Abstract
Adjunct therapy with the histone deacetylase inhibitor (HDACi) romidepsin increases plasma viremia in HIV patients on combination antiretroviral therapy (cART). However, a potential concern is that reversing HIV latency with an HDACi may reactivate the virus in anatomical compartments with suboptimal cART concentrations, leading to de novo infection of susceptible cells in these sites. We tested physiologically relevant romidepsin concentrations known to reactivate latent HIV in order to definitively address this concern. We found that romidepsin significantly inhibited HIV infection in peripheral blood mononuclear cells and CD4+ T cells but not in monocyte-derived macrophages. In addition, romidepsin impaired HIV spreading in CD4+ T cell cultures. When we evaluated the impact of romidepsin on quantitative viral outgrowth assays with primary resting CD4+ T cells, we found that resting CD4+ T cells exposed to romidepsin exhibited reduced proliferation and viability. This significantly lowered assay sensitivity when measuring the efficacy of romidepsin as an HIV latency reversal agent. Altogether, our data indicate that romidepsin-based HIV eradication strategies are unlikely to reseed a latent T cell reservoir, even under suboptimal cART conditions, because romidepsin profoundly restricts de novo HIV infections.
INTRODUCTION
Combination antiretroviral therapy (cART) greatly reduces HIV disease-related mortality but does not cure HIV infection. During cART, transcriptionally silent HIV persists in resting CD4+ T cells as a latent HIV reservoir (1). If cART is interrupted, viral replication is reinitiated from this reservoir. The resumption of viral replication typically manifests clinically as rebound in plasma viremia accompanied by a rapid decline in CD4+ T cells. Thus, lifelong cART is essential for continued virus suppression. The primary goal of HIV eradication therapies is to eliminate the latent HIV reservoir such that cART can be interrupted without viral rebound (2). Currently, activating HIV from latency such that viral cytopathic effects or the host immune system kills the infected cells is under investigation as a curative strategy. Key to the success of such strategies, collectively referred to as “kick and kill” approaches, is the ability to effectively reverse HIV latency in vivo (3).
The mechanisms by which HIV establishes latency are complex and include enzymatic processes that affect the chromatin organization of the HIV-promoter region, one of the key determinants of virus transcriptional activity (4–7). Histone deacetylation mediated by histone deacetylases (HDAC) leads to structural changes in chromatin that inhibit transcription (2, 6, 8). Conversely, histone deacetylase inhibitors (HDACi) turn on gene transcription by promoting acetylation of lysine residues on histones leading to chromatin relaxation (9). There are four classes of HDAC into which the 18 described enzymes have been categorized (10). Relevant to HIV eradication strategies, class I HDAC are particularly important for maintaining HIV latency (10, 11). HDACi, including romidepsin and panobinostat, may serve as latency reactivating agents (LRA) via direct interference with HDAC maintained latency (9, 12). Romidepsin exhibits inhibitory activity in the lower nanomolar range against class I HDAC and panobinostat exhibits activity against class I/II HDAC (13). Consistent with the close interactions between HDAC and the maintenance of HIV latency, HDACi have the ability to reactivate and induce HIV expression from latently infected cells in both ex vivo and in vivo studies (9, 14–16).
Recently, Lucera et al. reported that supraphysiological concentrations (200 nM) of romidepsin increased the susceptibility of CD4+ T cells to HIV infection (17). These data, as well as those of others (18), raise the concern that inducing latent HIV in anatomical compartments with suboptimal cART concentrations (19) could lead to the infection of new target cells and reseeding of the latent reservoir. To determine the likelihood of such paradoxical outcomes, we comprehensively analyzed the impact of HDACi on de novo virus infection employing a broad panel of ex vivo experimental systems. We found that romidepsin-based HIV eradication strategies are unlikely to reseed the latent reservoir because this drug profoundly restricted de novo HIV infections. Further, resting CD4+ T cells exposed to romidepsin exhibited reduced proliferation and viability in the viral outgrowth assay, which led to significantly lower assay sensitivity when measuring the efficacy of romidepsin as an HIV latency reversal agent.
MATERIALS AND METHODS
Source and isolation of primary human cells.
Healthy blood donor buffy coat fractions were obtained from the Department of Clinical Immunology Blood Bank at Aarhus University Hospital, Aarhus, Denmark. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll density separation. CD4+ T cells were enriched by negative depletion from PBMCs using EasySep human CD4+ T cell enrichment kit (catalog no. 19052; Stem Cell Technologies) or CD4+ T cell isolation kit (catalog no. 130-096-533; Miltenyi Biotec) according to the respective manufacturer's protocol. Monocyte-derived macrophages (MDMs) were generated from monocytes purified from PBMCs by plastic adherence, with subsequent culture for 7 days in RPMI growth medium (RPMI 1640, 50,000 IU of penicillin, 50,000 μg of streptomycin, 10% fetal calf serum, 1% glutamine, interleukin-2 [IL-2; 20 U/ml]) supplemented with 10 ng of macrophage colony-stimulating factor/ml and 1 ng of granulocyte-macrophage colony-stimulating factor/ml (both from PeproTech). Resting CD4+ T cells were enriched from cryopreserved PBMCs by negative depletion via a two-step protocol as previously described (20). Briefly, the first step was to enrich CD4+ T cells from PBMCs using Miltenyi CD4+ T cell isolation kit (catalog no. 130-096-533) according to the manufacturer's protocol. The second step was to further enrich for resting CD4+ T cells via depletion of cells expressing CD69, CD25, or HLA-DR (Miltenyi CD69 Microbeads kit II, catalog no. 130-092-355; CD25 Microbeads II, catalog no. 130-092-983; HLA-DR Microbeads, catalog no. 130-046-101). All cell incubations were performed at 37°C unless otherwise noted. The purity of enriched cells populations was assessed by flow cytometry using either a LSR Fortessa (BD Biosciences) or a FACSVerse (BD Biosciences) flow cytometer and FlowJo software (Tree Star).
Viability assay for TZM-bl cells, PBMCs, and activated CD4+ T cells.
The viability of TZM-bl cells (21–25) and primary human cells was assessed after exposure to romidepsin (Selleckchem) or panobinostat (Selleckchem) (26–28). We tested a range of drug doses that spanned the physiologically relevant doses of each drug (i.e., 40 nM romidepsin [14] and 30 nM panobinostat [15, 29]) and, in select experiments, subphysiological doses of romidepsin were included along with medium controls to facilitate the observation of dose-dependent effects on HIV infection.
