Abstract
Cancer cells are exposed to adverse conditions in the tumor microenvironment, and utilize post-transcriptional control mechanisms to re-program gene expression in ways that enhance cell survival. Stress granules and processing bodies are RNA-containing granules that contribute to this process by modulating cellular signaling pathways, metabolic machinery, and stress response programs. This review examines evidence implicating RNA granules in the pathogenesis of cancer and discusses their potential as targets for anticancer therapies.
Keywords: Cancer, Stress Granules, P-bodies, post-transcriptional regulation, apoptosis, stress response
INTRODUCTION
Nascent mRNAs bind to an array of RNA-binding proteins (RBPs) and microRNAs that conspire to determine their fates. These transcripts are transported from the nucleus to the cytoplasm as a part of compositionally diverse complexes known as messenger ribonucleoprotein particles (mRNPs). The composition of these exported mRNPs determines whether transcripts are immediately translated or transported to specific subcellular regions for storage and/or localized translation. These mRNPs are also subject to quality control mechanisms that promote the degradation of transcripts that contain errors that could lead to the production of incomplete protein products (reviewed in [1, 2]).
There is an imperfect correlation between levels of mRNA and levels of the proteins they encode. Up to two-thirds of this variation can be attributed to post-transcriptional mechanisms that modulate mRNA stability and translation [3, 4]. Proteins that mediate post-transcriptional control bind to cis elements that are typically found in 5′- and 3′-untranslated regions of individual transcripts. These regulatory elements can be unique to a single RNA molecule (allowing targeted regulation of this mRNA) or be found in a subset of mRNAs encoding functionally related proteins (coordinated regulation). Individual RBPs can influence several aspects of mRNA metabolism, including mRNA decay/stabilization, subcellular localization or translation rate. The availability of these RBPs is under the control of cell-intrinsic and extracellular cues. The RBP components of RNP complexes assembled on mRNA transcripts coordinately determine mRNA stability, localization and translation to allow precise and dynamic control over protein synthesis. The reader is referred to several recent reviews on post-transcriptional regulation of gene expression by microRNAs and RBPs for more detailed information [5–7].
The intricate RBP network that regulates mRNA stability, localization and translation is spatially regulated by the assembly of stress granules and processing bodies, mRNP-containing cytoplasmic granules that influence multiple aspects of cell metabolism, especially during changing conditions. Perturbations in RNA granule functions lead to pathological phenotypes observed in multiple neurodegenerative, immunological and infectious diseases [8–11]. Involvement of RNA granules in cancer initiation and progression is an emerging concept in tumor biology, and is the subject of this review.
COMPLEX LIFE OF mRNA
Cytoplasmic mRNAs that pass quality control are modified with 5’-caps (m7G) and 3’-poly(A) tails that are major determinants of mRNA stability. The eukaryotic decay machinery relies on ribonucleolytic activities that remove/hydrolyze the 5′-cap structure (decapping activity) and shorten or remove the poly(A) tail (deadenylation activity) to allow 5′-3′- or 3′-5′-exonucleolytic mRNA degradation, respectively [12].
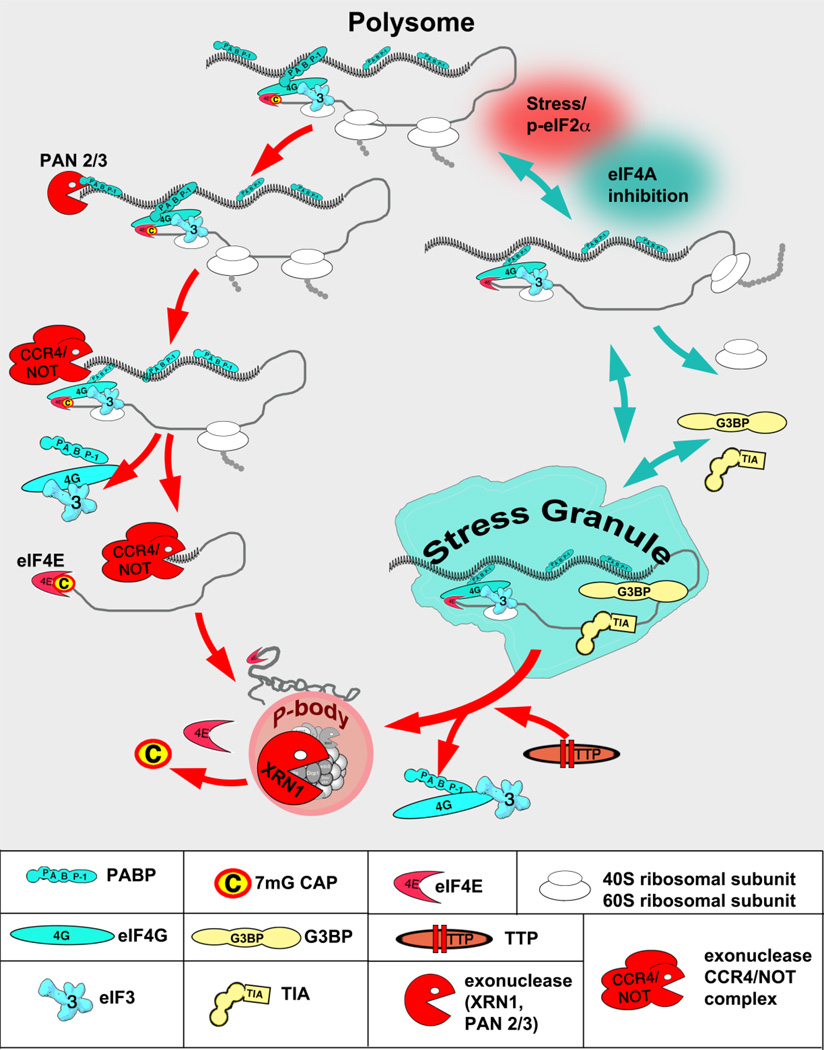
Typically, bulk mRNA decay is initiated by deadenylation [13], which is performed by one of three deadenylases that reside in multi-subunit complexes containing regulatory proteins [14]. These deadenylases include the PAB-specific ribonuclease 2 and 3 (PAN2/3) complex, the CAF1/CCR4/NOT complex and a homodimer of poly(A) ribonuclease (PARN). The CAF1/CCR4/NOT complex often cooperates with PAN2/PAN3 or specific RBPs bound to selected mRNAs to remove poly(A) tails [15]. By removing the poly(A) tail from targeted transcripts, these deadenylase complexes also displace polyadenylate-binding protein (PABP), which is a major mRNA stabilization factor [12]. Following deadenylation, the decapping enzymes that remove m7G from the 5′-ends of mRNA are activated (Figure 1). A decapping complex consisting of DCP1A and DCP2 decapping enzymes in association with the regulatory factors EDC3 (enhancer of decapping-3) and Hedls (also referred to as Ge-1 and Edc4) are responsible for hydrolysis of the cap structure [16]. Under steady-state conditions, the cap is protected from decapping by bound cap-binding eIF4F complex (consisting of translation initiation factors eIF4E, eIF4A, and eIF4G). The eIF4G scaffold protein communicates with the poly(A) tail through direct binding to PABP molecules, and deadenylation disrupts this communication (Figure 1). Following deadenylation/decapping, transcripts are degraded by the 5′-3′-exoribonuclease XRN1 or by exosome-associated 3′-5′-exoribonucleases RRP44 (hDIS3) or PM-SCL75 (RRP45, EXOSC10). In some cases, decay of specific mRNAs is initiated by endoribonucleolytic cleavage within its body followed by degradation in both 5′-3′ and 3′-5′ directions [17].
Figure 1. RBP-RNA granules interplay on the target mRNA.
