Summary
Human intestinal stem cell and crypt dynamics remain poorly characterized because transgenic lineage-tracing methods are impractical in humans. Here, we have circumvented this problem by quantitatively using somatic mtDNA mutations to trace clonal lineages. By analyzing clonal imprints on the walls of colonic crypts, we show that human intestinal stem cells conform to one-dimensional neutral drift dynamics with a “functional” stem cell number of five to six in both normal patients and individuals with familial adenomatous polyposis (germline APC−/+). Furthermore, we show that, in adenomatous crypts (APC−/−), there is a proportionate increase in both functional stem cell number and the loss/replacement rate. Finally, by analyzing fields of mtDNA mutant crypts, we show that a normal colon crypt divides around once every 30–40 years, and the division rate is increased in adenomas by at least an order of magnitude. These data provide in vivo quantification of human intestinal stem cell and crypt dynamics.
Graphical Abstract
Highlights
-
•
Somatic mtDNA mutations for quantitative lineage tracing in human intestine
-
•
Human intestinal stem cells evolve according to a neutral drift process
-
•
Loss of APC causes an increase in the stem cell loss/replacement rate
-
•
Intestinal crypts divide once every 30–40 years in the normal human colon
Baker et al. examine the in vivo stem cell biology of human colonic crypts. They reveal that each crypt contains a small number of functional stem cells and that stem cell division is predominantly symmetric and also quantify perturbation of stem cell architecture within adenomas. Additionally, they measure the division rate of human colonic crypts as once every 30–40 years in the healthy colon and demonstrate that the crypt division rate is increased 10-fold within small adenomas.
Introduction
The intestinal crypt has become the archetypal model for quantitative understanding of adult stem cell dynamics and regulation. Intestinal crypts are finger-like invaginations of the intestinal mucosa lined by a monolayer of epithelial cells. Genetic lineage-tracing experiments in mice have shown that stem cell self-renewal can be characterized by the dynamics of a single population of equipotent intestinal stem cells located at the crypt base, which divide frequently to produce the specialized cell progeny that occupy the upper portion of the crypt (Barker et al., 2007; Sangiorgi and Capecchi, 2008). Quantitative analysis has revealed that the stem cells are in continual neutral competition with each another to retain a privileged position within the crypt base niche; on average, each stem cell division results in loss and replacement of an individual stem cell lineage (Kozar et al., 2013; Lopez-Garcia et al., 2010; Snippert et al., 2010). However, the fruitful transgenic approach cannot be used in humans. Consequently, it has remained uncertain if and how the biology of the human intestinal crypt is mirrored by its murine counterpart.
Crypts themselves also represent a dynamic evolving population. A low level of crypt fission (the bifurcation of a crypt to produce two daughter crypts) is observed in the adult intestine (Cheng et al., 1986a; Totafurno et al., 1987; Wong et al., 2002), and the healing response following epithelial damage increases the proportion of crypts in fission (Cheng et al., 1986a, 1986b; Miyoshi et al., 2012; Park et al., 1995; Totafurno et al., 1987). Quantitative analysis of genetic lineage-tracing studies in mice revealed the low basal rate of crypt fission in the normal murine colon is increased 30-fold by oncogenic KRAS mutation (Snippert et al., 2014). Moreover, it is the fission of a transformed crypt, rather than the aberrant growth of cells per se, that drives the initial growth of colorectal adenomas (Preston et al., 2003; Thirlwell et al., 2010; Wong et al., 2002). Despite the central importance of crypt fission in the initiation of colon cancer, the evolutionary dynamics of the human intestinal crypt population remain obscure.
Here, we have measured the clonal evolution of stem cell populations within the human colon by using naturally occurring somatic mtDNA mutations to trace clonal lineages; this methodology circumvents the need to externally label cells in order to trace their progeny. Our analysis exploits the stereotypic architecture of the intestinal crypt to resolve the temporal dynamics of clone evolution.
