Cytological and biochemical analyses show that Ylr072w, here renamed Lipid transfer at contact site 1 (Ltc1), is a sterol transport protein localized to both ER–mitochondria and ER–vacuole contact sites in partnership with the organelle-specific components Tom70/81 and Vac8, respectively.
Abstract
Organelle contact sites perform fundamental functions in cells, including lipid and ion homeostasis, membrane dynamics, and signaling. Using a forward proteomics approach in yeast, we identified new ER–mitochondria and ER–vacuole contacts specified by an uncharacterized protein, Ylr072w. Ylr072w is a conserved protein with GRAM and VASt domains that selectively transports sterols and is thus termed Ltc1, for Lipid transfer at contact site 1. Ltc1 localized to ER–mitochondria and ER–vacuole contacts via the mitochondrial import receptors Tom70/71 and the vacuolar protein Vac8, respectively. At mitochondria, Ltc1 was required for cell viability in the absence of Mdm34, a subunit of the ER–mitochondria encounter structure. At vacuoles, Ltc1 was required for sterol-enriched membrane domain formation in response to stress. Increasing the proportion of Ltc1 at vacuoles was sufficient to induce sterol-enriched vacuolar domains without stress. Thus, our data support a model in which Ltc1 is a sterol-dependent regulator of organelle and cellular homeostasis via its dual localization to ER–mitochondria and ER–vacuole contact sites.
Introduction
Organelle contact sites are critical for interorganellar lipid transfer, ion homeostasis, and organelle membrane dynamics. In yeast, a contact site is formed between the ER and mitochondria by the ER–mitochondria encounter structure (ERMES), composed of four subunits: outer membrane mitochondrial Mdm10 and Mdm34, cytosolic Mdm12, and integral ER Mmm1 (Kornmann et al., 2009; Stroud et al., 2011). Mdm34, Mdm12, and Mmm1 are members of the extended synaptotagmin domain family, which bind lipids (Toulmay and Prinz, 2012; Schauder et al., 2014), and consistently, ERMES has been implicated in lipid exchange between the ER and mitochondria (Kornmann et al., 2009). ERMES-marked ER–mitochondria contacts are also linked to mitochondrial division and mitochondrial DNA (mtDNA) segregation and mitophagy, suggesting they represent microdomains that coordinate cellular functions (Murley et al., 2013; Böckler and Westermann, 2014). Recently, additional mitochondrial contacts sites have been described whose functions are related to ERMES, including V-CLAMP, a contact between mitochondria and vacuoles (Elbaz-Alon et al., 2014; Hönscher et al., 2014) and the ER protein complex EMC, proposed to form an additional contact between ER and mitochondria (Lahiri et al., 2014). In the case of V-CLAMP and ERMES, these sites are coregulated. Thus, different contact sites between organelles are coordinated, but how this regulation is achieved remains unknown.
To understand the composition, mechanisms, functional scope, and modes of communication of organelle contact sites, we explored the environment of ERMES using proteomics and identified a conserved uncharacterized ER protein, Ylr072w, as an ERMES interactor, which belongs to a larger uncharacterized protein family. Based on cytological and biochemical analyses, we have renamed Ylr072w Lipid transfer at contact site 1 (Ltc1). Ltc1 localizes to both ER–mitochondria and ER–vacuole contact sites in partnership with the organelle-specific components Tom70/71 and Vac8, respectively. Our data suggest that Ltc1 functions to transport and/or sense sterols at contact sites to facilitate separate organelle-specific functions and support a model in which the dual organelle localization of Ltc1 is a means of coordinating the functions of mitochondria and vacuoles.
Results and discussion
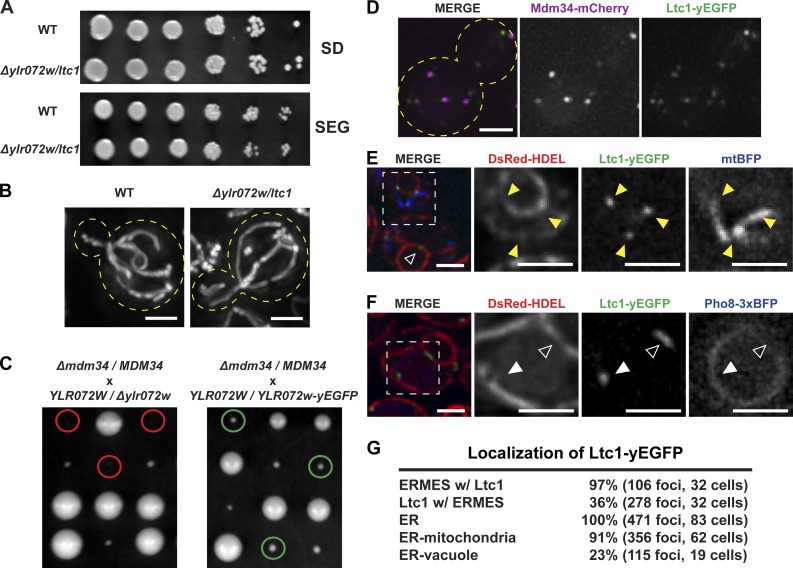
To explore the composition of ER–mitochondria contacts, we immunopurified GFP-tagged versions of ERMES subunits Mmm1, Mdm12, and Mdm34 from dithiobis succinimidyl propionate (DSP) cross-linked yeast extracts to identify interacting proteins by tandem mass spectrometry (IP-MS/MS). As shown in Table 1, proteomic analysis of ERMES subunit purifications identified multiple core ERMES complex components and the ERMES auxiliary subunit Gem1, consistent with previous observations (Kornmann et al., 2011; Stroud et al., 2011). In addition, we identified an uncharacterized protein, Ylr072w. Deletion of YLR072W from cells did not cause significant growth defects on media with fermentable (glucose) or nonfermentable (ethanol/glycerol) carbon sources (Fig. 1 A) nor defects in mitochondrial morphology (Fig. 1 B). However, deletion of YLR072W in combination with deletion of MDM34 caused a significant synthetic growth defect as compared with deletion of MDM34 alone, indicating that YLR072W has both a physical and functional relationship to ERMES (Fig. 1 C). We used this MDM34-dependent phenotype to determine that a YLR072W-yEGFP fluorescent protein fusion allele integrated at its endogenous locus was functional, as the growth of YLR072W-yEGFP Δmdm34 cells was similar to Δmdm34 cells (Fig. 1 C).
Table 1.
