ELAV-like family member 1, or CELF1, competes with another RNA-binding protein, HuR, to modulate MYC translation and plays an important role in the regulation of intestinal epithelial renewal.
Abstract
The mammalian intestinal epithelium is one of the most rapidly self-renewing tissues in the body, and its integrity is preserved through strict regulation. The RNA-binding protein (RBP) ELAV-like family member 1 (CELF1), also referred to as CUG-binding protein 1 (CUGBP1), regulates the stability and translation of target mRNAs and is implicated in many aspects of cellular physiology. We show that CELF1 competes with the RBP HuR to modulate MYC translation and regulates intestinal epithelial homeostasis. Growth inhibition of the small intestinal mucosa by fasting in mice was associated with increased CELF1/Myc mRNA association and decreased MYC expression. At the molecular level, CELF1 was found to bind the 3′-untranslated region (UTR) of Myc mRNA and repressed MYC translation without affecting total Myc mRNA levels. HuR interacted with the same Myc 3′-UTR element, and increasing the levels of HuR decreased CELF1 binding to Myc mRNA. In contrast, increasing the concentrations of CELF1 inhibited formation of the [HuR/Myc mRNA] complex. Depletion of cellular polyamines also increased CELF1 and enhanced CELF1 association with Myc mRNA, thus suppressing MYC translation. Moreover, ectopic CELF1 overexpression caused G1-phase growth arrest, whereas CELF1 silencing promoted cell proliferation. These results indicate that CELF1 represses MYC translation by decreasing Myc mRNA association with HuR and provide new insight into the molecular functions of RBPs in the regulation of intestinal mucosal growth.
INTRODUCTION
The epithelium of the mammalian intestinal mucosa undergoes a continual renewal process, characterized by active proliferation of stem cells localized near the base of the crypts and progression of these cells up the crypt–villus axis with cessation of proliferation and subsequent differentiation and apoptosis (Sato and Clevers, 2013; Xiao and Wang, 2014). This rapid self-renewal process is tightly controlled at multiple levels and highly regulated by a number of factors. In response to stress, rapid changes in gene expression patterns in intestinal epithelial cells (IECs) control cell division, migration, differentiation, and survival, thereby preserving epithelial integrity and homeostasis (Gunther et al., 2013; Sato and Clevers, 2013). Inhibition of intestinal mucosal growth occurs commonly in various critical disorders, particularly in patients who are supported with total parenteral nutrition after massive surgical operations (Wildhaber et al., 2003; Wang et al., 2010; Puleo et al., 2011; Gunther et al., 2013). Although the exact mechanisms that control gut epithelial homeostasis are not fully understood, posttranscriptional processes, especially altered mRNA turnover and translation, are shown to play an important role in the control of IEC proliferation and apoptosis under biological and pathological conditions (Xiao et al., 2013; Xiao and Wang, 2014; Cao et al., 2014; Liu et al., 2014). Changes in mRNA stability and translation are governed by two major types of trans-acting factors that directly interact with the mRNA: RNA-binding proteins (RBPs) and noncoding RNAs such as microRNAs (miRNAs) (Keene, 2007; Houseley and Tollervey, 2009; Zhuang et al., 2013). RBPs and miRNAs directly interact with cis elements on the mRNAs, frequently present at the 3′-untranslated regions (3′-UTRs), and regulate the stability and translation rates of target transcripts (Krol et al., 2010; Siomi and Siomi, 2010).
The RBP ELAV-like family member 1 (CELF1), also referred to as CUG-binding protein 1 (CUGBP1), contains three RNA recognition motifs through which it binds to specific mRNAs that often contain GU-rich elements (GREs) in the 3′-UTR or coding region (CR) (Vlasova and Bohjanen, 2008; Vlasova et al., 2008; Rattenbacher et al., 2010). In addition to its well-defined role as a regulator of splicing, the interaction of CELF1 with given mRNAs also enhances mRNA decay and represses mRNA translation (Vlasova et al., 2008; Xiao et al., 2011; Talwar et al., 2013), although in some instances CELF1 also promotes mRNA translation (Iakova et al., 2004; Chang et al., 2012). CELF1 was first discovered as an RBP that contributes to the pathogenesis of myotonic dystrophy (Brook et al., 1992), since CELF1 is overexpressed and interacts with GU-rich sequence of target mRNAs in patients with myotonic dystrophy (Wang et al., 2007; Ward et al., 2010; Cardani et al., 2013). Further studies have demonstrated that CELF1 is a multifunctional RBP that plays an important role in many cellular functions (Vlasova and Bohjanen, 2008; Talwar et al., 2013; Yu et al., 2013) and is implicated in several other human diseases, including cancer (Choi et al., 2007; Chettouh et al., 2013), fragile-X tremor/ataxia syndrome (Sofola et al., 2007), and liver dysfunction (Jones et al., 2012). Knockout of the CELF1 in mice is generally embryonic lethal, but the few CELF1-deficient mice that are born exhibit decreased cell viability, growth retardation, and spermatogenesis defects (Kress et al., 2007). Results obtained from in vitro experiments demonstrate that ectopic overexpression of CELF1 prevents apoptosis in HeLa cells (Rattenbacher et al., 2010) and alters the susceptibility of esophageal epithelial cells to chemotherapy-induced cell death (Chang et al., 2012).
Recently CELF1 has also emerged as a master regulator of gut epithelial homeostasis by modulating IEC proliferation, apoptosis, and cell-to-cell interaction (Xiao and Wang, 2014; Yang et al., 2014). CELF1 in IECs interacts with and inhibits Cdk4 (cyclin-dependent kinase 4) mRNA translation (Xiao et al., 2011), whereas decreased levels of endogenous CELF1 in Jnk2−/−-mice associate with crypt hyperplasia in the small intestine (Chung et al., 2014). CELF1 also represses translation of the tight junction protein occludin by increasing Occludin mRNA recruitment to processing bodies, resulting in gut epithelial barrier dysfunction (Yu et al., 2013). Moreover, expression of CELF1 in IECs is translationally repressed by miRNA miR-503 (Cui et al., 2012), providing insight into the regulation of one type of posttranscriptional regulator (an RBP) by another type of posttranscriptional regulator (an miRNA). In this paper, we report a novel finding showing that CELF1 competes with the RBP HuR to regulate MYC translation and thus modulates intestinal epithelial renewal. Fasting increases mucosal CELF1 in the small intestine, which associates with an increase in [CELF1/Myc mRNA] complex and Myc repression. In cultured IECs, CELF1 was found to bind the Myc 3′-UTR, and elevating CELF1 levels led to repression of MYC translation without affecting total Myc mRNA levels. Moreover, HuR competes with CELF1 for binding to the same Myc 3′-UTR element, but the two RBPs regulate MYC translation in opposite directions.
