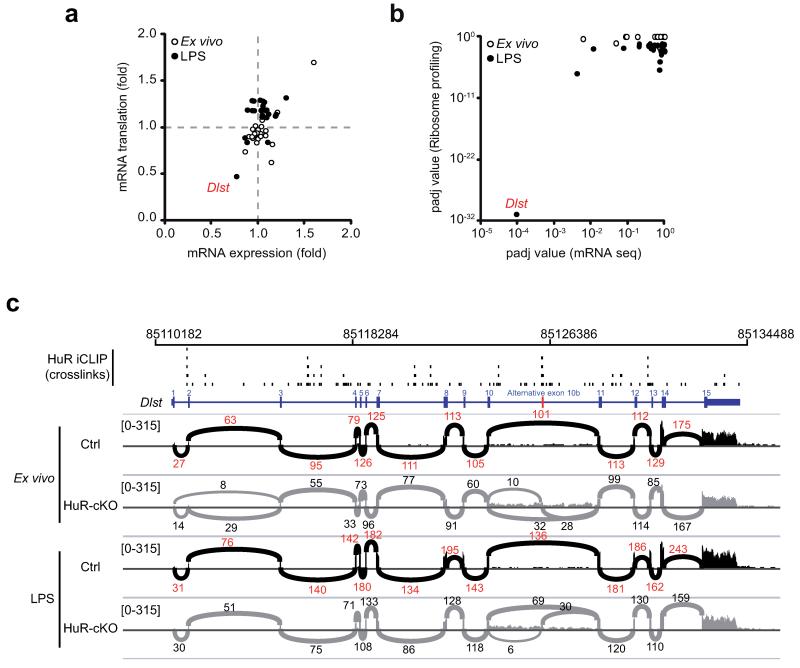
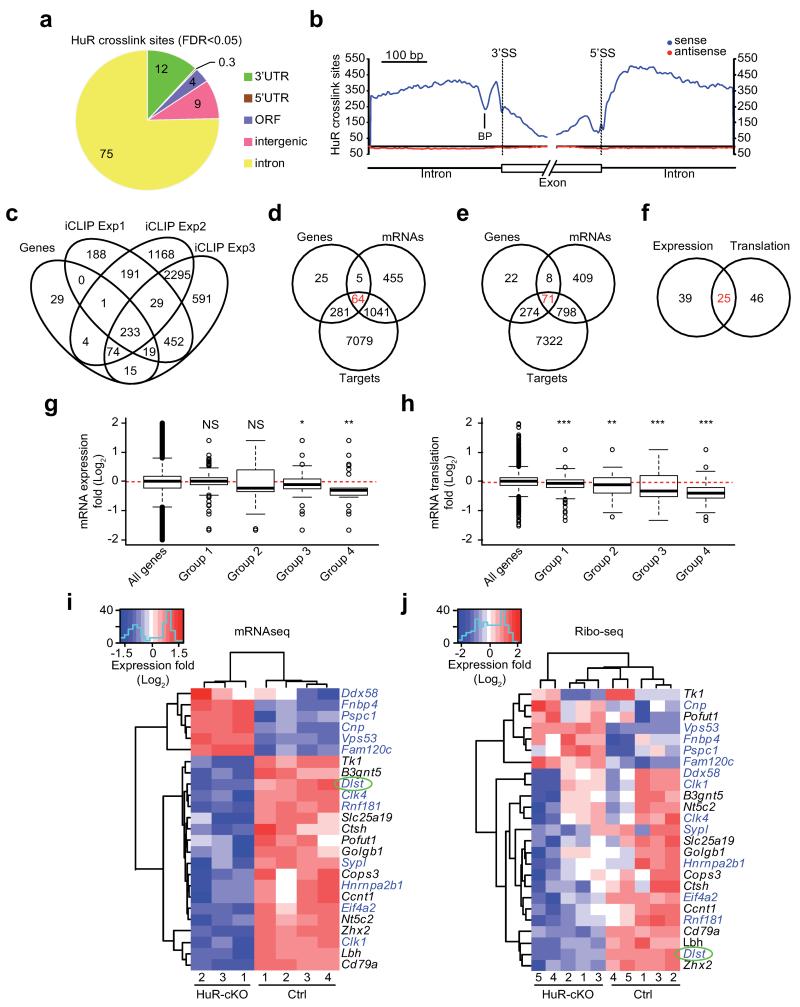
Differential expression analysis of mRNAseq and Ribo-Seq followed by pathway enrichment analysis indicated gene signatures related to different aspects of the cell cycle and energy metabolism were enriched in HuR-cKO B cells (Table 1). HuR deficiency influenced expression of genes related to glycolysis, TCA cycle and oxidative phosphorylation, three pathways increased in GC B cells and after B cell activation with LPS (Supplementary Fig. 4a, 4b and 4c). Blockade of these energy pathways using 2-deoxyglucose (2DG), CPI-613 or Oligomycin A decreased B cell survival, cell growth and cell proliferation after mitogen stimulation (Supplementary Fig. 4d and 4e). Analysis of the global fold-change in mRNA expression and translation of genes related to glycolysis, TCA cycle and oxidative phosphorylation showed no change when comparing ex vivo (unstimulated) control versus HuR-cKO B cells. By contrast, global expression and/or translation of these mRNAs were increased in LPS-activated HuR-cKO B cells, suggesting that HuR is only important for regulating the reprogramming of energy metabolism upon activation (Supplementary Fig. 4f). Data correlation between mRNAseq and Ribo-Seq of only those metabolic genes that are differentially translated in LPS-activated HuR-cKO B cells showed that all of them, with the exception of dihydrolipoamide S-succinyltransferase (Dlst), were modestly increased (Fig. 4a and Supplementary Fig. 4g, 4h and 4j). Dlst mRNA was increased in GC B cells when compared to naive B cells (Supplementary Fig. 4i), but its mRNA expression and translation was significantly reduced in LPS-activated HuR-cKO B cells (Fig. 4b).
Table 1. Pathway enrichment analysis.
Gene ontology analysis of Ribo-seq data from LPS-activated B cells performed using WebGestalt pathway enrichment analysis. The number of total and differentially expressed (DE) genes in HuR-cKO B cells compared to control (Ctrl) B cells is indicated. Gene sets contained a minimum of 6 genes and a hypergeometric test and multiple test correction (Benjamini-Hochberg) of p values was performed during the statistical analysis.
| Pathway Name | Total genes | DE genes | padj value |
|---|---|---|---|
| mRNA processing | 456 | 58 | 5.98*10−14 |
| miRNA regulation of DNA Damage Response | 69 | 14 | 6.54*10−06 |
| G1 to S cell cycle control | 65 | 12 | 8.63*10−05 |
| Senescence and Autophagy | 109 | 15 | 2.00*10−04 |
| Apoptosis | 93 | 13 | 5.00*10−04 |
| Androgen Receptor Signaling Pathway | 123 | 15 | 6.00*10−04 |
| Cytoplasmic Ribosomal Proteins | 78 | 11 | 1.20*10−03 |
| B Cell Receptor Signaling Pathway | 200 | 19 | 1.60*10−03 |
| Electron Transport Chain | 101 | 12 | 2.40*10−03 |
| Glycolysis and Gluconeogenesis | 50 | 8 | 2.60*10−03 |
| DNA Replication | 41 | 7 | 3.20*10−03 |
| PluriNetWork | 289 | 23 | 3.20*10−03 |
| IL-5 Signaling Pathway | 80 | 10 | 3.30*10−03 |
| TCA Cycle | 32 | 6 | 3.70*10−03 |
| T Cell Receptor Signaling Pathway | 143 | 14 | 3.70*10−03 |
| Adipogenesis | 133 | 13 | 5.10*10−03 |
| Alzheimers Disease | 77 | 9 | 6.50*10−03 |
| Toll Like Receptor signaling | 37 | 6 | 6.50*10−03 |
| Cell cycle | 95 | 10 | 8.10*10−03 |
| Amino Acid metabolism | 112 | 11 | 8.40*10−03 |
| MAPK signaling pathway | 165 | 14 | 9.60*10−03 |
Figure 4. Genes involved in energy metabolism are deregulated in HuR-deficient B cells.