Under steady-state conditions, mRNA is actively engaged into translation as a part of polysome and in the form of translationally-competent mRNP consisting of several translation initiation factors (such as cap-binding complex eIF4F and multi-subunit eIF3) and mRNA-bound RBPs (such as poly(A)-bound PABP). In response to cell-intrinsic and extracellular changes mRNA translation is arrested and transcript can be routed from the polysomes to SGs (blue arrows) or PBs (red arrows). Transcripts destined for assembly into PBs are first deadenylated by one or more mRNA deadenylases (PAN2/3 and CCR4/NOT are shown as examples). This causes the release of PABP molecules from the mRNA poly(A) tail, and the remodeling/decircularization of the mRNP which primes it for decapping. PB-associated decay enzymes subsequently may mediate mRNA decapping and decay within PBs. In contrast, transcripts assembled into SGs retain their poly(A) tails and associated PABP. Stresses, which may trigger phosphorylation of eIF2α (red shading) or not (such as inhibition of eIF4A, aqua shading), disrupt translational initiation of the mRNA, resulting in the runoff of ribosomes and the accumulation of stalled mRNPs. Binding of mRNAs by aggregation-prone RNA binding proteins (such as TIA-1, G3BP1) subsequently promotes its coalesce into SGs. Certain decay-promoting proteins (TTP, BRF1) can promote deadenylation and decay of SG-associated transcripts in tandem with promoting SG-PB fusion/interactions. Multiple additional proteins and signaling molecules associate with SGs or PBs once they are formed, depending on conditions.
Multiple other RBPs regulate recruitment of mRNA deadenylases/decapping enzymes in a transcript-specific manner. These RBPs bind to cis-elements found in the untranslated regions of selected mRNAs. A common example is the so-called ARE element (adenine and uridine-rich element) found in 3′-UTRs (Untranslated Regions) of short-lived RNA molecules [6]. AREs recruit numerous RBPs (ARE-BPs) that determine the stability of target transcripts. To date, more than 20 different ARE-RBPs have been identified. We will discuss ARE-BPs in the context of their association with RNA granules and possible roles in carcinogenesis.
Under steady-state conditions, recruitment of translation initiation complexes to the newly-exported mRNP promotes protein synthesis and the assembly of polysomes. The polysome-translated mRNA is circularized by interactions between eIF4G, a component of the cap-binding eIF4F complex, and 3′-poly(A) tail-bound PABP. This closed-loop configuration not only facilitates efficient translation by promoting the recycling of terminating ribosomes to the initiation complex, but also protects mRNA from degradation and ensures that only intact mRNAs are translated. Correspondingly, early views on the relationship between mRNA translation and mRNA decay were very simple: when mRNA is not translated, it is degraded. More recent data challenges this view and suggests a more sophisticated model in which RNA granules play a central role in regulating the assembly/disassembly of translating polysomes and the stability of specific mRNA transcripts (see Figure 1).
BASICS OF STRESS GRANULES AND PROCESSING BODIES
Although there are many types of RNA granules (e.g. Stress Granules (SGs), P-bodies (PBs), germ granules, neuronal granules, nuclear paraspeckles) [18], SGs and PBs are the most well understood and are closely associated with a variety of diseases including cancer. The classification of RNA granules is based on their composition (the presence of specific markers), subcellular localization (nuclear, cytoplasmic, axonal etc.), cell of origin (germ cells, neurons), response to stimuli (stress, viral infections), dynamic behavior and proposed functions (sites of mRNA storage/decay, stress response etc.) [9, 18].
PBs are cytoplasmic RNA granules that are enriched in the factors involved in mRNA degradation, mRNA surveillance, translational repression and RNA-mediated gene silencing. Initially they were discovered as “XRN1 foci” due to the presence of the exoribonuclease XRN1 in small granular structures within the cytoplasm of mammalian cells [19]. Later, other components of the RNA decay machinery such as decapping enzymes DCP1/DCP2 [20, 21] and decapping activators Hedls [22, 23], Edc3 [22], Pat1 [24, 25], LSm1–7 [20, 26, 27] and RCK (p54, DDX6) [28] were shown to co-localize with XRN1 foci. Further studies demonstrated that PBs are sites at which the key enzymes of cytoplasmic RNA degradation are concentrated (deadenylase complex CCR4/CAF1/NOT [29, 30] and its enhancer TOB2 [31], nonsense-mediated mRNA decay proteins UPF1, UPF2, UPF3, SMG5, SMG6 and SMG7 [32–34], ARE-mediated decay factors TTP, BRF1 and BRF2 [26, 35, 36]) and the RNAi machinery (GW182 [37], Argonautes [38, 39]). In addition, PBs contain translation regulation-associated factors such as eIF4E [40], eIF4E-T [41], CPEB [28], PCBP2 [42] and FASTK [26]. This composition determines the type of mRNAs deposited in PBs: these mRNAs seem to be primed for degradation (e.g. they lack poly(A) tails) [30]. However, although initially proposed to be sites for mRNA translational repression, storage or decay, the physical integrity of PBs is not required for global or specialized decay pathways [24, 27]. Many PB proteins exchange rapidly with the cytoplasmic pool and are not stable PB components whereas others (e.g. Dcp2) are very stable PB components [26, 40, 43, 44]. Decay enzymes are also present on polysomes, and transcript degradation can take place while mRNAs are associated with actively translating ribosomes [45, 46]. PBs are not sites for storage of selected mRNA degradation factors (such as Argonautes) because less than 10% of the cytoplasmic pool of these factors is concentrated at PBs [43, 44]. However, this does not rule out the possibility that PBs are storage sites for selected mRNAs [47] or protein factors [43]. It is also possible that PBs are sites of recycling/modification of decay factors.
SGs are cytoplasmic foci wherein translation initiation factors, 40S ribosomal subunits and diverse RBPs translationally-stalled mRNAs coalesce into discrete granules which recruit various signaling molecules [48–50]. SGs are non-membranous structures formed in response to different stress stimuli, typically through the stress-induced phosphorylation of eukaryotic initiator factor (eIF) 2 (eIF2) [51] (Figure 1). This results from activation of one or more stress-sensing serine/threonine kinases that phosphorylate serine residue 51 of the alpha subunit of eIF2 (eIF2α) [52], resulting in impaired translational initiation and consequent polysome disassembly (Figure 1). eIF2 is a component of the translation-competent eIF2/tRNAiMet/GTP ternary complex, an early initiation factor that mediates binding of initiator tRNAiMet to the 40S ribosomal subunit in a GTP-dependent manner [53]. To initiate mRNA translation, this ternary complex interacts not only with 40S ribosomal subunits but with a number of core translation initiation factors (such as multi-subunit eIF3, eIF4F complex, eIF5, eIF5B, eIF1, eIF1A) to form the 48S pre-initiation complex on the AUG start codon [54]. Productive assembly of the 48S complex on the start codon is followed by the joining of the large 60S ribosomal subunit to form a translation elongation-competent 80S ribosome. When eIF2α is phosphorylated, GDP-GTP exchange is inhibited, thus dramatically decreasing the levels of the active ternary complex, leading to the inhibition of translation initiation (reviewed in [53]). At the same time, protein synthesis is reprogrammed by the preferred translation of select mRNAs, including transcripts with upstream Open Reading Frames (uORFs) in the 5′ UTR. Such transcripts typically encode stress response proteins such as the transcription factor ATF4, to activate other pathways to activate long-term survival pathways [52].
Phosphorylation of eIF2α also leads to the assembly of SGs. Because translation elongation is not affected by eIF2α phosphorylation, ribosomes already engaged in translation “run-off” polysomes, converting them into non-canonical, translationally-stalled (so called 48S* complexes) pre-initiation complexes that lack selected initiation factors (such as eIF2 and eIF5) [55]. These 48S* complexes are the core constituents of SGs, but still require a subset of mRNA-associated RBPs to mediate SG assembly (Figure 1). The best characterized of these are TIA1 (T-cell internal antigen 1 [56]) and G3BP1 (Ras-GTPase-Activating Protein SH3-Domain-Binding Protein 1 [57]); these proteins contain aggregation-prone domains that mediate the coalescence of 48S* complexes into SGs [58]. Over-expression of G3BP1 or TIA1 is sufficient to drive (“nucleate”) SG formation even in the absence of stress, whereas siRNA-mediated knockdown of transcripts encoding these proteins significantly impairs with SG assembly despite phosphorylation of eIF2α and polysome disassembly. Moreover, multiple post-transcriptional modifications of various SG components contribute to SG assembly and disassembly (e.g. phosphorylation of G3BP1 at serine 149 impairs its ability to nucleate SGs [57]).