RESULTS
Fasting increases CELF1 and lowers MYC levels in small intestinal mucosa
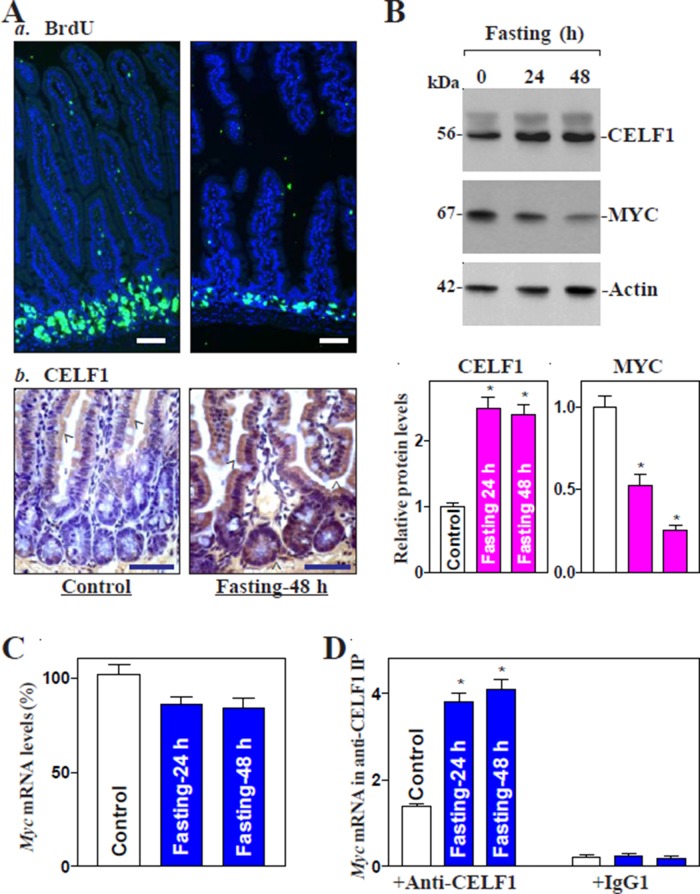
To determine the involvement of CELF1 in the regulation of intestinal mucosal growth, we used a mouse fasting model in this study, because it represents a physiological model of intestinal mucosal atrophy (Ito et al., 2010; Lalles and David, 2011). As reported previously (Xiao et al., 2013), fasting for 48 h inhibited small mucosal growth, as indicated by a reduction in the proliferating crypt cell population marked by incorporation of BrdU (and thus representing S-phase cells) (Figure 1A, a) and a decrease in the lengths of villi and crypts (Supplemental Figure 1). Importantly, the inhibition of small intestinal mucosal growth in fasted mice was associated with a significant increase in the levels of CELF1. CELF1 immunostaining (Figure 1A, b) and protein levels (Figure 1B) increased substantially in the small intestinal mucosa in fasted mice compared with control mice. CELF1 increase by fasting was paralleled by a decrease in the levels of MYC protein, although fasting only marginally reduced Myc mRNA levels (Figure 1C). In particular, fasting-induced intestinal mucosal atrophy was associated with an increase in CELF1 binding to Myc mRNA, as measured by ribonucleoprotein (RNP) immunoprecipitation (IP) assays using anti-CELF1 antibody under conditions that preserved RNP integrity (Figure 1D). The interaction of Myc mRNA with CELF1 was examined by isolating RNA from the immunoprecipitated material and subjecting it to reverse transcription followed by real-time quantitative PCR (RT-qPCR) analysis. The induction in levels of the [CELF1/Myc mRNA] complex occurred 24 h after fasting and remained elevated 48 h thereafter. We also examined changes in CELF1 association with Occludin mRNA, a known CELF1 target transcript (Yu et al., 2013), and found that fasting for 24 or 48 h did not increase the levels of the [CELF1/Occludin mRNA] complex (Supplemental Figure 2A). On the other hand, HuR association with Myc mRNA decreased significantly after fasting (Supplemental Figure 2B). These findings suggest that fasting increases CELF1 abundance in small intestinal mucosa and that induced [CELF1/Myc mRNA] association, along with reduction of the [HuR/Myc mRNA] complex, plays a role in MYC repression and subsequent mucosal atrophy.
FIGURE 1:
Fasting-induced intestinal mucosal atrophy associates with an increased CELF1 but decreased MYC. (A) Changes in cell proliferation as measured by BrdU labeling (a) and immunohistochemical staining of CELF1 (b) in small intestinal mucosa after fasting for 48 h. Green, BrdU, 1 h after injection, S-phase; brown, CELF1. (B) Changes in the levels of CELF1 and MYC proteins in control mice and mice fasted for 24 or 48 h. Top, representative immunoblots of CELF1 and MYC proteins; bottom, quantitative analysis of the immunoblotting signals as measured by densitometry. Values are the means ± SEM (n = 3). *, p < 0.05 compared with control. (C) Levels of Myc mRNA as measured by RT-qPCR analysis in the mucosa described in B. (D) Association of CUGBP1 with Myc mRNA in small intestinal mucosa as measured by RNP-IP/RT-qPCR analysis. After IP of RNA–protein complexes using either anti-CELF1 antibody (Ab) or control IgG1, RNA was isolated and measured with RT-qPCR analysis. The levels of Myc mRNA in IP material were normalized to the levels of Gapdh mRNA in each sample; values are the means ± SEM (n = 5).