Affinity purification of the ERMES complex identifies an uncharacterized protein, Ylr072w
| Identified protein | Average spectral counts | Description | CRAPome score | ||
| Mdm12 | Mdm34 | Mmm1 | |||
| Mmm1 | 13 | 30 | 71 | ERMES subunit | 0.00 |
| Mdm34 | 17 | 48 | 37 | ERMES subunit | 0.00 |
| Gem1 | 12 | 35 | 34 | Miro GTPase | 0.00 |
| Ylr072w | 0 | 17.5 | 5.5 | Uncharacterized | 0.00 |
| Mdm12 | 4 | 8 | 6 | ERMES subunit | 0.00 |
| Msc7 | 0 | 9 | 8 | ER protein | 0.00 |
| Tom71 | 0 | 7.5 | 3 | Mitochondrial protein import receptor | 0.00 |
| Pbn1 | 0 | 0 | 8 | Component of GPI-mannosyltransferase I | 0.00 |
| Pho84 | 0 | 1.5 | 4.5 | Abundant inorganic phosphate transporter | 0.00 |
| Vtc1 | 0 | 1 | 2 | Vacuolar transporter complex | 0.00 |
| Spt10 | 2 | 5 | 1.5 | Histone acetylase | 0.06 |
| Mdm10 | 8 | 26 | 20.5 | ERMES subunit | 0.12 |
| Myo2 | 0 | 3 | 1 | Type V myosin; mitochondria/vacuole transport | 0.18 |
| Lsp1 | 0 | 2.5 | 0 | Eisosome protein | 0.18 |
| Por1 | 6.5 | 22.5 | 17.5 | Mitochondrial porin | 0.35 |
| Fks1 | 0 | 0.5 | 3 | β-Glucan synthase component | 0.41 |
Functional GFP fusions to the ERMES proteins Mdm12, Mdm34, and Mmm1 encoded at their endogenous chromosomal loci were purified from yeast cell lysates with GFP antibodies. Purified proteins were identified by tandem mass spectrometry with a <1% decoy false discovery rate. As a control, mock purifications were performed from an isogenic strain background that did not express GFP. The average total spectrum counts assigned to each protein from two IPs for Mdm12 and Mmm1 and four IPs for Mdm34 are tabulated. We included in this table proteins that met the following criteria: (a) they were identified in duplicate sample experiments for Mdm12/Mmm1 or 3/4 IPs for Mdm34, and (b) their average spectral counts were at least 10-fold higher than in mock purifications. Tabulated are proteins that were identified in duplicate sample experiments and whose average spectral counts were at least 10-fold higher than in mock purifications. The “CRAPome” score for identified proteins is listed at right and is a measure of how likely the identified protein is the result of a nonspecific interaction. This number is derived from the Contaminant Repository for Affinity Purifications database, CRAPome, and is the proportion of mock purifications in which the protein is identified.
Figure 1.
Ylr072w/Ltc1 is localized to ER–mitochondria and ER–vacuole contact sites. (A) Deletion of YLR072W causes no significant growth defect on fermentable or nonfermentable carbon sources. Cells were grown to mid-log phase and plated on synthetic complete media containing 2% dextrose (SD) or 3% glycerol + 2% ethanol (SEG). (B) Deletion of YLR072W causes no significant change in mitochondrial morphology. Cells expressing mitochondria-targeted DsRed were grown to mid-log phase and imaged as described in “Fluorescence microscopy.” (C) Deletion of YLR072W causes a synthetic sick/lethal phenotype in Δmdm34 cells. Expression of Ylr072w-yEGFP restores Δmdm34 growth phenotype. Yeast diploids with the indicated genotypes were sporulated, subjected to tetrad dissection, and the resulting spore colonies were genotyped based on segregation pattern of markers. Red circles indicate inviable Δmdm34 Δylr072w cells. Green circles indicate viable Δmdm34 YLR072W-yEGFP cells. (D–F) Ylr072w localization was assessed in WT cells using Ylr072w-yEGFP integrated at its endogenous locus relative to ERMES marked by Mdm34-mCherry (D), ER and mitochondria marked by DsRed-HDEL and mitochondrial-targeted mtBFP, respectively (E), and ER marked by DsRed-HDEL and vacuoles marked by Pho8-3XBFP (F). Cells were grown to mid-log phase and imaged as described in “Fluorescence microscopy.” Yellow arrowheads in E mark ER–mitochondria contact sites, open white arrowheads in E and F mark NVJs, and closed white arrowheads in F mark ER–vacuole contacts. (G) Quantification of Ltc1 foci localization from D–F. Dashed lines in B and D demarcate cell boundaries. Dashed lines in E and F indicate enlarged regions shown as separate grayscale images to the right. Bars, 2 µm.
Ylr072w localized to punctate structures in cells, of which 35–40% colocalized with ERMES foci, marked by Mdm34-mCherry, suggesting that Ylr072w foci localize to ER–mitochondria contact sites (Fig. 1, D and G). However, Ylr072w foci formation was independent of ERMES, and we also observed ERMES foci formation in the absence of Ylr072w (Fig. S1, A and B). We directly examined whether Ylr072w marks ER–mitochondria contact sites by examining the localization of Ylr072w in cells relative to ER and mitochondria, using ER-targeted DsRed (DsRed-HDEL) and mitochondrial matrix–targeted BFP (mtBFP), respectively. We observed that a vast majority of Ylr072w foci (80–90%) were present at ER–mitochondria contact sites (Fig. 1, E [yellow arrowheads] and G). However, we also observed Ylr072w foci that were not localized near mitochondria (Fig. 1 E, open arrowhead). The localization of a proportion of nonmitochondrial Ylr072w foci to the nuclear envelope prompted us to examine their localization relative to nucleus–vacuole junctions (NVJs; Pan et al., 2000; Roberts et al., 2003; Millen et al., 2008). Relative to the ER and vacuoles, we observed that Ylr072w localized to both NVJs (Fig. 1 F, open arrowheads; and Fig. S1 C, open arrowheads) and other non-NVJ ER–vacuole contact sites (Fig. 1 F, closed arrowheads; and Fig. S1 C, closed arrowheads). Quantification of Ylr072w localization at ER–mitochondria contacts and ER–vacuole contacts yielded a proportion >100% (Fig. 1 G), which is likely the result of error introduced by the resolution limitation of fluorescence microscopy. In total, our data indicate that Ylr072w localizes to both ER–mitochondria and ER–vacuole contacts.
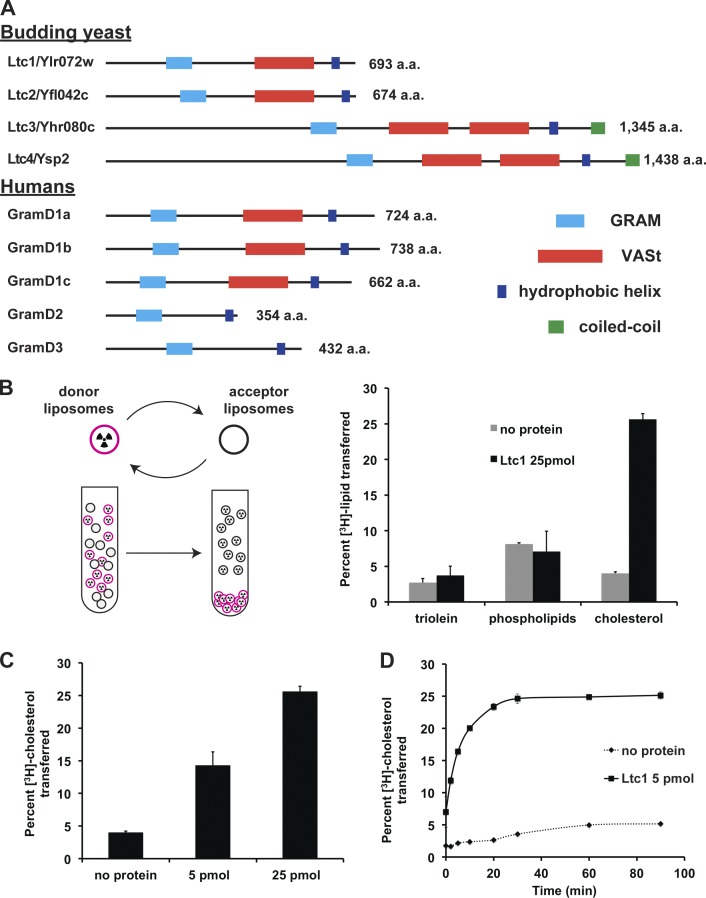
Phylogenetic analysis and structure prediction programs indicate that Ylr072w contains two predicted lipid-binding domains and is a member of a larger family with multiple homologues present in nearly all eukaryotes (Fig. 2 A). Both Phyre2 (Kelley and Sternberg, 2009) and iTasser (Yang et al., 2015) reveal a conserved N- to C-terminal domain architecture within the family: an unstructured N terminus, a GRAM domain structurally similar to pleckstrin homology (PH) domains (Begley et al., 2003), a previously unannotated StART-like domain also called a VASt domain (Khafif et al., 2014); and a C-terminal hydrophobic α helix. The VASt domain in this family shares no sequence similarity to StART domains, but is predicted to form a similar hydrophobic pocket that accommodates a lipid molecule (Khafif et al., 2014). We also include vertebrate GramD2 and GramD3 proteins in this family as phylogenetic analyses indicate they are derived from a common ancestor with GramD1a-c, suggesting that they arose through VASt domain loss. Based on these structural predictions and our observations, we rename YLR072W LTC1 for Lipid Transfer at Contact site 1 and the other members of the yeast family LTC2 (YFL042C), LTC3 (YHR080C), and LTC4 (YSP2/YDR326C). Although LTC1 is paralogous to LTC2, our physiological data suggest that it possesses nonredundant functions (Fig. 1 and see Fig. 4).