TZM-bl cells in cDMEM (Dulbecco modified Eagle medium, 50,000 IU of penicillin, 50,000 μg of streptomycin, 10% fetal calf serum, 1% glutamine) were seeded (5 × 103 cells/well) in 96-well plates, incubated for 24 h, and then primed (8 or 18 h) with romidepsin or panobinostat at the indicated range of concentrations (2-fold dilutions). After priming, the cells were washed with cDMEM and incubated for 48 h. TZM-bl cells were treated with trypsin, resuspended in 100 μl of cDMEM, and transferred to 96-well enzyme-linked immunosorbent assay (ELISA) plates, and 20 μl of MTS substrate (CellTiter 96 AQueous One Solution cell proliferation assay; Promega) was added to each well. Plates were incubated for 4 h, and formazan production was then analyzed with an ELISA plate reader (BioTek, ELx808). Cell viability was calculated according to the CellTiter protocol.
PBMCs or CD4+ T cells were stimulated in RPMI growth medium supplemented with phytohemagglutinin (PHA; 5 μg/ml) for 48 h. PHA-containing medium was replaced with fresh RPMI growth medium, and cells were incubated for 24 h. The PBMCs or CD4+ T cells were then seeded (2 × 105 cells/well) in 96-well plates and primed (8 or 18 h) with romidepsin or panobinostat at the indicated concentrations (2-fold dilutions). After priming, the cells were washed, resuspended in RPMI growth medium, and incubated for 48 h. Cells were resuspended in 100 μl of RPMI growth medium and assayed for formazan production as described above.
Single-round, VSVg-pseudotyped HIV infection assay.
Vesicular stomatitis virus G protein (VSVg)-pseudotyped HIV virions were produced by transfecting HEK293T cells with a packaging system consisting of four plasmids: pCCL-PGK-eGFP, pMD.2G, pRSV-REV, and pMDlg/p-RRE. Viral supernatants were harvested after 48 and 72 h and concentrated through a 20% sucrose cushion (25,000 × g for 2 h). Viral pellets were resuspended in phosphate-buffered saline and pooled, and aliquots were stored at −80°C.
PBMCs or enriched CD4+ T cells were stimulated in RPMI growth medium supplemented with PHA (5 μg/ml) for 48 h. The PHA-containing medium was replaced with fresh RPMI growth medium, and the PBMCs or enriched CD4+ T cells were incubated for 24 h. MDMs were grown as described above. Cells were seeded (2 × 105 cells/well) in 48-well plates with RPMI growth medium and pulsed for 2 h with romidepsin (10 or 40 nM) or panobinostat (30 nM). VSVg-pseudotyped HIV virions were then inoculated into the cell cultures, along with Polybrene (6 μg/ml), followed by incubation for an additional 6 h. The cells were then washed to remove unbound virions and resuspended in fresh RPMI growth medium and incubated for 48 h. The cells were then harvested by aspiration for the PBMCs and T cells or after EDTA treatment for the MDMs. Cells were incubated on ice with a Live/Dead fixable violet dead cell stain kit 405 (Life Technologies) according to the manufacturer's instructions. Data were collected using a LSR Fortessa flow cytometer. Viable cells exhibiting enhanced green fluorescent protein (eGFP) expression were characterized as infected.
HIV spreading assay.
Enriched CD4+ T cells were stimulated in RPMI growth medium supplemented with PHA (5 μg/ml) for 48 h. PHA-containing medium was replaced with fresh RPMI growth medium, and cells were incubated for 24 h. CD4+ T cells were pulsed for 2 h with romidepsin (10 or 40 nM) or panobinostat (30 nM). The cells were then infected with HIV-1HXB2 (30). After an additional 4 h of incubation, the HDACi and virus inoculum were removed, and the cells were resuspended in fresh RPMI growth medium. At the indicated time points, culture supernatants were harvested and evaluated for p24 antigen by ELISA to determine the replication kinetics of HIV as previously described (9).
Romidepsin and IFN-stimulated genes (ISGs) in PBMCs.
PHA-activated PBMCs were seeded (105 cells/well) into a conical 96-well plate and primed for 2 or 8 h with romidepsin (10 or 40 nM) or beta interferon (IFN-β; 1,000 IU/ml). After priming, RNA was collected from the cells using a High Pure RNA isolation kit (catalog no. 11828665001; Roche) according to the manufacturer's instructions. The RNA expression levels of 39 different genes were subsequently analyzed by using a Fluidigm 48-48 Multiplex Gene Array Biomark system, including TaqMan probes, according to the manufacturer's instructions.
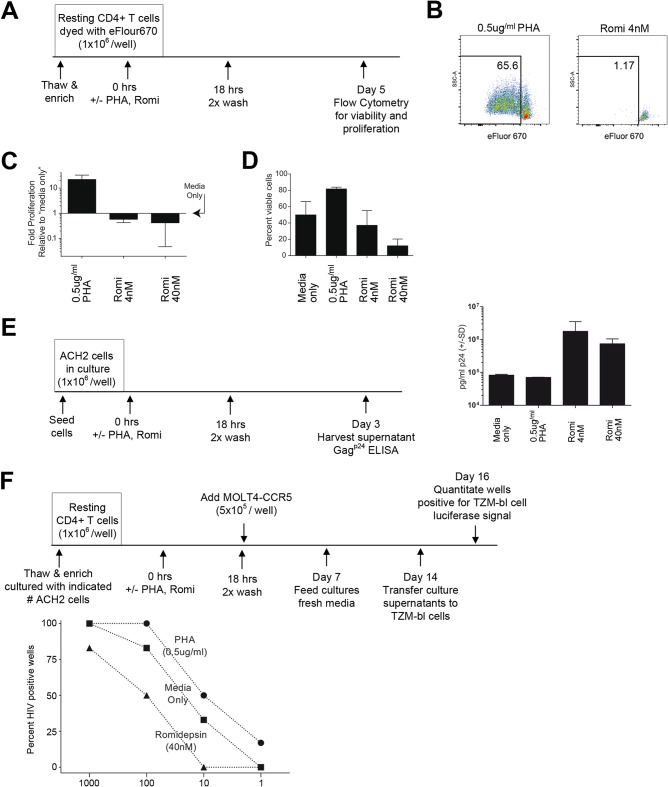
Resting CD4+ T cell proliferation and viability assay.