Myc 3′-UTR is a direct target of CELF1
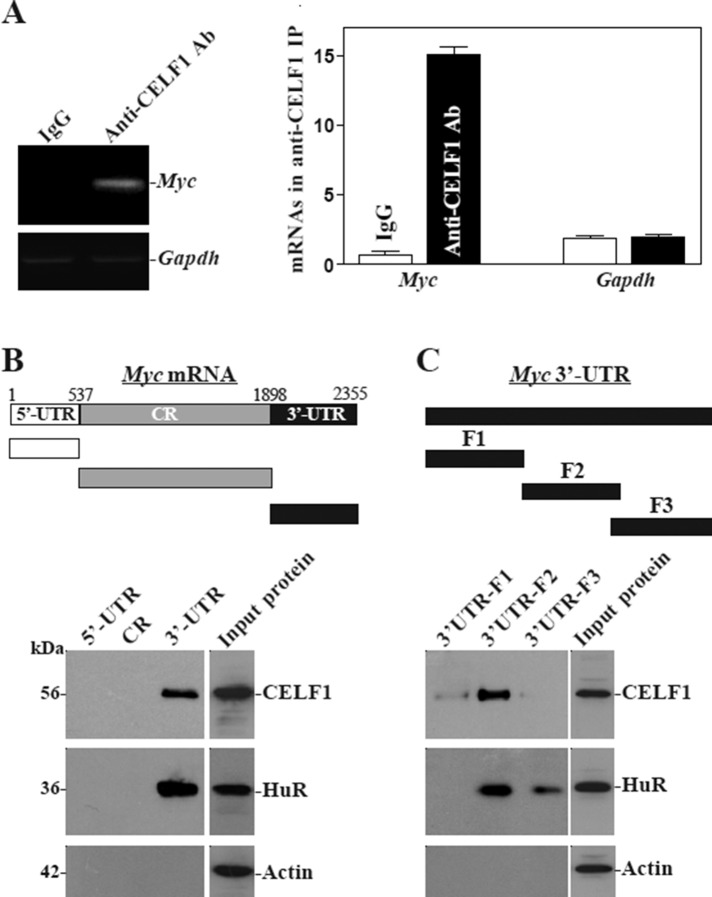
There are several computationally predicted hits of the CELF1 motif in the Myc 3′-UTR based on the reported CELF1-binding sequences (Tsuda et al., 2009; Rattenbacher et al., 2010), suggesting that CELF1 interacts with Myc mRNA via its 3′-UTR. Consistent with the findings obtained from small intestinal mucosal tissue (Figure 1D), CELF1 was also found to bind to Myc mRNA in cultured IEC-6 cells (Figure 2A). Myc PCR products were highly enriched in CELF1 samples compared with control immunoglobulin G1 (IgG1) samples. The enrichment of CDK4 PCR product was also examined and served as a positive control (unpublished data), since Cdk4 mRNA is a known target of CELF1 (Xiao et al., 2011), while the amplification of Gapdh PCR products, a nonspecific contaminating transcript (not a target of CELF1) encoding the housekeeping protein GAPDH, served to monitor the evenness of sample input, as reported previously (Zhang et al., 2009). [CELF1-Myc mRNA] associations were further tested by using biotinylated transcripts that spanned the Myc 5′-UTR, CR, or 3′-UTR (Figure 2B, schematic). Following incubation with cytoplasmic lysates, the interaction between the biotinylated Myc transcripts and CELF1 was examined by biotin pull down followed by Western blot analysis (Abdelmohsen et al., 2007; Liu et al., 2009). The Myc 3′-UTR transcripts readily associated with CELF1 (Figure 2B, bottom), but the Myc 5′-UTR or CR did not. In addition, HuR also formed complexes with the Myc 3′-UTR but not with the 5′-UTR and CR as reported previously (Liu et al., 2009). On the other hand, none of the Myc partial transcripts (5′-UTR, CR, 3′-UTR) was found to interact with β-actin, included here as a negative control. For further definition of the specific CELF1-binding regions in the Myc 3′-UTR, various partial biotinylated transcripts spanning the Myc 3′-UTR (spanning positions 1898–2355) were prepared (Figure 2C, schematic). As shown, CELF1 predominantly bound to the F2 (spanning positions 2044–2212, with predicted CELF1 binding sites, Supplemental Table 1) of the Myc 3′-UTR, but there was no detectable binding of CELF1 to fragments F1 (spanning positions 1899–2071) or F3 (spanning positions 2205–2355). Interestingly, HuR also predominantly interacted with fragment F2, while it only marginally associated with F3. These results indicate that CELF1 interacts with Myc mRNA via specific RNA segments within the F2 of the 3′-UTR.
FIGURE 2:
CELF1 binds the 3′-UTR of Myc mRNA via a GRE. (A) Association of endogenous CELF1 with endogenous Myc mRNA in IEC-6 cells. The levels of Myc mRNA in samples immunoprecipitated by using anti-CELF1 antibody (Ab) or control IgG1 were measured by RT-PCR (left) and RT-qPCR (right) analyses. Low-level amplification of Gapdh mRNA served to monitor the evenness in sample input. Values are the means ± SEM from triplicate samples. (B) Representative CELF1 and HuR immunoblots using the pull-down materials by biotinylated transcripts of the Myc 5′-UTR, CR, or 3′-UTR. Top, schematic representation of the biotinylated transcripts used in this study. Cytoplasmic lysates were incubated with 6 μg biotinylated Myc 5′-UTR, CR, or 3′-UTR, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. The presence of CUGBP1 or HuR in the pull-down material was assayed by Western blotting. β-Actin in the pull-down material was also examined and served as a negative control. (C) Binding of CELF1 or HuR to different fractions of 3′-UTR of the Myc mRNA. Top, schematic representation of the Myc 3′-UTR biotinylated transcripts. After incubation of cytoplasmic lysates with various fractions (F1–F3) of the Myc 3′-UTR, the resulting RNP complexes were pulled down, and the abundance of CELF1, HuR, and β-actin proteins in the pull-down material was examined.