(a) Analysis of the fold change in mRNA expression and mRNA translation (HuR-cKO/Ctrl) of those genes involved in cell energy pathways (Glycolysis and Gluconeogenesis, TCA Cycle and Electron Transport Chain) that are differentially translated in the absence of HuR (number of genes=25). mRNAseq and Ribo-seq libraries were generated in two independent experiments using LPS-activated splenic B cells from Elavl1fl/flMb1+/+ (Ctrl) and Elavl1fl/flMb1-Cre (HuR-cKO) mice (RNAseq, n=3-4 per group; Ribo-seq, n=5 per group). (b) Statistical significance of changes in mRNA expression and translation of the genes shown in a. P adjusted values (padj) were calculated after multiple correction of p values (Benjamini-Hochberg correction) obtained from DESeq analysis of RNAseq and Ribo-seq datasets. (c) Dlst mRNA splicing profiles in Ctrl and HuR-cKO B cells. Representative sashimi plots were generated in IGV. The exon number and read counts across each exon-exon junction are indicated for representative mRNAseq data from ex vivo and mitogen activated B cells. HuR iCLIP data for the Dlst locus collected from three independent experiments is shown as unique single nucleotide crosslink sites.
Dlst is one of the three subunits of the αKGDH enzymatic complex, which is essential for maintaining tricarboxylic acid (TCA) cycle flux and cell energy supply. In order to understand the role of HuR in Dlst mRNA regulation, we examined mRNAseq data and plotted the reads mapped across the Dlst locus as Sashimi plots (Fig. 4c). These mRNA splicing profiles showed that a single mRNA transcript was generated after RNA splicing in ex vivo and LPS-activated control B cells. In the absence of HuR, Dlst mRNA showed two alternative splicing events: intron 10 retention and alternative inclusion of a cryptic exon between exon 10 and 11. iCLIP data showed that HuR binds to several locations along Dlst RNA (Fig. 4c and Supplementary Fig. 5a-c). Peak calling analysis showed that HuR binds preferentially to introns, including the poly-pyrimidine tract found downstream the 3′ splice site of the cryptic exon present within intron 10 (Supplementary Fig. 5d). Taken together, these data demonstrate that HuR binding to Dlst pre-mRNA might promote Dlst mRNA expression and translation in HuR-cKO B cells. The modest change in translation of other components of cell energy pathways may reflect a compensatory mechanism.
HuR binding to introns modulates alternative intron usage
To gain a mechanistic insight into the role of HuR in mRNA splicing in B cells we further examined the HuR iCLIP data obtained from LPS-activated B cells. Analysis of unique read counts in all three iCLIP experiments showed that 75% of HuR-RNA crosslink sites were mapped to introns (Fig. 5a and Supplementary Fig. 5e and 5f). Visualisation of HuR crosslink sites close to the exon-intron boundaries indicated that HuR preferentially binds to introns, and showed a significant binding enrichment between the branch point and the 3′ splice site (Fig. 5b). These data suggested that HuR might be a splicing regulator in B cells, thus we studied whether HuR modulates pre-mRNA splicing by further analysis of mRNAseq data from LPS-activated B cells. Differential exon analysis using DEXSeq did not reveal significant changes in exon usage of protein coding transcripts in the absence of HuR, and failed to identify the alternative splicing events associated with Dlst mRNA (Supplementary Tables 1-5). Thus, we performed an intron-centric analysis of the mRNAseq data (Supplementary Fig. 6a), which showed that 530 introns belonging to 375 genes were differentially used in LPS-activated HuR-cKO B cells compared to control B cells (padj<0.1, Supplementary Fig. 6b). HuR was bound to 85% of these 375 genes in, at least, two of the three independent HuR iCLIP experiments (Fig. 5c). Dlst was found amongst these genes. Taken together data correlation from the intron-centric analysis and HuR iCLIP experiments identifies alternative intron usage in the absence of HuR.
Figure 5. HuR regulates intron usage in B cells.
(a) Proportion of unique HuR crosslink sites annotated to each genomic features. The sum of unique cDNA counts from three independent iCLIP experiments performed with LPS-activated B cells (Supplementary Fig. 5) was used for peak call analysis (FDR<0.05) and quantification of HuR crosslink sites. (b) RNA map of crosslink sites assessed at the exon-intron boundaries (BP=branch point; 5′ SS=5′ splice site; 3′ SS=3′ splice site). (c) Correlation between HuR binding and differential intron expression in HuR-cKO B cells. (d) Venn-diagram showing the relationship between genes with differential intron expression (Genes), differential mRNA expression (mRNAs) and mRNA bound by HuR (Targets). (e) Relationship between genes with differential intron expression (Genes), differential ribosome occupancy (mRNAs) and HuR binding (Targets). (f) Identification of mRNAs with differential intron expression which are both differentially expressed and differentially translated. (g) Global analysis of the fold change (HuR-cKO/Ctrl) (log2) in mRNA expression of all genes; genes with differential intron expression (group 1; n=375); HuR-targeted genes with differential intron usage and differential RNA abundance (group 2; n=64); HuR-targeted genes with differential intron usage and differential ribosome occupancy (group 3; n=71); and HuR-targeted genes with differential intron expression which are both differentially expressed and differentially translated (group 4; n=25). (h) Global analysis of the fold change (HuR-cKO/Ctrl) (log2) in ribosome occupancy in the groups described in g. Data in g and h is shown as box whisker plots showing the mean ± SD and 10-90 percentiles. Wilcoxon signed-rank test was performed between each group against all genes (* p<0.05, ** p<0.005). (i) Heatmap of the fold change (HuR-cKO/Ctrl) (log2) in mRNA expression of genes in group 4. (j) Heatmap showing the fold change (HuR-cKO/Ctrl) (log2) in ribosome occupancy of genes in group 4. Genes previously annotated as alternative spliced are highlighted in blue.