Results
Naturally Occurring Somatic Mutations to Trace Clonal Lineages
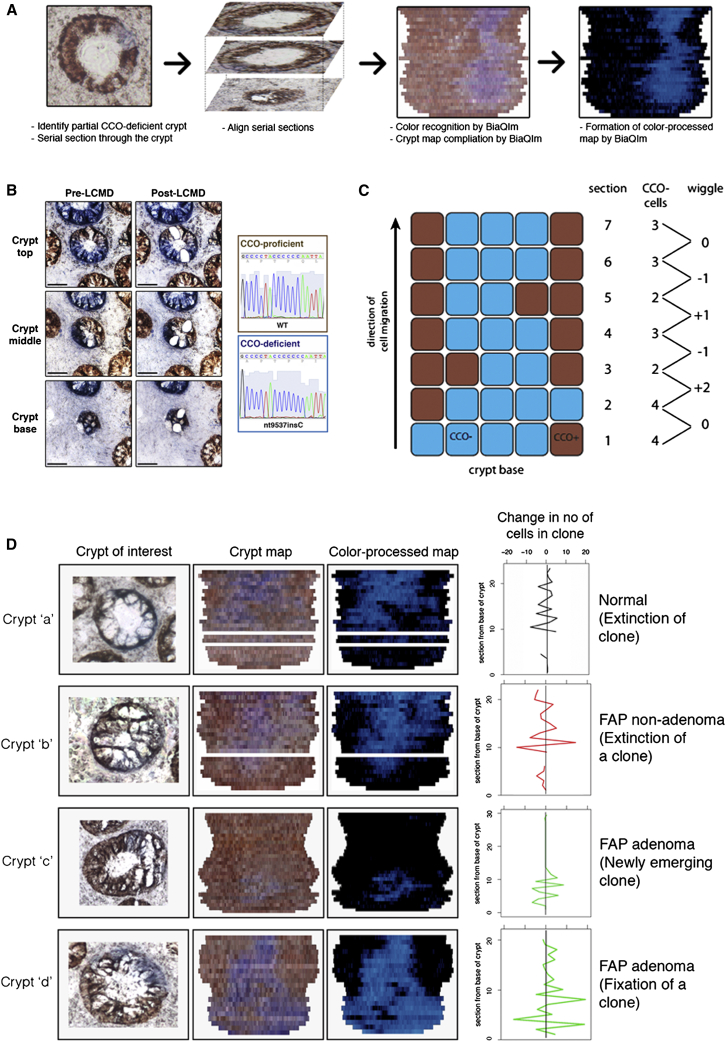
To trace clonal lineages in the human intestine, we performed enzyme-histochemistry for the activity of cytochrome c oxidase (CCO). Infrequent stochastic loss of CCO activity (CCO−) is observed in the human intestine and is attributed to an underlying somatic mitochondrial DNA (mtDNA) mutation (Taylor et al., 2003). mtDNA sequencing confirms that adjacent CCO− cells in the intestine are clonally derived (Fellous et al., 2009; Greaves et al., 2006; Gutierrez-Gonzalez et al., 2009; Taylor et al., 2003). CCO activity was assessed in en face serial sections of colonic mucosa (n = 9 patients; Table S1). Within each specimen there were crypts that contained only CCO-proficient (CCO+) cells, only CCO− cells, and a mixture of CCO− and CCO+ cells (“partially-mutated”) (Figure 1).
Figure 1.
Measurement of CCO-Deficient Clone Size and Migration
(A) Schematic diagram showing compilation of a “crypt map” by the BiaQIm software. Aligned serial sections are processed by the software to describe the position of CCO-deficient cells in the crypt (adapted from Fellous et al., 2009). Displayed crypt is a nonadenomatous crypt from an AFAP patient.
(B) Laser capture microdissection followed by sequencing of mtDNA in a partially CCO-deficient crypt. In this example, the CCO-deficient clone (blue staining) contains an insertion of a cytosine residue (nt9537insC), causing a frameshift in the gene encoding CCO subunit III. Displayed crypt is a nonadenomatous crypt from a patient with FAP. Scale bar represents 50 μm.
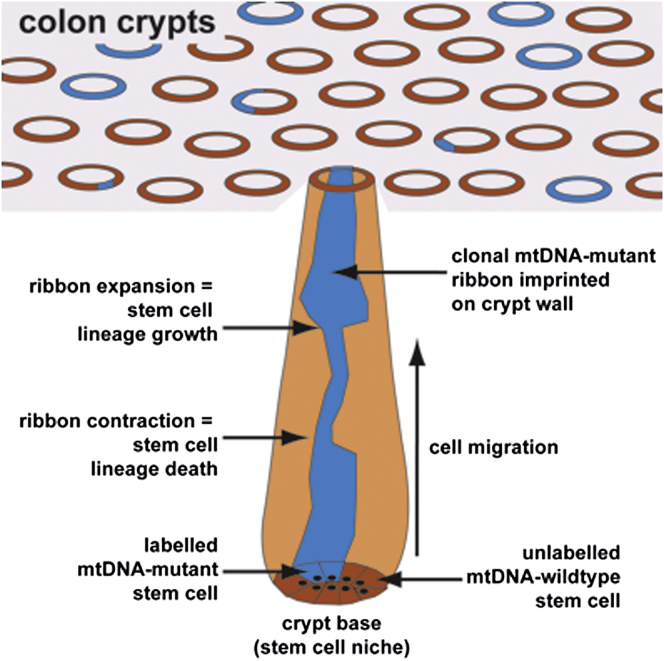
(C) Schematic diagram showing the expansion and contraction of a CCO-deficient population as it migrates from the crypt base. “Wiggles” of the CCO-deficient clone size are quantified by difference in the CCO− area between adjacent serial sections.
