SUMMARY
Mitochondrion plays essential roles in many aspects of biology, and its dysfunction has been linked to diverse diseases. Central to mitochondrial function is oxidative phosphorylation (OXPHOS), accomplished by respiratory chain complexes (RCCs) encoded by nuclear and mitochondrial genomes. How RCC biogenesis is regulated in metazoans is poorly understood. Here we show that Parkinson’s disease (PD)-associated genes PINK1 and Parkin direct localized translation of certain nuclear-encoded RCC (nRCC) mRNAs. Translationally-repressed nRCC mRNAs are localized in a PINK1/Tom20-dependent manner to mitochondrial outer membrane, where they are de-repressed and activated by PINK1/Parkin through displacement of translation repressors, including Pumilio and Glorund/hnRNP-F, a Parkin substrate, and enhanced binding of activators such as eIF4G. Inhibiting the translation repressors rescued nRCC mRNA translation and neuromuscular-degeneration phenotypes of PINK1 mutant, whereas inhibiting eIF4G had opposite effects. Our results reveal previously unknown functions of PINK1/Parkin in RNA metabolism and suggest new approaches to mitochondrial restoration and disease intervention.
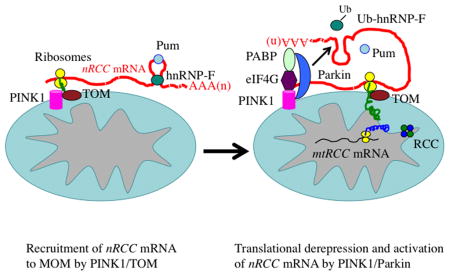
Graphical Abstract
INTRODUCTION
Mitochondria (mito) exert essential cellular functions, from bioenergetics and intermediary metabolism to ion homeostasis and apoptosis. Mito integrity is particularly important for neuromuscular (NM) tissues with high energy demand (Chan, 2006; Wallace, 2005). Other aspects of mito physiology are also important for these tissues. In neurons, mito help buffer Ca2+ influxes elicited by neuronal activity (Mattson et al., 2008; Saxton and Hollenbeck, 2012). It is thus not surprising that mito dysfunction has been linked to various neurological disorders (Chan, 2006; Schon and Przedborski, 2011; Wallace, 2005). The cause of mito dysfunction in most diseases, however, remains largely undefined.
OXPHOS is arguably the most fundamental mito function carried out by five RCCs whose subunits are dually encoded by the nuclear and mito genomes. The biogenesis and maintenance of RCCs require regulated expression of nRCC and mito -encoded RCC (mtRCC) genes. How this process is coordinated is not well understood. Subcellular targeting and local translation of mRNAs offers an effective mechanism to achieve spatially restricted protein synthesis (Balagopal and Parker, 2009; Besse and Ephrussi, 2008), making it highly pertinent to processes like RCC biogenesis. In yeast, mtRCC mRNAs are targeted close to the mito inner membrane (MIM) (McMullin and Fox, 1993), whereas certain nRCC mRNAs are recruited to the vicinity of mito outer membrane (MOM) (Kellems et al., 1975). Localized translation of nRCC and mtRCC mRNAs presumably ensures co-translational import and assembly of subunits into multimeric RCCs. Whether mito-resident regulatory factors are required for this process, and the in vivo significance of this process in metazoans, are largely unknown.
PD is an age-dependent degenerative condition caused primarily by dopaminergic neuron (DN) deficits. Mito dysfunction and OXPHOS impairment in particular has been profoundly implicated in PD pathogenesis (Henchcliffe and Beal, 2008). Strong genetic evidence supporting a mito etiology of PD came from the identification of a familial PD (FPD) gene PINK1 (Pten-induced kinase 1) encoding a mito -targeted Ser/Thr kinase (Valente et al., 2004). Genetic studies in Drosophila first established that PINK1 and another FPD gene product Parkin, an E3 ubiquitin ligase, act in a common pathway to maintain mito function (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). Recent studies have emphasized roles of PINK1 and Parkin in mitophagy (Narendra et al., 2010).
We previously observed OXPHOS impairment in Drosophila PINK1 model (Liu et al., 2011). Here we investigated the underlying molecular cause. Our results reveal a previously unknown mechanism of PINK1 action in regulating localized translation of select nRCC mRNAs, whereby PINK1 acts as a mito-resident regulatory factor to promote the MOM-targeting of nRCC mRNAs via the translocase of outer membrane (TOM) complex. PINK1-regulated nRCC mRNAs are translationally repressed in the cytosol. Upon recruitment to MOM, however, they are translationally derepressed and activated by PINK1 and Parkin. The in vivo significance of this process is supported by the phenotypic rescue of PINK1 mutant after inhibiting the translational repressors bound to nRCC mRNAs, including Pumillio (Pum), GW182, and Glorund (Glo), the fly homolog of hnRNP-F/H. We also show that PINK1 and Parkin cooperate to promote hnRNP-F/Glo ubiquitination, thus directly linking the PINK1/Parkin pathway to mRNA metabolism and translational control of OXPHOS.
RESULTS
PINK1 Regulates the mRNA Localization and Protein Abundance of Certain nRCC Genes in Drosophila
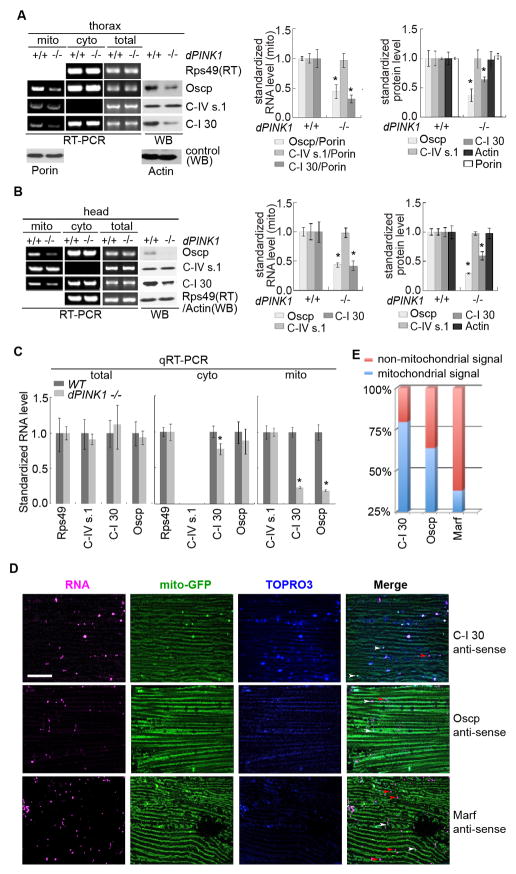
To understand the OXPHOS defects in PINK1 mutant, we examined the expression of RCC proteins in NM tissues of control and PINK1B9 mutant flies. Ten RCC (7 nRCC and 3 mtRCC) proteins were chosen for analysis. Levels of 4 nRCC proteins, including the 30 kD subunit of complex-I (C-I 30), OSCP subunit of C-V, core protein 2 of complex-III, and ND75 subunit of complex-I (Figure 1A, B; data not shown), were reduced, whereas other nRCC proteins tested were unaltered in PINK1 mutant (Figure S1A). Levels of 3 mtRCC proteins tested, cytochrome c oxidase subunit I (C-IV-s1) (Figure 1A, B), ATP synthase subunit 6, and cytochrome b (Figure S1A), were not changed. The levels of several nuclear-encoded, mito proteins, e.g., Porin (VDAC) and MCU, were also unaffected (Figure S1A). PINK1 appears to specifically regulate the expression of select nRCC proteins.
Figure 1. PINK1 Regulates mRNA Localization and Protein Expression of Select nRCC Genes in Drosophila.