HuR modulates mRNA expression and translation via splicing
Expression and translation analysis of all 375 genes with differential intron usage in HuR-cKO B cells (group 1) showed no differences globally (Supplementary Fig. 6c). Individually, 64 genes (group 2) out of these 375 were differentially expressed in LPS-activated HuR-cKO B cells and bound to HuR (Fig. 5d). A similar data correlation showed that 71 out of the 375 genes (group 3) were both differentially translated and bound to HuR (Fig. 5e). Only 25 of these genes (group 4) were both differentially expressed and translated in HuR-cKO B cells (Fig. 5f). When expression of genes in groups 1, 2 and 3 was analysed globally, no changes in mRNA abundance was observed when comparing HuR-cKO versus control B cells (Fig. 5g). By contrast, global translation of these mRNAs was significantly reduced in HuR-cKO B cells, suggesting that, even though HuR-dependent regulation of alternative splicing might not necessarily affect overall mRNA levels, HuR is required for mRNA translation (Fig. 5h). Global mRNA expression and translation of the genes in group 4 were both reduced by up to 50%. Closer examination at individual genes indicated that both mRNA expression and translation of 76% of genes in group 4 (19 out of 25) were reduced in the absence of HuR, including Dlst (Fig. 5i and 5j). In summary, differential intron usage analysis and its correlation with differential expression in mRNAseq and Ribo-Seq allowed us to discover the alternative intron events associated with HuR binding. Visualization of the read coverage revealed high complexity in mRNA splicing associated with these genes, detecting up to 8 different alternative splicing events associated with the solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 (Slc25a19) in the absence of HuR (Supplementary Fig. 6d and 6e). Genetic deletion of either Dlst or Slc25a19 has been previously associated with reduced αKGDH enzymatic activity and alterations in the TCA cycle23, 24.
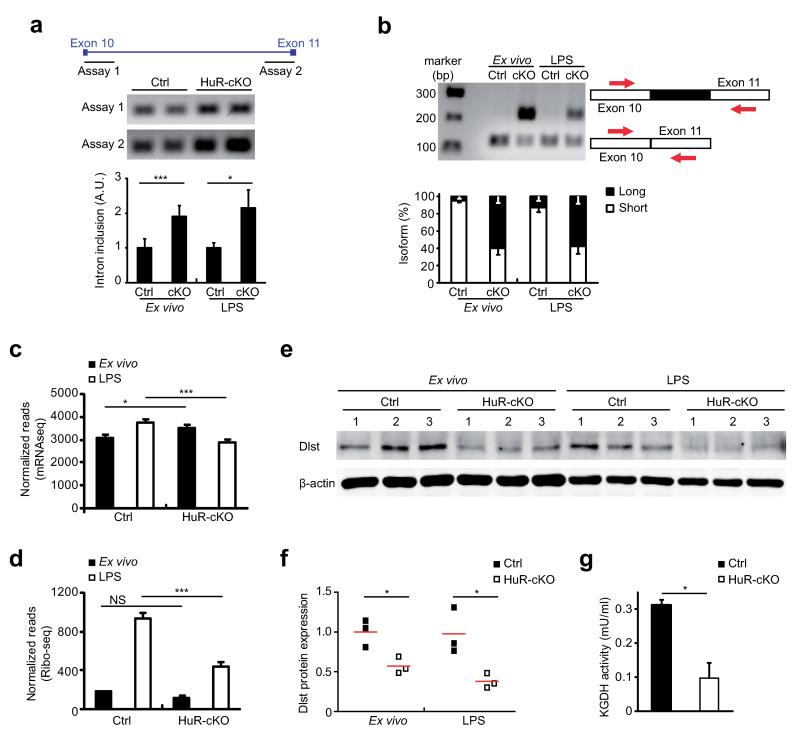
HuR regulates Dlst mRNA splicing
To understand the role of HuR in mRNA splicing, we selected Dlst mRNA for detailed analysis and evaluated usage of intron 10 with two RT-PCR assays. HuR-cKO B cells showed increased intron retention compared to control B cells (Fig. 6a). Read coverage analysis of the Dlst gene locus using mRNAseq data from HuR-cKO B cells identified a novel alternative exon splicing event located after exon 10. This alternative exon 10b was previously annotated in Ensembl as the start exon of predicted transcript ENSMUST00000165575, a transcript for which evidence of its existence is lacking. Detection of reads mapped across the exon 10/alternative exon 10b junction suggested that this was an alternative exon inclusion rather than an alternative first exon. RT-PCR validated this observation and indicated that over 50% of Dlst transcripts within HuR-cKO B cells included the alternatively spliced exon 10b (Fig. 6b). mRNAseq data showed little difference in Dlst mRNA expression between ex vivo or LPS-activated HuR-cKO and control B cells (Fig. 6c). However, the number of ribosome footprints mapped to Dlst mRNA was reduced by 2-fold in HuR-cKO B cells, indicating reduced mRNA translation (Fig. 6d). Visualization of ribosome footprints as Sashimi plots failed to identify ribosome-protected reads mapped to the alternative exon 10b, suggesting that transcripts containing this exon were not translated (Supplementary Fig. 7a). Consistent with this, inclusion of alternative exon 10b introduces several stop codons into the coding sequence (Supplementary Fig. 7b). Immunoblot analysis of B cell protein extracts from ex vivo and LPS activated B cells confirmed a 50% reduction in Dlst protein abundance in HuR-deficient B cells (Fig. 6e and 6f). Antibodies reactive with the N-terminus of Dlst could not detect any alternative protein isoforms produced as a consequence of exon 10b insertion in the absence of HuR (Supplementary Fig. 7c). Decreased Dlst protein expression was associated with a reduction in αKGDH enzymatic activity in HuR-cKO B cells (Fig. 6g). Taken together, HuR binding to Dlst intron 10 prevented alternative exon inclusion, thus guaranteeing Dlst mRNA translation into enzymatically active protein.
Figure 6. Alternative splicing of Dlst in HuR-cKO B cells reduces αKGDH enzymatic activity.
(a) Design summary of PCR primers, spanning exon 10/intron 10 junction and intron 10/exon 11 junction, used to detect intron inclusion in Elavl1fl/flMb1-Cre (HuR-cKO) B cells. Intron inclusion in ex vivo and LPS-activated B cells was quantified relative to Elavl1fl/flMb1+/+ (Ctrl) cells. (b) PCR across the exon 10/exon 11 junction. The percentage of alternative exon inclusion was calculated after gel density quantification of long (alternative exon included) and short (alternative exon skipped) amplicons. Data in a and b was from 6 (naïve cells) or 4 (LPS-treated cells) biological replicates collected in two independent experiments and is shown as mean values ± SD (unpaired t test; * p<0.05, *** p<0.0005). Images are representative. (c) Dlst reads measured by mRNAseq in ex vivo and LPS-activated B cells from Ctrl and HuR-cKO mice. Mean normalised read counts ± SD are shown (n=3-4 per group; differential expression analysis performed with DESeq; *padj<0.05, ***padj < 0.0005). (d) Quantification of ribosome footprints mapped to Dlst mRNA. Data from ex vivo and LPS-activated B cells is shown as mean normalised read counts ± SD (n=4-5 per group; differential expression analysis performed using DESeq2; ***padj < 0.0005). (e) Immunoblot analysis of Dlst protein. Total protein lysates were prepared from ex vivo and LPS-activated splenic B cells from Elval1fl/flMb1+/+ (Ctrl) and HuR-cKO mice in three independent experiments and loaded in the same gel. β-actin was detected as sample loading control. (f) Gel density quantification of Dlst protein abundance relative to β-actin (n=3 per group; unpaired t test; *p<0.05). (g) αKGDH enzymatic activity in total cell extracts from ex vivo B cells collected from individual Elavl1fl/flMb1+/+ (Ctrl.) and HuR-cKO mice (Mean ± SD; n=4 per group from two independent experiments; unpaired t test; *p<0.05).