Resting CD4+ T cells were incubated with eFluor-670 (catalog no. 65-0840-85; eBioscience) according to the manufacturer's instructions. Dyed resting CD4+ T cells (106 cells per well) were placed into a 24-well plate in cRPMI+IL-2 medium (RPMI 1640 with l-glutamine, 50,000 IU of penicillin, 50,000 μg of streptomycin, 10% fetal calf serum, IL-2 [10,000 U/ml], and conditioned medium from a mixed-lymphocyte reaction culture as described previously [31]) plus PHA (0.5 μg/ml) and/or romidepsin (4 or 40 nM). After an 18-h incubation, the cultures were washed twice (2-h interval) with cRPMI+IL-2 medium and then maintained in 2 ml of cRPMI+IL-2 medium for 4 additional days. Cells were incubated on ice with a Live/Dead Fixable Green Dead Cell Stain Kit 488 (L-23101; Life Technologies) according to the manufacturer's instructions. The data were collected using a FACSVerse flow cytometer. Viable cells exhibiting a dilution of eFluor-670 were characterized as having proliferated during the culture period. The fold proliferation was determined relative to the respective human donor “medium-only” culture. The percentage of viable cells represents the proportion of resting CD4+ T cells, which excluded the fixable green dead cell stain.
Activation of latent HIV in ACH2 cells.
ACH2 cells (12, 32, 33) were incubated for 18 h with or without PHA (0.5 μg/ml) or romidepsin (4 or 40 nM), washed twice (using a 2-h interval) with cRPMI+IL-2 medium, and then incubated in fresh cRPMI+IL-2 medium for 48 h. Viral p24 antigen was measured in culture supernatants by ELISA as an indicator of the ability of either romidepsin or PHA to induce virus production from this latent HIV cell model (9).
Modified viral outgrowth assay.
The viral outgrowth assay was performed essentially as described previously (34). Briefly, resting CD4+ T cells were isolated from healthy blood donor buffy coat fractions. On day 0, 106 resting CD4+ T cells per well from each donor were plated into a 24-well plate, together with the indicated number of ACH2 cells in cRPMI+IL-2 medium with PHA (0.5 μg/ml) or romidepsin (4 or 40 nM). After 18 h, the cultures were washed twice (using a 2-h interval) with cRPMI+IL-2 medium, and then the cultures were moved into six-well plates and fed with 5 × 105 MOLT-4/CCR5 cells in a final volume of 8 ml of cRPMI+IL-2 medium. On day 7, 6 ml of the culture volume was replaced with fresh cRPMI+IL-2 medium. On day 14, 200 μl of each culture supernatant was transferred to TZM-bl cells in duplicate, and the luciferase activity from these reporter cells was measured on day 16 as an indicator of HIV production within a given culture (15).
Statistics.
Comparisons of infectivity within a single-round infection assay were performed using Student t tests of each drug condition versus dimethyl sulfoxide (DMSO) controls. Differences in replication (using the area under the curve [AUC]) in the HIV spreading assay were evaluated by a one-way repeated-measures analysis of variance (ANOVA). Comparisons between modified viral outgrowth conditions were made using a two-way ANOVA wherein the sources of variation were either the latency reactivation treatment or the number of ACH2 cells per well.
RESULTS
Limited HDACi-induced cell death in primary human cells.
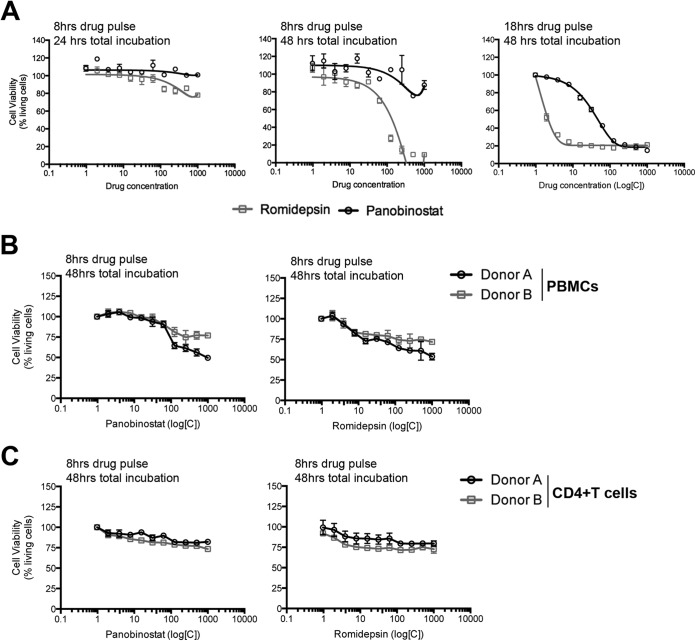
Our research focus was to determine the extent of romidepsin's impact on de novo HIV infection. As a control for the generalizability of our observations with romidepsin to other HDACi, we also evaluated the impact of panobinostat on de novo HIV infection in selected assays. To design the best strategies for evaluating the impact of HDACi on de novo viral infection, we first determined the toxicity of each drug in (i) HIV reporter TZM-bl cells, (ii) primary human PBMCs, and (iii) primary CD4+ T cells. The dose range examined included physiologically relevant doses of each drug (i.e., 40 nM romidepsin [14] and 30 nM panobinostat [15, 29]). Furthermore, we designed these analyses to detect potentially delayed toxic effects of drug exposure by measuring viability at both 24 and 48 h after removal of the drug (Fig. 1A, left and middle panels). Romidepsin and panobinostat treatment of TZM-bl cells resulted in only a minor impact on cell viability at 24 h, with the exception of the very high doses of romidepsin. In contrast, cell viability at 48 h was dramatically affected by romidepsin treatment, where cell death exceeded 90% at ≥100 nM (Fig. 1A, middle panel). Cell death was less pronounced for panobinostat, even at high doses (Fig. 1A, left and middle panels). In these experiments, TZM-bl cells were exposed to the HDACi for 8 h. Given that key ex vivo analyses have incorporated drug exposures with these HDACi of 18 h (34); we also evaluated 18 h of drug exposure in TZM-bl cells and observed a notably greater reduction in cell viability (Fig. 1A, right panel). As little as 7 nM romidepsin resulted in >80% of TZM-bl cells dying within 24 h. We observed ∼50% cell death at 30 nM panobinostat and 80% cell death at 125 nM. Next, to explore whether HDACi toxicity also occurred in primary human cells, we determined the viability of both PBMCs and CD4+ T cells exposed to romidepsin or panobinostat for a total of 8 h. We found that romidepsin-induced cell death in primary cells was less pronounced versus TZM-bl cells. Specifically, ca. 70 to 80% of PBMCs and CD4+ T cells remained viable for 48 h after exposure to the drug concentrations used in our subsequent virus infection assays (Fig. 1B and C).