A defining feature of SGs is their dynamic nature: SGs are quickly assembled under stress and rapidly disperse after the stress is removed. The effects of pharmacological agents that either disassemble polysomes (puromycin) or freeze ribosomes on mRNA transcripts to stabilize polysomes (cycloheximide) reveal that SGs are in a dynamic equilibrium with polysomes [56]. Fluorescence recovery after photobleaching (FRAP) studies directly show that mRNPs within SGs rapidly shuttle in and out of SGs [26, 44, 56, 59]. SGs also recruit multiple enzymes and signaling molecules including kinases and phosphatases, scaffolding and adaptor proteins, ubiquitin-modifying enzymes, RNA helicases, ribonucleases, ribosyl-, glucosyl- and methyl-transferases. As signaling centers, SGs transiently alter multiple signaling pathways to ensure cell adaptation to stress (reviewed in [50]).
Despite their many differences, SGs and PBs share some protein and RNA components and are often observed in the close proximity to one another [26]. Like SGs, PBs can be induced by stress conditions and contain translationally-stalled mRNAs that can resume translation and be converted into polysomes [29, 56, 60]. Several proteins found in both PBs and SGs are known to regulate translation or stability of transcripts and include the cap-binding factor eIF4E [26, 40], the RNA helicase RCK/p54 (DDX6) [28], RNAi-associated Argonaute proteins [39, 44, 61, 62], the ARE-BPs TTP, BRF1 [26] and HuR [63], the translational silencer YB-1 (Y-box binding protein 1) [64], and CPEB [28]. Some proteins such as TTP and BRF1 promote docking between SGs and PBs which may allow mRNPs to move from one granule to the other [26, 35, 65] (Figure 1). Although SGs and PBs can fuse, each can also form independently of the other [26, 66, 67]. For more detailed overview of RNA granules, their components and functions in different organisms we refer the readers to recent reviews [9, 48, 50, 68–70].
SURVIVING THE HOSTILE CANCER MICROENVIRONMENT: ROLE OF RNA GRANULES
RNA granules contain hundreds of different molecules. Their direct roles in cancer metabolism, regulation of gene expression in cancer cells, and cancer cell adaptation to tumor microenvironments are yet to be uncovered. Here, we discuss some examples of PB- and SG-associated proteins that have been implicated in cancer cell physiology.
Stress responses and Cancer Microenvironment
The integrated stress response (ISR) is a program that receives information from different “stress sensors” and coordinates cellular adaptation to stress [71, 72]. Among these stress sensors are serine/threonine kinases that phosphorylate eIF2α [73]. They include GCN2 (which senses amino acid levels and is activated by amino acid deprivation [74]), HRI (which monitors oxidative stress/ROS levels and activated by heme deprivation in erythroid cells [75]), PERK (senses endoplasmic reticulum (ER) stress [76, 77]) and PKR (activated in response to UV exposure, viral infections and heat shock [78]). As eIF2α phosphorylation is the main trigger of SG formation [51], SG formation is part of the ISR that promotes adaptation and survival during changing local environmental conditions.
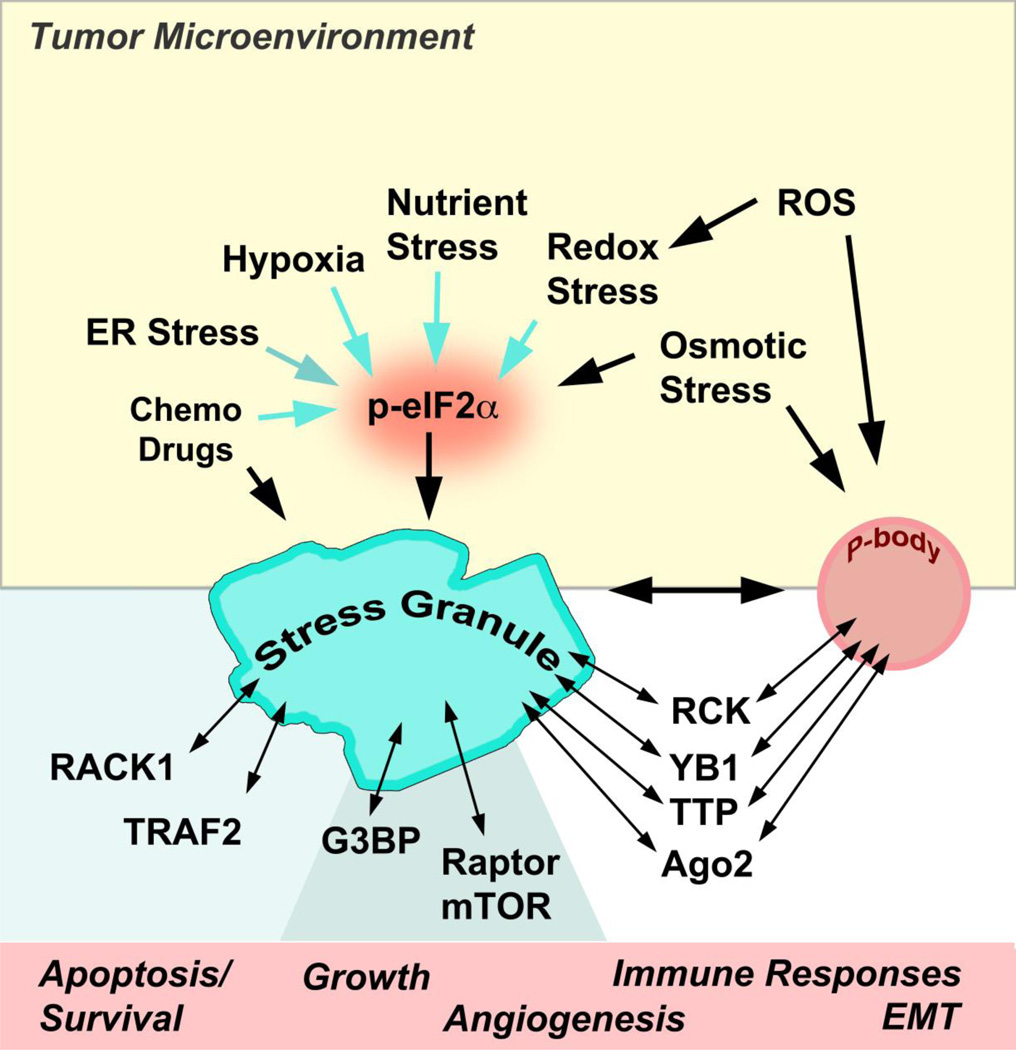
Responding and adapting to stress is important in both cancer development and the tumor response to anticancer therapies (Figure 2). During solid tumor development, cancers can quickly outgrow the existing vasculature, as high metabolic demands of rapidly proliferating cancer cells can out-strip the availability of oxygen and nutrients. This exposes the cancer cells to hypoxia, hyperosmolarity, and nutrient starvation [79]. Although tumors counter this by stimulating neovascularization (formation of new blood vessels), the new blood vessels are often leaky and disorganized. Moreover, the high proliferative rate and dysregulation of protein synthesis in cancer cells cause endoplasmic reticulum (ER) stress, a consequence of protein misfolding and overload in ER due to imbalanced protein synthesis and improper protein folding within secretory pathways [80]. All these condition create a hostile microenvironment that affects various aspects of tumor growth and metabolism (Figure 2).
Figure 2. RNA granules and tumor microenvironment.