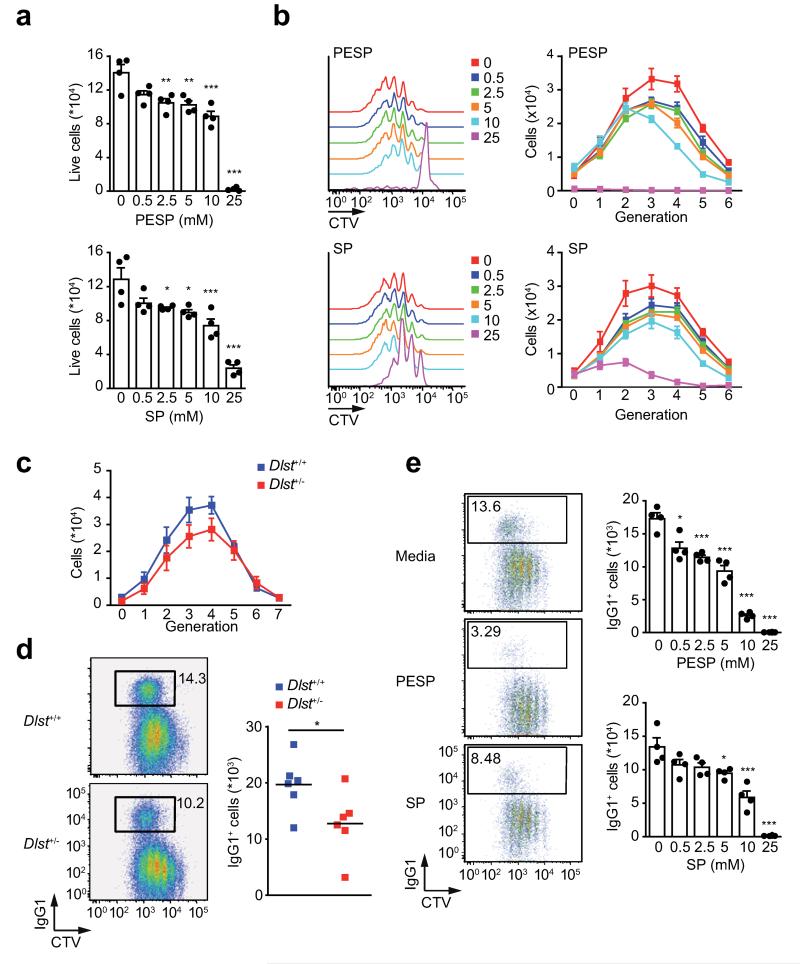
αKGDH is required for B cell proliferation and Ig CSR
Dlst, as part of the αKGDH complex, is highly regulated in order to maintain the flux through the TCA cycle. Inhibition of the αKGDH enzymatic activity using αKG analogs (succinyl phosphonate (SP) and phosphonoethyl ester of succinyl phosphonate (PESP)) significantly reduced B cell viability (Fig. 7a). Analysis of B cell proliferation following LPS + IL-4 stimulation showed that αKGDH inhibition reduced proliferation in a dose-dependent manner (Fig. 7b and 7c). Further analysis of CSR showed that the number of IgG1+ cells was significantly reduced both after chemical inhibition of αKGDH enzymatic activity or in B cells with a single copy of the Dlst gene (Dlst+/− B cells) (Fig. 7d and 7e). Thus, αKGDH enzymatic activity is required for in vitro B cell survival, proliferation and CSR.
Figure 7. αKGDH enzymatic activity is required for B cell survival and proliferation.
(a) Viability of splenic B cells activated with LPS + IL-4 (96 h) in the presence of the indicated doses of succinyl phosphonate (SP) or phosphonoethyl ester of succinyl phosphonate (PESP). Data from one of the two independent experiments performed is shown as mean ± SD (n=4 per group; unpaired t tests comparing control samples against each different dose; *p<0.05, **p<0.01, ***p<0.001). (b) Representative CellTrace™ Violet (CTV) dye profiles in the presence of SP and PESP. The number of cells in each generation was calculated based on dye dilution. (c) In vitro proliferation of Dlst+/+ and Dlst+/− splenic B cells after activation with LPS + IL-4 for 96 hours analysed by CellTrace™ Violet dye dilution as described in b. Splenic B cells were isolated from 6 different mice and treated separately in two independent experiments. Data are shown as the mean number of cells per generation ± SD. Two way ANOVA test was performed (p=0.0162). (d) Quantification of the number of IgG1+ cells in cultures described in c. Representative plots show CellTrace™ Violet dye dilution versus IgG1 surface expression. Unpaired Student’s t test was performed for statistical analysis of the data (n=6; *p<0.05). (e) Analysis of CSR in the cultures in a. Representative plots and the mean number of IgG1+ cells ± SD from one of the two independent experiments are shown (n=4 per group; unpaired Student’s t test comparing no treated cells against the other conditions; *p<0.05, **p<0.01, ***p<0.001).
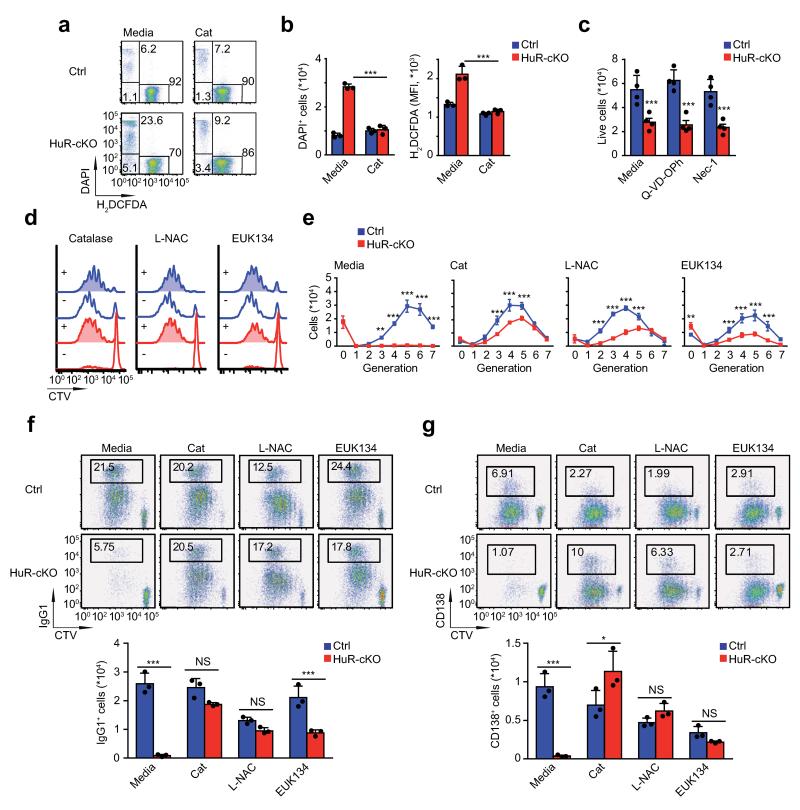
ROS scavengers rescue B cell proliferation and Ig CSR
Analysis of HuR-cKO B cell cultures showed that cell viability was reduced in the absence of HuR (Fig. 8a and 8b). Viability of HuR-cKO B cells could not be rescued by blocking caspase-dependent apoptosis or programed necrosis using Q-VD-OPh or necrostatin-1 respectively, but was reversed by the H2O2 scavenger catalase (Fig. 8b and c). Consistent with this, the fluorescence of the ROS reporter dye H2DCFDA showed that viable HuR-cKO B cells produced larger amounts of ROS compared to Mb1-Cre+ controls. Addition of catalase reduced H2DCFDA fluorescence, indicating diminished ROS concentration (Fig. 8b).