(A) RT-PCR and western blot (WB) analyses of RCC mRNAs and proteins in PINK1 thoracic muscle. mRNAs or proteins prepared from total, cyto, or mito fractions were used. Mito-bound nRCC mRNA levels were normalized with Porin protein level, total and cyto mRNA levels with rps49, and protein levels with actin (n=3 experiments). Since the expression of the mtRCC gene C-IV s1is not altered by PINK1, it is used later to normalize RNA and protein levels of nRCC genes.
(B) RT-PCR and WB of RCC mRNAs and proteins in PINK1 head tissues. Experiments were done as in A (n=3 experiments).
(C) qRT-PCR of RCC mRNAs in total and subcellular fractions from PINK1 muscle tissues (n=3–4 experiments).
(D, E) RNA FISH of nRCC mRNAs and control marf mRNA in wild type muscle. Mito and nuclei were labelled with mito-GFP and TOPRO3, respectively. Arrows: RNA particles colocalizing (white) or not colocalizing (red) with mito. Scale bar: 25μm. Bar graph in E shows quantification of RNA signals colocalizing with mito (150 signals from 3 individual animals).
Error bar: SEM; *, p<0.05 in Student’s t-tests.
See also Figure S1.
We next examined the mechanisms underlying the selective reduction of nRCC proteins in PINK1 mutant. Semi-quantitative RT-PCR and quantitative real-time RT -PCR (qRT-PCR) revealed that total mRNA levels for the affected proteins were largely unaffected (Figure 1A–C). To test whether localized translation might be involved, we analyzed nRCC mRNA distribution in subcellular fractions. In the cytosolic (cyto) fraction, nRCC mRNA levels were comparable between control and mutant (Figure 1A–C). Intriguingly, a fraction of the mRNAs for the affected nRCC subunits was found in Percoll gradient-purified mito (Figure 1A–C), the purity of which was verified by the absence of other membrane structures such as the ER and endosomes (Figure S1B). RNase A-treatment of purified mito degraded the nRCC mRNAs, but not mtRCC RNAs residing in the matrix (Figure S1C), suggesting that the nRCC mRNAs are MOM-bound. Fluorescent in situ hybridization (FISH) confirmed the mito localization of select nRCC mRNAs (Figure 1D, E; Figure S1D), and transmission EM analysis revealed the association of ribosomes with muscle MOM, suggesting ongoing translation on MOM (Figure S1E). Strikingly, correlating with the change of protein levels, levels of MOM-bound nRCC mRNAs, but not mtRCC mRNAs, were significantly reduced in NM tissues of PINK1 mutant (Figure 1A–C). In contrast, in the intestine, which was phenotypically normal in PINK1 mutant, the mRNA localization and protein expression of those nRCC genes were unaltered (Figure S1F). PINK1 thus regulates the mRNA localization and protein expression of select nRCC genes in a tissue-specific manner. Due to the importance of C-I to PD in general (Schapira, 2010), and C-I 30 to PINK1 function in particular (Wu et al., 2013), we focused on C-I 30 mRNA in later analyses.
PINK1 Regulates the mRNA Localization and Protein Expression of Select nRCC Genes in Human DNs
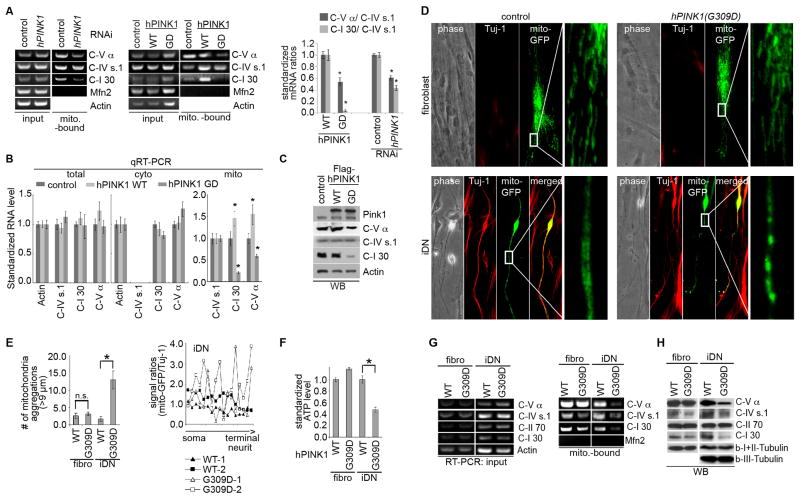
We next tested possible conservation of PINK1 function in nRCC mRNA regulation. In HEK293T cells, the nRCC mRNAs homologous to those regulated by PINK1 in flies were found in highly purified mito (Figure 2A, B; S2A). Mito localization of mammalian C-I 30 mRNA was further confirmed using the MS2-GFP/MS2-bs tagging system (Bertrand et al., 1998) (Fig. S2B, C). Moreover, in PINK1 siRNA-treated cells, levels of MOM-bound nRCC mRNAs were significantly reduced, whereas mtRCC RNA (C-IV s1) was unaffected (Figure 2A, S2D). Similar to PINK1 RNAi, expression of pathogenic PINK1(G309D) (Hoepken et al., 2007) greatly reduced levels of MOM-bound nRCC mRNAs, but not mtRCC RNA (Figure 2A, B), suggesting that PINK1(G309D) acts in a dominant-negative manner. Not all nRCC mRNAs or mRNAs for nuclear-encoded mito proteins were MOM-bound (Figure 2A, S2D). Correlating with the change in abundance of MOM-bound, but not total, mRNAs of select nRCC genes, their protein levels were also reduced when PINK1 was inhibited (Figure 2C). Thus, the function of PINK1 in nRCC mRNA localization and protein expression is conserved in human cells.
Figure 2. PINK1 Regulates mRNA Localization and Protein Expression of Select nRCC Genes in Mammalian Cells.
(A) Semi-quantitative RT-PCR of mito-bound nRCC mRNAs in HEK293 cells with hPINK1 knocked down or transfected with PINK1-WT or PINK1-G309D. Knockdown efficiency is shown in Figure S2D. Bar graph shows normalized mito-bound nRCC mRNA levels (n=3 experiments).
(B) qRT-PCR of RCC mRNAs in total and subcellular fractions of HEK293 cells transfected with PINK1-WT or PINK1-G309D (n=3 experiments).
(C) WB of RCC proteins in HEK293 cells transfected with PINK1-WT or PINK1-G309D.
(D) Mitochondrial morphology in fibroblasts or iDNs from control subject (WT) and PINK1(G309D) patient.
(E) Quantification of mito size in the periphery of fibroblasts and iDNs (left), and signal intensity ratios of mito-GFP versus Tuj-1 in iDN neurites (right). For each sample at least 4 cells were analyzed (*p<0.05).
(F, G) ATP level (F) and nRCC mRNA and protein expression (G) in control (WT) and PINK1(G309D) fibroblasts and iDNs (*p<0.05, n=4).
See also Figure S2.
To validate these findings in disease -relevant neurons, we used induced DNs (iDNs) trans-differentiated from patient skin fibroblasts (Caiazzo et al., 2011). Expression of DN-related markers and DN-like transcriptomes were confirmed in iDNs (Figure S2E–G). The functionality of iDNs was validated by rescue of locomotor deficit in 6-OHDA-lesioned rats upon transplantation (Figure S2H). Comparison of PINK1(G309D) patient iDNs with control iDNs revealed disease-specific abnormal mito distribution and morphology in neuronal processes (Figure 2D, E), and reduced ATP levels (Figure 2F). Importantly, reduced mito-association of select nRCC mRNAs and reduced protein levels were observed in patient iDNs (Figure 2G, H). Patient fibroblasts, however, did not show significant change in mito distribution, morphology (Figure 2D, E), ATP production (Figure 2F), or nRCC protein expression (Figure 2G, H), indicating that the PINK1(G309D) mutation exerts DN-specific effects on mito function.