Figure 2.
Ylr072w/Ltc1 is a member of a conserved protein family and catalyzes sterol transport between membranes. (A) Ltc1 family members in budding yeast and humans, also identified by Khafif et al. (2014). Members were identified by BLAST searches with the amino acid sequence of Ylr072w/Ltc1. Proteins with significant sequence similarity were queried for shared predicted secondary structure and domains with Phyre2 and i-TASSER (Kelley and Sternberg, 2009; Yang et al., 2015). In yeast, the paralogues Ltc1/Ltc2(Yfl042c) and Ltc3(Yhr080c)/Ltc4(Ysp2) arose from a genome duplication event. (B–D) Ltc1 selectively transports sterol between membranes (B; n = 4 for cholesterol and n = 2 for triolein and phospholipids) in a concentration (C; n = 4)- and time (D; with protein n = 3; control without protein n = 1)-dependent manner and were performed as depicted in the schematic in B and as described in Materials and methods. Errors bars represent standard deviation.
Figure 4.
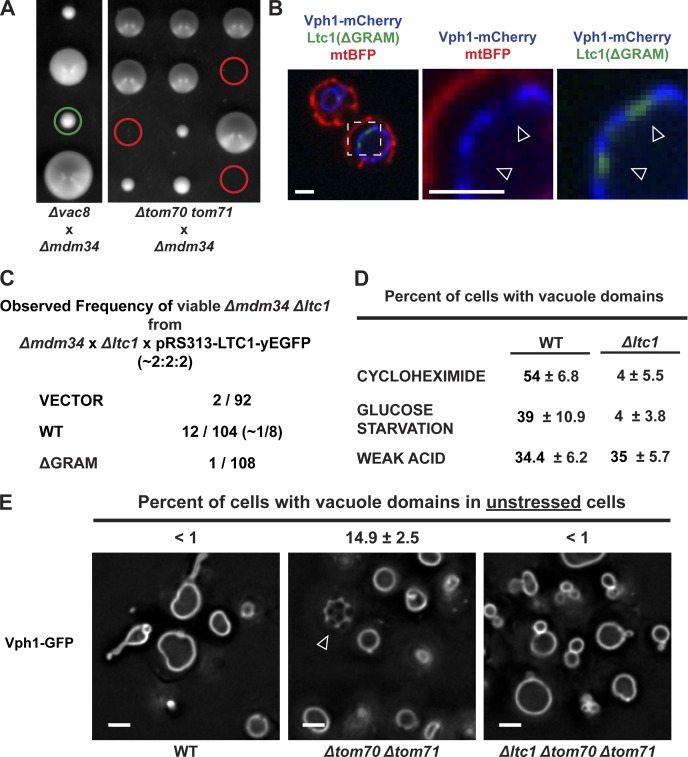
The distribution of Ltc1 to ER–mitochondria and ER–vacuolar contacts regulates Ltc1-dependent separable mitochondrial and vacuolar functions. (A) The localization of Ltc1 to ER–mitochondria, but not to ER–vacuole, contacts is essential in Δmdm34 cells. Diploid yeast heterozygous for the indicated deletions were sporulated and analyzed by tetrad dissection as described in “Fluorescence microscopy.” The green circle indicates viable Δvac8 Δmdm34 cells (the colonies immediately above and below are Δvac8 and WT, respectively). Red circles indicate nonviable Δtom70 Δtom71 Δmdm34 cells. (B) The Ltc1 GRAM domain is required for the mitochondrial localization of Ltc1. Cells expressing Ltc1(ΔGRAM)-yEGFP, Vph1-yEmCherry (vacuoles), and mtBFP (mitochondria) were grown to mid-log phase and imaged as described in “Fluorescence microscopy.” Open arrowheads indicate the localization of Ltc1 to Vph1-depleted regions on vacuoles. Dashed lines denote the enlarged region shown as two-color images to the right of the merged, three-color image. (C) The mitochondrial localization of Ltc1 is essential in Δmdm34 mutants. Diploid yeast heterozygous for Δmdm34 and Δltc1 and harboring a yeast centromeric plasmid pRS313 containing Ltc1-yEGFP or Ltc1(ΔGRAM) were sporulated and analyzed by tetrad dissection as described in “Fluorescence microscopy.” The expected frequency of viable Δmdm34 Δltc1 mutants for pRS313 plasmids containing a WT Ltc1 is 1/8 spore colonies. The observed frequency for WT Ltc1 was 12/104 (∼1/8), whereas the observed frequency for Ltc1(ΔGRAM) and an empty vector control were 1/108 and 2/92, respectively. (D) Ltc1 is required for vacuole membrane domain formation in response to cycloheximide and glucose starvation. WT and Δltc1 cells expressing Vph1-yEGFP were grown exponentially in synthetic complete media for at least 12 generations and then subjected to the indicated treatments and imaged as described in “Fluorescence microscopy.” Quantification represents triplicate biologically independent experiments. (E) Shifting the proportional localization of Ltc1 to ER–vacuole contacts is sufficient to induce vacuole membrane domain formation under normal growing conditions. WT, Δtom70 Δtom71, and Δltc1 Δtom70 Δtom71 cells were grown for at least 12 generations in nutrient-replete media, and domain formation was assessed using Vph1-yEGFP as described in “Fluorescence microscopy.” Quantification represents triplicate biologically independent experiments. The arrowhead indicates a vacuole with Vph1-GFP–labeled domains. Bars, 2 µm.
To characterize Ltc1 molecular features, we expressed and purified the predicted soluble domain of Ltc1 (amino acids 1–590, 6xHis-Ltc1(ΔTM)) from Escherichia coli. Size exclusion chromatography coupled with multiangle light scattering indicated that recombinant Ltc1 is a monomer (73,690 ± 0.283% D). To test whether Ltc1 facilitates lipid transfer, we used an established in vitro lipid transfer assay (Schulz et al., 2009), in which dense sucrose-loaded donor liposomes containing one of three different types of radiolabeled lipids, phospholipids, triolein (a triglyceride), or cholesterol, were incubated with lighter, unlabeled acceptor liposomes in the presence and absence of Ltc1(ΔTM) and subsequently separated by centrifugation (Fig. 2 B). Radioactive cholesterol, but not triolein or major classes of phospholipids, was transferred to acceptor liposomes in an Ltc1-dependent manner (Fig. 2 B), and transport efficiency was dependent on Ltc1(ΔTM) concentration and time (Fig. 2, C and D). Thus, Ltc1 selectively transports sterols between membranes in vitro, suggesting that it functions as a sterol transfer protein and/or sensor in vivo.