Figure 8. ROS scavengers rescue HuR-deficient B cells from cell death.
(a) Representative plots showing H2DCFDA and DAPI staining of Mb1-Cre (Ctrl) and HuR-cKO B cells after four hours of in vitro culture in the absence or presence of recombinant catalase (Cat, 20 U/ml). (b) Quantitation of the number of DAPI+ cells and the MFI of H2DCFDA in DAPI− cells in the cultures described in a (Mean ± SD, n=3 per group). (c) Effect of Q-VD-OPh (3 μM) and Necrostatin-1 (Nec-1, 3 μM) on in vitro B cell survival (Mean number of DAPI− cells ± SD, n=4 per group). (d) Representative CellTrace™ Violet dye profiles of LPS + IL-4 stimulated Mb1-Cre (Ctrl.) and HuR-cKO B cells cultured in complete RPMI media supplemented with catalase (20 U/ml), L-N-acetyl-cysteine (L-NAC, 10 mM) or EUK-134 (5 μM) for 96 hours. (e) Number of live cells in each cell generation of the proliferation profile shown in d (Mean ± SD, n=3 per group). (f) Representative plots and the mean number of IgG1+ cells ± SD. (g) Representative plots and the mean number of CD138+ cells ± SD is shown. Data shown in each figure panel is from one of the at least three independent experiments performed. Unpaired Student’s t tests were performed for statistical analysis of the data (*p<0.05,**p<0.01,***p<0.001).
HuR-cKO B cells failed to proliferate when cultured with mitogens in vitro. Media supplementation with catalase as well as with ROS scavengers like sodium pyruvate, N-acetyl cysteine and EUK134 enhanced the proliferation of HuR-cKO B cells in response to a variety of stimuli (Fig. 8d and supplementary Fig. 8). Quantitation of the number of B cells recovered showed that HuR-cKO B cells undergo a similar number of cell divisions; however the number of cells in each division was reduced, indicating that ROS scavengers did not completely restore proliferation (Fig. 8e). ROS scavengers led to a recovery in the percentage and number of IgG1+ and CD138+ (ASC) HuR-cKO B cells (Fig. 8f and 8g). However NAC and EUK134 marginally inhibited the yield of IgG1+ and CD138+ cells in control cultures. Taken together, our data indicate that HuR-cKO B cells contained elevated amounts of ROS and these compromised B cell survival. Mitigating ROS allowed the HuR-cKO B cells to proliferate, undergo CSR and form ASC in vitro.
DISCUSSION
Here we show that HuR is dispensable for B cell development, but that the GC response and the production of class-switched antibodies following immunisation with T-independent antigens are highly dependent upon HuR. Our in vitro experiments indicate that HuR may control B cell metabolism upon activation to limit ROS production, but further validation in the context of the GC response is required.
The regulation of intracellular ROS is important during lymphocyte activation25. Increased ROS are found in activated B cells taken from virally infected mice26. B cells defective in HVCN1 make less ROS and have attenuated antibody responses27, and antioxidant administration to mice increased B cell apoptosis and decreased proliferation26. However, lack of ROS production in gp91 deficient mice has been linked to modestly increased responses to TI-2 antigens28. Thus a general picture is emerging in B cells in which ROS production can be separated in space and time. Low levels of ROS have mitogenic properties whereas high levels will induce cell death29, 30. H2O2 produced by NAPDH oxidases after BCR crosslinking exerts a short-lived function as inhibitor of protein tyrosine and lipid phosphatases31, thus enhancing signal transduction. BCR-associated ROS production early after antigen encounter may not be essential for B cell proliferation, however, mitochondrial-mediated ROS production is required to sustain BCR-mediated activation28, 32. Interestingly, in vitro exposure to H2O2 can revive signalling by the BCR on GC B cells33. The mechanism proposed by Wheeler et al. involves enhancement of the PI3K-mTORC axis for metabolic reprogramming after lymphocyte activation34,35. Our global transcriptome analysis, comparing naive versus GC B cells and ex vivo versus LPS-stimulated B cells, reinforces previous observations that both glycolysis and oxidative phosphorylation are increased after B cell activation2, 36, as part of a metabolic program taking place both in GC B cells 37 and in vitro stimulated B cells to promote survival and proliferation, and in which post-transcriptional regulation by HuR is a critical component.
The presence of hydroxyl radicals also enhances deamination of cytosine to uracil, a reaction that is necessary for CSR38. CSR requires control of ROS levels by DNA damage sensing proteins such as p53 and Atm. P53 limits ROS production in B cells and promotes switching to IgG2a39. Atm expression during DSB decreases ROS levels via regulation of cell energy pathways. Atm−/− mice have an impaired antibody response and elevated ROS that is restored by treatment with NAC40. Furthermore, the Atm-specific inhibitor KU-55933 decreases cellular ATP levels by reducing TCA cycle intermediates and oxidative respiration41. Thus we suggest that reduced energy metabolism during CSR may limit cell cycle progression and prevent the accumulation of ROS until DSB are repaired. By sustaining the TCA cycle, HuR ensures provision of the reducing equivalents required for ATP synthesis and/or ROS scavenging.
Evidence has accumulated to demonstrate increased glycolysis is part of a metabolic switch during activation of immune cells1. Although the TCA cycle is the main provider of reducing equivalents for oxidative phosphorylation, its regulation remains largely unexplored during lymphocyte activation. We have used CPI-613 to inhibit PDH and αKGDH, two key enzymes of the TCA cycle, resulting in a block in B cell growth and proliferation. Recently, it has been reported that CPI-613 induces an oxidative burst responsible for αKGDH enzymatic activity inhibition42. This observation underscores the importance of regulating ROS production during B cell activation and is strengthened by our finding that phosphonate analogues of αKG43 reduce B cell viability and proliferation. The number of IgG1+ cells recovered after in vitro B cell activation in the presence of 10 mM PESP is reduced by five-fold even though cell proliferation is less affected, being consistent with our results from Dlst+/− B cells, suggesting that B cells undergoing CSR are more susceptible to ROS.