FIG 1.
Differential impact of HDACi in immortalized versus primary human cells. (A to C) The viability of TZM-bl cells (A) and primary human cells (B and C) was assessed by flow cytometry after the cells were pulsed with either romidepsin or panobinostat for 8 or 18 h and then incubated for an additional 24 or 48 h, as indicated in each subpanel. Panel A presents compiled results from (two) experiments performed in triplicate. Panels B and C present compiled data for PBMCs and CD4+ T cells, in triplicate measures, respectively, from two human donors. Error bars indicate the standard deviations (SD). Gray squares indicate romidepsin; black circles indicate panobinostat.
Romidepsin inhibits VSVg-pseudotyped HIV infection of CD4+ T cells.
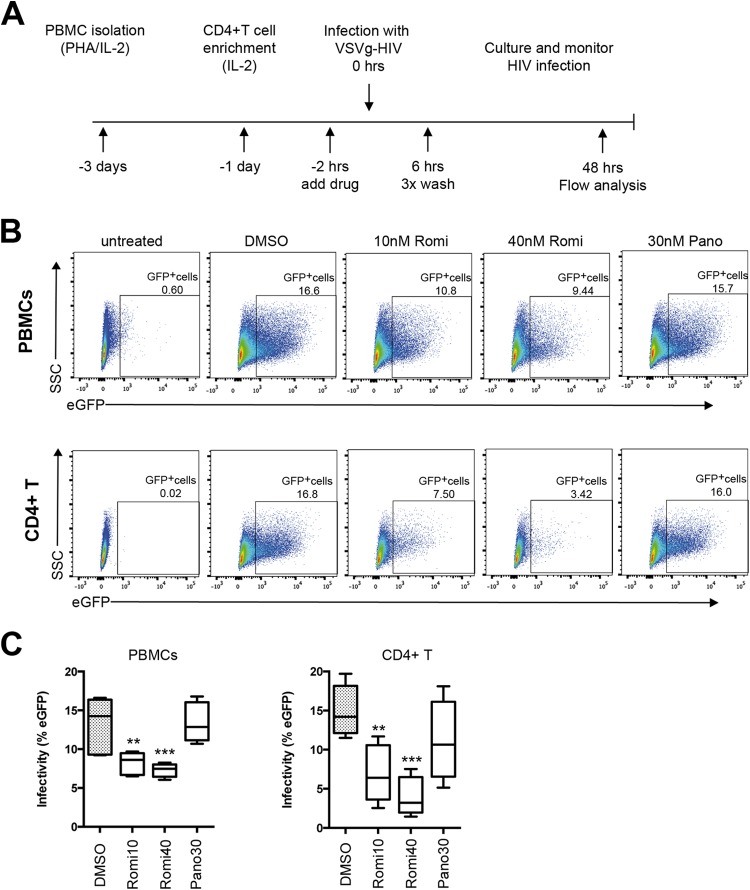
To understand the impact of HDACi on de novo viral infection, we performed a series of primary human cell experiments to measure de novo infection following HDACi treatment. In the first line of experiments, IL-2/PHA-activated PBMCs were primed with increasing amounts of either romidepsin or panobinostat for 2 h prior to incubation with VSVg-pseudotyped single-round HIV virions expressing eGFP (Fig. 2A). The live cells in each culture were examined using flow cytometry to quantitate the proportion of eGFP positive cells as a measure of infectivity (Fig. 2B). We found that priming cells with panobinostat did not alter the proportion of infected PBMCs (Fig. 2B and C); however, romidepsin significantly reduced the infectivity of VSVg-pseudotyped HIV at a relatively low concentration (10 nM, P ≤ 0.01; 95% confidence interval [CI] = 1.874 to 8.240), as well as at a more physiologically relevant concentration (40 nM, P ≤ 0.001; 95% CI = 2.845 to 9.211) (Fig. 2B and C). Collectively, our experiments with activated PBMCs showed a substantial reduction in infectivity after exposure to HDACi, although this experimental approach did not permit cell lineage-specific data interpretation.
FIG 2.
Single-round HIV infection of primary human cells with VSVg-pseudotyped virus was inhibited by romidepsin. (A) Schematic representation of the experimental approach. (B) Representative flow cytometry data highlight the reduced infection that results from romidepsin treatment. The percentages of infected, eGFP-positive cells are presented with each dot plot. (C) Compiled data for PBMCs and for CD4+ T cells from four human donors are shown (means ± the standard errors of the mean). **, P < 0.01; ***, P < 0.001.
To examine whether the observed inhibition of infectivity by romidepsin specifically affected CD4+ T cells and/or macrophages, we sorted CD4+ T cell populations from activated PBMCs prior to HDACi priming and generated MDMs. Similar to the results described above, significantly fewer primary CD4+ T cells were infected after priming with 10 or 40 nM romidepsin (P ≤ 0.01 [95% CI = 3.017 to 13.32] and P ≤ 0.0001 [95% CI = 5.910 to 16.22], respectively) (Fig. 2C). In contrast, MDMs were generally permissive for infection by the VSVg-pseudotyped single-round HIV virions regardless of HDACi treatment (see Fig. S1 in the supplemental material). Specifically, we observed >20% infection in DMSO-, romidepsin-, and panobinostat-treated MDMs derived from one of the donors whose CD4+ T cells exhibited profoundly reduced infection after exposure to romidepsin. Together, these data indicate that de novo infection of activated CD4+ T cells by VSVg-pseudotyped HIV was inhibited by romidepsin.