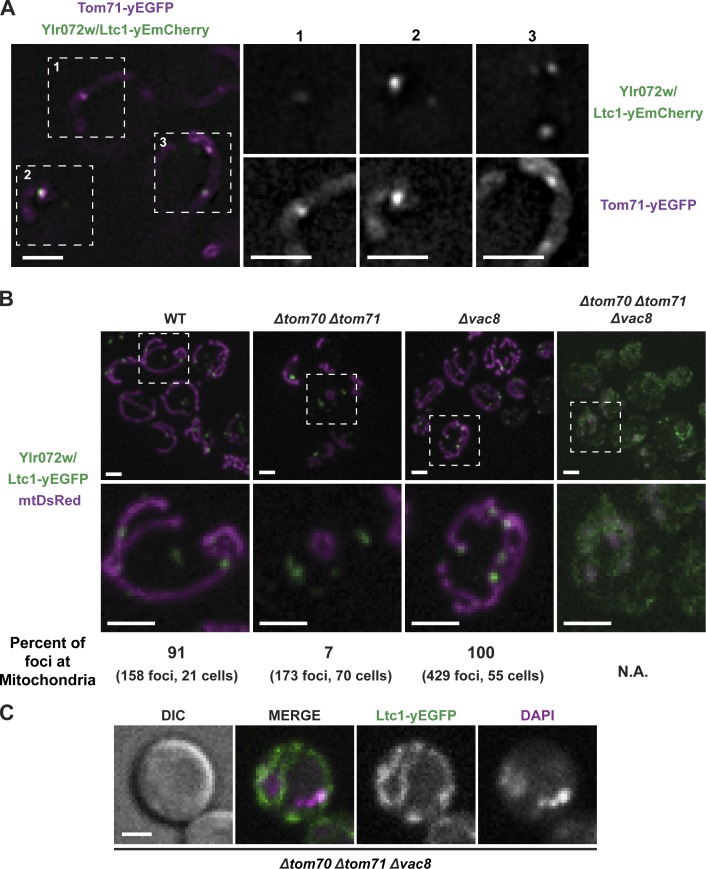
To examine the molecular basis for Ltc1’s dual localization, we immunopurified Ltc1-yEGFP from DSP cross-linked yeast whole cell lysates using anti-GFP antibodies. Consistent with our cytological and proteomic data (Fig. 1 and Table 1), we identified the ERMES subunit Mdm34 and the ERMES-associated protein Gem1 (Table 2, wild type [WT]). Based on the number of spectral counts, the most abundant proteins identified in Ltc1 purifications were the paralogous mitochondrial preprotein import receptors, Tom70/71 (Table 2, WT; Schlossmann et al., 1996; Schmidt et al., 2010), also identified in the ERMES complex subunits purifications (Table 1). Localization of a functional Tom71-yEGFP fusion relative to Ltc1 in cells revealed that, in addition to a uniform labeling of the mitochondrial outer membrane, Tom71 was present in focal structures that colocalized with Ltc1 (Fig. 3 A and Fig. S1 A). We tested whether Tom70/71 is required for Ltc1’s localization to mitochondria. In Δtom70 Δtom71 cells, Ltc1 was localized to foci, but in contrast to WT cells, a vast majority of these were not localized to mitochondria (Fig. 3 B) and instead were at ER–vacuole contacts (Fig. S1 C). Tom70/71 also functions redundantly to target the F-box protein Mfb1 to mitochondria (Kondo-Okamoto et al., 2008). Although not detected in our Ltc1 purifications, we tested whether localization of Ltc1 to mitochondria was Mfb1 dependent. In contrast to deletion of TOM70 and TOM71, deletion of MFB1 did not alter the localization pattern of Ltc1 relative to WT (Fig. S1 D). Thus, these data indicate a major and perhaps direct role of Tom70/71 in the localization of Ltc1 to ER–mitochondria contact sites.
Table 2.
Ylr072w/Ltc1 interacts with proteins on mitochondria and vacuoles
| Identified protein | Average spectral counts | Description | CRAPome score | |
| WT | Δtom70 Δtom71 | |||
| Ylr072w/Ltc1 | 280.5 | 242.5 | – | 0.00 |
| Tom71 | 53 | 0 | Mitochondrial protein import receptor | 0.00 |
| Tom70 | 13 | 0 | Mitochondrial protein import receptor | 0.00 |
| Vac8 | 11.5 | 22.5 | Multifunctional vacuolar protein | 0.00 |
| Gem1 | 10.5 | 0 | Miro GTPase | 0.00 |
| Mdm34 | 6 | 0 | ERMES subunit | 0.00 |
| Leu9 | 3 | 1 | Leucine biosynthesis; mitochondrial | 0.00 |
| Npr1 | 2 | 7 | Protein kinase; substrate of TORC1 | 0.00 |
| Erj5 | 2 | 1 | ER-localized J protein | 0.00 |
| Ybt1 | 1.5 | 4 | Vacuole-localized ABC transporter | 0.00 |
| Mam33 | 0 | 12.5 | Mitochondrial matrix acidic protein | 0.00 |
| Pdr10 | 0 | 5.5 | ABC transporter | 0.00 |
| Erg6 | 6 | 7 | Ergosterol biosynthesis | 0.11 |
| Kog1 | 1.5 | 5.5 | Subunit of TORC1 | 0.11 |
| Mnp1 | 0 | 7.5 | Mitochondrial nucleoid protein | 0.17 |
| Vph1 | 4.5 | 14 | Vacuolar ATPase subunit | 0.22 |
Ylr072w/Ltc1-yEGFP was purified from yeast WT or Δtom70 Δtom71 cell lysates, and proteins were identified by tandem mass spectrometry with a <1% false discovery rate. Control mock purifications were performed from isogenic strain backgrounds that did not express yEGFP. The average total spectral counts assigned to each protein from two independent experiments are tabulated. Tabulated are proteins that were identified in duplicate sample experiments and whose average spectral counts were at least 10-fold higher than in mock purifications. The “CRAPome” score for identified proteins is listed at right and was measured as in Table 1.
Figure 3.
Localization of Ltc1 to ER–mitochondria and ER–vacuole contact sites requires Tom70/71 and Vac8, respectively. (A) Ltc1, marked by Ltc1-yEmCherry, colocalizes with Tom71, marked by Tom71-yEGFP, in foci on the mitochondrial outer membrane. Cells expressing Ltc1-yEmCherry and Tom71-yEGFP were grown to mid-log phase and imaged as described in “Fluorescence microscopy.” Dashed lines and numbers denote enlarged regions shown as separate grayscale images of each channel to the right of the merged image. (B) Vac8 and Tom70/71 are required for Ltc1 foci localization to vacuoles and mitochondria, respectively. Cells of the indicated genotypes expressing Ltc1-yEGFP and mtDsRed were grown to mid-log phase and imaged as described in “Fluorescence microscopy.” Images represent a maximum intensity z-projection. Dashed lines denote enlarged regions shown below. (C) Ltc1 is an ER membrane–associated protein. Δtom70 Δtom71 Δvac8 cells expressing Ltc1-yEGFP were grown to mid-log phase and treated with 1 µg/ml DAPI for 20 min to label the nucleus, washed twice in SD media, and imaged as described in “Fluorescence microscopy.” Extranuclear DAPI-stained structures are likely mtDNA. Bars, 2 µm.