Notably, Dlst+/− B cells do not recapitulate the in vitro phenotype of HuR-cKO B cells. This likely reflects as yet unidentified roles for other targets of the HuR-splicing program that are important during B cell activation. Amongst the 375 genes identified as HuR dependent is Slc25a19 depletion of which also reduces αKGDH enzymatic activity23.
The αKGDH complex has three subunits: α-oxoglutarate dehydrogenase (Ogdh), dihydrolipoamide S-succinyltransferase (Dlst), and dihydrolipoamide dehydrogenase (Dld). Dlst is a haploinsufficient gene and heterozygote mice show increased ROS production and cell death in the brain44. The APP670/671 mutation of Dlst in humans reduced enzyme activity and causes Alzheimer’s disease associated with mitochondrial dysfunction and cell death45. Decreased Dlst diminishes the provision of reducing equivalents and succinyl-CoA required for GTP synthesis by matrix substrate-level phosphorylation46. Increased ROS levels in the absence of HuR may be due increased mRNA translation of genes of the oxidative respiratory chain, but reduced matrix substrate-level phosphorylation may additionally contribute to ROS production by increasing ADP levels47. Furthermore, αKGDH activity generates •O2− which acts as a negative regulator of αKGDH enzymatic activity48, 49. Thus, oxidative stress may promote cell death due to inhibition of flux through the TCA cycle50.
Our novel approach of integrating global analysis of mRNA expression and translation comparing control and HuR-cKO primary B cells reveals a role for HuR in the regulation of the metabolism after B cell activation. We have applied a novel bioinformatics approach to identify differential intron usage based on read counting within intron bins defined as windows between annotated exons. We have identified 530 introns from 375 genes that are differentially expressed in the absence of HuR. HuR loss is associated with aberrant intron inclusion which compromises, in case of Dlst, mRNA abundance and translation, but may also affect the isoform of translated proteins. A quantitative analysis of full length transcripts and alternative protein isoforms in the presence and absence of HuR should address this. We suggest that HuR, like other splicing regulators, promotes the correct splicing of pre-mRNA and provides a quality control mechanism for the transcriptome. In the context of B cell activation, HuR depletion promotes an imbalance in energy metabolism that leads to the fatal accumulation of ROS and impairs B cell proliferation and CSR.
ONLINE METHODS
Reagents, antibodies and oligonucleotides
Information is detailed in Supplementary Table 6, 7 and 8.
Mouse strains and animal procedures
Mice on the C57BL/6 background used in this study are: CD79atm1(cre)Reth 51 and Elavl1tm1Dkon 16. Dlst+/− mice were generated by Lexicon Pharmaceuticals, Inc. (The Woodlands, Texas)24. Immunisations were performed by intraperitoneal administration of Sheep Red Blood Cells (SRBCs, 2 × 108 cells/mouse), NP-Ficoll (25 μg), NP-LPS (25 μg) or alum-precipitated NP-KLH (100 μg). For in vivo experiments, the number of animals was decided based on preliminary studies using small cohorts to assess the magnitude of the changes (2-3 mice per genotype). 6 to 10 mice (12-16 weeks old) were considered a sufficient number of mice in final studies to reach sufficient statistical power analysis. No animals were excluded due to a lack of responsiveness to immunisation. Randomization, but not experimental blinding, was performed in these studies. All animal procedures at the Babraham Institute were approved by the Babraham Institute AWEEC, the UK Home Office. Local Ethical Committee for Animal Experiments of KU Leuven, Belgium approved experimentation with Dlst+/− mice under license number 000/2014.
ELISA and ELISPOT assays
Serum immunoglobulins and NP-specific antibodies were detected by ELISA52. NP-specific antibody end point titers were used as a measure of relative concentration. NP-specific antibody secreting B cells (ASC) were detected by ELISPOT assays performed as previously described9.
Flow cytometry
Analysis of B cell populations was performed using specific antibodies for cell surface markers. B cell population analysed in bone marrow were: Pre-Pro-B cells - B220lo IgM− CD25− c-kit+ CD19−; Pro-B cells - B220lo IgM− CD25− c-kit+; Pre-B cells - B220lo IgM− CD25+ CD19lo; immature-B cells - B220+ IgM+ IgD−; and mature-B cells - B220+ IgM+ IgD+. B cell subsets identified in spleen were: T1 - B220+ CD93− CD23− IgMhi; T2 - B220+ CD93− CD23+ IgMhi; T3 - B220+ CD93− CD23+ IgM+; MZ - B220+ CD93+ CD21hi CD23−, FO - B220+ CD93+ CD21+ CD23+ and GC B cells - B220+CD95+PNA+.
Fixable Viability Dye eFluor® 780 or DAPI was used to test cell viability. For intracellular staining of HuR, the BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit was used. In vitro B cell proliferation was tested by flow cytometry after the labelling of ex vivo B cells with the CellTrace™ Violet dye. ROS levels in B cells were measured by using OxyBURST® Green H2DCFDA (1 μM) added into the B cell culture 30 min before cell analysis by flow cytometry.
Calcium flux analysis
Purified LN B cells were loaded with Fluo-4 AM (3 μM) in PBS + 0.5% BSA (5 × 106 cells/ml) for 30 min at RT in the dark. After washing, cells were incubated in 0.5% BSA/PBS containing 1 mM CaCl2 for further 30 min to allow de-esterification of intracellular Fluo-4 AM ester. Changes in fluorescence emission (at 494 nm) after surface Ig crosslinking were measured using a PerkinElmer LS 55 Fluorescence spectrometer.
B cell isolation and cell culture
B cells from spleen or peripheral lymph nodes were isolated by using the B Cell Isolation Kit from Miltenyi Biotec. B cells were cultured in RPMI 1640 Medium (Dutch Modification) plus 5% FCS, antibiotics, 2 mM L-glutamine and β-mercaptoethanol (5 μM). ROS scavengers, Q-VD-OPh, Necrostatin-1 (Nec-1), 2-deoxy-glucose (2-DG), CPI-613, Oligomycin A, Succinyl phosphonate (SP) and Phosphonoethyl ester of succinyl phosphonate (PESP) were added to the cell culture media when indicated in the figure legend. Mitogen activation of B cells was carried out using either LPS (E.coli 0127:B8), αCD40 (3/23 clone) or αIgM (B7.6 clone) in the presence or not of IL4 and IL5.
Protein separation and Western Blot analysis
Total cell extracts were prepared by incubating cells in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.1% SDS and 0.5% Sodium deoxycholate) supplemented with protease inhibitors. 10% polyacrylamide-SDS gels were loaded with 20 μg of total protein extracts per lane. Gel electrophoresis and protein transfer to nitrocellulose membranes was performed prior detection of HuR, Dlst and β-actin by Western blot using specific antibodies and enhanced chemiluminescence or IRDye detection.
RNA extraction and RT-PCR assays
Total RNA extraction from purified B cells was performed using TRIzol (LifeTech). 1 μg of RNA was treated with DNase I prior reverse transcription into cDNA. Analysis of Dlst alternative splicing events were performed by RT-PCR using specific primers (table S3). Intron retention was quantified by qPCR whereas alternative exon inclusion was quantified after gel densitometry of the PCR products. qPCR assays were performed using Platinum® SYBR® Green qPCR SuperMix (Life Technologies). mRNA expression of Dlst and Elavl genes was quantified by the ΔΔCT method (comparative threshold cycle) and normalised to the expression of 18S rRNA.