(D) Representative examples of crypt maps. The left column represents en face images of the crypts of interest, the middle column the resulting crypt maps, and the right represents the color-processed maps (blue, CCO− cells; black, CCO+ cells). White lines represent missing sections. The graph displays the change in the number of CCO− cells between adjacent sections. Examples of newly emerging clones (crypt “c”) and clones that were putatively in the process of becoming extinct (crypts “a” and “b”) were observed.
Deviations in Clone Size along the Crypt Axis Reveal Stem Cell Dynamics
We reconstructed the cellular composition of partially mutated crypts using serial sections and BiaQIm (http://www.deconvolve.net/bialith/BAQIFeatures.htm) image-processing software (Figure 1A). As previously reported (Fellous et al., 2009; Graham et al., 2011; Humphries and Wright, 2008), CCO− cells typically formed contiguous ribbons along the length of the crypt that were confirmed as de novo clonal populations (Figure 1B). In some cases, the width of these ribbons varied considerably along the crypt length (Figures 1A, 1D, and Figure S1; Movies S1 and S2). The intestinal crypt acts as a conveyor belt: cells are produced at the crypt base and migrate upward along the crypt axis before being shed into the lumen days later (Wright and Alison, 1984). Therefore, we reasoned that the “wiggles” in the width of the CCO− ribbon along the crypt axis represented a temporal record of the dynamic evolution of the CCO− stem cell population at the crypt base (Figure 1C). In other words, the CCO− ribbon records the clonal evolution of functioning stem cells at the crypt base, but we note that there may be many more cells within the crypt that have the potential to function as a stem cell. Specifically, we supposed that symmetric division of a CCO− stem cell that resulted in the replacement of a neighboring CCO+ stem cell with a CCO− stem cell (expansion of the CCO− clone) would increase the ribbon width, whereas replacement of a CCO− stem cell by a CCO+ stem cell (loss) would decrease the ribbon width. To verify this concept in a situation where the ground-truth was known, we analyzed ribbon evolution in an established computational model of the colonic crypt (van Leeuwen et al., 2009; see the Supplemental Experimental Procedures) and verified that the changes in the ribbon width along the crypt axis reflected the temporal dynamics of cell division at the crypt base (Figures S1A–S1C).
To quantify stem cell dynamics in vivo, we measured the deviation in ribbon width (Figure 1D) between successive en face serial sections in partially mutated normal crypts (n = 11) from three patients with sporadic colorectal cancer. Deviations were quantified by changes in the proportion of blue to brown staining cells between successive sections (using both manual and automated measurements; Figures S1D and S1E) and expressed in terms of cell numbers (Figure S2 and Table S1). The distribution of the deviations was approximately symmetric around zero (skewness = 0.3; KS-test p = 1; Figure 2A), indicating that the expansions and contractions of the ribbon widths were balanced, and therefore implying that the clonal evolution of the crypt base stem cells constituted a “neutral drift” type process. This also verified CCO deficiency as a neutral marker. Consistent with neutral drift dynamics, we observed CCO− populations located only in the lower portion of the crypt (a CCO− clone), ribbons disconnected from the crypt base (clone extinction), and examples of clone fixation where all cells become CCO− (Figures 1D and S1F). Led by analogous studies of intestinal stem cell neutral drift dynamics in transgenic mice (Kozar et al., 2013; Lopez-Garcia et al., 2010; Snippert et al., 2010), we expected that the temporal evolution of the number of functioning stem cells in the CCO− clone would follow a one-dimensional random walk, and consequently that the temporal evolution in CCO− ribbon width along the crypt axis would be described by a simple one-dimensional diffusion process (see the Supplemental Experimental Procedures). We note that this simple model describes the stem cell dynamics