CELF1 inhibits MYC translation by interacting with the Myc 3′-UTR
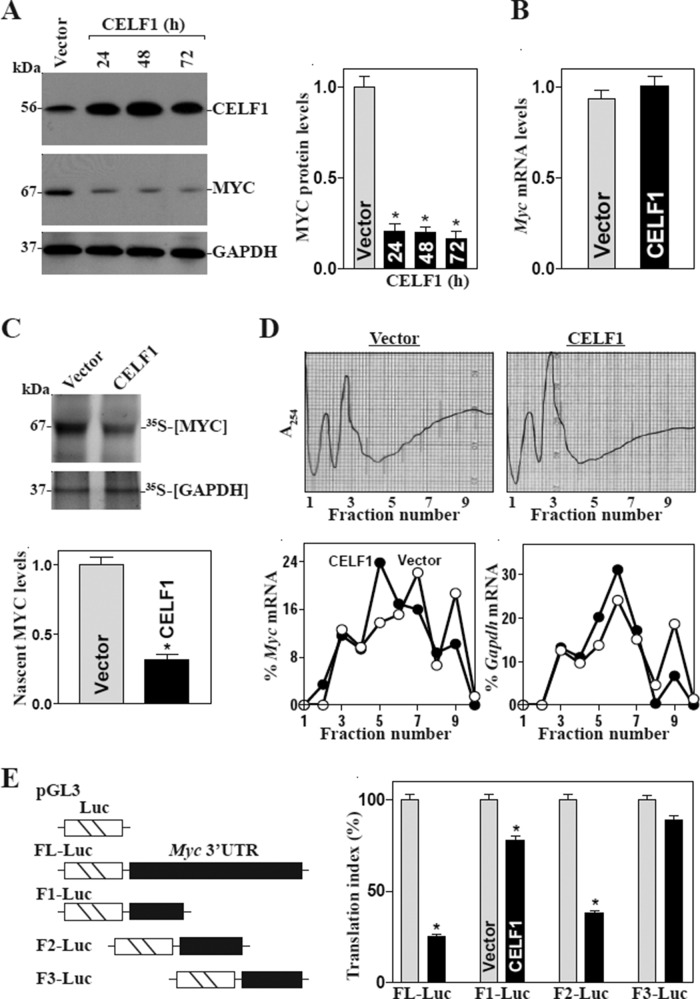
To examine the functional consequences of CELF1 interactions with Myc mRNA, we first determined the effect of overexpressing the wild-type Celf1 gene on MYC abundance in IECs. As shown in Figure 3A, transient transfection with the CELF1 expression vector increased CELF1 protein expression levels approximately fivefold relative to cells transfected with the empty vector. Increased CELF1 abundance was associated with a potent inhibition of MYC expression; the reduction in MYC levels likely occurred at the translation level, since ectopic CELF1 overexpression did not decrease the levels of total Myc mRNA (Figure 3B) but repressed the rate of nascent MYC protein (Figure 3C). To further define the role of CELF1 in the regulation of MYC translation, we examined the relative distribution of Myc mRNA in individual fractions from polyribosome gradients after CELF1 overexpression. Although increasing the levels of CELF1 did not affect global polysomal profiles (Figure 3D, top), the abundance of Myc mRNA associated with actively translating components of the gradient (fractions 7–10) decreased dramatically in CELF1-transfected cells, where a significant shift of Myc mRNA was observed toward low-translating parts of the gradient (fraction 5; Figure 3D, bottom). In contrast, Gapdh mRNA, which is not a target of CELF1 and encodes the housekeeping protein GAPDH, distributed similarly in both groups. To examine whether the translational effect of CELF1 on Myc mRNA was exerted through GREs, we used a firefly luciferase (FL) reporter gene construct containing the GRE present in the Myc 3′-UTR, and negative control vector pGL3-Luc (Figure 3E, schematic). A plasmid expressing Renilla luciferase (RL) was also cotransfected as an internal control for normalization of firefly luciferase. For distinguishing translational output from changes in mRNA turnover, the luciferase activities were normalized to luciferase mRNA levels (RL mRNA, FL mRNA) to assess the translational efficiency (the “translation index”). Ectopic CELF1 overexpression was found to decrease the levels of luciferase reporter gene activity when cells were transfected with the FL-Luc (containing full-length Myc 3′-UTR) or F2-Luc but not with the F3-Luc. When cells were transfected with the F1-Luc, increasing the levels of CELF1 just slightly reduced the reporter gene activity.
FIGURE 3:
CELF1 overexpression inhibits MYC translation via the Myc 3′-UTR. (A) Changes in the levels of MYC after ectopic CELF1 overexpression. Cells were transfected with the vector expressing CELF1 or control empty vector; protein levels were assessed by Western blot analysis at various times after the transfection. Left, representative immunoblots of CELF1 and MYC proteins; right, quantitative analysis of the immunoblotting signals as measured by densitometry. Values are the means ± SEM (n = 3). *, p < 0.05 compared with vector. (B) Levels of Myc mRNA 48 h after transfection, as measured by RT-qPCR analysis. Data were normalized to Gapdh mRNA levels, and values are shown as the means ± SEM (n = 3). (C) Newly translated MYC protein in cells treated as described in B. Cells were incubated with l-[35S]methionine and l-[35S]cysteine for 20 min; this was followed by IP by using anti-MYC antibody, resolving immunoprecipitated samples by SDS–PAGE, and transferring for visualization of signals by using a PhosphorImager. The translation of housekeeping control GAPDH was measured similarly. (D) Distributions of Myc and Gapdh mRNAs in each gradient fraction of polysomal profile in cells described in B. Top, polysomal profiles in cells described in B. Nuclei were pelleted, and the resulting supernatants were fractionated through a 10–50% liner sucrose gradient. Bottom, the levels of Myc and Gapdh mRNAs in different fractions as measured by RT-qPCR analysis and plotted as a percentage of the levels of total Myc mRNA (left) and Gapdh mRNA (right). (E) Changes in MYC translation efficiency as measured by Myc 3′-UTR luciferase reporter assays. Left, schematic of plasmids: control (pGL3-Luc); chimeric firefly luciferase–full-length Myc 3′-UTR (FL-Luc); and luciferase–various Myc 3-UTR fractions (F). Right, levels of activities of luciferase reporters containing Myc 3′-UTR or its different fractions. Forty-eight hours after the Luc reporters or pGL3-Luc (negative control) were cotransfected with a Renilla luciferase reporter, luciferase activity was measured using the Dual Luciferase Assay System. For measurement of translational changes, the ratio of firefly luciferase to Renilla luciferase was further normalized to the levels of Firefly and Renilla mRNAs. The values were expressed as means ± SEM (n = 3). *, p < 0.05 compared with cells transfected with control vector.