Immunofluorescence microscopy
Ex vivo or mitogen- activated splenic B cells were cultured for 20 minutes on cover slips coated with 0.01% Poly-L-Lysine prior detection of HuR by immunofluorescence microscopy as previously described53. Images from three independent experiments were captured with an Olympus FV1000 System and processed using Image-J.
Individual-nucleotide resolution Cross-Linking and ImmunoPrecipitation (iCLIP)
iCLIP experiments were performed as described 21, using protein lysates from splenic B cells activated with LPS for 48 hours (n=3). Briefly, splenic B cells were irradiated with UV- light to crosslink protein-RNA interactions (150 mJ/cm2, Stratalinker 2400). After washing cells with ice-cold PBS, total cell extracts were obtained using RIPA buffer and sonication (x3). Extract supernatants was treated with RNase I (0.167 U/ml) for 3 minutes at 37°C. Immunoprecipitation of HuR-RNA complexes was performed using 2 μg. of αHuR mAb coupled to protein G dynabeads. B cell extracts from HuR-cKO B cells were used as a negative control. After washing twice with high-salt buffer (50 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS and 0.5% sodium deoxycholate) and once with PNK washing buffer (20 mM Tris-HCl pH 7.4, 10 mM MgCl2, 0.2% Tween-20), one tenth of the sample were labelled with 32P-ATP using PNK, whereas an RNA Linker was ligated to the rest of the sample after RNA dephosphorilation. RNA-protein complexes were separated by electrophoresis (SDS-Page) and transfer to nitrocellulose membranes. HuR-RNA complexes higher than 55 KDa were marked after visualization by autoradiography. RNA extraction was performed by incubating the nitrocellulose fragment at 37°C for 10 minutes with proteinase K in PK buffer (100 mM This-Cl pH 7.5, 50 mM NaCl and 10 mM EDTA). 200 μl of PK buffer containing urea (7 M) was added to each sample and further incubated at 37°C for 20 minutes. RNA was isolated by phenol/clorophorm extraction and ethanol precipitation. RNA was retro transcribed into cDNA using RCLIP primers and SuperScript III reverse transcriptase. After cDNA purification using 6% TBE-urea gels, cDNA was circularised and amplified by PCR using Solexa P5/P7 primers. cDNA libraries from three independent experiments were prepared and analysed. RCLIP primers included a 7 bases long barcode at the 5′end (three known bases plus four unknown nucleotides). Additional two bases (AT) were added to the 3′end. Barcodes allowed us to multiplex different cDNA libraries for Illumina sequencing and identify PCR duplicate reads.
High throughput sequencing and library preparation
mRNAseq libraries were obtained using TruSeq Stranded mRNA Sample Prep Kit (Illumina Inc). Splenic B cells from individual HuRf/f control or HuR-cKO mice were independently processed for RNA extraction (ex vivo samples, n=4 per genotype) or were stimulated with LPS for 48 hours in RPMI media plus 1 mM NaPyr (n=3-4 per genotype).
Previously published mRNAseq libraries were used to analyse the expression of genes involved in glycolysis, TCA cycle and electron transport in naive and GC B cells (GEO deposition number - GSE47705)54.
Ribosome footprinting profiling (Ribo-Seq) assays were performed using ARTseq™ Ribosome Profiling Kit (Epicentre, Illumina). Ex vivo or LPS-activated B cells (n=4-5 per genotype and condition) were treated with cyclohexamide (CHX, 100 μg/ml) three minutes prior cell extract preparation. cDNA libraries were sequenced using either GAIIX (iCLIP) or HTSeq2000 (mRNAseq and Ribo-Seq) Illumina technology. The type of sequencing performed was: iCLIP, 40 bp SE; mRNAseq, 100 bp SE; and Ribo-seq, 50 bp SE.
Bioinformatics and statistics
Mapping and peak call analysis of HuR iCLIP data was performed as described previously 21. Briefly, RCLIP primers were used to generate the cDNA libraries from HuR iCLIP experiments. Data demultiplexing involved identification of the three known bases of the 5′ barcode. Quality analysis of sequencing data was done using FastQC. Data analysis was done in the iCount pipeline. First, the four unknown bases of the 5′ barcode were used to remove PCR duplicate reads. 3′ barcode (AT) was used to define the length of the reads. Both parameters were important to identify unique reads. Then, reads were trimmed to remove 5′ and 3′ barcodes along with any adaptor sequence before mapping to the genome using Bowtie. Data was originally annotated to mm9 genome, but was lifted over to mm10 genome using Galaxy. Once genome annotation was completed, nucleotide −1 was located and used for data visualization in UCSC genome viewer. This base is the HuR- crosslink binding site. The number cDNA counts associated to a single HuR- crosslink site was quantified base on read length and unique 5′ barcode sequence. Peak call analysis was performed to identify unique HuR-crosslink binding sites with a FDR<0.05. Peak call analysis was performed in iCount by genome wide analysis of 30 base windows defined after annotation at single nucleotide resolution of HuR-RNA interactions (15 nucleotides flanking a cross link site). Peak enrichment analysis and FDR calculation was performed allowing 100 permutations and multiple hypothesis testing to correct for multiple comparisons.
Raw sequencing data from mRNAseq and Ribo-Seq was demultiplexed, FastQ analysed and mapped to mm10 genome annotation using Tophat. Differential gene expression analysis of mRNAseq and Ribo-Seq datasets was performed using DESeq and DESeq2 packages55. Briefly, data was adjusted to a negative binomial distribution after correction by library size factor. Differential expression analysis, p value determination and Benjamini-Hochberg multiple test correction of the p values were performed to obtain final padj values. DEXSeq package was used for exon and intron usage analysis56. First, we define exons and introns by using the Ensembl exon annotation for protein coding transcripts (GRCm38.72) and we independently counted reads within the exon and intron bins by using HTSeq. Read counting along exon bins was required in order to estimate the library size factors. We then used these size factors to normalise the intron bins and assessed differential intron usage with DEXSeq. A hypergeometric statistical test followed by Benjamini-Hochberg correction of the p- values was performed for pathway enrichment analysis57, 58. UCSC and IGV were used to visualise read coverage (see Supplemental Information). Sequencing data was visualised in UCSC genome viewer. Read coverage of mRNAseq and Ribo-Seq data was visualised in the Integrated Genome Browser (IGV).
Unpaired t tests or Mann-Whitney tests were performed for statistical analysis of non-sequencing data. More details about sample size and the statistical test performed in each experiment are indicated in each figure legends.
Data deposition
High throughput sequencing data was deposited in Gene Expression Omnibus (GEO). Identification numbers are: GSE62129 for mRNAseq, GSE62134 for Ribo-seq and GSE62148 for iCLIP.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Bates, D. Sanger, N. Evans, S. Walker, K. Tabbada, G. Morgan, R. Walker and the Babraham Institute Biological Support Unit for expert technical assistance.