with a degree of detail appropriate to our data, and has equivalent scaling behavior to the more complex model previously used to understand cellular dynamics at the crypt base (Ritsma et al., 2014). In particular, if a CCO− clone has a total of n(t) functional stem cells at time t, the mean-square change in cell number is predicted to vary linearly with time as 〈n(T + t) − n(T)〉2T = 2λt, where the “diffusion constant,” λ, defines the functional stem cell loss/replacement rate, and 〈…〉T denotes an average over time. Applied to the measured clonal imprint on the crypt, this translates to a mean-square change in ribbon width of:
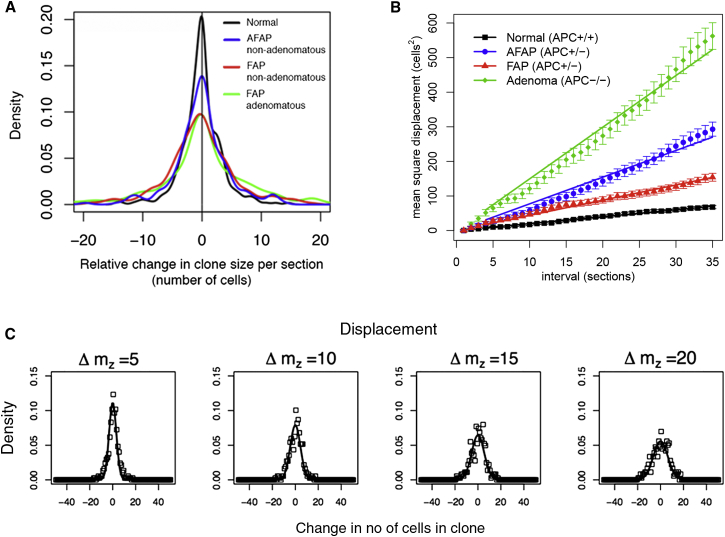
where ƒ defines the ratio of the functional stem cell number at the crypt base to the number of cells in a circumferential crypt section, ρ = 12 μm is the section thickness, ν is the average cell migration rate along the crypt axis, and mz indexes the z-stack coordinate of the serial sections. The experimental data confirmed the predicted linear relationship for M(mz) (Pearson’s R = 1; p < 0.001; Figure 2B). Additional quantitative analysis showed agreement of the distribution of ribbon widths along the crypt axis with the predicted time-dependence (Figure 2C, see the Supplemental Experimental Procedures). Altogether, these results show that the stem cells in human colon also follow a process of one-dimensional neutral drift dynamics.
Figure 2.
Assessment of Crypt Base Cell Number and Proliferative Fraction
(A) Smoothed density plot showing change in the relative clone size between sequential sections (“wiggle”). The symmetry around zero indicates that clonal contraction is balanced by equally frequent clonal expansion. The flatter and broader distribution in FAP, AFAP, and adenomatous crypts as compared to normal indicates more frequent, larger fluctuations in clone size.
(B) Mean squared difference in CCO− ribbon width as a function of distance along the crypt axis. Normal, FAP, AFAP, and adenomatous crypts all showed the linear relationship predicted by a one-dimensional neutral drift dynamics. Error bars denote SE.
(C) Distributions of the change in CCO− ribbon width as a function of displacement along the crypt axis in the normal colon. The measured distribution of changes in ribbon width (points) and the distributions predicted by the mathematical model (see the Supplemental Experimental Procedures) parameterized with the fitted value of the diffusion coefficient (lines) show good agreement. Columns represent the displacement measured in sections along on the crypt axis (Δmz).
From these results, we then sought to estimate the functional stem cell loss/replacement rate. From the analysis of bromodeoxyuridine incorporation data (Potten et al., 1992; see the Supplemental Experimental Procedures and Figure S2A), we could infer an average migration speed of around ν ≈ 4 μm/hour in the lower half of the crypt (where most of our histological sections were collected), consistent with previous pulse-chase labeling studies using tritiated thymidine incorporation (Lipkin et al., 1963). However, to determine the ratio, ƒ, we require an estimate of the functional stem cell number, n. Previous studies of mouse intestine have determined an average of n = 5, far smaller than the number of Lgr5+ crypt base columnar cells (Kozar et al., 2013). Here, noting that the functional stem cell loss/replacement rate must be bound by the cell division rate at the crypt base (e.g., we require λ ≤ 1 per cell division), which is estimated to be around once per 2–3 days for human colon (Potten et al., 1992), we can deduce a minimum ratio of around ƒ = 5. With an average of 23 (95% quantile 6–30) cells per crypt circumference (Figures S2B–S2D), a figure similar to that found in mouse colon, this is consistent with a maximum functional stem cell number of around n = 6, approximately a factor of two smaller than the average number of cells at the crypt base (mean = 10 cells; 95% quantile 6–15; Table 1 and Figures S2E–S2G). Because we expect some of the functional stem cell divisions to result in asymmetric fate outcome, it is likely that this figure represents a small overestimate.
Table 1.