Our results further showed that CELF1 silencing by transfection with small interfering RNA (siRNA) targeting the Celf1 mRNA (siCELF1) resulted in an increase in MYC expression. These specific siCELF1 nucleotides were designed to reduce Celf1 mRNA with high specificity and efficacy and low toxicity (Xiao et al., 2011; Cui et al., 2012). The levels of CELF1 protein decreased by > 80% at 48 h after the transfection with siCELF1, whereas the levels of MYC protein increased by approximately twofold compared with those in cells transfected with C-siRNA (Figure 4A). Decreasing the levels of endogenous CELF1 by siCELF1 induced MYC expression by enhancing its translation, because CELF1 silencing did not alter the levels of total Myc mRNA (Figure 4B), but it enhanced nascent synthesis of MYC protein (Figure 4C) and increased the Myc 3′-UTR luciferase reporter gene activity (Figure 4D). These results indicate that CELF1 inhibits MYC translation by directly interacting with its 3′-UTR.
FIGURE 4:
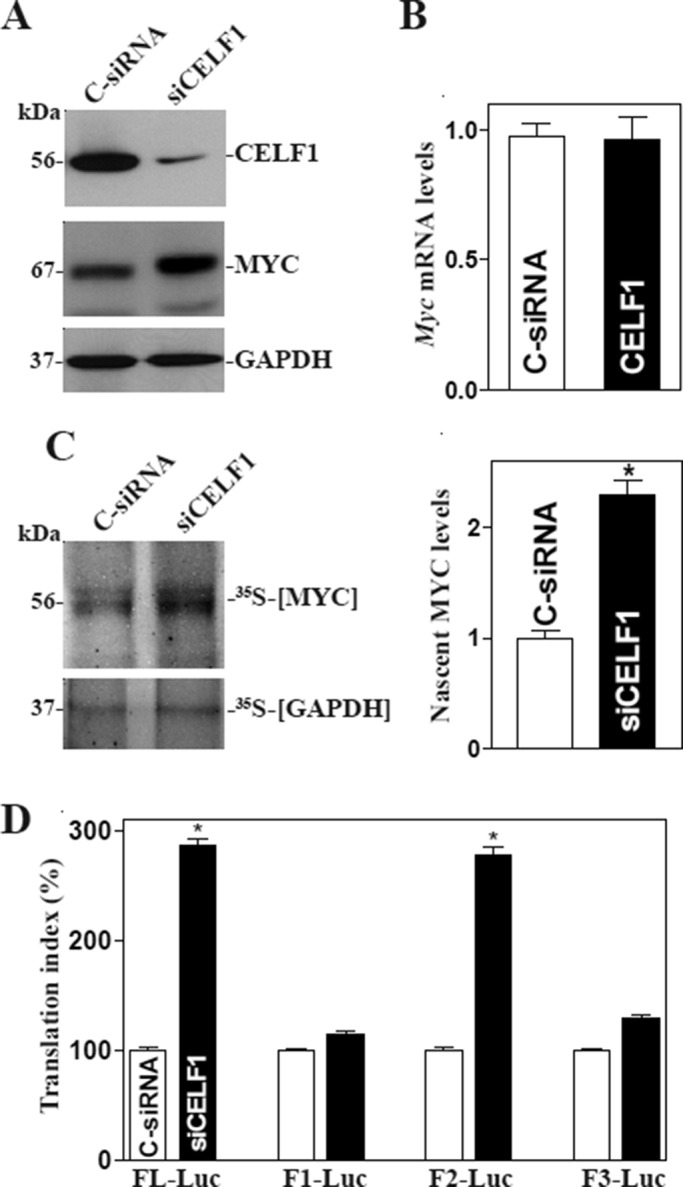
CELF1 silencing enhances MYC translation. (A) Representative immunoblots of CELF1 and MYC proteins. Forty-eight hours after cells were transfected with either siRNA targeting the CELF1 mRNA CR (siCELF1) or control siRNA (C-siRNA), whole-cell lysates were harvested for Western blot analysis. (B) Levels of Myc mRNA as measured by RT-qPCR analysis in cells described in A. The data were normalized to Gapdh mRNA levels and are shown as the means ± SEM of data from triplicate experiments. (C) Newly translated MYC protein in cells described in A as measured by [35S]methionine/[35S]cysteine incorporation assays. Left, immunoblots; right panel, quantitative analysis of the immunoblotting signals as measured by densitometry. Values are the means ± SEM (n = 3). *, p < 0.05 compared with C-siRNA. (D) Changes in MYC translation efficiency as measured by using Myc 3′-UTR luciferase reporter assays in cells described in A. Twenty-four hours after cells were transfected with the Luc-Myc-3′-UTR or pGL3-Luc, the levels of luciferase activity were examined and normalized to the mRNA levels to calculate the translation efficiencies.