PINK1 Functionally Interacts with Pum and the TOM/TIM Complex to Regulate the Localization and Translation of nRCC mRNAs
The tight correlation of levels of select nRCC proteins with their mRNAs in the mito fraction, but not their mRNAs in the cyto fraction which constitute the majority of total mRNAs (Figure S3A) and were unchanged in PINK1 mutant (Figure 1C), suggests that the MOM-bound nRCC mRNAs are translationally active, whereas those in the cyto are repressed. Indeed, we observed cytoplasmic ribosomes associated with MOM (Figure S1E), and detected cytoplasmic translation factors in highly purified mito from fly tissues or human cells (Figure S3B). Moreover, polysome analysis of cyto and mito fractions indicated that PINK1-regulated nRCC mRNAs are translationally more engaged in the latter fraction (Figure S3C), a notion supported by in vitro translation assays (Figure S3D). Consistently, PINK1-regulated nRCC proteins are present almost exclusively in the mito fraction, whereas ND42 and Tom20, which are synthesized in the cyto and subsequently imported into mito, are present in both the cyto and mito fractions (Figure S3E).
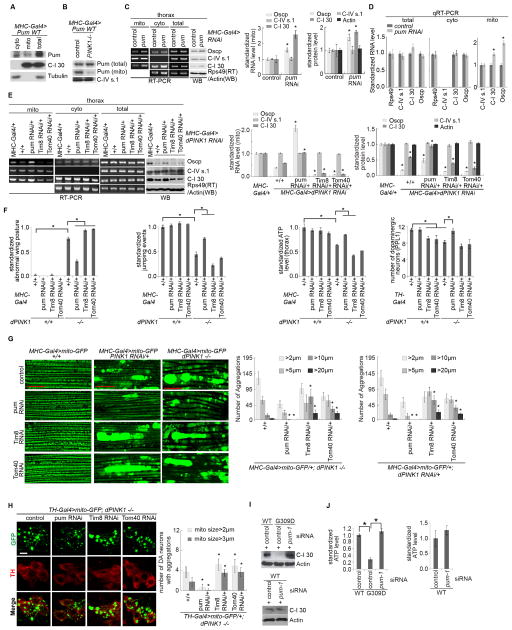
To probe the molecular mechanism of translational control of nRCC mRNAs, we tested the role of the translational repressor Pum (Quenault et al., 2011). Pum is present in both the cyto and mito fractions in fly tissues (Figure 3A) and mammalian cells (Figure S3F). The mito localization of Pum appeared to be regulated byPINK1, as the a mount of mito-bound Pum, but not total Pum, was reduced in PINK1 mutant tissues (Figure 3B) or PINK1(G309D)-transfected HEK293 cells (Figure S3G). To test whether the mito localization of Pum depends on its binding to mRNA, we treated purified mito with RNase A and observed significantly reduced mito-association of Pum (Figure S3H). Moreover, an RNA binding-deficient form of Pum (Friend et al., 2012) was compromised in mito localization (Figure S3I), suggesting that Pum may be recruited to MOM via binding to mRNAs.
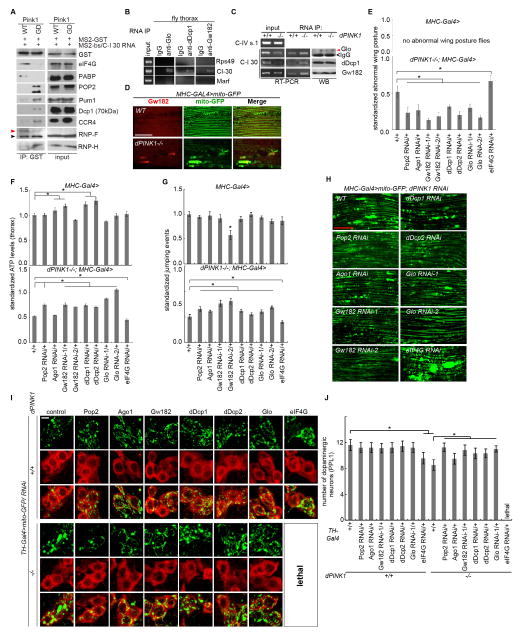
Figure 3. Involvement of Pum and TOM/TIM Complex in the MOM Localization and Translation of Select nRCC mRNAs.
(A) WB showing the presence of Pum in the mito fraction of Mhc-Gal4>Pum-WT muscle. C-I 30 serves as mito marker and Actin as loading control.
(B) Effects of PINK1 mutation on the MOM recruitment of Pum.
(C) RT-PCR and WB showing the effect of Pum RNAi on the MOM localization and protein expression of nRCC mRNAs (n=3 experiments).
(D) qRT-PCR showing the effect of Pum RNAi on RCC mRNA levels in the total and subcellular fractions (n=3–4 experiments).
(E) RT-PCR and WB showing the effects of Pum, Tom40, or Tim8 RNAi on the defective MOM localization and translation of nRCC mRNAs in PINK1 RNAi flies (n=3 experiments).
(F, G, H) Effects of Pum, TOM40, or TIM8 RNAi on the wing posture, jumping ability, ATP level, and DN loss (F), and on the mito aggregation defect caused by PINK1 mutation in flight muscle (G) or DNs (H) (n=3 experiments, 20–25 flies per genotype per experiment; For DN counting, 7–8 animals were assayed). Scale bar (G): 25μm.
(I, J) Effects of Pum-1 RNAi on C-I 30 protein expression (I) and ATP level (J) in control and PINK1(G309D) iDNs (n=3 experiments).
Error bar: SEM; *, p<0.05 in Student’s t-tests.
See also Figure S3.
We next tested the role of Pum in nRCC mRNA translation. Pum RNAi moderately increased mito-bound nRCC mRNAs and their protein levels in fly muscle tissues (Figure 3C, 3D, S3J). Similar effect was observed in HEK293 cells (Figure S3K). Importantly, Pum RNAi effectively restored levels of MOM-bound nRCC mRNAs and their proteins in PINK1 RNAi (Figure 3E) or PINK1 mutant (Figure S3L) muscle tissues.
Since all nRCC proteins require passage through the TOM/TIM complex to enter mito, we asked whether the MOM-targeting and translation of nRCC mRNAs is linked to nascent nRCC protein import. For this purpose, we knocked down components of the TOM/TIM complexes. Inhibition of Tom40 or Tim8 strongly exacerbated PINK1 mutant effects on the MOM-targeting and translation of nRCC mRNAs (Figure 3E, S3L). Independent RNAi lines for each gene produced similar effects (Figure S3N). The MOM-targeting and translation of nRCC mRNAs thus appears to be linked to protein import during RCC biogenesis.
PINK1 has been shown to regulate mito fission/fusion dynamics (Yang et al., 2008) and motility (Liu et al., 2012). However, loss- or gain-of-function of genes involved in fission/fusion (Drp1, Marf) or motility (Miro) had no obvious effect on the levels of MOM-bound nRCC mRNAs, although for some unknown reason the protein levels of C-V (OSCP) and C-I 30 were reduced in Drp1-DN or Miro overexpression (OE) conditions (Figure S3O). Thus, defects in mito dynamics or motility cannot explain the effect of PINK1 on select nRCC mRNA localization or translation, and PINK1 appears to play a specific role in this process.