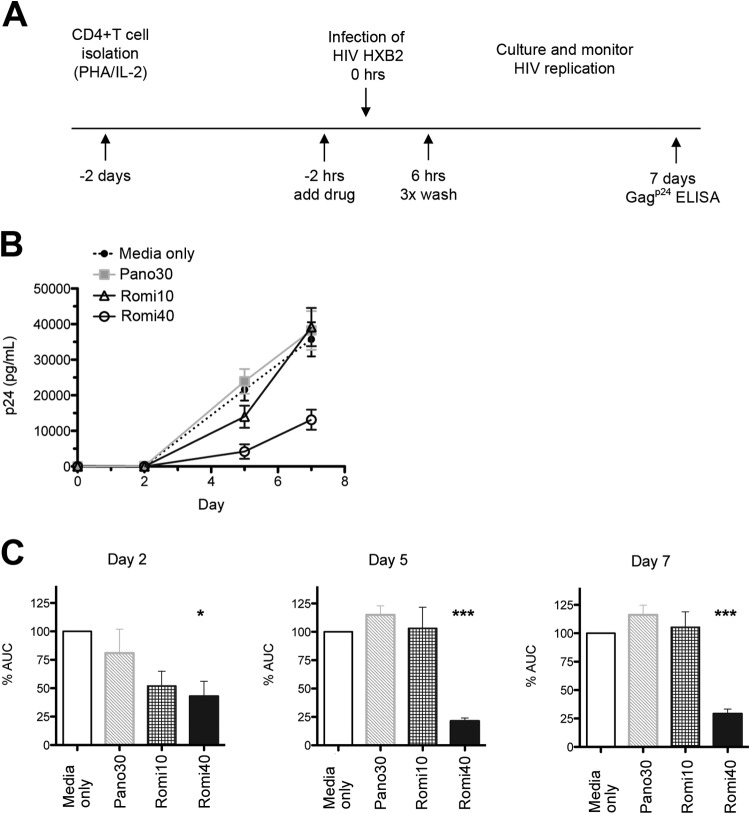
Romidepsin inhibits wild-type HIV infection of CD4+ T cells.
Next, we examined whether the mechanism of inhibition was specific to the pH-dependent fusion that characterizes VSVg-pseudotyped virions or whether wild-type HIV envelope fusion was similarly affected by treatment. To do this, HDACi-primed activated CD4+ T cells were challenged with replication-competent HIV-1HXB2—a highly pathogenic, CXCR4-tropic, laboratory-adapted viral isolate (30) (Fig. 3A). CD4+ T cell cultures primed with 10 nM romidepsin exhibited significantly reduced infection after 2 days in culture, as measured by Gagp24 in supernatants (P ≤ 0.05) (Fig. 3B and C). Similar effects were observed for romidepsin at 40 nM (P ≤ 0.05) but not for 30 nM panobinostat. By day 5, reduced HIV replication was only observed in the cultures primed with 40 nM romidepsin (P ≤ 0.001). This pronounced effect was maintained through 7 days of the culture, despite the fact that the drug pulse was relatively short in duration (Fig. 3B and C). The exposure of activated CD4+ T cells to physiologically relevant doses of romidepsin significantly decreased viral spreading, and thus the effect of romidepsin on de novo HIV infection is not dependent on the viral route of entry.
FIG 3.
Romidepsin significantly inhibits HIV replication in primary CD4+ T cells. (A) Schematic representation of the experimental approach. (B) Delayed HIV replication kinetics in activated CD4+ T cells observed after 40 nM romidepsin treatment from five human donors (means ± the standard errors of the mean). (C) From the curves in panel B, the AUC was calculated for each indicated time. AUC values for each condition and donor were normalized to the medium only control of each respective donor. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
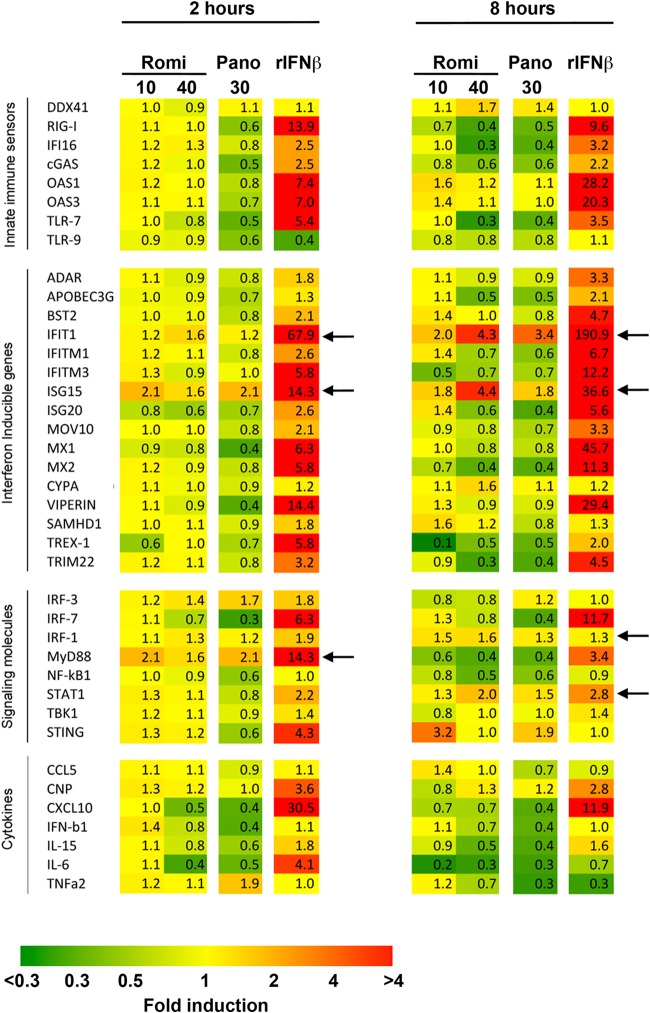
Romidepsin selectively activates ISGs in PBMCs.
To gain further insights into a potential mechanism for the HIV inhibition exhibited by romidepsin, we investigated the gene profile in PBMCs challenged with either romidepsin or panobinostat. We found that nearly all tested innate immune sensors, as well as other genes examined, were affected by 2 or 8 h of panobinostat exposure (e.g., the downregulation of MX1, Viperin, APOBEC3G, NF-κB, and IRF7) (Fig. 4). In contrast, whereas the expression of most genes was unchanged after 2 h of exposure to romidepsin, 8 h of 40 nM romidepsin exposure led to major alterations in the gene profile (Fig. 4, right panel). Expression was downregulated for most of the examined genes; however, there were three notable exceptions: IFIT1, ISG15, and STAT1. The expression of each of these genes was increased after 8 h of exposure to 40 nM romidepsin (Fig. 4, right), a finding indicative of differential regulation of innate restriction factors important for viral confinement.
FIG 4.