Additional proteins were identified in Ltc1 purifications, including the vacuolar protein Vac8, which based on total spectral counts was the most abundant vacuolar protein present (Table 2, WT). Vac8 has diverse functions in autophagy, vacuolar transport, and in the formation of NVJs (Wang et al., 1998; Veit et al., 2001; Subramanian et al., 2006; Tang et al., 2006). Of note is that Tom70/71 and Vac8 have TPR repeats and Armadillo repeats, respectively, which form protein–protein interaction platforms (Wang et al., 1998; Wu and Sha, 2006; Grove et al., 2008; Li et al., 2009), suggesting that similar to Tom70/71 for mitochondria, Vac8 may function to localize Ltc1 to vacuoles. Consistently, in Δvac8 cells, Ltc1 localized to foci exclusively at ER–mitochondria contact sites (Fig. 3 B and Fig. S1 E). Vac8 is required for the formation of NVJs through a direct interaction with the outer nuclear envelope protein Nvj1 (Pan et al., 2000). Thus, we tested whether the Ltc1 localization to ER–vacuole contacts was dependent on NVJ formation. We observed that Ltc1-yEGFP localization was unaffected in Δnvj1 cells (Fig. S1 E), indicating that Ltc1-dependent ER–vacuolar contact is distinct from Nvj1-dependent NVJ contact sites. In cells harboring deletions in TOM70/71 and VAC8, Ltc1 foci were absent, and instead, Ltc1 was localized diffusely throughout the ER (Fig. 3 C), consistent with Ltc1 being an ER membrane–associated protein. In contrast, Ltc1 formed foci localized to vacuoles in Δnvj1 Δtom70 Δtom71 cells, similar to its localization in Δtom70 Δtom71 cells (Fig. S1 F). Together our data indicate that Tom70/71 and Vac8 are partners of Ltc1 in the formation of ER–mitochondria and ER–vacuolar contact sites/tethers, respectively, and suggest that they may regulate the relative distribution of Ltc1 to these distinct organelle contacts.
To test whether the dual localization of Ltc1 is a means for organelle communication, we first asked whether Ltc1 has distinct functions at mitochondria and vacuoles. Consistent with this, we found that deletion of TOM70 and TOM71, but not VAC8, produced a synthetic lethal/sick phenotype in combination with deletion of MDM34 (Fig. 4 A). However, given that Tom70 and Tom71 function in mitochondrial protein import, it is possible that the synthetic lethality/sickness observed in Δtom70 Δtom71 Δmdm34 cells is not a specific consequence of defects in Ltc1 localization to mitochondria. Thus, we searched for cis-regulators of Ltc1 localization to interrogate Ltc1 organelle-specific functions.
Ltc1 contains a GRAM domain that in other contexts controls the cellular localization of proteins via protein and/or phosphoinositide lipid interactions (Doerks et al., 2000; Begley et al., 2003). We asked whether the GRAM domain of Ltc1 determines its localization to ER–mitochondria and/or ER–vacuolar contacts. Ltc1(ΔGRAM) (deletion of amino acids 145–360) localized exclusively to ER–vacuole contact sites, a pattern similar to that observed in Δtom70 Δtom71 cells (Fig. 4 B). Thus, the GRAM domain is an essential determinant of Ltc1’s localization to ER–mitochondria contacts, although we do not presently understand the molecular basis for GRAM-dependent Ltc1 mitochondrial localization. Transformed plasmids containing full-length Ltc1-yEGFP, but not Ltc1(ΔGRAM)-yEGFP or an empty vector, rescued the synthetic lethal/sick interaction phenotype of Δltc1 Δmdm34 cells (Fig. 4 C). Together, these observations indicate that Ltc1-dependent functions at ER–mitochondria and ER–vacuole contact sites are nonredundant and separable.
In response to glucose starvation, cycloheximide treatment, or weak acid stress, the vacuole membrane rearranges into ergosterol-enriched (marked by Ivy1) and ergosterol-depleted (marked by Vph1) lipid domains (Toulmay and Prinz, 2013). Given that Ltc1 possesses sterol transport activity in vitro, we examined whether it is required for ergosterol-dependent vacuolar lipid domain formation in cells. Vacuolar domain formation was severely defective in Δltc1 cells after glucose starvation and cycloheximide treatment, but was unaffected in response to weak acid stress as compared with WT cells (Fig. 4 D). These observations are consistent with the in vitro ergosterol transport activity of Ltc1 and suggest that Ltc1 functions at ER–vacuole contacts in the stress-dependent reorganization of vacuolar membranes. These results also support the idea that different stresses mediate vacuolar domain formation through different mechanisms (Toulmay and Prinz, 2013).
We asked whether the relative localization of Ltc1 to ER–mitochondria and ER–vacuole contacts alters organelle-specific Ltc1 functions. Specifically, we increased the proportion of Ltc1 localized to the ER–vacuole contact sites and examined vacuolar domain formation in unstressed cells (Fig. 4, B and E; and Fig. S2). We accomplished this by deleting TOM70 and TOM71 or by replacing Ltc1 with Ltc1-ΔGRAM and assessed vacuolar domain formation based on patterned Vph1 localization in cells after exponential growth for over a dozen generations in nutrient-replete media. Under these conditions, vacuolar domains were readily observed in Δtom70 Δtom71 and Ltc1-ΔGRAM Δltc1 cells, but not in WT or Δltc1 Δtom70 Δtom71 cells (Fig. 4, B and E; and Fig. S2). These observations demonstrate that Ltc1 functions specifically at the ER–vacuole contact to control vacuole membrane domain formation and that shifting the localization of Ltc1 to ER–vacuole contact sites is sufficient to induce vacuolar membrane domain formation under normal growing conditions. In addition, we observed that Ltc1-ΔGRAM localization in Δltc1 cells was restricted to ergosterol-enriched domains (Fig. 4 B, open arrowheads; and Fig. S2). The exclusive localization of Ltc1 to ergosterol regions in the absence of its GRAM domain suggests that the VASt domain harbors the ergosterol binding/transport activity of Ltc1.
Our data show that ER membrane–associated Ltc1 is capable of transporting sterols and forming ER–mitochondria and ER–vacuole membrane contacts by partnering with the organelle-specific components Tom70/71 and Vac8. We envision that at these organelle contacts, Ltc1 acts as dynamic organizer of local membrane lipid composition rather than, or in addition to, mediating bulk sterol transfer in cells. Regulation of local lipid composition by Ltc1 may promote the formation of specialized domains, an extreme example of which is the stress-induced microscale vacuolar domains in yeast. Ltc1-induced lipid domains could act cooperatively to regulate the localization of proteins that, together with a selective lipid milieu, would act as regulators of organelle biogenesis and signaling.