Altered Pum and TOM/TIM Activities Modulate PINK1 Mutant Effects on NM Tissue Function and Integrity in Drosophila
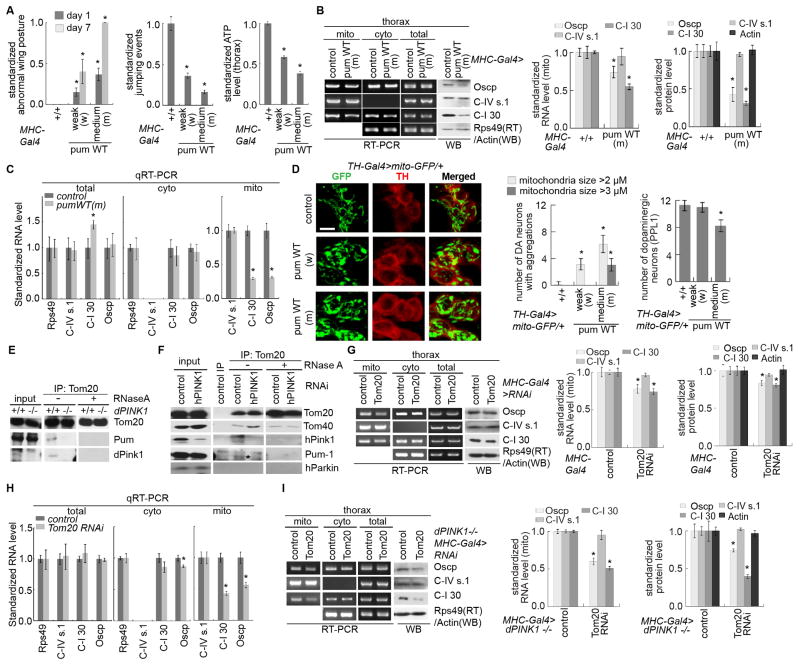
We next assessed the effects of Pum and Tom/Tim on PINK1 function at the organismal level. While Pum RNAi had little effect on NM tissue function or integrity on its own, it effectively rescued the wing posture, jumping ability, ATP level, and DN survival defects of PINK1 B9 mutant (Figure 3F) or PINK1 RNAi flies (Figure S3M). In addition, Pum RNAi rescued the abnormal mito morphology caused by PINK1 inactivation (Figure 3G, H). Consistent with Pum performing a conserved function, a portion of mammalian Pum-1 was localized to mito (Figure S3F), and Pum-1 RNAi rescued the reduced nRCC protein expression and ATP production in PINK1(G309D) iDNs (Figure 3I, J).
In contrast to the rescuing effect of Pum RNAi, Tim8 or Tom40 RNAi exacerbated PINK1 mutant phenotypes in muscle (Figure 3F, G, S3M) and DNs (Figure 3H). Moreover, Tim8 or Tom 40 RNAi, but not Pum RNAi, caused mild mito aggregation and DN loss (Figure S4A, B), emphasizing the importance of TOM/TIM to DN health. To further probe the function of the TOM/TIM complex, we inhibited additional components. RNAi of Tom7 and Tom20, but not Tim10 or Tim13, enhanced PINK1 mutant phenotypes (Figure S4C). Since the knockdown efficiencies of the various RNAi transgenes were similar (Figure S4D), their differential genetic interactions with PINK1 suggest that components of the TOM/TIM complex do not function equivalently in the PINK1 pathway. We further probed the genetic epistasis between PINK1 and TOM in gain-of-function studies. PINK1 mutant phenotypes were specifically rescued by Tom20 OE (Figure S4E); reciprocally, PINK1 OE rescued Tom20 RNAi effects (Figure S4F). These results, together with their mutual enhancement of mutant phenotypes shown later, suggest that PINK1 and Tom20 may act at the same level, e.g., by working together in a complex, rather than at different steps of a linear pathway.
We also examined Pum OE effects. While strong Pum OE with the muscle-specific Mhc-Gal4 driver was lethal, weak or medium Pum OE lines produced viable progenies with muscle defects (Figure 4A, S4G). Using the medium Pum OE line (Pum -m), we found that its phenotypic effects correlated well with impaired MOM-targeting and translation of nRCC mRNAs (Figure 4B, C). In DNs, abnormal mito aggregation and neuronalloss (Figure 4D) similar to that seen in PINK1mutant were observed in TH-Gal4>Pum-m flies. PINK1 OE failed to rescue Pum-m OE effects (Figure S4H). This result, together with the rescue of PINK1 mutant phenotypes by Pum RNAi (Figure 3), suggests that Pum may act downstream of PINK1.
Figure 4. Effects of Pum Overexpression and a TOM20-PINK1 Complex on the Localization and Translation of nRCC mRNAs.
(A) Effects of Pum OE on wing posture, jumping ability, and ATP levels. Weak (Pum-w) and moderate (Pum-m) Tg lines were used (n=3 experiments, 20–25 flies per genotype per experiment).
(B) RT-PCR and WB showing the effect of Pum-m OE on the MOM localization and translation of nRCC mRNAs (n=3 experiments).
(C) qRT-PCR showing the effect of Pum-m OE on RCC mRNA levels in the total and subcellular fractions (n=3 experiments).
(D) Effects of Pum-w and Pum-m OE on mito morphology and survival of DNs (n=7–8 flies). Scale bar: 5μm.
(E) RNA-dependent interaction of Tom20 with PINK1 or Pum as detected by co-IP using fly muscle extracts.
(F) Tom20 interacting proteins as detected by co-IP. Extracts made from CCCP-treated HEK293 cells were mock treated or treated with RNase A before IP. *: Pum-1 signal.
(G) RT-PCR and WB showing the effect of Tom20 RNAi on the MOM localization and translation of nRCC mRNAs in fly muscle (n=3 experiments).
(H) qRT-PCR showing the effect of Tom20 RNAi on RCC mRNA levels in the total and subcellular fractions (n=3–4 experiments).
(I) RT-PCR and WB showing the effect of Tom20 RNAi on the defective localization and translation of nRCC mRNAs inPINK1 mutant (n=3 experiments).
Error bar: SEM; *, p<0.05 in Student’s t-tests.
See also Figure S4.
PINK1-TOM20 Interaction Promotes the MOM-Targeting and Translation of nRCC mRNAs
We sought to elucidate the molecular basis of the genetic interactions between TOM and PINK1, especially between Tom20 and PINK1. Tom20 is a key component of the protein import system that serves as a receptor for precursor proteins containing mito targeting sequences (MTS). Recently, PINK1 was found to associate with the TOM complex in depolarized mito in mammalian cells (Lazarou et al., 2012), although its physiological role was unclear. We hypothesized that Tom20 helps anchor translating nRCC mRNAs and associated ribosomes/RNPs to the MOM, presumably through binding to MTS of nascent nRCC proteins during co-translational import, and that PINK1 may facilitate this process. To test this model, we analyzed Tom20 immunoprecipitate (IP) prepared from fly muscle (Figure 4E) and HEK293T cells (Figure 4F). To stabilize endogenous PINK1, we briefly subjected flies or cultured cells to mild CCCP treatment (Narendra et al., 2010). We found that PINK1 and Pum, but not Parkin, associated with Tom20. The association of PINK1 or Pum with Tom20 was dramatically reduced by RNase A treatment (Figure 4E, F), whereas Tom20-Tom40 interaction was not affected (Figure 4F). The RNA-dependency of the above interactions prompted us to test the involvement of nRCC mRNAs. RNA-IP of fly muscle (Figure S4I) or HEK293 cells (Figure S4J) showed that the levels of nRCC mRNAs associated with Tom20 were reduced when PINK1 was inhibited. Supporting a critical role of Tom20 -PINK1 interaction in the localization and translation of nRCC mRNAs, Tom20 RNAi inhibited the MOM-targeting and translation of nRCC mRNAs (Figure 4G, H). Moreover, Tom20 RNAi enhanced the reduced MOM-targeting and translation of nRCC mRNAs (Figure 4I), mito aggregation (Figure S4L, M), and muscle degeneration (Figure S4C) effects ofPINK1 mutation, whereas Tom20 OE had opposite effects (Figure S4E, S4K).