Collective contributions from the different stress-associated conditions and surrounding cellular environment result in the unique niche where cancer cells exist, known as the tumor microenvironment (yellow). The tumor microenvironment changes dynamically during tumor progression, which requires constant adaptation of cancer cells to the changing intracellular and extracellular conditions. Hypoxia and nutrient starvation force cells to alter their metabolism/cellular energetics ultimately causing chronic ROS production that has multiple effects on cancer cell (patho)physiology. Chronic ER stress results from the imbalance between increased protein synthesis due to the high demands on cancer cell proliferation and ER capacity often. These stresses, often augmented by chemotherapy, trigger formation of PBs/SGs, spatial manifestations of ISR (Integrative Stress Response) that orchestrates cellular adaptation to changing conditions. The integration and balanced management of these cancer-associated stresses through SGs and PBS modulate multiple physiological responses (bottom red panel) including activation of anti-apoptotic, pro-survival mechanisms, enhanced tumor growth, modulation of immune responses, promotion of angiogenesis and/or activation of a novel developmental regulatory program/transformation referred to as the “epithelial-mesenchymal transition” (EMT). Multiple RNA-binding proteins with signaling properties (Ago2, RCK, TTP, YB-1, G3BP) and signaling molecules (RACK1, TRAF2, Raptor, mTOR) dynamically associate with SGs and PBs (double-headed arrows). They may transduce signals incoming from tumor microenvironment (yellow) to the physiological responses (bottom red panel), thus mediating both short- and long-term adaptations critical to tumor growth and metastasis. For other details, see text.
Cancer cells adapt to this environment by uncoupling glycolysis from oxidative phosphorylation, a process facilitated by HIF1alpha (reviewed in [81]), which results in a high level of oxidative stress resulting from production of reactive oxygen species (ROS) and requires increased anti-oxidant capacity (reviewed in [82]). The altered glycolytic metabolism of cancer cells [81] is characterized by high mitochondrial membrane potential, elevated leakage of electrons, and production of O2− superoxide, which mitochondrial and cytoplasmic superoxide dismutases (SODs) metabolize into the longer-lived metabolite hydrogen peroxide, which acts as second messenger to activate ROS scavenging pathways (reviewed in [83]). Chronically elevated ROS depletes cellular anti-oxidants (reduced NADH/NADPH, peroxiredoxin, and glutathione), altering redox-dependent cell signaling [84] and activating several antioxidant defense pathways, including NRF2/KEAP1, HIF1, HSF and p53/myc (reviewed in [82]). The term “oxidative stress” is often equated with production of “reactive oxygen species” or ROS, but these are not precisely identical. ROS are small reactive molecules including superoxide, hydroxyl free radicals, and hydrogen peroxide (H2O2) that result from leaky or malfunctioning mitochondria or are produced enzymatically. ROS levels are balanced by antioxidants and ROS scavengers; when ROS production exceeds the antioxidant capacity, stress ensues.
The direct stress sensors of redox imbalance are largely cysteine residues in regulatory proteins such as KEAP1/NRF2 and HSF1. Arsenicals directly bind to reduced cysteines in proteins, and inactivate perhaps 200 enzymes, including key anti-oxidant enzymes (glutathione reductase, glutathione peroxidase, thioredoxin reductase and peroxidase), signaling kinases and phosphatases, key metabolic enzymes (pyruvate dehydrogenase) and signaling enzymes (reviewed in [85]). Increased ROS levels associated with arsenite treatment may be produced by cellular metabolism (mitochondrial electron transport; NAD redox reactions, etc) but accumulate because arsenite inactivates the protective antioxidant systems that would normally conteract the imbalance caused by ROS. Sodium arsenite was first chemical found to potently induce SGs [51], and remains the most effective and consistent agent used to induce SG assembly. Induction of SGs by sodium arsenite [86] or hypoxia [87] can promote resistance to apoptosis. Although poorly understood, this may require both short-term suppression of apoptosis, and long-term adaption mediated by transcription factors such as NRF2 and HIF1alpha. SG-mediated, short-term responses may involve sequestration/inactivation of proapoptotic factors (such as RACK1 and TRAF2) in SGs [87, 88] or sequestration of mRNAs encoding pro-apoptotic proteins [89]. Treatment of diverse cancer cell lines with oxidative stress agents including sodium arsenite [56], H2O2 [90], or selenite [91] induces different types of SGs. Sodium arsenite activates the eIF2α kinase HRI which phosphorylates eIF2alpha, which is necessary and sufficient to induce SGs under normal culture conditions [56]. In contrast, although H2O2 also induces eIF2α phosphorylation, it triggers SGs through an eIF2a-independent pathway but requires eIF4E [90]. Selenite induces SG assembly through a combination of mechanisms, but the SGs it induces appear to lack several pro-survival molecules found in arsenite-induced SGs ([91] and see below).
The connection between SGs, ROS and redox imbalance is complex. A recent study [92] suggested that the balance between G3BP1 and one of its binding partners Ubiquitin-specific protease 10 (USP10) coordinately regulates ROS levels and inhibits apoptosis when both proteins are colocalized in SGs (induced by arsenite stress), but not in response to hydrogen peroxide, which did not induce SGs under the conditions they employed. This study did not determine what type of “arsenite-induced” ROS was produced [92], and did not consider differences between arsenite and H2O2 (reviewed in [85]), so this study is difficult to evaluate. An important earlier study clearly demonstrated that the balance between NAD(P)H and nonoxidized glutathione during stress conditions regulated both arsenite and G3BP1-induced SG formation [93], suggesting that redox stress rather than ROS is key to G3BP-mediated SG regulation. Another role for G3BP1 in redox regulation may involve its translational repression of the catalytic beta-subunit of mitochondrial H+-ATP synthase [94], suggesting that G3BP1-mediated translational repression is mediated by SGs, and that this repression may be relieved by release of the mRNA from the granule to sites proximal to the mitochondria [94]. G3BP1/USP10 are linked to autophagy [95], and other studies link each molecule independently to cancer: G3BP1 is important in cancer cell proliferation, migration and resistance chemotherapy [96], whereas USP10 stabilizes p53 [95] and SIRT6 [97] to suppress tumor formation.
SG formation and chemotherapy drugs
Several studies have linked the survival of cancer cells to the formation of SGs in response to chemotherapeutic treatments. Bortezomib (PS-341/Velcade), an FDA-approved drug that targets the 26S proteasome, displays significant anti-tumor activity against mantle cell myeloma and multiple myeloma. In contrast, many types of solid tumors are refractory to bortezomib treatment and resistant to bortezomib-induced apoptosis. Fournier et al. showed that bortezomib potently induces the formation of SGs in colon cancer (Caco), cervical cancer (HeLa) and lung cancer (Calu-I) cell lines [98]. Similarly to MG132 (another proteasome inhibitor used in chemotherapy) [99], bortezomib triggers phosphorylation of eIF2α promoting SG formation [98]. Interestingly, these drugs activate different eIF2α kinases: MG132 activates GCN2 [100] while bortezomib activates HRI [98]. Upon HRI depletion, bortezomib fails to trigger SG assembly and cancer cells undergo efficient apoptosis correlating with the lack of pro-survival anti-apoptotic SGs. In addition, CUGBP1 (CUG triplet repeat RNA-binding protein 1) mediates recruitment of p21 (WAF1/CIP1) mRNA into SGs upon proteasome inhibition by MG132 or bortezomib [101]. p21 is a stress-responsive cyclin-dependent kinase inhibitor with tumor suppression functions that rely on cell cycle arrest or apoptosis induction. CUGBP1-mediated localization of p21 mRNA into SGs stabilizes this mRNA and promotes resistance to bortezomib-induced apoptosis. Conversely, knocking down CUGBP1 promotes bortezomib-induced apoptosis [101].