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC BB/J001457/1 and BB/J00152X/1). K.F. was supported by the National Health and Medical Research Council, Australia (KF Fellowship 516786) and the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. K.Z. was supported by an EMBL EIPOD fellowship. VB work with the phosphonate analogs of 2-oxo acids was supported by the Russian Science Foundation (grant number 14-15-00133). J.U. and T.C. were supported by the Slovenian Research Agency (grant J7-5460).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caro-Maldonado A, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair D, Dufort FJ, Chiles TC. Protein kinase Cbeta is critical for the metabolic switch to glycolysis following B-cell antigen receptor engagement. The Biochemical journal. 2012;448:165–169. doi: 10.1042/BJ20121225. [DOI] [PubMed] [Google Scholar]
- 4.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Seminars in immunology. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nature immunology. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov P, Anderson P. Post-transcriptional regulatory networks in immunity. Immunological reviews. 2013;253:253–272. doi: 10.1111/imr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nature immunology. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 8.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 9.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 11.de Yebenes VG, Bartolome-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunological reviews. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertossi A, et al. Loss of Roquin induces early death and immune deregulation but not autoimmunity. The Journal of experimental medicine. 2011;208:1749–1756. doi: 10.1084/jem.20110578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadri N, Lu JY, Badura ML, Schneider RJ. AUF1 is involved in splenic follicular B cell maintenance. BMC immunology. 2010;11:1. doi: 10.1186/1471-2172-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee N, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Molecular cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Molecular cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Katsanou V, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Molecular and cellular biology. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yiakouvaki A, et al. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. The Journal of clinical investigation. 2012;122:48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubin MM, et al. Conditional knockout of the RNA-binding protein HuR in CD4(+) T cells reveals a gene dosage effect on cytokine production. Mol Med. 2014;20:93–108. doi: 10.2119/molmed.2013.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh M, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. The Journal of clinical investigation. 2009;119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papadaki O, et al. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 21.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindhurst MJ, et al. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15927–15932. doi: 10.1073/pnas.0607661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. Mice deficient in dihydrolipoyl succinyl transferase show increased vulnerability to mitochondrial toxins. Neurobiology of disease. 2009;36:320–330. doi: 10.1016/j.nbd.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nature immunology. 2002;3:1129–1134. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 26.Crump KE, Langston PK, Rajkarnikar S, Grayson JM. Antioxidant treatment regulates the humoral immune response during acute viral infection. Journal of virology. 2013;87:2577–2586. doi: 10.1128/JVI.02714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capasso M, et al. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nature immunology. 2010;11:265–272. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards SM, Clark EA. BCR-induced superoxide negatively regulates B-cell proliferation and T-cell-independent type 2 Ab responses. European journal of immunology. 2009;39:3395–3403. doi: 10.1002/eji.200939587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buttke TM, Sandstrom PA. Redox regulation of programmed cell death in lymphocytes. Free radical research. 1995;22:389–397. doi: 10.3109/10715769509147548. [DOI] [PubMed] [Google Scholar]
- 30.Bertolotti M, et al. B- to plasma-cell terminal differentiation entails oxidative stress and profound reshaping of the antioxidant responses. Antioxidants & redox signaling. 2010;13:1133–1144. doi: 10.1089/ars.2009.3079. [DOI] [PubMed] [Google Scholar]
- 31.Singh DK, et al. The strength of receptor signaling is centrally controlled through a cooperative loop between Ca2+ and an oxidant signal. Cell. 2005;121:281–293. doi: 10.1016/j.cell.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler ML, Defranco AL. Prolonged production of reactive oxygen species in response to B cell receptor stimulation promotes B cell activation and proliferation. J Immunol. 2012;189:4405–4416. doi: 10.4049/jimmunol.1201433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil AM, Cambier JC, Shlomchik MJ. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 2012;336:1178–1181. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fruman DA, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 35.Yang K, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell metabolism. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heise N, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. The Journal of experimental medicine. 2014;211:2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labet V, Grand A, Cadet J, Eriksson LA. Deamination of the radical cation of the base moiety of 2′-deoxycytidine: a theoretical study. Chemphyschem: a European journal of chemical physics and physical chemistry. 2008;9:1195–1203. doi: 10.1002/cphc.200800154. [DOI] [PubMed] [Google Scholar]
- 39.Guikema JE, et al. p53 represses class switch recombination to IgG2a through its antioxidant function. J Immunol. 2010;184:6177–6187. doi: 10.4049/jimmunol.0904085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito K, et al. Regulation of reactive oxygen species by Atm is essential for proper response to DNA double-strand breaks in lymphocytes. J Immunol. 2007;178:103–110. doi: 10.4049/jimmunol.178.1.103. [DOI] [PubMed] [Google Scholar]
- 41.Zakikhani M, et al. Alterations in cellular energy metabolism associated with the antiproliferative effects of the ATM inhibitor KU-55933 and with metformin. PloS one. 2012;7:e49513. doi: 10.1371/journal.pone.0049513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuart SD, et al. A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer & metabolism. 2014;2:4. doi: 10.1186/2049-3002-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunik VI, et al. Phosphonate analogues of alpha-ketoglutarate inhibit the activity of the alpha-ketoglutarate dehydrogenase complex isolated from brain and in cultured cells. Biochemistry. 2005;44:10552–10561. doi: 10.1021/bi0503100. [DOI] [PubMed] [Google Scholar]
- 44.Shi Q, Chen HL, Xu H, Gibson GE. Reduction in the E2k subunit of the alpha-ketoglutarate dehydrogenase complex has effects independent of complex activity. The Journal of biological chemistry. 2005;280:10888–10896. doi: 10.1074/jbc.M409064200. [DOI] [PubMed] [Google Scholar]
- 45.Gibson GE, et al. Alpha-ketoglutarate dehydrogenase in Alzheimer brains bearing the APP670/671 mutation. Annals of neurology. 1998;44:676–681. doi: 10.1002/ana.410440414. [DOI] [PubMed] [Google Scholar]
- 46.Kiss G, et al. The negative impact of alpha-ketoglutarate dehydrogenase complex deficiency on matrix substrate-level phosphorylation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27:2392–2406. doi: 10.1096/fj.12-220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lardy HA, Wellman H. Oxidative phosphorylations; role of inorganic phosphate and acceptor systems in control of metabolic rates. The Journal of biological chemistry. 1952;195:215–224. [PubMed] [Google Scholar]
- 48.Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan CL, et al. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. The Journal of biological chemistry. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2000;20:8972–8979. doi: 10.1523/JNEUROSCI.20-24-08972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doody GM, et al. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nature immunology. 2001;2:542–547. doi: 10.1038/88748. [DOI] [PubMed] [Google Scholar]
- 53.Diaz-Munoz MD, Osma-Garcia IC, Iniguez MA, Fresno M. Cyclooxygenase-2 deficiency in macrophages leads to defective p110gamma PI3K signaling and impairs cell adhesion and migration. J Immunol. 2013;191:395–406. doi: 10.4049/jimmunol.1202002. [DOI] [PubMed] [Google Scholar]
- 54.Hogenbirk MA, et al. Differential programming of B cells in AID deficient mice. PloS one. 2013;8:e69815. doi: 10.1371/journal.pone.0069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome research. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.