Measured Diffusion Coefficients and Crypt Physiology Parameters
| APC Status | D Measured Diffusion Coefficient (95% SE) | Mean Number of Cells at Crypt Base (95% Quantile) | Average Percent Cells Ki67+ at Crypt Base (95% Quantile) | Mean Cells in Crypt Circumference (95% Quantile) | v Average Velocity of Cell Migration in Lower Half of Crypt (μm/hr); Reference |
|---|---|---|---|---|---|
| WT | 1.000 (±0.014) | 10 (6–15) | 9 (0–25) | 23 (16–30) | 4; Lipkin et al., 1963 |
| APC−/+ AFAP | 3.86 (±0.11) | 10 (6–13) | 10 (0–23) | 31 (24–41) | 1 (estimated) |
| APC−/+ FAP | 2.20 (±0.04) | 11 (8–14) | 13 (0–36) | 34 (27–44) | 1; Lightdale et al., 1982 |
| APC−/− | 7.47 (±0.19) | 16 (8–26) | 27 (16–49) | 49 (30–98) | 1 (estimated) |
APC Mutations Alter Stem Cell Dynamics
APC is the critical tumor suppressor gene in the colon (Lamlum et al., 2000), and loss of normal APC function is suggested to alter stem cell dynamics (Kim et al., 2004; Vermeulen et al., 2013). To quantify the effect of APC mutation on stem cell dynamics in the human colon, we examined the temporal evolution of CCO− clones in partially CCO-deficient but nondysplastic crypts from patients with familial adenomatous polyposis (FAP), and attenuated-FAP (AFAP). These patients have a germline loss-of-function mutation in the APC gene (Miyoshi et al., 1992a), and adenoma growth is initiated when the remaining wild-type allele is lost or mutated (Miyoshi et al., 1992b). In both nondysplastic FAP and AFAP crypts (APC−/+, n = 22 crypts from six patients) and dysplastic crypts from adenomas (APC−/−, n = 10 crypts from two patients), analysis of the balance of expansion versus contraction events in CCO− ribbons (Figure 2A) indicated the stem cells underwent neutral evolution (APC−/+ FAP skewness = −0.41 KS-test, p = 0.9; AFAP skewness = 0.76 p = 0.7; APC−/− skewness = −0.08, p = 0.3). The linear dependence of the mean-square displacement, M(mz), for both APC−/+ crypts and APC−/− adenomatous crypts (Figure 2B) confirmed neutral drift continued to occur within the stem cell compartment of APC-mutant crypts. As with normal crypts, there was good agreement of the distribution of ribbon widths along the crypt axis with the predicted time-dependence (Figure S2H). Taking the functional stem cell number to scale in proportion to the number of cells at the crypt base and the circumference of the crypt (Figures S2B–S2G); i.e., assuming that the ratio ƒ remains fixed, we found that the apparent increase in the speed of clone dispersion around the crypt circumference is approximately accounted for by the measured decrease in the cell migration speed along the crypt axis in both FAP and AFAP patients (Table 1), leading to loss/replacement rates comparable to those of normal tissue. In contrast, the measured increase in the dispersion of the clonal ribbons around the crypt circumference in adenomas is not compensated by the reduction in the migration speed, and translated to an acceleration of the loss/replacement rate by a factor of approximately two. This proportionate increase was consistent with the assessment of the pattern of CpG island methylation at a neutral locus, which showed decreased within-crypt methylation pattern diversity in adenomatous versus AFAP/FAP crypts, in line with an increased stem cell loss/replacement rate in adenomas (Figures S3A and S3B). Faster stem cell replacement leads to the suppression of methylation pattern diversity, because the methylation pattern borne by the “dominant” clone will be more rapidly propagated, and hence methylation pattern diversity that arises because of de novo methylation changes in different stem cells is suppressed. Furthermore, the increase in the rate of stem cell loss/replacement was in line with previous measurements showing disruption of mitotic spindle orientation in adenomas compared to normal colon (Quyn et al., 2010).
Evolutionary Dynamics of Colonic Crypts in Normal and Neoplastic Colon
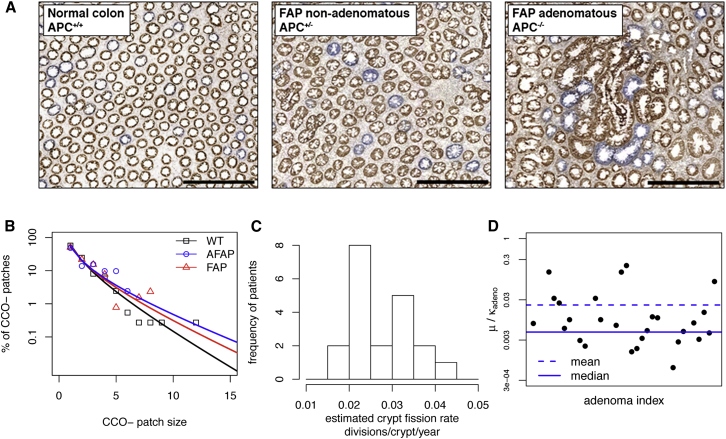
We next sought to define the evolutionary dynamics of the crypt population. Wholly mutant CCO− crypts are often found in clonal patches consisting of two or more CCO− crypts (Figure 3A), and the mean patch size increases with age (Greaves et al., 2006), suggesting that crypt fission occurs at a baseline rate throughout life. To estimate the crypt fission rate, we counted the patch size distribution of CCO− crypts in normal epithelium from resection specimens from 20 patients aged between 42 and 85 years who underwent resection for colorectal cancer (ncrypts = 136,899 crypts assessed; nCCO− = 5,313). We supposed that patch evolution followed a simple birth process defined by the crypt fission rate κ (Snippert et al., 2014; see the Supplemental Experimental Procedures), and fitted the predicted patch size distribution to that observed for each patient using a maximum likelihood approach (Figure 3B). The estimated crypt fission rate varied significantly between patients with a mean of κWT = 0.028 (σ = 0.007), indicating an average crypt cycle length of 36 years (Figure 3C). This is significantly longer than the previous heuristic estimate of 9–18 years (Totafurno et al., 1987).