Romidepsin treatment leads to the upregulation of select ISG transcription. PBMCs were primed with the indicated concentration of HDACi for either 2 h (left) or 8 h (right). After priming, the cells were analyzed using a Fluidigm multiplex gene array. The cells were primed with recombinant interferon beta (rIFN-β) as a positive control for altered gene expression. The fold changes in transcription relative to baseline are presented in each box of the grid. In the heat map, yellow hues indicate stable transcription relative to baseline, green hues indicate reduced transcription relative to baseline, and red hues indicate increased transcription relative to the baseline. Representative data from one of two human donors are shown. Arrows indicate the fold changes consistently above 1.5.
Romidepsin inhibits proliferation of resting CD4+ T cells and reduces the sensitivity of viral outgrowth assays.
Given our observations that romidepsin reduced de novo HIV infection and virus spread in culture, we investigated the impact of this drug specifically on resting CD4+ T cells. We chose these cells because they are the primary cell type included in the viral outgrowth assays used to quantitate the size of the latent reservoir in HIV patients (20, 31, 34–39). The impact of romidepsin on the proliferation of magnetically enriched, HIV-negative blood donor-derived resting CD4+ T cells was profound. We first confirmed a previously reported observation (40) that exposure to romidepsin interferes with T cell proliferation when combined with mitogenic stimulation (see Fig. S2 in the supplemental material). However, it was essential that we also evaluate the impact of romidepsin (18-h drug exposure) in the absence of mitogenic stimulation to more closely approximate the conditions utilized in viral outgrowth assays (34) (Fig. 5A). We found that both the proliferation (Fig. 5B and C) and cell viability (Fig. 5D) of resting CD4+ T cells were dramatically reduced by 18 h of exposure to romidepsin.
FIG 5.
Romidepsin decreases the proliferative capacity and viability of resting CD4+ T cells, leading to reduced sensitivity for the viral outgrowth assay. (A) Schematic representation of each experimental approach. (B to D) Flow cytometry data reveal that resting CD4+ T cell proliferation (B and C) and viability (D) are negatively impacted in response to romidepsin treatment. Viable cells exhibiting a dilution of eFluor-670 were defined as having proliferated during the culture period. The fold proliferation was determined relative to the respective human donor “medium only” culture from three human donors (means ± the SD) (C), and the percentage of viable cells represents the proportion of resting CD4+ T cells, which excluded the fixable green dead cell stain from three human donors (means ± the SD) (D). Error bars indicate the SD. (E) Culture supernatant Gagp24 quantities indicate that romidepsin, but not PHA, readily reactivates latent HIV in ACH2 cells (triplicate measures; means ± the SD). (F) Romidepsin treatment reduced the sensitivity of the viral outgrowth assay relative to medium control conditions (e.g., viral outgrowth from the condition recapitulating 100 IUPM was detected in 50% of the wells treated with romidepsin versus 83% for the medium control). Resting CD4+ T cells (106 per well) were cultured with the indicated numbers of ACH2 cells (n = 6 to 8 wells per data point [2 to 4 wells per human donor]). The results were analyzed by using a two-way ANOVA wherein the sources of variation were either the latency reactivation treatment (P = 0.0002) or the number of ACH2 cells per well (P = 0.012).
Latent HIV infection in resting CD4+ resting T cells is infrequent (∼1 infectious unit per million resting CD4+ T cells tested [IUPM]). Viral outgrowth assays are extended-duration (∼14 days), stimulatory cultures of resting CD4+ T cells that are optimized for the quantitation of the latent HIV reservoir (20, 31, 34–39). To test our hypothesis that romidepsin's broad impact on cell viability, cell proliferation, and de novo HIV infection could alter the sensitivity of viral outgrowth assays and to experimentally overcome the limitation posed by the fact that latency events in cART-suppressed HIV patients are rare, we established a modified viral outgrowth assay with 106 resting CD4+ T cells per culture from HIV-negative blood donors spiked with a defined number of ACH2 cells. ACH2 cells are a model for HIV latency (12, 32, 33), where cells can be stimulated to produce HIV progeny virions by romidepsin exposure (as with latently infected resting CD4+ T cells [14]), but not the mitogenic stimulator PHA (unlike latently infected resting CD4+ T cells [Fig. 5E]). Three different treatment conditions were evaluated in this modified viral outgrowth system: medium only, 0.5 μg of PHA, and 40 nM romidepsin (Fig. 5F). When we tested the typical patient condition of 1 IUPM, we observed viral outgrowth in only 17% (1 of 6) of PHA-treated wells tested and 0% (0 of 6) in the medium-only or romidepsin (0 of 8) wells. When a 10-fold-higher IUPM was stimulated, we observed 0% viral outgrowth in the romidepsin wells (0 of 8), whereas 33% of the medium-only wells (2 of 6) and 50% of the PHA-stimulated cultures (3 of 6) exhibited viral outgrowth. When 100 IUPM was evaluated, only 50% of the romidepsin cultures (4 of 8) versus 83% of the medium-only wells (5 of 6) and 100% of the PHA wells (6 of 6) were positive for viral outgrowth. Even at 1,000 IUPM, not all wells treated with romidepsin (5 of 6) were positive for viral outgrowth, as was the case for both medium control (6 of 6) and PHA-treated (6 of 6) wells. We conclude from these data that romidepsin reduced the sensitivity of the viral outgrowth assay and that romidepsin's induction effects are obscured within this assay with the typical patient condition of 1 IUPM.
DISCUSSION
The rationale for the incorporation of HDACi into HIV eradication strategies is that epigenetic modulation of molecular mechanisms blocking transcription of integrated HIV DNA can reactivate HIV expression in resting infected memory CD4+ T cells and disrupt latency (29). Numerous in vitro, ex vivo, and clinical studies have shown that HDACi reactivate latent HIV infection (9, 12–16, 37, 38, 40–48). However, a theoretical concern is that HDACi-induced viral production in cART sanctuary sites could result in de novo HIV infections and reseeding of the latent reservoir (17, 18). In the present study, we addressed this concern by comprehensively exploring the impact of romidepsin on HIV infection. We found that this effective LRA profoundly restricts de novo HIV infections of primary human T cells.