RNA-dependent Association of PINK1 with eIF4G and Other Components of the Translation Initiation Complex
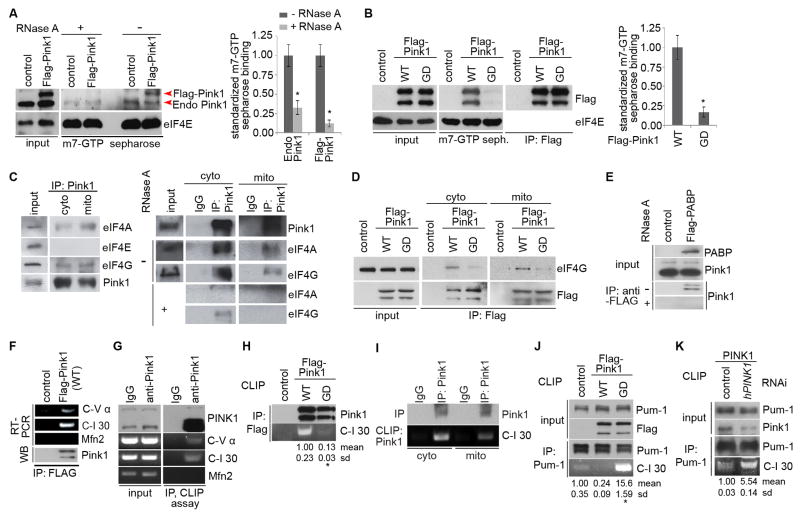
We next probed the mechanism by which PINK1 regulates nRCC mRNA translation. We found that PINK1 associated with mRNA 5′ cap structure, as detected by m7-GTP-sepharose chromatography (Figure 5A). Importantly, the G309D mutation impaired such association (Figure 5B). The association of PINK1 with m7-GTP-sepharose was RNA-dependent, whereas m7-GTP-sepharose binding by eIF4E, a translation initiation complex (TIC) component that directly binds to 5′ cap, was RNA-independent (Figure 5A), suggesting that PINK1 may associate with mRNA 5′ cap indirectly through the TIC. Indeed, endogenous PINK1 exhibited RNA-dependent associations with TIC components eIF4A and eIF4G, which occurred in the mito and cyto fractions (Figure 5C). It is worth pointing out that the association between PINK1 and eIF4G was not eliminated by RNase A treatment (Figure 5C), and PINK1 did not associate with eIF4E, suggesting certain degree of specificity and directness of PINK1-eIF4G interaction. Human genetics studies recently implicated eIF4G as a candidate FPD gene (Chartier-Harlin et al., 2011). We tested whether the PINK1-eIF4G association might be altered in disease condition and found that the G309D mutation attenuated PINK1-eIF4G interaction (Figure 5D). PINK1 also associated RNA-dependently with polyA binding protein (PABP), a protein known to interact with the TIC to promote translation (Figure 5E). Together, these data support a role of PINK1 in promoting translation through RNA-dependent and possibly RNA-independent associations with the TIC.
Figure 5. PINK1 Binds to nRCC mRNAs and RNA-dependently Associates with the TIC.
(A, B) m7-GTP sepharose binding showing RNA-dependent association of endogenous PINK1 (control sample) and exogenous PINK1 (Flag-PINK1) with the TIC (A), and compromised binding of PINK1(G309D) with the TIC (B). eIF4E exhibits RNA-independent m7-GTP binding. Bar graphs show data quantification (n=3 experiments).
(C) RNA-dependent associations between PINK1 and TIC components in the cyto and mito fractions in co-IP assays.
(D) Compromised association between PINK1-G309D and eIF4G.
(E) RNA-dependent association of endogenous PINK1 with PABP.
(F) RNA-IP showing PINK1 binding to nRCC mRNAs but not Mfn2 mRNA in HEK293 cells.
(G) Direct binding of PINK1 to nRCC mRNAs but not Mfn2 mRNA in CLIP assays.
(H) Reduced C-I 30 mRNA binding by PINK1-G309D compared to PINK1-WT in CLIP assays.
(I) PINK1 binding to C-I 30 mRNA in both the mito and cyto fractions in CLIP assays.
(J, K) CLIP assays showing competition between PINK1 and Pum-1 in binding to C-I 30 mRNA in HEK293 cells transfected with PINK1-WT or PINK1-G309D (J), or with PINK1 shRNA (K).
See also Figure S5.
To further demonstrate the role of PINK1 in regulating the translation of mito-bound nRCC mRNAs, we made a translational reporter in which the synthesis of a luciferase-C-I 30 fusion protein is under the control of the 5′ and 3′ UTRs of C-I 30 mRNA. Combining in vitro transcribed reporter mRNA with purified mito from PINK1-WT transfected HEK293T cells led to stimulated translation of the reporter (Figure S5A). Using the same assay, we found that purified mito from PINK1 or parkin mutant fly tissues or PINK1−/− knockout mice tissues translated the reporter mRNA at a lower efficiency than WT mito (Figure S5B–D). Collectively, these data support a role of PINK1 in stimulating the translation of MOM-bound C-I 30 mRNA.
PINK1 Binds to nRCC mRNAs
We sought to elucidate the molecular basis of the RNA-dependency of PINK1 association with TIC. RNA-IP analysis revealed PINK1 association with nRCC mRNAs (Figure 5F). To test whether PINK1 directly binds mRNAs, we performed cross-linking and immunoprecipitation (CLIP) assays (Ule et al., 2005). Endogenous PINK1 directly bound to C-I 30 and C-V α mRNAs but not Mfn2 mRNA (Figure 5G). Moreover, compared to PINK1-WT, PINK1(G309D) bound to C-I 30 mRNA poorly (Figure 5H), supporting the importance of nRCC mRNA-binding to PINK1 function. CLIP assays performed on mito fraction showed that PINK1 binds to MOM-localized C-I 30 mRNA (Figure 5I).
Pum also exhibited C-I 30 mRNA -binding activity in CLIP assays (Figure 5J). In HEK293 cells transfected with PINK1 -WT, mRNA binding by Pum was significantly weakened. In contrast, nRCC mRNA binding by Pum was enhanced in PINK1(G309D) transfected cells or PINK1 RNAi cells (Figure 5J, K). These data suggest that PINK1 negatively impacts the nRCC mRNA-binding activity of Pum. We tested whether this effect might be mediated by a PINK1/Pum physical interaction. Our repeated co-IP assays were unable to detect such interaction (Figure S5E).
PINK1 Promotes Translational Derepression and Activation of nRCC mRNAs on MOM
Data presented earlier suggest that the MOM-localized nRCC mRNAs are translationally repressed in the cyto before reaching MOM, where they are reactivated by PINK1. To dissect the molecular mechanism, we generated a construct expressing C-I 30 mRNA tagged with MS2 binding sites, allowing purification of C-I 30 RNPs using GST-MS2. To specifically monitor events occurring on MOM-bound C-I 30 mRNAs, we used mito for RNP purification. Analysis of mito C-I 30 RNPs purified from PINK1-WT or PINK1-G309D transfected cells showed that PINK1-WT increased the association of eIF4G and PABP with C-I 30 mRNA (Figure 6A). Intriguingly, a number of translational repressors, including Dcp1, POP2, Pum-1, hnRNP-F, and hnRNP-H, were enriched in the C-I 30 RNPs purified from PINK1-G309D transfected cells (Figure 6A). In the case of hnRNP-F, we observed a modified form in PINK1-WT transfected cells. These results suggest that PINK1 may translationally derepress nRCC mRNAs on MOM by displacing and/or modifying translation repressors and further stimulate translation by promoting activator association.
Figure 6. PINK1 De-represses and Activates the Translation of MOM-Bound nRCC mRNAs.
(A) WB of mito C-I 30 mRNA-RNPs purified from HEK293 cells transfected with PINK1-WT or PINK1-G309D. Red arrow: a higher MW species of hnRNP-F.
(B) RNA-IP of wild type thoracic mito showing association of Glo, Dcp1, and GW182 with C-I 30 mRNA. Rps49 and marf serve as negative controls.
(C) RNA-IP showing increased binding of translation repressors to C-I 30 mRNA in PINK1 mutant.
(D) Immunofluorescence staining showing increased GW182 localization to swollen mito in PINK1 mutant muscle. Scale bar: 25μm.