Although the physiological roles of vacuole membrane domains is poorly understood (Toulmay and Prinz, 2013; Wang et al., 2014), our proteomic data suggest that the Target-Of-Rapamycin Complex 1 (TORC1) subunit Kog1 as well as the downstream TORC1 target Npr1 are in close proximity to Ltc1 at the vacuole (Table 2). Thus, one possible function of Ltc1-dependent domain formation may be in TORC1 regulation. Consistent with this possibility, Gtr2, a Rag GTPase component of the EGO (Exit from G0) complex, a conserved upstream regulator of TORC1 (Dubouloz et al., 2005; Bonfils et al., 2012), is also localized with Ltc1 and Ivy1, a protein with a putative BAR-like domain, in the ergosterol-enriched vacuolar domain (Toulmay and Prinz, 2013). Domain formation at the vacuole may thus function similar to TORC2 regulation at the plasma membrane, where the BAR domain activators Slm1/2 are dynamically partitioned between the eisosome and TORC2 membrane domain compartments to regulate TORC2 activity in response to stress (Olivera-Couto et al., 2011; Berchtold et al., 2012; Niles et al., 2012). The exact molecular role of Vac8 in Ltc1-mediated vacuolar domain formation as well as whether Ltc1 affects and/or coordinates Vac8’s known functions in NVJ formation, vacuolar motility, and autophagy (Wang et al., 1998; Tang et al., 2006) remain to be determined.
Our data suggest Ltc1 possesses a separate lipid-linked function at mitochondria as the simultaneous loss of Ltc1 and ERMES functions produces a synthetic lethal/sick growth phenotype. Loss of ERMES subunits alters the levels of mitochondrial lipids relative to WT, including ergosterol (Tan et al., 2013). Thus, we speculate that Ltc1 function at mitochondria, similar to its function at vacuoles, is related to sterols. However, although sterols are important for the maintenance of mitochondrial morphology in yeast, their exact functions remain poorly described (Altmann and Westermann, 2005). In mammals, however, the nonvesicular transport of sterols from ER to mitochondria has been specialized for cell type–specific steroid hormone production. A mammalian homologue of Ltc1, GramD1b (Fig. 2 A), is highly expressed in steroidogenic Leydig cells (McDowell et al., 2012) and in the adrenal glands (Rosenbloom et al., 2015), suggesting that it may function in this capacity.
Our observations raise the possibility that Ltc1 via Tom70/71 might play an important regulatory role in mitochondrial biogenesis specific to Tom70/71 clients, which are the metabolite carrier proteins of the inner mitochondrial membrane (Hines et al., 1990; Söllner et al., 1990; Steger et al., 1990; Schlossmann et al., 1996). Cytosolic kinases, such as the nutrient-sensing kinase PKA, regulate Tom70/71 through phosphorylation (Schmidt et al., 2011), which may be a means of regulating Ltc1–Tom70/71 interactions in addition to regulating protein import. The exact function of Ltc1 at mitochondria and whether the canonical function of Tom70/71 in protein import is integrated with Ltc1 remain outstanding questions.
Although Ltc1 functions at mitochondria and vacuoles are separate, we show that increasing the proportion of Ltc1 at ER–vacuole contacts is sufficient to induce ergosterol-enriched vacuolar domains under normal growth conditions. Thus, we propose that the dual localization of Ltc1 to mitochondria and vacuolar contacts serves as a unique mode of interorganellar communication. Although the physiological pathways that regulate the relative distribution of Ltc1 at these distinct contacts are not known, it is likely that such coordination will be important for nutrient sensing and signaling (Hönscher et al., 2014) and cellular replicative aging (Hughes and Gottschling, 2012), which both involve extensive cross-talk between mitochondria and the vacuole.
Materials and methods
Plasmid and strain construction
All Saccharomyces cerevisiae strains constructed in this study are based on the W303 (ade2–1; leu2–3; his3–11, 15; trp1–1; ura3–1; can1–100 [Rothstein, 1983]) genetic background and are listed in Table S1. Strains were constructed by mating and/or by direct transformation using the lithium acetate method. Correct targeting of chromosomal integrations was confirmed by PCR. Fluorescent protein fusion alleles were derived from plasmids described in Sheff and Thorn (2004; yEGFP and mCherry) and derivatives described in Lackner et al. (2013; yEmCherry).
To generate pRS313::LTC1-yEGFP, genomic DNA from the W303 LTC1-yEGFP yeast strain was used to amplify a PCR product containing LTC1-yEGFP plus its native promoter (250 bp 5′ of start codon) and the entire ADH1 terminator used in the yEGFP::HIS cassette, which was then cloned into linearized pRS313 (Sikorski and Hieter, 1989) using a Gibson isothermal assembly reagent (New England Biolabs, Inc.) according to the manufacturer’s instructions. This was then used to generate pRS313::LTC1(ΔGRAM)-yEGFP by isothermal assembly of PCR products that when recombined would remove the nucleotides encoding amino acids 145–360 of Ltc1. To generate 6xHis-tagged Ltc1(ΔTM) for expression in E. coli, the DNA encoding amino acids 1–590 of the protein was amplified from yeast genomic DNA and cloned directionally into pET15b using 5′-XhoI/3′-BamHI sites. pVT100U-mtTagBFP was constructed by synthesizing yeast codon–optimized TagBFP de novo and replacing GFP in pVT100U-mtGFP (Westermann and Neupert, 2000) with 5′-KpnI/3′-XhoI sites.
pYES-mitoTagBFP, which drives galactose-inducible expression of mitoTagBFP from a GAL1 promoter (Murley et al., 2013); pYX142-mtDsRed, a CEN/ARS plasmid driving mitochondrial matrix–targeted DsRed (mtDsRed) expression from a TDI promoter (Westermann and Neupert, 2000); pKW1803 (DsRed-HDEL), which expresses a fusion protein containing the Kar2 signal sequence, DsRed, and an HDEL retention sequence (Madrid et al., 2006); and pRS305 3xBFP-Pho8, an integrating plasmid driving expression of 3xBFP-Pho8 from a PGK1 promoter (Graef et al., 2013), are all previously described in more detail.
Fluorescence microscopy
Cells were grown to log phase (OD600 ∼0.6–1.0) at 30°C in the appropriate synthetic medium to select for plasmids, concentrated by centrifugation, deposited directly on a glass slide, and then sealed under a #1.5 coverslip using nail polish. Cells in Fig. 1 E were grown in synthetic medium containing 2% galactose to drive expression of mtTagBFP from pYES-mtTagBFP; otherwise, cells were grown in synthetic medium with 2% glucose. Cells were imaged at 25°C.
Cells in Fig. 1 (B and D–F), Fig. S1 (A and C), Fig. 3 A, and Fig. 4 E were imaged on a DeltaVision Real-Time microscope (IX70 DeltaVision; Olympus) using a 60× 1.40 NA objective lens (Olympus) and a 100-W mercury lamp (Applied Precision). Light microscopy images were collected using an integrated, cooled charge-coupled device (CCD)–based camera (CoolSNAP HQ; Photometrics) equipped with an Interline Chip (Sony). Datasets were processed using softWoRx’s (Applied Precision) iterative, constrained three-dimensional deconvolution method to remove out-of-focus light.
Cells depicted or analyzed in Fig. 3 B, Fig. 4 B, Fig. S1 (B and D–F), and Fig. S2 were imaged using the spinning disc module of a Marianas SDC Real-Time 3D Confocal-TIRF microscope (Intelligent Imaging Innovations, 3i) fit with a Yokogawa spinning disk head, a 63× 1.40 NA (Olympus; Fig. S2) or 100× 1.46 NA objective (Olympus; Fig. 3 B, Fig. 4 B, Fig. S1 [B, D, and E], and Fig. S2), and EMCCD camera.
For stress treatments in Fig. 4 D, cells were grown for >12 generations to mid-log phase (to ensure no residual vacuole domains from cells in the colony) in synthetic complete (SC) media containing dextrose as a carbon source (SD) and harvested by centrifugation. They were then resuspended in an equal volume of SD media containing 60 mM sodium acetate, pH 5.2, 50 µg/ml cycloheximide, or SC media without glucose. For glucose starvation, cells were washed three times with water before resuspending them in an equal volume of SC-glucose. All treatments lasted 3 h. Cells were imaged on the DeltaVision wide-field deconvolution microscope described above. Experiments were performed in independent biological replicates.