After demonstrating that the physiologically relevant dosages of the drugs that we used were not overtly cytotoxic (Fig. 1), we challenged primary human cells with romidepsin and then infected the cells with either VSVg-pseudotyped HIV or highly pathogenic wild-type HIV, which in both cases were virus models with robust infectivity rates (Fig. 2 and 3). To our surprise, we observed that de novo infection of activated PBMCs and activated CD4+ T cells by VSVg-pseudotyped HIV was inhibited by physiological drug levels of romidepsin. In contrast, when we performed a similar experiment with donor-matched MDM, we observed no appreciable HDACi effect on infection by these VSVg-pseudotyped HIV virions, a finding which agrees with recent findings presented by Campbell et al. (18). When activated CD4+ T cells were treated with romidepsin and then infected with wild-type HIV, we observed a significant reduction in viral spread within the cell culture. Taken together, these two observations reveal that the mechanism of inhibiting de novo HIV was not envelope dependent. However, further investigations are needed to dissect the differences in infectivity observed between romidepsin-treated CD4+ T cells and MDMs.
Next, we used Fluidigm gene expression analyses to begin to elucidate whether the mechanism of inhibiting de novo HIV is dependent upon an HDACi-induced alteration in the innate antiviral activities of the target cells. To do this, we utilized a multiplex gene array (49) designed to investigate mRNA levels for (i) molecules involved in innate immune sensing pathways, (ii) ISGs, (iii) innate immunity signal transduction mediators, and (iv) cytokines related to innate immune responses. We noted that after 8 h of exposure to 40 nM romidepsin the expression of IFIT1, ISG15, and STAT1 in PBMCs were significantly upregulated (Fig. 4, right panel). This is important because of the known antiviral effects for each of these proteins. Specifically, the members of the family of IFITs, including IFIT1, are known to be upregulated after IFN treatment, viral infections, and pathogen-associated molecular pattern recognition (50–52). ISG15 is a ubiquitin-like protein shown to protect cells in numerous viral infection models, but for HIV it is believed mainly to impair viral release (53, 54). STAT1 activation can often be correlated with cellular proinflammatory, antiproliferative, and apoptotic activities (55). Although this analysis focused on a subset of genes involved in innate immunity and did not address other potential mechanisms of virus inhibition, the observation that romidepsin-induced increases in expression for these genes suggests that HDACi inhibition of de novo HIV infections could be mediated through HDACi-altered innate immunity versus direct interference with virus replication.
In selected assays, we included panobinostat as a control to assess the generalizability of our romidepsin-related findings to other HDACi. Romidepsin and panobinostat target class I and class I/II HDAC, respectively (13). These two HDACi had similar cell viability profiles in activated, primary PBMCs and CD4+ T cell cultures (Fig. 1). When we examined the impact of these two HDACi on infection with VSVg-pseudotyped HIV and wild-type HIV, we observed a dose-dependent effect with romidepsin wherein the most profound inhibition of de novo infection was observed at the highest tested dose (40 nM). The dose of panobinostat (30 nM) evaluated here was matched to the concentration of panobinostat that was attained in vivo with the dosing strategy used in a recent clinical trial (9, 15). At this dose (and also at the supraphysiological dose of >100 nM [data not shown]), panobinostat did not exhibit the same level of inhibition of de novo HIV infection that we observed for romidepsin (Fig. 2 and 3).
The cells that compose the latent HIV reservoir are relatively infrequent (20, 31, 34–39). Because positive signals detected by viral outgrowth assays are rare, we hypothesized that romidepsin's broad impacts on cell viability, cell proliferation, and de novo HIV infection could alter the sensitivity of this assay. Such an outcome could lead to an underestimation of efficacy when this assay is utilized ex vivo to evaluate the function of a potential LRA such as romidepsin. Our modified viral outgrowth assay was designed with defined IUPM values to allow us to test this hypothesis. Two relevant comparisons were incorporated into the experiment. The “PHA versus medium only” comparison was included to evaluate whether resting CD4+ T cells that were activated by PHA would in turn activate ACH2 cell HIV production, whereas the “medium only versus romidepsin” comparison permitted us to discern whether romidepsin reduced the ability of this assay to detect viral outgrowth from latently infected cells or not. Our results showed that ex vivo romidepsin exposure profoundly impacted the ability of the viral outgrowth assay to quantitate the latent HIV reservoir for the typical patient condition of 1 IUPM. Thus, these data indicate that caution should be exercised when determining whether a given molecule is capable of reactivating latent HIV infection from HIV patient-derived resting CD4+ T cells after ex vivo stimulation with potential LRA. We further demonstrate that the inhibition of HIV spreading in cell culture that results from romidepsin treatment creates a technical barrier for the use of virus spreading assays, such as the viral outgrowth assay, for assessing the potential of romidepsin to reverse HIV latency.
In conclusion, we show that romidepsin inhibits HIV infection in an envelope-independent manner. Furthermore, we provide evidence that this inhibition could result from a romidepsin-mediated enhancement in antiviral innate immunity. Altogether, our data indicate that reseeding of the latent reservoir is an unlikely outcome of a romidepsin-based HIV eradication strategy. Translation of our ex vivo findings to the clinic may best be facilitated through the use of an animal model of HIV persistence wherein ART sanctuaries may exist (1). The in vivo testing of adjunct therapy with romidepsin as an HIV eradication strategy in such a model is warranted to fully characterize the likelihood of adjunct romidepsin treatment to enhance the risk of HIV infection and reservoir reseeding during a kick-and-kill HIV eradication therapy.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by the Lundbeck Foundation (K.L.J. and M.R.J.), Aarhus University Research Foundation (M.R.J.), Danish Strategic Research Council grant 0603-00521B (L.Ø.), and Danish Research Council grant 12-133887 (O.S.S.). The funders had no role in study design, data collection, and analysis, decision to publish, or writing of the manuscript. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells (John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc.), ACH-2 cells (Thomas Folks), and MOLT-4/CCR5 cells (Masanori Baba, Hiroshi Miyake, and Yuji Iizawa).
We thank Lene Svinth Jøhnke, Ane Kjeldsen, and the flow cytometry core facilities at Aarhus University Institute of Biomedicine for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00574-15.