HuR competes with CELF1 to bind Myc mRNA
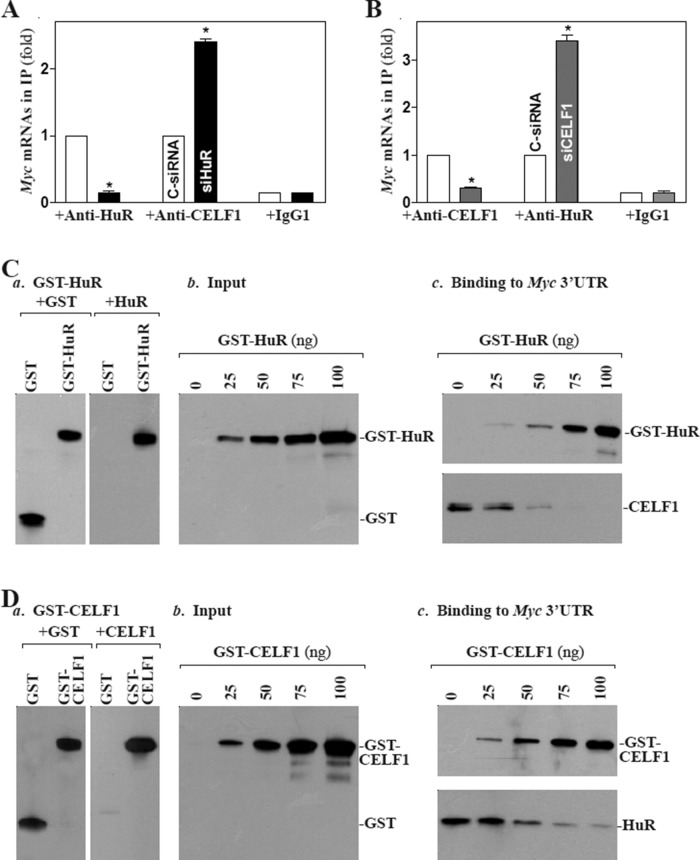
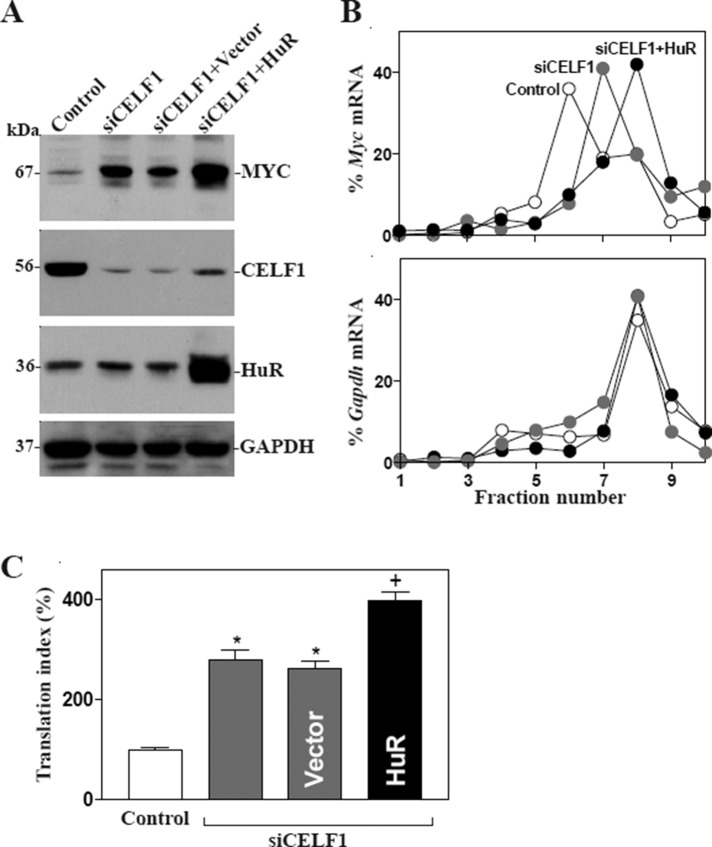
We recently demonstrated that HuR enhances MYC translation (Liu et al., 2009), whereas target deletion of HuR in IECs causes small intestinal mucosal atrophy as a result of inactivation of the Wnt signaling pathway (Liu et al., 2014). As both HuR and CELF1 predominantly bound to the F2 of the Myc 3′-UTR (Figure 2C), we postulated that HuR and CELF1 might compete for interaction with Myc mRNA, thus jointly regulating MYC translation. To test this possibility, we first examined the effect of altering HuR levels on CELF1 binding to Myc mRNA. As shown in Figure 5A, HuR silencing by transfection with siRNA targeting the HuR mRNA (siHuR) decreased the levels of [HuR/Myc mRNA] complex, but it increased the amount of Myc mRNA associated with CELF1, although it had no effect on total Myc mRNA levels as reported previously (Liu et al., 2009; unpublished data). In contrast, CELF1 silencing not only decreased the amount of Myc mRNA associated with CELF1 but also increased the levels of Myc mRNA bound to HuR (Figure 5B). Second, we investigated the competitive binding of HuR and CELF1 to Myc mRNA by examining the effect of adding purified glutathione S-transferase (GST)-HuR (Figure 5C, a) or GST-CELF1 fusion proteins to in vitro binding reactions of Myc 3′-UTR with HuR and CELF1. When increasing concentrations of GST-HuR were added to the binding reaction (Figure 5C, b), its interaction with CELF1 was reduced (Figure 5C, c). Neither HuR nor CELF1 binding to the Myc 3′-UTR was affected by GST being added to the binding reaction (unpublished data). Conversely, increasing the concentrations of GST-CELF1 in the binding reaction mixture decreased, in a concentration-dependent manner, Myc 3′-UTR association with HuR (Figure 5D). Third, we determined the role of competitive binding of HuR and CELF1 in the regulation of MYC translation. Our results showed that CELF1 silencing and ectopic HuR overexpression together stimulated MYC translation synergistically, since the levels of MYC protein expression in cells cotransfected by siCELF1 and HuR expression vector were higher than those observed in cells transfected with siCELF1 alone (Figure 6A). Moreover, ectopic HuR overexpression in CELF1-silenced cells induced the abundance of Myc mRNA associated with actively translating fractions (fractions 8–9) in the polyribosome gradients (Figure 6B) and also resulted in additional increases in the levels of Myc 3′-UTR luciferase reporter gene activity (Figure 6C). Together these results indicate that HuR and CELF1 competitively bind to the Myc 3′-UTR and modulate MYC translation in opposite directions; HuR promoted MYC translation, CELF1 repressed it.