Interestingly, another chemotherapeutic agent, the antimetabolite 5-Fluorouracil (5-FU), also induces the assembly of SGs [102]. 5-FU is used for treatment of various types of cancers including breast, colorectal, head and neck cancers [103]. 5-FU cytotoxicity is caused by interference with enzymatic activities of thymidylate synthase and by its incorporation into DNA and RNA intermediates. 5-FU incorporation into RNA (5-FU-containing RNA metabolites) causes activation of PKR kinase [104], phosphorylation of eIF2α [104] and assembly of SGs that sequester RACK1, an important regulator of apoptosis [102]. RACK1-positive SGs are prosurvival and anti-apoptotic (see below). Other chemotherapeutic agents known to incorporate into RNA (such as 6-thioguanine and 5-azacytidine) also trigger assembly of SGs [102]. In contrast, DNA-incorporating chemotherapeutic metabolites (such as trifluorothymidine and gemcitabine) do not trigger SG formation but, surprisingly, potently increase the number of PBs in cancer cells [102]. The biological significance of these observations is not clear, although recently PBs have been implicated in the cellular response to hypoxia. Under hypoxic conditions, p54/Rck (a component of both PBs and SGs) regulates mRNA translation of the hypoxia inducible factor HIF-1α, a major regulator of cellular response to oxygen fluctuation [105]. SGs play a role in the resistance of cancer cells to radiation therapy and hypoxia by the regulation of HIF-1α and HIF-1α-regulated transcripts [89]; whether p54/Rck association with PBs/SGs is required for such regulation is to be determined.
Sodium selenite, a selenium-containing chemotherapeutic agent, promotes ROS production and selective apoptosis of cancer cells. It triggers ER stress resulting in phosphorylation of eIF2α and ROS-dependent formation of SGs in prostate cancer (DU145) and osteosarcoma (U2OS) cell lines [91]. Selenite-induced SGs lack translation initiation factor eIF3b, RACK1 and importin α1/β1, components of “canonical” sodium arsenite-induced SGs. The phosphorylation of eIF2α is dispensable for selenite-induced assembly of SGs; instead selenite relies on the action of eIF4E-binding protein 1 (4EBP1) for SG assembly and 4EBP1 depletion compromises its ability to assemble SGs. In response to selenite treatment, 4EBP1 disrupts the assembly of translationally-competent eIF4F complexes, resulting in the assembly of SGs. Importantly, selenite-induced SGs are functionally distinct from canonical SGs in that they promote cell death instead of cell survival in response to stress. This difference is at least partially attributed to the 4EBP1-mediated contribution to the cytotoxicity of selenite [91]. All these data suggest that SGs might be a part of the program by which cancer cells resist chemotherapy treatments and provide a basis for potential targeting of SGs in cancer.
Phosphorylation of eIF2α (p-eIF2α) is a key trigger of SG assembly and ISR, but p-eIF2α independent pathways of SG assembly also exist. Synthetic and natural compounds that induce SG assembly without eIF2α phosphorylation have been described, some of which are particularly relevant to cancer biology. One of them is 15-deoxy-Δ(12, 14)-prostaglandin J2 (15d-PGJ2), a natural lipid inflammatory mediator belonging to a class of prostaglandins with diverse roles in cancer and inflammatory diseases. 15d-PGJ2 binds to the helicase eIF4A and inhibits eIF4A/eIF4G interactions, resulting in translation inhibition and assembly of SGs [106]. In leukemia and colorectal cancer cells, 15d-PGJ2 induces apoptosis and proliferative arrest [107]. The role of SGs in 15d-PGJ2-mediated apoptosis is proposed to be a consequence of global translational arrest, but the exact molecular details are lacking. Further studies of pro-apoptotic and anti-proliferative properties of 15d-PGJ2 are required to determine whether this compound is of a potential use for anti-cancer therapy.
At least three other drugs that target helicase eIF4A and induce SGs show promise as anti-cancer therapeutics: Pateamine A (PatA) [108, 109], hippuristanol [110] and silvestrol [111]. PatA is a metabolite isolated from the sea sponge Mycale specious that binds eIF4A [108, 109], stimulates its helicase and ATPase activities leading to the disruption of eIF4A/eIF4G interaction [108, 109, 112], reduction of translationally-competent eIF4F complex levels and assembly of SGs [100, 113]. PatA and its synthetic derivatives show potent anti-proliferative and proapoptotic activities in many tumor cell lines in vitro [109, 114, 115] and on melanoma xenograft models in vivo [115]. Silvestrol, a compound belonging to the class of rocaglates and isolated from plant Aglaia foveolata, stimulates the RNA binding and helicase activities of eIF4A leading to the sequestration of eIF4A from the eIF4F complex [111, 116] and thus inducing SG assembly [117] in a p-eIF2a-independent manner, similar to PatA [108, 109, 112]. Silvestrol is a potent inducer of apoptosis, effective inhibitor of cancer cell proliferation in vitro, and effective suppressor of tumor growth in xenograft models of breast, prostate and hepatocellular cancers [111, 117, 118]. Hippuristanol, a steroid isolated from coral Isis hippuris, is an allosteric inhibitor of eIF4A that reduces RNA binding, ATPase and helicase activities [110, 119] and thus promotes formation of SGs [100]. Hippuristanol induces cell cycle arrest and apoptosis in adult T-cell leukemia (ATL) cells and shows synergistic effects with other anti-tumor agents against diverse human leukemia and lymphoma cell lines and in the Eu-Myc lymphoma mouse model [120, 121]. Although all three eIF4A-targeting agents are promising anti-cancer drugs, the mechanisms underlying their anti-proliferative effects are still elusive. eIF4A is required for the translation of mRNAs with highly structure 5’ -UTRs, which encode many proteins involved in carcinogenesis, therefore its inactivation and consequent SG formation may regulate the translation/ subcellular localization of such transcripts. Future studies relying on genome-wide approaches such as ribosome and/or polysome profiling correlated with subcellular localization of selected mRNA targets could dissect mechanisms of SG-inducing chemotherapy agents in cancer.
mTOR, SGs and cancer
SGs may regulate the activity and localization of the mammalian target of rapamycin (mTOR) kinase, a key regulator of cell metabolism and growth, which is widely implicated in cancer [122]. mTOR regulates cell growth and mRNA translation in response to insulin signaling, growth factors and amino acid availability. mTOR is the catalytic subunit of two structurally distinct complexes associated with different subcellular compartments: mTORC1 (composed of mTOR, regulatory-associated protein of mTOR (Raptor), mammalian lethal with SEC13 protein 8 (MLST8) and associated non-core components PRAS40 and DEPTOR) and mTORC2 (composed of mTOR, rapamycin-insensitive companion of mTOR (Rictor), MLST8 and mammalian stress-activated protein kinase interacting protein 1 (mSIN1)). mTORC2 is an important regulator of the cytoskeleton, while mTORC1 constitutes an energy/nutrient/redox sensor coupling metabolism to protein synthesis through the regulation of phosphorylation of eIF4E-binding proteins (4EBPs), the key regulators of eIF4E functions (reviewed in [123, 124]).
In human cells, osmotic or oxidative stress promotes the recruitment of mTOR and its mTORC1-specific partner Raptor to SGs; their targeting to SGs is controlled by DYRK3 (dual specificity tyrosine-phosphorylation regulated kinase 3) [125]. Oxidative stress affects mTORC1 activity via both inhibitory and stimulatory inputs. While redox-induced hyperactivation of mTORC1 is mediated by the inhibition of the upstream tuberous sclerosis complex 1/2 (TSC1-TSC2) [126], redox-induced inhibition of mTORC1 is mediated by astrin, a binding partner of Raptor [127]. In response to redox and metabolic stresses, astrin inhibits Raptor:mTOR association by binding and sequestering Raptor in SGs, thus inhibiting mTORC1 assembly and activation [127]. Consequently, SG-mediated regulation of mTORC1 activity leads to changes in cellular translation by regulating eIF4F complex assembly. In addition, mTOR-regulated recruitment of TIA1/TIAR proteins into SGs selectively modulates translation of mRNAs with 5'-terminal oligopyrimidine tracts (5'TOP) [49, 128]. Specifically, mTORC1 cooperates with GCN2 kinase to regulate translation of 5'TOP mRNAs under conditions of nutrient starvation and stress [128]. This subclass of mRNAs predominantly encodes protein biosynthesis factors such as translation factors and ribosomal proteins [129]. As cancer cells have elevated needs for protein synthesis, the interplay between SGs, mTOR activity/localization and translational control of 5′ TOP mRNAs in the stressful tumor microenvironment is of vital importance for cancer cell metabolism.