REFERENCES
- 1.Eisele E, Siliciano RF. 2012. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG. 2012. HIV: shock and kill. Nature 487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 4.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun TW, Churchill M, Mascio MD, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O'Doherty U, Palmer S, Penicaud MC, Peterlin M, Poli G, Routy JP, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Lint CV, Verdin E, Woolfrey A, Zaia J, Barre-Sinoussi F. 2012. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durand CM, Blankson JN, Siliciano RF. 2012. Developing strategies for HIV-1 eradication. Trends Immunol 33:554–562. doi: 10.1016/j.it.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl V, Josefsson L, Palmer S. 2010. HIV reservoirs, latency, and reactivation: prospects for eradication. Antivir Res 85:286–294. doi: 10.1016/j.antiviral.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Archin NM, Sung JM, Garrido C, Soriano-Sarabia N, Margolis DM. 2014. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol 12:750–764. doi: 10.1038/nrmicro3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colin L, Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen TA, Schmeltz-Sogaard O, Brinkmann C, Wightman F, Lewin SR, Melchjorsen J, Dinarello C, Ostergaard L, Tolstrup M. 2013. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccines Immunother 9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. 2009. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol 83:4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. 2009. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS 23:1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wightman F, Lu HK, Solomon AE, Saleh S, Harman AN, Cunningham AL, Gray L, Churchill M, Cameron PU, Dear AE, Lewin SR. 2013. Entinostat is a histone deacetylase inhibitor selective for class 1 histone deacetylases and activates HIV production from latently infected primary T cells. AIDS 27:2853–2862. doi: 10.1097/QAD.0000000000000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber K, Doyon G, Plaks J, Fyne E, Mellors JW, Sluis-Cremer N. 2011. Inhibitors of histone deacetylases: correlation between isoform specificity and reactivation of HIV type 1 (HIV-1) from latently infected cells. J Biol Chem 286:22211–22218. doi: 10.1074/jbc.M110.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, Hesselgesser J, Irrinki A, Murry JP, Stepan G, Stray KM, Tsai A, Yu H, Spindler J, Kearney M, Spina CA, McMahon D, Lalezari J, Sloan D, Mellors J, Geleziunas R, Cihlar T. 2014. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog 10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Østergaard L, Søgaard OS. 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 16.Søgaard O, Graversen M, Leth S, Brinkmann C, Kjær A-S, Olesen R, Denton P, Nissen S, Sommerfelt M, Rasmussen T, Østergaard L, Tolstrup M. 2014. The HDAC inhibitor romidepsin is safe and effectively reverses HIV-1 latency in vivo as measured by standard clinical assays. 20th International AIDS Conference (AIDS 2014), Melbourne, Australia. [Google Scholar]
- 17.Lucera MB, Tilton CA, Mao H, Dobrowolski C, Tabler CO, Haqqani AA, Karn J, Tilton JC. 2014. The histone deacetylase inhibitor vorinostat (SAHA) increases the susceptibility of uninfected CD4+ T cells to HIV by increasing the kinetics and efficiency of postentry viral events. J Virol 88:10803–10812. doi: 10.1128/JVI.00320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell GR, Bruckman RS, Chu YL, Spector SA. 2015. Autophagy induction by histone deacetylase inhibitors inhibits HIV type 1. J Biol Chem 290:5028–5040. doi: 10.1074/jbc.M114.605428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. 2013. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information. 2015, posting date PubChem Compound Database: CID 6918837. Panobinostat compound summary. National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 27.National Center for Biotechnology Information. 2015. PubChem Compound Database: CID 5352062. Romidepsin compound summary. National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 28.Atadja P. 2009. Development of the pan-DAC inhibitor panobinostat (LBH589): successes and challenges. Cancer Lett 280:233–241. doi: 10.1016/j.canlet.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen TA, Tolstrup M, Winckelmann A, Ostergaard L, Sogaard OS. 2013. Eliminating the latent HIV reservoir by reactivation strategies: advancing to clinical trials. Hum Vaccines Immunother 9:790–799. doi: 10.4161/hv.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratner L, Fisher A, Jagodzinski LL, Mitsuya H, Liou RS, Gallo RC, Wong-Staal F. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses 3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 31.Siliciano JD, Siliciano RF. 2005. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304:3–15. [DOI] [PubMed] [Google Scholar]
- 32.Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A 86:2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clouse KA, Powell D, Washington I, Poli G, Strebel K, Farrar W, Barstad P, Kovacs J, Fauci AS, Folks TM. 1989. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol 142:431–438. [PubMed] [Google Scholar]
- 34.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 36.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 37.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, Bandarenko N, Schmitz JL, Bosch RJ, Landay AL, Coffin JM, Margolis DM. 2008. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS 22:1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RB, O'Connor R, Mueller S, Foley M, Szeto GL, Karel D, Lichterfeld M, Kovacs C, Ostrowski MA, Trocha A, Irvine DJ, Walker BD. 2014. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T lymphocytes. PLoS Pathog 10:e1004287. doi: 10.1371/journal.ppat.1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR. 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remoli AL, Marsili G, Battistini A, Sgarbanti M. 2012. The development of immune-modulating compounds to disrupt HIV latency. Cytokine Growth Factor Rev 23:159–172. doi: 10.1016/j.cytogfr.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Rigby L, Muscat A, Ashley D, Algar E. 2012. Methods for the analysis of histone H3 and H4 acetylation in blood. Epigenetics 7:875–882. doi: 10.4161/epi.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savarino A, Mai A, Norelli S, El Daker S, Valente S, Rotili D, Altucci L, Palamara AT, Garaci E. 2009. “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 6:52. doi: 10.1186/1742-4690-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choudhary SK, Archin NM, Margolis DM. 2008. Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4+ T cells. J Infect Dis 197:1162–1170. doi: 10.1086/529525. [DOI] [PubMed] [Google Scholar]
- 48.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 49.Jang JS, Simon VA, Feddersen RM, Rakhshan F, Schultz DA, Zschunke MA, Lingle WL, Kolbert CP, Jen J. 2011. Quantitative miRNA expression analysis using Fluidigm microfluidics dynamic arrays. BMC Genomics 12:144. doi: 10.1186/1471-2164-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkar SN, Sen GC. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther 103:245–259. [DOI] [PubMed] [Google Scholar]
- 51.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. 2012. Global landscape of HIV-human protein complexes. Nature 481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valente ST, Gilmartin GM, Mott C, Falkard B, Goff SP. 2009. Inhibition of HIV-1 replication by eIF3f. Proc Natl Acad Sci U S A 106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A 103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morales DJ, Lenschow DJ. 2013. The antiviral activities of ISG15. J Mol Biol 425:4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Decker T. 1999. Introduction: STATs as essential intracellular mediators of cytokine responses. Cell Mol Life Sci 55:1505–1508. doi: 10.1007/s000180050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.