FIGURE 5:
HuR competitively represses [Myc mRNA-CELF1] association. (A) Changes in binding of Myc mRNA to CELF1 and HuR as detected by RNP-IP/RT-qPCR analysis in cells 48 h after transfection with siHuR or C-siRNA. Values are means ± SEM from triplicate samples. *, p < 0.05 compared with cells transfected with C-siRNA. (B) Binding of Myc mRNA to CELF1 and HuR 48 h after transfection with siCELF1 or C-siRNA. (C) Effect of GST-HuR added to the binding reaction on association of HuR or CELF1 with the Myc 3′-UTR: (a) GST-HuR fusion protein identified by anti-GST antibody (left) or recognized by anti-HuR antibody (right); (b) protein input in the binding reaction mixture; and (c) interactions of HuR and CELF1 with the Myc 3′-UTR. Various concentrations of GST-HuR were used; the levels of binding complexes were detected by pull-down assays. Three independent experiments were performed showing similar results. (D) Effect of GST-CELF1 on association of HuR or CELF1 with the Myc 3′-UTR: (a) GST-CELF1 fusion protein; (b) protein input in the binding reaction mixture; and (c) binding of HuR and CELF1 to the Myc 3′-UTR.
FIGURE 6:
CELF1 silencing and HuR induction increase MYC translation synergistically. (A) Representative immunoblots of MYC, CELF1, and HuR in cells 48 h after transfection with siCELF1 alone or cotransfection with siCELF1 and the HuR expression vector. (B) Distributions of Myc (top) and Gapdh (bottom) mRNAs in each gradient fraction of polysomal profile in cells described in A. (C) Changes in MYC translation efficiency as measured by Myc 3′-UTR luciferase reporter assays. Values were expressed as means ± SEM of data from three separate experiments. *,+, p < 0.05 compared with cells transfected with control or siCELF1-trasfected cells, respectively.
Polyamines regulate MYC abundance by altering the levels of CELF1
Polyamines (spermidine and spermine and their precursor putrescine) are organic cations found in all eukaryotic cells (Casero and Marton, 2007; Pegg and Casero, 2011) and are essential for maintenance of gut epithelial integrity (Wang and Johnson, 1994; Wang et al., 2010; Guo et al., 2002; Liu et al., 2003, 2005). The supply of polyamines to cells dividing in the crypts is essential for normal intestinal mucosal growth, as it increases MYC expression, whereas polyamine depletion inhibits epithelial renewal through down-regulation of MYC. In this study, we examined whether polyamines regulate MYC expression by altering CELF1. Consistent with our previous studies (Xiao et al., 2007; Zou et al., 2006), inhibition of ornithine decarboxylase (ODC, a key enzyme for polyamine biosynthesis) by treatment with DFMO (d, l-α-difluoromethylornithine) for 4 d almost totally depleted polyamines, as putrescine and spermidine were undetectable and spermine was decreased by ∼50% (unpublished data). Polyamine depletion by DFMO increased CELF1 abundance, and this induction was completely prevented by exogenous putrescine given together with DFMO (Figure 7A, top). The levels of [CELF1-Myc mRNA] complex were also increased in polyamine-deficient cells (Figure 7B), which was associated with a decrease in MYC expression (Figure 7A, middle). Furthermore, CELF1 silencing by transfection with siCELF1 in polyamine-deficient cells (Figure 7C) not only prevented the increased CELF1-Myc mRNA association (Figure 7D), but it also promoted MYC translation, as shown by an increase in the levels of MYC protein (Figure 7E) and its 3′-UTR luciferase reporter gene activity (Figure 7F). On the other hand, CELF1 silencing also enhanced formation of the [HuR-Myc mRNA] complex in polyamine-deficient cells (Supplemental Figure 3). These findings strongly suggest that polyamines regulate CELF1 negatively and that elevated CELF1 contributes to repression of MYC translation in polyamine-depleted cells.
FIGURE 7:
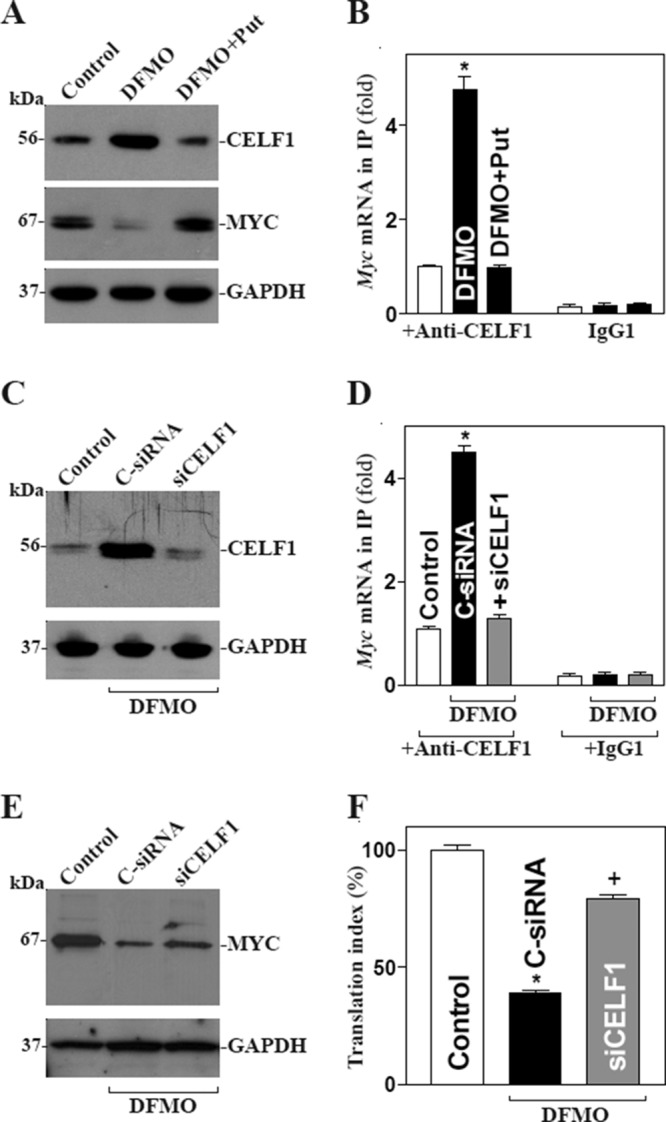
Polyamine depletion represses MYC translation by inducing the association of CELF1 with Myc mRNA. (A) Representative immunoblots of CELF1 and MYC after polyamine depletion. Cells were exposed to DFMO alone or DFMO plus putrescine (Put) for 4 d; the levels of CELF1 and MYC proteins were examined by Western blot analysis. (B) Association of endogenous CELF1 with endogenous Myc mRNA as measured by RNP-IP/RT-qPCR analysis in cells described in A. Values are means ± SEM from three separate experiments. *, p < 0.05 compared with control cells and cells exposed to DFMO plus Put. (C) Representative immunoblots of CELF1 after CELF1 silencing. Cells were exposed to DFMO for 2 d and then transfected with siCELF1 or C-siRNA. Whole-cell lysates were harvested 48 h after the transfection in the presence of DFMO. (D) CELF1/Myc mRNA association in cells described in C. Values are means ± SEM of data from three separate experiments. *,+, p < 0.05 compared with control or cells transfected with siCELF1, respectively. (E) Changes in expression of MYC protein in cells described in C. (F) Changes in MYC translation efficiency as measured by Myc 3′-UTR luciferase reporter assays.