(E, F, G) Effects of translation repressor and eIF4G RNAi on wing posture (E), ATP level (F), and jumping activity (G) of PINK1 mutant. RNAi alone effects are shown on the top graphs (n=3 experiments, 20–25 flies per genotype per experiment).
(H–J) Effects of translation repressor and eIF4G RNAi on muscle mito morphology in Mhc>PINK1-RNAi flies (H), and on DN mito morphology (I) and survival (J) in PINK1 mutant (n=7–8 flies tested). Scale bar (H): 25μm; (I): 5μm.
Error bar: SEM; *, p<0.05in Student’s t-tests.
See also Figure S6.
We next tested the in vivo relevance of the above findings. The fly homologues of hnRNP-F/H, known as Glo (Kalifa et al., 2006), and Dcp1 were found to associate with C-I30 mRNA in muscle tissues (Figure 6B). Moreover, in PINK1 mutant more C-I 30 mRNA was bound by Glo and Dcp1 in the mito fraction (Figure 6C). Binding of repressors to MOM -bound C-I 30mRNA was also observed in HEK293 cells (Figure S6A), and increased repressor binding to nRCC mRNAs was found in PINK1−/− mouse midbrain tissues (Figure S6B). Considering that Dcp1 is a core component of P –bodies (Franks and Lykke-Andersen, 2008), cytoplasmic foci involved in mRNA turnover or repression, we tested association of other P-body components with C-I 30 mRNA. GW182, a conserved key component of P-body, also bound to C-I 30 mRNA (Figure 6B, C), and GW182-positive foci were found residing in the immediate vicinity of mito in PINK1 mutant but not wild type muscle tissues (Figure 6D), suggesting that nRCC mRNAs might be targeted to P-bodies in PINK1 mutant condition. We further tested the functional relationships between PINK1 and the translational repressors. Knockdown of the translation repressors effectively rescued PINK1 loss-of-function phenotypes, whereas eIF4G knockdown had opposite effect (Figure 6E–J). The phenotypic effects of these genetic interactions correlated well with effects on C-I 30 protein expression (Figure S6C). Thus, both the positive and negative regulators associated with C-I 30 mRNA are important effectors in the PINK1 pathway of nRCC mRNA regulation.
Parkin Cooperates with PINK1 to Promote hnRNP-F/Glo Ubiquitination and nRCC mRNA Translation
Parkin was previously shown to act downstream of PINK1 (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). We tested the role of Parkin in translational control of nRCC mRNAs. Parkin associated with Pum -1 in HEK293 cells (Figure 7A). This association raised the possibility that Parkin may mediate the effect of PINK1 in recruiting Pum to mito. Contrary to this expectation, mito-bound Pum was increased in parkin mutant (Figure S7A), suggesting that not only Parkin is not required for the MOM localization of Pum, it may act at a later step to displace Pum from MOM-localized nRCC mRNAs to relieve translation al repression. Supporting a role for Parkin in translational regulation, we found that transfected (Figure 7B) and endogenous Parkin (Figure 7C) associated with 5′ cap in an RNA-dependent fashion, that pathogenic mutations disrupted this association (Figure 7D), that Parkin associated with TIC component PABP (Figure 7E), and that Parkin stimulated the translation of the C -I 30 reporter (Figure S5D). As in PINK1 mutant, the levels of MOM-bound nRCC mRNAs and their proteins were reduced in parkin mutant (Figure 7F, G). However, unlike in PINK1 case, parkin mutant phenotypes were not rescued by Pum RNAi (Figure S7B). Moreover, unlikePINK1 -OE, Parkin OE was able to rescue Pum OE effects on nRCC mRNA translation and tissue integrity (Figure 7H, S7C), suggesting that Parkin may function downstream of Pum.
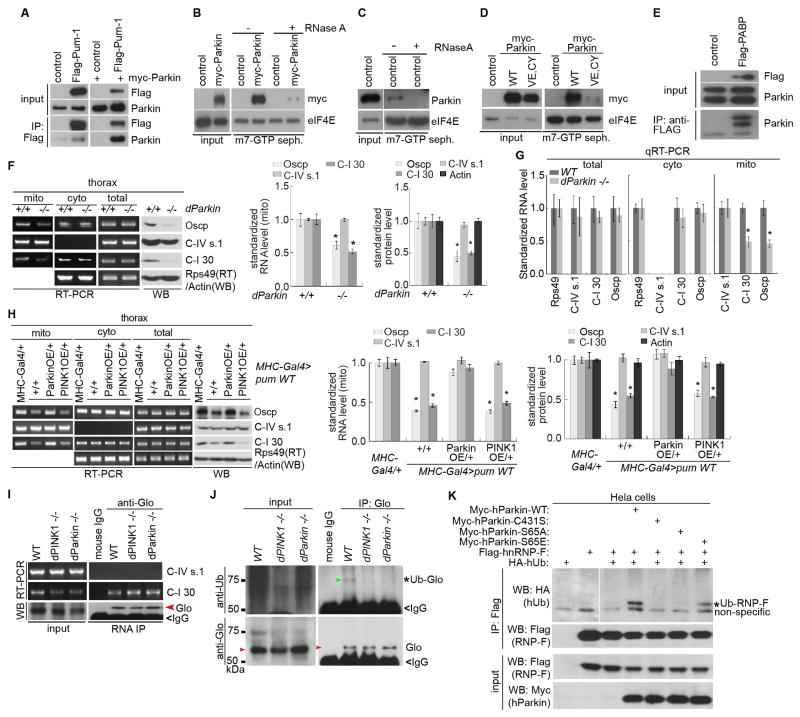
Figure 7. Parkin Cooperates with PINK1 to Promote Glo/hnRNP-F Ubiquitination and nRCC mRNA Translation.
(A) Association of endogenous (left) and exogenous (right) Parkin with Pum-1 in co-IP assays.
(B, C) m7-GTP sepharose binding showing RNA-dependent association of myc-Parkin (B) and endogenous Parkin (C) with TIC.
(D) m7-GTP sepharose binding showing impaired association of Parkin (V56E, C212Y) with the TIC.
(E) Association of endogenous Parkin with exogenous PABP in co-IP assays.
(F) RT-PCR and WB showing the effects of parkin mutation on the MOM-targeting and translation of nRCC mRNAs (n=3 experiments).
(G) qRT-PCR showing the effect of parkin mutation on RCC mRNA levels in the total and subcellular fractions (n=3 experiments).
(H) RT-PCR and WB showing effects of Parkin or PINK1 OE on the defective MOM-targeting and translation of nRCC mRNAs caused by Pum-m OE (n=3 experiments).
(I) RNA-IP showing the effect of parkin mutation on C-I 30 mRNA binding by Glo.
(J) Effect of PINK1 and parkin mutations on Glo ubiquitination in fly muscle. Glo IP from control or mutant tissues was probed with anti-Ub. Red arrow: Glo; Green arrow: Ub-Glo.
(K) Effects of WT and mutant forms of hParkin on hnRNP-F ubiquitination. Hela cells transfected with the indicated constructs were subjected to hnRNP-F IP, followed by WB. *: Ub-hnRNP-F. Left two lanes of the top panel are from a different gel and used to show that the band below Ub-hnRNP-F is non-specific.
Error bar: SEM; *, p<0.05in Student’s t-tests.
See also Figure S7.