SG-associated RBPs and regulation of their target cancer-relevant mRNAs
The cap-binding protein eIF4E is a component of the eIF4F complex that integrates information from major receptor signaling pathways and orchestrates cell proliferation and cell death/survival. eIF4E expression and activity are elevated in ~30% of all cancers, and its overexpression is correlated with poor prognosis, especially in hematological malignancies. In fact, eIF4E is an oncogene itself, and even modest over-expression of eIF4E enhances cancer cell proliferation and resistance to cell death (reviewed in [130]). While eIF4E is a general regulator of cap-dependent translation, a specific subset of mRNAs is especially sensitive to the levels of this factor [52, 131]. These eIF4E–sensitive transcripts encode cancer-related protein kinases, transcription factors, cell cycle and apoptosis regulators, growth factors and cytokines. Some proteins that activate eIF4E function (e.g., Akt, Myc and Ras) also increase the transcription of eIF4E (e.g., c-Myc). Importantly, eIF4E cooperates with c-Myc to promote cancer progression by inhibiting apoptosis [132, 133]. While localization of eIF4E into SGs and PBs is well documented, it remains to be determined whether eIF4E-sensitive mRNA targets are preferentially recruited to these granules in response to extra- and intra-cellular stimuli.
RACK1, another SG component, is a highly conserved intracellular adaptor protein belonging to the Trp-Asp (WD) repeat protein family. It is a scaffolding protein that interacts with a vast array of signaling molecules including kinases and phosphatases, membrane receptors, G proteins, ion channels and apoptosis-related molecules [134]. RACK1 is a component of the 40S ribosomal subunit, binds eIF3, and regulates mRNA translation and quality control [135]. Up-regulation of RACK1 mRNA and protein expression is observed in many cancers (lung, gastric, breast cancers, gliomas) [136]. It is proposed to both suppress and promote cancer cell proliferation via its interactions with different partners that influence various signaling pathways. In one case, RACK1 interacts with the pro-apoptotic proteins Fem1b and BIM to inhibit cancer cell apoptosis. Sequestration of RACK1 into SGs has a negative impact on the stress-activated p38 and JNK (c-Jun N-terminal kinase)/MAPK pathways causing inhibition of apoptosis. However, RACK1 also can exert pro-apoptotic effects by interacting with apoptosis-related proteins such as BAX, a member of BCl-2 family [136]. The extent to which RACK1 interactions with pro- and anti-apoptotic molecules is regulated by stress-induced RACK1 localization into SGs, and whether RACK1 binding to 40S subunits and/or eIF3 is required for its targeting to SGs is presently unknown.
HuR (ELAV1) and TTP (Tristetraprolin) are among the best-characterized ARE-BPs with multiple functions in cancer. Both proteins are components of SGs, but exhibit opposite effects on their target mRNAs: while HuR generally stabilizes transcripts and regulates their translation, TTP actively promotes degradation of mRNA targets. The shared targets TTP and HuR include mRNAs encoding inflammatory cytokines, cell growth factors, angiogenesis-, apoptosis- and differentiation-related factors. Surveys of TTP expression in cancer cell lines and human tumors suggest that TTP functions as tumor suppressor [137], and its levels negatively correlate with tumor progression in breast and prostate cancer [138]. In a v-H-ras-dependent mast cell tumor model, where tumor development in vivo is dependent on the induction of autocrine IL-3 secretion, over-expression of TTP reduces levels of IL-3 mRNA and suppresses tumor growth [139]. In epithelial EpRas cells, knock down of TTP promotes epithelial-to-mesenchymal transition (EMT) and metastasis [140]. A microRNA frequently deregulated in hematological, cholangiocytic and lung cancers (miR-29a) dampens expression of TTP. Consequently, over-expression of miR-29a in EpRas cells leads to suppression of TTP levels followed by disruption of epithelial polarity, EMT and metastasis [140]. Similarly, over-expressed TTP impairs Myc-induced lymphomagenesis and abolishes the transformed state by disabling Myc-mediated proliferative responses [141]. While down-regulation of TTP is observed in tumors of different origin (e.g., cancers of the breast, colon, esophagus, kidney, lung, pancreas, prostate, skin, stomach, and thyroid) up-regulation of HuR is often observed in cancer [142]. In pancreatic and colon cancers, high HuR levels correlate with high levels of VEGF, potent angiogenic factor, and poor prognosis. In mouse xenograft models, over-expression of HuR in carcinoma cells lead to the development of larger tumors, while HuR depletion reduces the tumor size [142–144]. While SGs regulate TTP/HuR localization and activities, the importance of these relationships to cancer development is still unclear.
YB-1 and LIN28 (LIN28A) are multifunctional RBPs belonging to the family of cold shock domain (CSD) proteins [145]. Both factors shuttle between the nucleus and cytoplasm and localize to SGs and/or PBs under stress. Mutations in the CSD of these proteins affect RNA binding and localization into SGs/PBs [64, 146]. While YB-1 is highly expressed in both adult and developing tissues, expression of LIN28 is highest in embryonic tissues and stem cells. Both YB-1 and LIN28 are overexpressed in a wide range of solid tumors and hematological malignancies (reviewed in [147, 148]). Although these proteins are closely related, their functions seem to be non-overlapping. LIN28 regulates gene expression by two mechanisms: 1) blocking the processing of the let-7 family of microRNAs, and 2) enhanced translation of mRNAs encoding metabolic enzymes, splicing factors and cyclins (reviewed in [149]). YB-1 efficiently stabilizes its mRNA targets and can either inhibit or stimulate their translation [147]. In cancer, LIN28-mediated down-regulation of let-7 microRNAs, which have tumor suppressor activity, leads to up-regulation of let-7 mRNA targets that encode pro-proliferative, oncogenic and anti-apoptotic factors. Lin28 overexpression is sufficient to transform NIH/3T3 cells while depletion of Lin28 in human leukemia cells increases let-7 levels and reduces cancer cell proliferation [150]. The role of YB-1 in cancer is even more complex. YB-1 inhibits translation of mRNAs associated with control of cellular proliferation and the epithelial phenotype but simultaneously facilitates activation of a mesenchymal gene expression program. YB-1-mediated EMT is associated with preferential translation of mRNAs encoding EMT-inducing proteins such as Snail1, Twist and others [151]. Overall this results in the paradoxical down-regulation of cancer cell proliferation and simultaneous increased mobility and invasiveness as well as survival in anchorage-independent conditions and metastasis.
Both angiogenin (ANG) and its inhibitor RNH1 (also known as ribonuclease inhibitor, RI) are components of SGs [152] and have well-characterized roles in cancer and angiogenesis (reviewed in [153]). ANG is a secreted ribonuclease (RNase) with angiogenic properties that is over-expressed in many cancers. It stimulates cancer cell proliferation by enhancing ribosomal RNA (rRNA) transcription and promotes cancer cell survival during stress [154]. Several stressors induce ANG translocation from the nucleus into the cytoplasm, where it cleaves transfer RNAs (tRNAs) in the anticodon loop to produce tRNA-derived stress-induced RNA (tiRNAs) [155, 156]. Selected tiRNAs inhibit mRNA translation by interfering with eIF4F complex functions, and promoting the assembly of pro-survival SGs in an eIF2α phosphorylation-independent manner that requires YB-1 [157–159]. The RNase activity of ANG is absolutely required for ANG-mediated angiogenesis, enhanced survival and proliferation [153]. RNH1 binds ANG and inhibits tRNA cleavage, rRNA transcription, and cell migration, but promotes apoptosis. Down-regulation of RNH1 promotes tumor growth and cancer cell proliferation, migration and metastasis (bladder, prostate and hematological cancers, melanoma) while up-regulation suppresses melanoma growth and metastasis [160–162]. While molecular details of RNH1-dependent suppression are still unclear, available data suggest that RNH1 regulates transcription and translation of EMT-associated transcription factors Twist, Snail, Slug and ZEB1 [161].