Figure 3.
Analysis of CCO-Deficient Patch Size
(A) Representative images of CCO enzyme histochemistry in normal (patient age = 64 years), FAP nonadenomatous (67 years), and FAP adenomatous (67 years) colonic tissue. The majority of adjacent CCO− crypts (“patches”) are clonally derived. Scale bar represents 500 μm.
(B) Distribution of CCO− patch sizes (points) with the maximum-likelihood fit of the crypt fission model with crypt fission rate κ (lines) in a normal patient, and in patients with FAP or AFAP. The fitting procedure is detailed in the Supplemental Experimental Procedures.
(C) Histogram of estimated crypt fission rates (κ divisions/year) for wild-type. The mean fission rate was κWT = 0.028 divisions/crypt/year showing a significant variation in rate between patients.
(D) Distribution of the ratio of CCO− induction to the crypt fission rate (μ/κadeno) as estimated for adenomatous crypts. A broad range of values was observed.
APC has been suggested to play a critical role in crypt fission regulation (Wasan et al., 1998). To quantitatively assess the impact of APC disruption, we counted CCO− patch size distributions in the nondysplastic colons of six patients with either FAP or AFAP (ncrypts = 40,839; nCCO− = 1,411) and also within dysplastic adenomas in these patients (n = 36 adenomas; ncrypts = 1,174; nCCO− = 138). In nondysplastic FAP and AFAP colon, the mean estimated crypt fission rate was κAFAP = 0.028 (σ = 0.004), and κFAP = 0.034 (σ = 0.007), which was slightly higher but comparable with normal colon. To estimate the crypt fission rate within adenomas required an independent estimate of adenoma age (Supplemental Experimental Procedures and Figure S3D). By showing that the distribution of adenoma sizes was consistent with boundary-driven expansion, we could infer the median value of the ratioμ/κadeno = 0.005 (σ = 0.05), where μ defined the rate at which adenomatous crypts became CCO-deficient (Figure 3D). If μ is comparable between normal and adenomatous crypts (μ = 0.001 crypt−1year−1; Figure S3E), then this corresponds to an order of magnitude increase in the fission rate within adenomas and is consistent with the previously reported increased numbers of crypts in adenomas (Wasan et al., 1998; Wong et al., 2002). We note that there is a large degree of variability between adenomas in the estimated value of κadeno, and that we exclusively analyzed very small adenomas that could potentially be growing more slowly than their larger and more advanced counterparts. Correspondingly, assessment of methylation pattern diversity showed lower diversity among clonally related adenomatous crypts as compared to clonally related AFAP/FAP crypts (Figures S3A and S3C).
Discussion
Lineage tracing studies using transgenic mouse models have provided significant insight into the dynamics of murine intestinal stem cells, but the significance of these findings for the human intestine has remained in question. Here we have shown that the combination of naturally occurring somatic mutations coupled with the unique anatomy of the intestinal crypt provides a platform from which the evolutionary record of the human intestinal stem cell compartment can be inferred. We have shown that the clonal evolution of human intestinal stem cells is a neutral process that mimics that observed in the murine crypt, and also inferred that human crypts house a similarly small number of functional stem cells to their murine counterparts (Kozar et al., 2013), despite the total number of cells being 10-fold greater in human crypts. Loss of normal APC function is usually the cause of neoplasia in the colon (Lamlum et al., 2000); our data from neoplastic crypts imply that the tumor-suppressive mechanism of APC is a consequence of its direct role in regulating stem cell dynamics.
We have measured a low basal rate of crypt fission occurring throughout life in the adult human colon; similarly to the murine small bowel (Li et al., 1994), human colonic crypts typically divide at most once or twice during a lifetime. Loss of APC function causes an order-of-magnitude increase in the crypt fission rate and demonstrates a critical role for APC in regulating growth. We note that aberrant crypt fission underpins intestinal neoplasia, and as such, crypts are the units of selection in the intestine. Thus, by measuring the crypt fission rate, we measured a fundamental evolutionary parameter of the human colon, which facilitates quantification of the age and rate of growth of neoplastic lesions.
Finally, we note that our methods represent a general toolkit to quantify in vivo human stem cell dynamics in any tissue with a stereotypic architecture centered around a stem cell niche. Consequently, our methodology is directly applicable for understanding the origins and evolution of epithelial disease such as Barrett's esophagus and intestinal metaplasia in the stomach.