Images were manipulated in Photoshop (Adobe). In some images found in this paper, linear adjustments were made to brightness and contrast.
Immunopurification and tandem mass spectrometry
Genes encoding ERMES subunits (MDM12, MDM34, and MMM1) were tagged at their C terminus with GFP at their native chromosomal locus. GFP fusion proteins were immunopurified from whole-cell lysates as previously described (Lackner et al., 2013). Cells were grown overnight to log phase (OD600 = 1) in YPD medium (2% glucose, 2% peptone, and 1% yeast extract supplemented with adenine and tryptophan), harvested by centrifugation, washed once with distilled water, and resuspended 1:1 with IPLB buffer (20 mM Hepes, pH 7.4, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 0.6 M sorbitol) containing protease inhibitor cocktail Set 1 (PIC; EMD Millipore). Cell suspensions were flash-frozen dropwise in liquid nitrogen. Frozen cells were lysed in a freezer mill (Spex 6970 EFM) and using three lysing periods of 2 min at speed “7” with 2 min of chilling between each. Lysates were stored at −80°C. Frozen cell lysates were thawed in a room temperature water bath with additional PIC, then clarified of unlysed cells and debris by centrifugation at 500 g for 5 min. Clarified lysates were treated with a reversible cross-linker DSP (Thermo Fisher Scientific) at a concentration of 1 mM for 30 min at 4°C, nutating at 50 rpm. The reaction was quenched by addition of Tris, pH 7.5, to 100 mM and incubation on ice for 10 min. Membranes were solubilized by addition of digitonin to a final concentration of 1% and nutating at 4°C at 50 rpm. Lysates were clarified by centrifugation at 12,000 g for 10 min at 4°C. Clarified lysates were incubated with 50 µl μMACs monoclonal mouse anti-GFP magnetic microbeads (Miltenyi Biotec) incubated at 4°C for 30 min. μMACS columns were equilibrated in IPLB + 1% digitonin + 1× PIC. Lysates and anti-GFP beads were flown over the column, and bound beads were washed three times with 800 µl IPLB + 0.1% digitonin + PIC and then twice more with 500 µl IPLB. On-bead digestion with trypsin was achieved by applying 25 µl of elution buffer 1 (EB1; 2 M Urea, 50 mM Tris-HCl, pH 7.5, 1 mM DTT, and 5 µg/ml Trypsin) to the column and incubating for 30 min. Digested proteins were eluted by adding 2× 50 µl EB2 (2 M Urea, 50 mM Tris-HCl, pH 7.5, and 5 mM chloroacetamide). Eluted proteins were incubated overnight at 25°C and then quenched by addition of 1 µl trifluoroacetic acid (TFA). Purified proteins were identified by tandem mass spectrometry in the University of California, Davis Proteomics Core. Mock purifications from cells that did not express GFP were used as a control for nonspecific interactions.
Ltc1-yEGFP was immunopurified using a modified protocol. Cells were grown in YPD and harvested routinely by centrifugation as above. Cells were then resuspended in a buffered solution (RIPA; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA). Suspended cells were flash-frozen dropwise in liquid nitrogen and lysed in a freezer mill. Clarified lysates were then cross-linked with DSP and quenched as above. Lysates were then solubilized by adding detergents to a final concentration of 1% sodium deoxycholate, 1% NP-40, and 0.1% SDS. Lysates were clarified and incubated with anti-GFP μMACs beads as above. Bound anti-GFP beads were washed three times with RIPA + 0.1% sodium deoxycholate, 0.1% NP-40, 0.01% SDS + PIC, and then twice more with RIPA (no detergents or PIC). On-bead digest, protein elution, and MS/MS were performed as above.
Protein purification
Hexahistidine-tagged Ltc1 was purified from BL21 E. coli containing RIPL plasmid (encoding nonabundant tRNAs) and pET15b::YLR072W(aa1–590). Cultures were grown to mid-log phase in LB media at 37°C and then shifted to 18°C for overnight expression in 0.5 mM IPTG. Cells were harvested routinely by centrifugation, resuspended in a buffer (50 mM Hepes, pH 8, 500 mM NaCl + 350 µg/ml PMSF) and lysed in a microfluidizer, upon which Triton X-100 was added to 0.1%. Insoluble proteins and debris were removed by centrifugation at 35,000 rpm in a Beckman 45Ti rotor for 45 min. The soluble protein fraction was incubated with Ni-IDA resin and washed twice with buffer (50 mM Hepes, pH 8, 500 mM NaCl, 5 mM imidazole, and 0.1% Mega-8 [Dojindo Molecular Technologies Inc.]). Ni-IDA resin was loaded onto an FPLC column and proteins were eluted with 250 mM imidazole. Peak fractions were pooled, concentrated, and then run on a size-exclusion chromatography column (Superdex 200) in a buffer (50 mM Hepes, pH 8, 500 mM NaCl, 0.1% Mega-8, and 0.01% BME), and peak fractions were pooled and concentrated to ∼1 mg/ml protein.
Lipid transfer assays
Liposomes were prepared as described previously (Schulz et al., 2009) except that EDTA was omitted from the buffers. In brief, lipids dissolved in chloroform were mixed and dried under nitrogen, rehydrated in a buffer, subjected to five freeze–thaw cycles, and extruded through a 0.4-µm-pore-size track-etched Nuclepore membrane (Whatman) using a mini extruder (Avanti Polar Lipids, Inc.). The liposomes contained PC/PE/total yeast lipids (80:10:10 mol%). Egg PC and Egg PE were purchased from Avanti Polar Lipids, Inc., and total yeast lipids were obtained by extraction from WT cells in mid-logarithmic growth phase in synthetic complete medium and total lipid phosphate was determined (Bartlett, 1959). Some heavy (sucrose-filled) liposomes were made so that 1 ml liposomes contained either 3 µCi [3H]cholesterol (61 pmol) or [3H]triolein (50 pmol; American Radiolabeled Chemicals). To prepare heavy liposome with radiolabeled phospholipids, total yeast lipids were obtained from a strain grown for 4 h in a medium containing 5 µCi/ml [3H]palmitate (American Radiolabeled Chemicals).
Lipid transfer assay contained 40 µl of 1 mM heavy liposomes, 40 µl of 1 mM light liposomes, 10 µl of 1% Mega-8, and 10 µl protein. The reactions were incubated at 30°C and stopped by transfer to an ice bath, and 10 µl of 1 mg/ml Proteinase K (Sigma-Aldrich) was added. After 30 min on ice, the samples were centrifuged at 16,000 g for 10 min at 4°C, and the radioactivity in 80 µl of the supernatant was determined by scintillation counting. The amount of radioactivity in the supernatant from a reaction prepared without light liposomes and protein was subtracted from other reactions to calculate the amount of radioactivity transferred to light liposomes.
Online supplemental material
Fig. S1 depicts results from experiments further characterizing the localization of Ltc1 and the factors that determine it; for example, we show that Ltc1 localization to ER–mitochondria contact sites is ERMES independent and that localization to ER–vacuole contact sites is independent of NVJs, formed by Nvj1. Fig. S2 depicts a z-series through a yeast cell that is expressing Vph1-mCherry and Ltc1(ΔGRAM)-yEGFP, showing their anticorrelated localization pattern with respect to the vacuole. Table S1, included as a separate Excel file, lists strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502033/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Michael Paddy in the University of California, Davis (UC Davis) MCB-CBS Imaging Facility for advice with fluorescence microscopy. We also thank Dr. Brett Finney and Mr. Darren Weber from the UC Davis Proteomics Core Facility for mass spectrometric analysis of protein samples. Finally, we thank members of the Nunnari, Prinz, and Powers laboratories for stimulating discussion.