We next searched for potential target(s) of Parkin mediating its effect on nRCC mRNA translation. A number of observations led us to focus on Glo. First, Glo RNAi effectively rescued PINK1 mutant (Figure 6E–J), whereas Glo OE had opposite effect (Figure S7D, E). Second, in PINK1 and parkin mutants, more nRCC mRNAs bound to Glo in the mito fraction (Figure 7I). Third, Glo physically associated with Parkin in fly tissues (Figure S7F). Fourth, we observed modification of hnRNP-F by PINK1-WT in C-I 30 RNP (Figure 6A). We tested whether Parkin might ubiquitinate Glo. A Ub-positive, higher molecular weight species consistent with mono-ubiquitinated Glo was detected in Glo-IP prepared from wild type but not parkin or PINK1 mutant flies, suggesting that PINK1 and Parkin regulate Glo ubiquitination (Figure 7J). Interestingly, in PINK1 mutant expressing a phospho-mimetic, but not a non-phosphorylatable forms of Parkin with the PINK1 phosphorylation site mutated (Shiba-Fukushima et al., 2014), Glo ubiquitination was restored (Figure S7G), indicating that PINK1 acts through Parkin to regulate Glo ubiquitination. Ubiquitination of hnRNP-F by PINK1/Parkin was also observed in mammalian cells, in which the WT (Parkin-WT) or phospho-mimetic (Parkin-S65E), but not E3-dead (Parkin-C431S) or non-phosphorylatable (Parkin-S65A) forms of Parkin, induced hnRNP-F mono-ubiquitination (Figure 7K). These data support that Glo/hnRNP-F is a direct substrate mediating the effect of PINK1/Parkin on nRCC mRNA translation. Glo or hnRNP-F protein level was not obviously changed by altered PINK1 or Parkin activities (Figure 7I–K), consistent with mono-ubiquitination generally not affecting protein stability. We hypothesize that ubiquitination of Glo alters the structure of the repressor complexes and their binding to nRCC mRNAs. Supporting this notion, repressor binding to C-I 30 mRNA was increased in parkin mutant, but reduced in PINK1 mutant upon Parkin OE (Figure S7H, I).
DISCUSSION
We report a role of the PINK1/Parkin pathway in regulating the MOM-targeting and translation of select nRCC mRNAs. Since similar effects were observed in vivo in flies and in mammalian cells, including human iDNs, this newly identified function is likely to be fundamental to PINK1/Parkin biology and PD pathogenesis. Combined with the synthesis of mtRCC proteins near the MIM, localized translation of nRCC mRNAs and co-translational protein import on MOM allow efficient assembly of the multi-subunit RCCs. As the PINK1/Parkin-regulated nRCC proteins and the mtRCC proteins are extremely hydrophobic, their coupled synthesis and assembly on mito membrane avoids potential misfolding or aggregation in the cytosol or matrix. Defects in this process could contribute to the mito etiology of PD and possibly other age-related disorders. Our results further suggest that pharmacological agents targeting the key players involved in localized nRCC mRNA translation may offer rational therapeutics for PD.
Our results implicate PINK1 in the localization of nRCC mRNAs to MOM. PINK1 may act in this process through its binding to nRCC mRNAs and through its interaction with Tom20. Tom20/TOM may act in the initial recruitment of nRCC mRNA RNPs from the cytosol to MOM, analogous to the targeting of ER-associated mRNAs, stalled ribosomes, and the SRP to the ER. An alternative, though not mutually exclusive, mechanism of nRCC mRNA targeting to MOM may involve RNA-binding proteins. Previous studies in yeast implicated Tom20 and a Pum homolog Puf3p in directing the mito localization of some nRCC mRNAs (Eliyahu et al., 2010). We found that the sole Pum in flies is localized to mito in PINK1- and RNA binding-dependent manners. Moreover, Pum inhibition increased, whereas Pum OE decreased, nRCC mRNA abundance on MOM, arguing against a positive role of Pum in directing nRCC mRNA localization. The primary role of Pum in nRCC mRNA regulation in metazoans is likely translational repression and its effect on nRCC mRNA abundance on MOM is likely due to links between mRNA translatability and stability (Balagopal and Parker, 2009; Besse and Ephrussi, 2008). Thus, although the process of nRCC mRNA localization to MOM is conserved from yeast to humans, the underlying mechanisms likely differ between yeast and metazoans. Consistently, while PINK1 acts as a key regulator of nRCC mRNA targeting and translation in flies and mammals, there is no counterpart of PINK1 in yeast.
Our results support direct roles of PINK1/Parkin in translational control of nRCC mRNAs. Several observations support this conclusion. First, our CLIP assays showed that PINK1 directly binds to nRCC mRNAs and competes with Pum for nRCC mRNA binding. Second, PINK1 physically associates with mRNA 5′ cap structures and components of the TIC, including eIF4A, eIF4G, and PABP, in an RNA-dependent manner, and at least in the case of eIF4G, RNA-independent interaction may also occur. PINK1 also positively regulates the association of eIF4G and PABP to C-I 30 mRNA. Of particular interest is PINK1’s relationship with eIF4G, mutations in which have been tentatively linked to autosomal dominant PD. Our results support that defective translational control may play a broader role in PD pathogenesis (Lu et al., 2014). Third, our results indicate that PINK1/Parkin actively participate in the de-repression of nRCC mRNAs by displacing the translational repressor hnRNP-F/Glo from nRCC mRNAs. The association of other repressors, such as Pum, Dcp1, GW182, and POP2, with nRCC mRNA was also affected in PINK1 or parkin mutants, although the detailed molecular mechanism remains to be elucidated. Given that Dcp1 and GW182 are components of the P-body (Franks and Lykke-Andersen, 2008), our results suggest that P-bodies and possibly stress granules, related cytoplasmic structures in the turnover or storage of translationally stalled mRNAs (Balagopal and Parker, 2009), may be intimately linked to PINK1/Parkin pathogenesis. Fourth, PINK1 and Parkin promote mono-ubiquitination of hnRNP-F/Glo to displace hnRNP-F/Glo and other repressors from nRCC mRNAs. hnRNPs function in distinct RNP complexes to control broad aspects of RNA physiology. Glo was identified as a protein binding to nanos mRNA (Kalifa et al., 2006), and it may repress translation at multiple steps (Andrews et al., 2011). A role for Glo in regulating translation at the initiation and post-initiation steps would conform to the proposed roles of PINK1/Parkin in a multi-step regulation of localized nRCC mRNA translation. Although PINK1 can also interact with the TOM complex, it is unlikely that the effect of PINK1/Parkin on mRNA translation is mediated by TOM, since the interaction between PINK1 and TOM is transient and weaker than the interaction between PINK1 and the translation machinery, and a direct role for TOM in mRNA translation is not known. Instead, our data support a direct role of PINK1/Parkin in translational control.
Our findings broaden the in vivo function of the PINK1/Parkin pathway in mito regulation. Recent studies have highlighted the roles of PINK1 and Parkin in mitophagy. Mitophagy and nRCC mRNA regulation may represent distinct stages of mito quality control (MQC) by PINK1/Parkin. Being highly active in metabolism, mito inevitably accumulate oxidative damages to constituents such as the RCCs during their lifetime. PINK1/Parkin may help repair mildly damaged mito, for example via localized translation of nRCC mRNAs to boost OXPHOS. As for severely damaged mito, PINK1/Parkin may segregate them from healthy mito, transport them to appropriate cellular locations, and target them for removal. It will be interesting to test whether and how these different phases of MQC are connected.
One important implication of this study is that OXPHOS, a seemingly constitutive and housekeeping function, is tightly controlled at the level of nRCC mRNA localization and translation in metazoan NM tissues. It is likely that this process is dynamically regulated under stress, aging, or disease conditions. Another important implication of this study is that defective translational control plays broader and multifaceted roles in neurodegenerative disease. Previous studies implicated excessive protein synthesis in the pathogenesis of dominantly inherited neurodegeneration such as LRRK2-associated PD (Gehrke et al., 2010; Imai et al., 2008), whereas the current study implicates reduced synthesis of nRCC proteins in PINK1/Parkin-associated recessive PD. Whether these diseases are caused by converging pathogenic pathways, and how defects in translation, a supposedly ubiquitous cellular process, leads to cell type-selective degeneration, are important questions for future studies.