Several other SG- and/or PB-associated proteins can contribute to carcinogenesis. eIF5A, a hypusine-modified factor involved in various aspects of mRNA metabolism, promotes polysome disassembly and assembly of SGs [163]. The hypusine modification affects eIF5A-associated effects on polysomes and SGs [163] and is required for eIF5A-mediated promotion of tumor growth in pancreatic cancer [164]. The conserved RNA-dependent helicase Ded1/DDX3, which works both as an activator and repressor of translation, is a SG-associated protein [165, 166] with anti-apoptotic properties that promotes transformation by modulating hypoxic responses in breast cancer [167] or by regulation of Rac1 mRNA translation and Rac1-mediated signaling in medulloblastomas [168]. Finally, Argonautes, key effectors of the RNAi pathway (reviewed in [169]), are components of both SGs and PBs that possess multiple cellular functions that are de-regulated in cancer cells. Their contribution to cancer initiation and progression via small RNA pathways is well documented and is a matter of several recent reviews [170–172].
We have described here only some RNA granule-associated proteins and their proposed roles in cancer. Note, however, that a number of studies trying to determine SG function(s) rely on deletion or mutation of one or more SG-associated proteins. Distinguishing between the functional effects of deletion/mutation of one or more SG-associated proteins on cell metabolism from the ancillary effects that these proteins exert on SGs (preventing/inducing) is a consistent problem in the field. Nonetheless, direct or indirect interactions between these and other proteins and apoptosis/signaling-related molecules suggest that the crosstalk between RNA granules and apoptotic machinery regulates the balance between cell survival and cell death. While the precise mechanistic details of crosstalk between RNA granules and apoptotic machineries are still elusive, the current evidence supports the notion that RNA granules could act as important signaling/regulatory centers exploited by cancer cells to allow their survival in hostile tumor microenvironments, as well as response to chemotherapy and radiotherapy.
CONCLUSIONS AND PERSPECTIVES
Over the past several years, substantial research has shown that RNA granules are not merely sites of mRNA storage or degradation. Instead, they are flexible and versatile regulators of gene expression, and act as signaling hubs to influence multiple aspects of cell metabolism central to carcinogenesis and metastasis. While the exact mechanisms by which RNA granules impact the development and progression of cancer are largely unknown, it is clear that the post-transcriptional mechanisms mediated by RNA granule-associated proteins and RNAs play major roles in adaptation to stress, cancer cell proliferation and tumor growth.
An intriguing aspect of RNA granule assembly, in particular SG assembly, is their responsiveness to chemotherapeutic drugs. As discussed above, some drugs promote the assembly of non-canonical SGs whose composition is different than that of classical SGs. Inclusion or exclusion of signaling molecules such as RACK1 or TRAF2 has profound effects on cancer cell viability, e.g. selenite-induced SGs promote cell death whereas 5-FU-induced SGs promote cell survival. Their assembly often relies on different signaling, e.g. selenite-induced SG assembly requires eIF4EBP1-mediated remodeling of the eIF4F complex and is independent of eIF2α phosphorylation. A precise understanding of the mechanisms by which chemotherapeutic drugs induce SG assembly could identify new targets for cancer therapy.
Many questions remain, and a number of areas require further studies. Cancer cells within tumors are very heterogeneous. Do these tumor cell subpopulations assemble SGs in response to stress and/or chemotherapeutic drugs with different efficiency? Do cancer-associated mutations in key signaling molecules influence assembly of prosurvival or pro-apoptotic SGs? What are the key mRNA targets and proteins that are assembled into RNA granules in response to chemotherapy treatment? Developing a detailed spatiotemporal knowledge of the specific functions of SG subtypes, their composition and their regulation by different signaling molecules represent a major challenge, and a thorough understanding of the roles of RNA granules in cancer cell metabolism may identify the next important targets for cancer therapy.
HIGHLIGHTS.
Stress Granules and PBs house cancer-associated RNA-binding proteins
Many Stress Granule-associated proteins are aberrantly expressed in cancer
SG and PB-associated proteins alter gene expression and cancer initiation/progression
Chemotherapy drugs modulate assembly of Stress Granules
SGs and PBs are potential targets and biomarkers for cancer therapy
ACKNOWLEDGMENTS
We thank the members of the Anderson's and Ivanov's research groups for the productive discussions and helpful comments. This work was supported by National Institutes of Health grant (CA168872 and GM111700, P.A.) and a Research Development Grant from the Muscular Dystrophy Association (ID158521, P.I.) and ALS Association (N7W220, P.I.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fasken MB, Corbett AH. Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol. 2005;12:482–488. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 4.Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Molecular systems biology. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebauer F, Preiss T, Hentze MW. From cis-regulatory elements to complex RNPs and back. Cold Spring Harbor perspectives in biology. 2012;4:a012245. doi: 10.1101/cshperspect.a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov P, Anderson P. Post-transcriptional regulatory networks in immunity. Immunological reviews. 2013;253:253–272. doi: 10.1111/imr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 8.Bloch DB, Nobre RA, Yang WH. GW/P-bodies and autoimmune disease. Advances in experimental medicine and biology. 2013;768:61–70. doi: 10.1007/978-1-4614-5107-5_5. [DOI] [PubMed] [Google Scholar]
- 9.Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA biology. 2014:11. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd RE. Regulation of stress granules and P-bodies during RNA virus infection. Wiley interdisciplinary reviews RNA. 2013;4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanderweyde T, Youmans K, Liu-Yesucevitz L, Wolozin B. Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology. 2013;59:524–533. doi: 10.1159/000354170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 13.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley interdisciplinary reviews RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 15.Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochimica et biophysica acta. 2013;1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Ling SH, Qamra R, Song H. Structural and functional insights into eukaryotic mRNA decapping. Wiley interdisciplinary reviews RNA. 2011;2:193–208. doi: 10.1002/wrna.44. [DOI] [PubMed] [Google Scholar]
- 17.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 19.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 21.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Diez J. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochimica et biophysica acta. 2007;1773:1786–1792. doi: 10.1016/j.bbamcr.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5'-3' decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 29.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoecklin G, Anderson P. In a tight spot: ARE-mRNAs at processing bodies. Genes Dev. 2007;21:627–631. doi: 10.1101/gad.1538807. [DOI] [PubMed] [Google Scholar]
- 37.Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 40.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E–transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E–binding protein 4E–T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aizer A, Kalo A, Kafri P, Shraga A, Ben-Yishay R, Jacob A, Kinor N, Shav-Tal Y. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci. 2014;127:4443–4456. doi: 10.1242/jcs.152975. [DOI] [PubMed] [Google Scholar]
- 48.Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19:R397–R398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Ivanov P, Kedersha N, Anderson P. Stress puts TIA on TOP. Genes Dev. 2011;25:2119–2124. doi: 10.1101/gad.17838411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 53.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 61.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. Journal of virology. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273–3284. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13:704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1224. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–4727. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harbor perspectives in biology. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 70.Stoecklin G, Kedersha N. Relationship of GW/P-bodies with stress granules. Advances in experimental medicine and biology. 2013;768:197–211. doi: 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 72.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Advances in nutrition. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cellular and molecular life sciences : CMLS. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 76.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 77.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 78.Srivastava SP, Kumar KU, Kaufman RJ. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 79.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer cell. 2014;25:563–573. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 81.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Frontiers in pharmacology. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nature reviews Drug discovery. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 83.Costa A, Scholer-Dahirel A, Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Seminars in cancer biology. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Wall SB, Oh JY, Diers AR, Landar A. Oxidative modification of proteins: an emerging mechanism of cell signaling. Frontiers in physiology. 2012;3:369. doi: 10.3389/fphys.2012.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen S, Li XF, Cullen WR, Weinfeld M, Le XC. Arsenic binding to proteins. Chemical reviews. 2013;113:7769–7792. doi: 10.1021/cr300015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 88.Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 90.Emara MM, Fujimura K, Sciaranghella D, Ivanova V, Ivanov P, Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2alpha phosphorylation. Biochem Biophys Res Commun. 2012;423:763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujimura K, Sasaki AT, Anderson P. Selenite targets eIF4E–binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 2012;40:8099–8110. doi: 10.1093/nar/gks566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, Fujii M. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol Cell Biol. 2013;33:815–829. doi: 10.1128/MCB.00763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cande C, Vahsen N, Metivier D, Tourriere H, Chebli K, Garrido C, Tazi J, Kroemer G. Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J Cell Sci. 2004;117:4461–4468. doi: 10.1242/jcs.01356. [DOI] [PubMed] [Google Scholar]