A. Murley was supported by National Institutes of Health (NIH) training grant 5T32GM007377-34; J. Nunnari is supported by NIH grants R01GM062942, R01GM097432, and R01GM106019.
J. Nunnari is on the Scientific Advisory Board of Mitobridge, Inc. The authors declare no other competing financial interests.
Footnotes
Abbreviations used in this paper:
- DSP
- dithiobis succinimidyl propionate
- ERMES
- ER–mitochondria encounter structure
- NVJ
- nucleus–vacuole junction
- WT
- wild type
References
- Altmann K., and Westermann B.. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 16:5410–5417. 10.1091/mbc.E05-07-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett G.R. 1959. Colorimetric assay methods for free and phosphorylated glyceric acids. J. Biol. Chem. 234:469–471. [PubMed] [Google Scholar]
- Begley M.J., Taylor G.S., Kim S.A., Veine D.M., Dixon J.E., and Stuckey J.A.. 2003. Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol. Cell. 12:1391–1402. 10.1016/S1097-2765(03)00486-6 [DOI] [PubMed] [Google Scholar]
- Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T.C., and Loewith R.. 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14:542–547. 10.1038/ncb2480 [DOI] [PubMed] [Google Scholar]
- Böckler S., and Westermann B.. 2014. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell. 28:450–458. 10.1016/j.devcel.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., and De Virgilio C.. 2012. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 46:105–110. 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Doerks T., Strauss M., Brendel M., and Bork P.. 2000. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25:483–485. 10.1016/S0968-0004(00)01664-9 [DOI] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., and De Virgilio C.. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell. 19:15–26. 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., and Nunnari J.. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 24:2918–2931. 10.1091/mbc.E13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T.Z., Cortajarena A.L., and Regan L.. 2008. Ligand binding by repeat proteins: natural and designed. Curr. Opin. Struct. Biol. 18:507–515. 10.1016/j.sbi.2008.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines V., Brandt A., Griffiths G., Horstmann H., Brütsch H., and Schatz G.. 1990. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9:3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Hughes A.L., and Gottschling D.E.. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 492:261–265. 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., and Sternberg M.J.. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Khafif M., Cottret L., Balagué C., and Raffaele S.. 2014. Identification and phylogenetic analyses of VASt, an uncharacterized protein domain associated with lipid-binding domains in Eukaryotes. BMC Bioinformatics. 15:222 10.1186/1471-2105-15-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N., Shaw J.M., and Okamoto K.. 2008. Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 9:63–69. 10.1038/sj.embor.7401113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Osman C., and Walter P.. 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum–mitochondria connections. Proc. Natl. Acad. Sci. USA. 108:14151–14156. 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria–cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S., Chao J.T., Tavassoli S., Wong A.K., Choudhary V., Young B.P., Loewen C.J., and Prinz W.A.. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12:e1001969 10.1371/journal.pbio.1001969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Qian X., Hu J., and Sha B.. 2009. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284:23852–23859. 10.1074/jbc.M109.023986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A.S., Mancuso J., Cande W.Z., and Weis K.. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371. 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell E.N., Kisielewski A.E., Pike J.W., Franco H.L., Yao H.H., and Johnson K.J.. 2012. A transcriptome-wide screen for mRNAs enriched in fetal Leydig cells: CRHR1 agonism stimulates rat and mouse fetal testis steroidogenesis. PLoS ONE. 7:e47359 10.1371/journal.pone.0047359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen J.I., Pierson J., Kvam E., Olsen L.J., and Goldfarb D.S.. 2008. The luminal N-terminus of yeast Nvj1 is an inner nuclear membrane anchor. Traffic. 9:1653–1664. 10.1111/j.1600-0854.2008.00789.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B.J., Mogri H., Hill A., Vlahakis A., and Powers T.. 2012. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA. 109:1536–1541. 10.1073/pnas.1117563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Couto A., Graña M., Harispe L., and Aguilar P.S.. 2011. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell. 22:2360–2372. 10.1091/mbc.E10-12-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., and Goldfarb D.S.. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141. 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43:D670–D681. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R.J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202–211. 10.1016/0076-6879(83)01015-0 [DOI] [PubMed] [Google Scholar]
- Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., and Reinisch K.M.. 2014. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 510:552–555. 10.1038/nature13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlossmann J., Lill R., Neupert W., and Court D.A.. 1996. Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J. Biol. Chem. 271:17890–17895. 10.1074/jbc.271.30.17890 [DOI] [PubMed] [Google Scholar]
- Schmidt O., Pfanner N., and Meisinger C.. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11:655–667. 10.1038/nrm2959 [DOI] [PubMed] [Google Scholar]
- Schmidt O., Harbauer A.B., Rao S., Eyrich B., Zahedi R.P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., and Meisinger C.. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 144:227–239. 10.1016/j.cell.2010.12.015 [DOI] [PubMed] [Google Scholar]
- Schulz T.A., Choi M.G., Raychaudhuri S., Mears J.A., Ghirlando R., Hinshaw J.E., and Prinz W.A.. 2009. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 187:889–903. 10.1083/jcb.200905007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Pfaller R., Griffiths G., Pfanner N., and Neupert W.. 1990. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 62:107–115. 10.1016/0092-8674(90)90244-9 [DOI] [PubMed] [Google Scholar]
- Steger H.F., Söllner T., Kiebler M., Dietmeier K.A., Pfaller R., Trülzsch K.S., Tropschug M., Neupert W., and Pfanner N.. 1990. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J. Cell Biol. 111:2353–2363. 10.1083/jcb.111.6.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud D.A., Oeljeklaus S., Wiese S., Bohnert M., Lewandrowski U., Sickmann A., Guiard B., van der Laan M., Warscheid B., and Wiedemann N.. 2011. Composition and topology of the endoplasmic reticulum–mitochondria encounter structure. J. Mol. Biol. 413:743–750. 10.1016/j.jmb.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Subramanian K., Dietrich L.E., Hou H., LaGrassa T.J., Meiringer C.T., and Ungermann C.. 2006. Palmitoylation determines the function of Vac8 at the yeast vacuole. J. Cell Sci. 119:2477–2485. 10.1242/jcs.02972 [DOI] [PubMed] [Google Scholar]
- Tan T., Ozbalci C., Brügger B., Rapaport D., and Dimmer K.S.. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126:3563–3574. 10.1242/jcs.121244 [DOI] [PubMed] [Google Scholar]
- Tang F., Peng Y., Nau J.J., Kauffman E.J., and Weisman L.S.. 2006. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 7:1368–1377. 10.1111/j.1600-0854.2006.00458.x [DOI] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2012. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125:49–58. 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2013. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202:35–44. 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M., Laage R., Dietrich L., Wang L., and Ungermann C.. 2001. Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 20:3145–3155. 10.1093/emboj/20.12.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206:357–366. 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.X., Catlett N.L., and Weisman L.S.. 1998. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 140:1063–1074. 10.1083/jcb.140.5.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., and Neupert W.. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- Wu Y., and Sha B.. 2006. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13:589–593. 10.1038/nsmb1106 [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y.. 2015. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 12:7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.