Experimental Procedures
Normal colon tissue samples were collected at University College Hospital, London, under multicenter ethical approval (07/Q1604/17 and 11/LO/1613). FAP tissue was collected at the Academic Medical Centre, Amsterdam, in accordance with national ethics guidelines on tissue procurement (local protocol 12-543). Tissue preparation and assessment of CCO activity and mtDNA sequencing (Taylor et al., 2003), construction of crypt maps (Fellous et al., 2009), and analysis of methylation patterns (Graham et al., 2011) was performed as previously described. Assessment of the number of cells at the crypt base was performed on 3 μm hematoxylin-and-eosin stained sections, and Ki67 expression assessed on serial sections with the Novocastra rabbit polyclonal antibody (dilution 1:400).
Details of computational and mathematical analyses are provided in the supplemental materials. Briefly, computational analysis of the crypt was performed in the CHASTE simulation framework (Fletcher et al., 2012; van Leeuwen et al., 2009), and mathematical analysis was based on a refinement of the previously described model of neutral drift processes in intestinal crypts (Lopez-Garcia et al., 2010; Snippert et al., 2010).
Acknowledgments
This study was supported by Cancer Research UK (to A.-M.B. and N.A.W.), the Medical Research Council (to B.C. and S.A.C.M.), the Engineering and Physical Sciences Research Council (to A.G.F.), Microsoft Research (to A.G.F.), the National Institute for Health Research University College London Hospitals Biomedical Research Centre (to M.R.J.), the Dutch Cancer Research Foundation (to M.J.), the Wellcome Trust (to B.D.S.), and Higher Education Funding Council for England (to T.A.G.).
Published: August 7, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
Serial sections along crypt axis (“z stack”) stained for CCO activity from two crypts. Variation in the proportion of crypt circumference occupied by CCO− cells is apparent.
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Cheng H., Bjerknes M., Amar J., Gardiner G. Crypt production in normal and diseased human colonic epithelium. Anat. Rec. 1986;216:44–48. doi: 10.1002/ar.1092160108. [DOI] [PubMed] [Google Scholar]
- Cheng H., McCulloch C., Bjerknes M. Effects of 30% intestinal resection on whole population cell kinetics of mouse intestinal epithelium. Anat. Rec. 1986;215:35–41. doi: 10.1002/ar.1092150106. [DOI] [PubMed] [Google Scholar]
- Fellous T.G., McDonald S.A.C., Burkert J., Humphries A., Islam S., De-Alwis N.M.W., Gutierrez-Gonzalez L., Tadrous P.J., Elia G., Kocher H.M. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cells. 2009;27:1410–1420. doi: 10.1002/stem.67. [DOI] [PubMed] [Google Scholar]
- Fletcher A.G., Breward C.J., Jonathan Chapman S. Mathematical modeling of monoclonal conversion in the colonic crypt. J. Theor. Biol. 2012;300:118–133. doi: 10.1016/j.jtbi.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Graham T.A., Humphries A., Sanders T., Rodriguez-Justo M., Tadrous P.J., Preston S.L., Novelli M.R., Leedham S.J., McDonald S.A.C., Wright N.A. Use of methylation patterns to determine expansion of stem cell clones in human colon tissue. Gastroenterology. 2011;140:1241–1250. doi: 10.1053/j.gastro.2010.12.036. e1–e9. [DOI] [PubMed] [Google Scholar]
- Greaves L.C., Preston S.L., Tadrous P.J., Taylor R.W., Barron M.J., Oukrif D., Leedham S.J., Deheragoda M., Sasieni P., Novelli M.R. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl. Acad. Sci. USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez L., Deheragoda M., Elia G., Leedham S.J., Shankar A., Imber C., Jankowski J.A., Turnbull D.M., Novelli M., Wright N.A., McDonald S.A. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J. Pathol. 2009;217:489–496. doi: 10.1002/path.2502. [DOI] [PubMed] [Google Scholar]
- Humphries A., Wright N.A. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- Kim K.-M., Calabrese P., Tavaré S., Shibata D. Enhanced stem cell survival in familial adenomatous polyposis. Am. J. Pathol. 2004;164:1369–1377. doi: 10.1016/S0002-9440(10)63223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar S., Morrissey E., Nicholson A.M., van der Heijden M., Zecchini H.I., Kemp R., Tavaré S., Vermeulen L., Winton D.J. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell. 2013;13:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Lamlum H., Papadopoulou A., Ilyas M., Rowan A., Gillet C., Hanby A., Talbot I., Bodmer W., Tomlinson I. APC mutations are sufficient for the growth of early colorectal adenomas. Proc. Natl. Acad. Sci. USA. 2000;97:2225–2228. doi: 10.1073/pnas.040564697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Q., Roberts S.A., Paulus U., Loeffler M., Potten C.S. The crypt cycle in mouse small intestinal epithelium. J. Cell Sci. 1994;107:3271–3279. doi: 10.1242/jcs.107.12.3271. [DOI] [PubMed] [Google Scholar]