- 94.Ortega AD, Willers IM, Sala S, Cuezva JM. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J Cell Sci. 2010;123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- 95.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang H, Shao RG. [G3BP: a promising target for cancer therapy] Yao xue xue bao = Acta pharmaceutica Sinica. 2010;45:945–951. [PubMed] [Google Scholar]
- 97.Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, Kong S, Ye J, Gao B, Fang D. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell reports. 2013;5:1639–49. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fournier MJ, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer cell international. 2010;10:12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–26018. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, Mazroui R. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaehler C, Isensee J, Hucho T, Lehrach H, Krobitsch S. 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res. 2014;42:6436–6447. doi: 10.1093/nar/gku264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature reviews Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 104.Garcia MA, Carrasco E, Aguilera M, Alvarez P, Rivas C, Campos JM, Prados JC, Calleja MA, Esteban M, Marchal JA, Aranega A. The chemotherapeutic drug 5-fluorouracil promotes PKR-mediated apoptosis in a p53-independent manner in colon and breast cancer cells. PLoS One. 2011;6:e23887. doi: 10.1371/journal.pone.0023887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saito K, Kondo E, Matsushita M. MicroRNA 130 family regulates the hypoxia response signal through the P-body protein DDX6. Nucleic Acids Res. 2011;39:6086–6099. doi: 10.1093/nar/gkr194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim WJ, Kim JH, Jang SK. Anti-inflammatory lipid mediator 15d–PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 2007;26:5020–5032. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shin SW, Seo CY, Han H, Han JY, Jeong JS, Kwak JY, Park JI. 15d–PGJ2 induces apoptosis by reactive oxygen species-mediated inactivation of Akt in leukemia and colorectal cancer cells and shows in vivo antitumor activity. Clin Cancer Res. 2009;15:5414–5425. doi: 10.1158/1078-0432.CCR-08-3101. [DOI] [PubMed] [Google Scholar]
- 108.Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 110.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 111.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA, Jr, Pelletier J. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. The Journal of clinical investigation. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chemistry & biology. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 113.Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 114.Hood KA, West LM, Northcote PT, Berridge MV, Miller JH. Induction of apoptosis by the marine sponge (Mycale) metabolites, mycalamide A and pateamine. Apoptosis. 2001;6:207–219. doi: 10.1023/a:1011340827558. [DOI] [PubMed] [Google Scholar]
- 115.Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, Zhao N, Marsh J, Agoulnik S, Twine N, Parent L, Chen Z, Shie JL, Jiang Y, Zhang H, Du H, Boivin R, Wang Y, Romo D, Littlefield BA. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Molecular cancer therapeutics. 2009;8:1250–1260. doi: 10.1158/1535-7163.MCT-08-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chambers JM, Lindqvist LM, Webb A, Huang DC, Savage GP, Rizzacasa MA. Synthesis of biotinylated episilvestrol: highly selective targeting of the translation factors eIF4AI/II. Organic letters. 2013;15:1406–1409. doi: 10.1021/ol400401d. [DOI] [PubMed] [Google Scholar]
- 117.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, Porco JA, Jr, Pelletier J. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kogure T, Kinghorn AD, Yan I, Bolon B, Lucas DM, Grever MR, Patel T. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS One. 2013;8:e76136. doi: 10.1371/journal.pone.0076136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun Y, Atas E, Lindqvist LM, Sonenberg N, Pelletier J, Meller A. Single-molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure. 2014;22:941–948. doi: 10.1016/j.str.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 120.Cencic R, Robert F, Galicia-Vazquez G, Malina A, Ravindar K, Somaiah R, Pierre P, Tanaka J, Deslongchamps P, Pelletier J. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood cancer journal. 2013;3:e128. doi: 10.1038/bcj.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochemical pharmacology. 2011;81:713–722. doi: 10.1016/j.bcp.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 122.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K–mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 126.Yoshida S, Hong S, Suzuki T, Nada S, Mannan AM, Wang J, Okada M, Guan KL, Inoki K. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J Biol Chem. 2011;286:32651–32660. doi: 10.1074/jbc.M111.238014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Klasener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, Nitschke R, Kuehn EW, Jonker JW, Groen AK, Reth M, Hall MN, Baumeister R. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 128.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–2068. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 130.Polunovsky VA, Bitterman PB. The cap-dependent translation apparatus integrates and amplifies cancer pathways. RNA biology. 2006;3:10–17. doi: 10.4161/rna.3.1.2718. [DOI] [PubMed] [Google Scholar]
- 131.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 133.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell communication and signaling : CCS. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gandin V, Senft D, Topisirovic I, Ronai ZA. RACK1 Function in Cell Motility and Protein Synthesis. Genes & cancer. 2013;4:369–377. doi: 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li JJ, Xie D. RACK1, a versatile hub in cancer. Oncogene. 2014 doi: 10.1038/onc.2014.127. [DOI] [PubMed] [Google Scholar]
- 137.Carrick DM, Blackshear PJ. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Archives of biochemistry and biophysics. 2007;462:278–285. doi: 10.1016/j.abb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 138.Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stoecklin G, Gross B, Ming XF, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 140.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, Cleveland JL. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–574. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 143.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Abdelmohsen K, Tominaga-Yamanaka K, Srikantan S, Yoon JH, Kang MJ, Gorospe M. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mihailovich M, Militti C, Gabaldon T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 146.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA biology. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 147.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry Biokhimiia. 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 148.Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. The international journal of biochemistry & cell biology. 2013;45:973–978. doi: 10.1016/j.biocel.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 149.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell stem cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, Sorensen PH. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 152.Pizzo E, Sarcinelli C, Sheng J, Fusco S, Formiggini F, Netti P, Yu W, D'Alessio G, Hu GF. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126:4308–4319. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. Journal of cellular physiology. 2012;227:2822–2826. doi: 10.1002/jcp.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li S, Hu GF. Angiogenin-mediated rRNA transcription in cancer and neurodegeneration. Int J Biochem Mol Biol. 2010;1:26–35. [PMC free article] [PubMed] [Google Scholar]
- 155.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fu P, Chen J, Tian Y, Watkins T, Cui X, Zhao B. Anti-tumor effect of hematopoietic cells carrying the gene of ribonuclease inhibitor. Cancer gene therapy. 2005;12:268–275. doi: 10.1038/sj.cgt.7700742. [DOI] [PubMed] [Google Scholar]
- 161.Pan X, Xiong D, Yao X, Xin Y, Zhang L, Chen J. Up-regulating ribonuclease inhibitor inhibited epithelial-to-mesenchymal transition and metastasis in murine melanoma cells. The international journal of biochemistry & cell biology. 2012;44:998–1008. doi: 10.1016/j.biocel.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 162.Yao X, Li D, Xiong DM, Li L, Jiang R, Chen JX. A novel role of ribonuclease inhibitor in regulation of epithelial-to-mesenchymal transition and ILK signaling pathway in bladder cancer cells. Cell and tissue research. 2013;353:409–423. doi: 10.1007/s00441-013-1638-2. [DOI] [PubMed] [Google Scholar]
- 163.Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS ONE. 2010;5:e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fujimura K, Wright T, Strnadel J, Kaushal S, Metildi C, Lowy AM, Bouvet M, Kelber JA, Klemke RL. A Hypusine-eIF5A–PEAK1 Switch Regulates the Pathogenesis of Pancreatic Cancer. Cancer Res. 2014;74:6671–6681. doi: 10.1158/0008-5472.CAN-14-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F–mRNA complex. Mol Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 167.Bol GM, Raman V, van der Groep P, Vermeulen JF, Patel AH, van der Wall E, van Diest PJ. Expression of the RNA helicase DDX3 and the hypoxia response in breast cancer. PLoS One. 2013;8:e63548. doi: 10.1371/journal.pone.0063548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2014 doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 169.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 171.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in molecular medicine. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 172.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Frontiers in genetics. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]