EXPERIMENTAL PROCEDURES
Drosophila Genetics
Fly culture and crosses were performed according to standard procedures and were raised at indicated temperatures. The UAS-Tom20, UAS-Tom40 and UAS-Tom70 transgenic flies were generated by BestGene Inc. Other fly stocks were obtained from VDRC or Bloomington Drosophila stock centers or from other investigators.
RT-PCR, qRT-PCR, CLIP Assays, and Purification of Mito-Associated mRNAs
For RT-PCR analysis, total RNAs were extracted from cell cultures, fly tissues, or adult mouse midbrain (P27) using an RNeasy® Mini kit, and one-step RT-PCR was performed using an RT-PCR kit. For qRT-PCR analysis, purified RNA were mixed with primers and Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) and PCR products detected by StepOnePlus real-time PCR system (Applied Biosystems). CLIP assay was performed under conditions modified from a published method (Ule et al., 2005). Intact mito from in vitro cell cultures, fly heads, fly thoraces, fly intestinal, and mouse midbrain (P7) tissues were purified at described previously (Kristian et al., 2006) and mito-associated mRNAs were extracted as described above.
Immunohistochemistry, RNA FISH, and TEM analysis
Immunohistochemical analyses of mito morphology of adult fly brains and muscle tissues were essentially done as described (Wu et al., 2013). For immunohistochemical analysis of mitochondrial morphology of adult fly brains and muscle tissues, male flies at around 5 days of age and raised at 29 degree were used. For analysis of DA neuron number in the various genetic backgrounds, male flies at around 14 days of age and raised at 29 degree were used. For RNA FISH in fly muscle, a protocol modified from a previously published method was used. Digoxigenin-labeled riboprobes for C -I 30, Oscp and Marf were generated using a PCR -based method. For TEM analysis, adult male flies at around 5 days of age and raised at 29°C were used. Samples were further sectioned and prepared for TEM analysis by the Cell Sciences Imaging Facility of Stanford University School of Medicine.
DNA Cloning, IP, RNP purification, m7-GTP Sepharose Affinity Chromatography, Western Blotting, RNAi, and Translational Reporter Assays
DNA cloning, IP, Western blotting, RNAi, and luciferase-based translational reporter assays were performed essentially as described (Gehrke et al., 2010). For RNP purification, cells were transfected with MS2-GST, C-I 30-MS2-bs, and PINK1 constructs. Sixty hours post-transfection, cells were UV-crosslinked, mito purified, and mito extracts subjected to GST-pull down and WB analyses. For IP and m7-GTP sepharose affinity chromatography, extracts made from HEK293T cells, fly tissues prepared from 5 day-old flies raised at 29 degree, or mouse midbrain (P7) tissue were prepared in lysis buffer [50 mM Tris-HCl, pH7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 0.5 mM DTT, 60 mM β-glycerolphosphate, 1 mM sodium vanadate, 20 mM NaF, and Complete protease inhibitor cocktail (Roche)] and subjected to IP using the indicated antibodies, or pull-down using m7-GTP sepharose beads at 4°C for 4 hours. For WB analysis of fly samples, male flies at around 5days of age and raised at 29 degree were used.
iDN Culture
Fibroblasts were seeded at 50% confluence onto matrigel-coated wells and infected with concentrated lentiviral particles expressing human TFs. Lentiviral particles were mixed with MEF media containing 8 μg/ml polybrene. 24 hours post-infection the medium was replaced with MEF medium containing 2 μg/ml doxycycline to activate TF expression. Medium was gradually changed to N3 medium (DMEM/F-12 medium containing 25 μg/ml insulin, 50 μg/ml transferrin, 30 nM sodium selenite, 20 nM progesterone, 100 nM putrescine, 2 μg/ml doxycycline) and cells were cultured for 2–3 weeks before immunohistochemical and biochemical analyses.
Behavioral Assays and ATP Measurement
Wing posture, flight and jumping ability assays on adult flies, and measurements of ATP contents in thoracic muscle were performed as described previously (Wu et al., 2013). For all wing posture assays, male flies at 1-day and 13~15-day old and aged in 29°C were used, except for the Mhc-Gal4>Pum-m OE flies, which were assayed at day 1 and day 7, as noted in Figure 4A. For the jumping and flight ability assays, male flies at 3–5 days of age raised at 29°C were used. For each genotype at least three independent experiments were performed for these assays. For ATP measurement in mammalian cells, in vitro culture d cells trypsinized from 3.7 cm 2 culture plates were collected and transferred into a new 1.5 ml reaction tube, centrifuged at 1,000 g at 4°C, and washed once with ice cold 1x PBS before ATP measurement.
Statistical Analysis
Statistical significance of all data were evaluated by unpaired Student’s t-tests. Error bars represent standard error of the mean (SEM).
See Extended Experimental Procedures for additional details.
Supplementary Material
Acknowledgments
We thank Drs. J Lykke-Andersen, B Cullen, M Gorospe, C-Y Huang, Y Imai, J Keene, J Kimble, and Open Biosystems for plasmids, Drs. H Bellen, J Chung, Y-N Jan, M Guo, W Saxton, K Zinsmaier, P Verstreken, A Whitworth, the VDRC, and the Bloomington Drosophila Stock Centers for fly stocks, the Cell Sciences Imaging Facility of Stanford University for TEM analysis, and Bestgene Inc. for help with making transgenic flies. Special thanks go to S Liu, J Gaunce and G Silverio for technical supports, and members of the Lu lab for discussions. Supported by NIH NS084412, NS083417 (Lu), NS082938(Guo), CIRM-RS1-00215-1 (Guo), and Fellowships from Stanford University School of Medicine and PDF (Z W).
Footnotes
AUTHOR CONTRIBUTIONS
SG and ZW designed the study, performed the experiments, analysed data, wrote the manuscript, and contributed equally. MK, YS, and GA provided key reagents for the iDN study. S. Guo participated in the design of the iDN study and provided key reagents. BL conceived and supervised the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews S, Snowflack DR, Clark IE, Gavis ER. Multiple mechanisms collaborate to repress nanos translation in the Drosophila ovary and embryo. RNA. 2011;17:967–977. doi: 10.1261/rna.2478611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol Cell Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat Clinic Pract Neurol. 2008;4:600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- Hoepken HH, Gispert S, Morales B, Wingerter O, Del Turco D, Mulsch A, Nussbaum RL, Muller K, Drose S, Brandt U, et al. Mitochondrial dysfunction, peroxidation damage and changes in glutathione metabolism in PARK6. Neurobiol Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian T, Hopkins IB, McKenna MC, Fiskum G. Isolation of mitochondria with high respiratory control from primary cultures of neurons and astrocytes using nitrogen cavitation. J Neurosci Methods. 2006;152:136–143. doi: 10.1016/j.jneumeth.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al. Parkinson’s Disease-Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Acin-Perez R, Geghman KD, Manfredi G, Lu B, Li C. Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc Natl Acad Sci U S A. 2011;108:12920–12924. doi: 10.1073/pnas.1107332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Gehrke S, Wu Z. RNA metabolism in the pathogenesis of Parkinsons disease. Brain Res. 2014 doi: 10.1016/j.brainres.2014.03.003. pii: S0006-8993(14)00329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin TW, Fox TD. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem. 1993;268:11737–11741. [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125:2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Complex I: inhibitors, inhibition and neurodegeneration. Exp Neurol. 2010;224:331–335. doi: 10.1016/j.expneurol.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode) generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Inoshita T, Hattori N, Imai Y. PINK1-mediated phosphorylation of Parkin boosts Parkin activity in Drosophila. PLoS Genet. 2014;10:e1004391. doi: 10.1371/journal.pgen.1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Sawada T, Shiba K, Liu S, Kanao T, Takahashi R, Hattori N, Imai Y, Lu B. Tricornered/NDR kinase signaling mediates PINK1-directed mitochondrial quality control and tissue maintenance. Genes Dev. 2013;27:157–162. doi: 10.1101/gad.203406.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.