- Lightdale C., Lipkin M., Deschner E. In vivo measurements in familial polyposis: kinetics and location of proliferating cells in colonic adenomas. Cancer Res. 1982;42:4280–4283. [PubMed] [Google Scholar]
- Lipkin M., Bell B., Sherlock P. Cell Proliferation Kinetics in the Gastrointestinal Tract of Man. I. Cell Renewal in Colon and Rectum. J. Clin. Invest. 1963;42:767–776. doi: 10.1172/JCI104769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C., Klein A.M., Simons B.D., Winton D.J. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y., Ando H., Nagase H., Nishisho I., Horii A., Miki Y., Mori T., Utsunomiya J., Baba S., Petersen G. Germ-line mutations of the APC gene in 53 familial adenomatous polyposis patients. Proc. Natl. Acad. Sci. USA. 1992;89:4452–4456. doi: 10.1073/pnas.89.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y., Nagase H., Ando H., Horii A., Ichii S., Nakatsuru S., Aoki T., Miki Y., Mori T., Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum. Mol. Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Ajima R., Luo C.T., Yamaguchi T.P., Stappenbeck T.S. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–113. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Goodlad R.A., Wright N.A. Crypt fission in the small intestine and colon. A mechanism for the emergence of G6PD locus-mutated crypts after treatment with mutagens. Am. J. Pathol. 1995;147:1416–1427. [PMC free article] [PubMed] [Google Scholar]
- Potten C.S., Kellett M., Rew D.A., Roberts S.A. Proliferation in human gastrointestinal epithelium using bromodeoxyuridine in vivo: data for different sites, proximity to a tumour, and polyposis coli. Gut. 1992;33:524–529. doi: 10.1136/gut.33.4.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston S.L., Wong W.-M., Chan A.O.-O., Poulsom R., Jeffery R., Goodlad R.A., Mandir N., Elia G., Novelli M., Bodmer W.F. Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res. 2003;63:3819–3825. [PubMed] [Google Scholar]
- Quyn A.J., Appleton P.L., Carey F.A., Steele R.J.C., Barker N., Clevers H., Ridgway R.A., Sansom O.J., Näthke I.S. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Ritsma L., Ellenbroek S.I., Zomer A., Snippert H.J., de Sauvage F.J., Simons B.D., Clevers H., van Rheenen J. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507:362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Snippert H.J., Schepers A.G., van Es J.H., Simons B.D., Clevers H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014;15:62–69. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.W., Barron M.J., Borthwick G.M., Gospel A., Chinnery P.F., Samuels D.C., Taylor G.A., Plusa S.M., Needham S.J., Greaves L.C. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirlwell C., Will O.C.C., Domingo E., Graham T.A., McDonald S.A.C., Oukrif D., Jeffrey R., Gorman M., Rodriguez-Justo M., Chin-Aleong J. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–1454. doi: 10.1053/j.gastro.2010.01.033. e1–e7. [DOI] [PubMed] [Google Scholar]
- Totafurno J., Bjerknes M., Cheng H. The crypt cycle. Crypt and villus production in the adult intestinal epithelium. Biophys. J. 1987;52:279–294. doi: 10.1016/S0006-3495(87)83215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen I.M.M., Mirams G.R., Walter A., Fletcher A., Murray P., Osborne J., Varma S., Young S.J., Cooper J., Doyle B. An integrative computational model for intestinal tissue renewal. Cell Prolif. 2009;42:617–636. doi: 10.1111/j.1365-2184.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., Morrissey E., van der Heijden M., Nicholson A.M., Sottoriva A., Buczacki S., Kemp R., Tavaré S., Winton D.J. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- Wasan H.S., Park H.S., Liu K.C., Mandir N.K., Winnett A., Sasieni P., Bodmer W.F., Goodlad R.A., Wright N.A. APC in the regulation of intestinal crypt fission. J. Pathol. 1998;185:246–255. doi: 10.1002/(SICI)1096-9896(199807)185:3<246::AID-PATH90>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wong W.-M., Mandir N., Goodlad R.A., Wong B.C.Y., Garcia S.B., Lam S.-K., Wright N.A. Histogenesis of human colorectal adenomas and hyperplastic polyps: the role of cell proliferation and crypt fission. Gut. 2002;50:212–217. doi: 10.1136/gut.50.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright N.A., Alison M. Clarendon; Oxford: 1984. The Biology of Epithelial Cell Populations. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serial sections along crypt axis (“z stack”) stained for CCO activity from two crypts. Variation in the proportion of crypt circumference occupied by CCO− cells is apparent.