Induced CELF1 causes G1-phase growth arrest in IECs
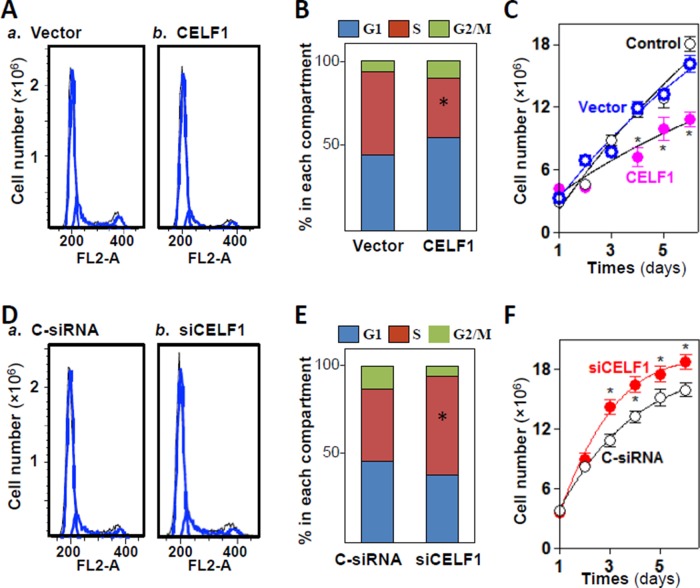
To study the in vitro functions of CUGBP1, we examined changes in IEC proliferation after ectopic CELF1 overexpression or CELF1 silencing. Ectopic overexpression of CELF1 using an expression vector resulted in G1-phase enrichment, with an increase in G1-phase cells, a reduction in S-phase cells, and a reduction in the total number of cells overexpressing CELF1 (Figure 8, A–C). In contrast, CELF1 silencing by transfection of siCELF1 enhanced cell proliferation, with a decrease in G1-phase cells, an increase in S-phase cells, and elevated cell numbers (Figure 8, D–F). Increasing or decreasing CELF1 did not directly affect cell death, as there were no apparent differences in cell viability, morphology, or caspase-3 cleavage after modulation of CELF1 levels (unpublished data). Together these findings indicate that CELF1 functions as a repressor of IEC proliferation and is implicated in the regulation of intestinal epithelial homeostasis.
FIGURE 8:
CELF1 results in G1-phase growth arrest. (A) Flow cytometric analysis of cell cycle distribution in Caco-2 cells transfected with CELF1 expression vector for 48 h. Black line: area; blue line: curve fit; FL2-A: DNA content. (B) The relative G1, S, and G2/M compartments calculated from data described in A. Values are the means of three separate experiments. *, p < 0.05 compared with vector. (C) Changes in cell numbers at different times after transfection with the CELF1 expression vector or control vector. Values are the means ± SEM (n = 6). *, p < 0.05 compared with control and cells transfected with control vector. (D and E) Flow cytometric analysis of cell cycle distribution in cells 48 h after transfection with siCELF1 or C-siRNA. *, p < 0.05 compared with C-siRNA. (F) Changes in cell numbers after CELF1 silencing. Values are the means ± SEM (n = 6). *, p < 0.05 compared with C-siRNA.
DISCUSSION
The transcription factor MYC is crucial for maintaining normal gut mucosal growth and for stimulating epithelial regeneration after injury (Liu et al., 2003, 2005; Wang, 2007). MYC expression is tightly controlled at multiple levels, both transcriptional and posttranscriptional. Although the transcriptional events that regulate MYC expression are well documented (Marcu et al., 1992), the contribution of posttranscriptional regulation of MYC is far less well understood (Wall et al., 2008; Kim et al., 2009). Our previous study showed that HuR enhances MYC translation through Chk2-dependent HuR phosphorylation, and this process is tightly regulated by cellular polyamines (Liu et al., 2009). In this study, we further discovered that Myc 3′-UTR is the target of CELF1 and that association of CELF1 with Myc mRNA represses MYC translation but does not affect Myc mRNA levels. Interestingly, HuR and CELF1 were found to bind the same region of the Myc 3′-UTR in a competitive manner: HuR silencing increased CELF1 association with the Myc 3′-UTR, while CELF1 silencing increased HuR association with Myc mRNA. Conversely, HuR overexpression displaced CELF1 from the Myc 3′-UTR, while CELF1 overexpression displaced HuR from the Myc 3′-UTR. Given CELF1 elevation by fasting in mice was associated with an inhibition of both MYC expression and gut mucosal growth, these findings provide insight into the mechanism that controls MYC expression at the posttranscriptional level and also highlight the important role of CELF1-mediated MYC repression in the pathogenesis of intestinal mucosal atrophy.
The results reported here indicate that CELF1 interacted with the 3′-UTR of Myc mRNA but not with the 5′-UTR or CR in IECs. Although the Myc 3′-UTR does not contain typical canonical GREs such as UGUUUGUUUGU, there are several GU repeats and CUG repeats in the Myc 3′-UTR, which were also recently recognized as the GRE and interacted with CELF1 (Tsuda et al., 2009; Rattenbacher et al., 2010). Through the use of various ectopic reporters bearing partial transcripts spanning the Myc 3′-UTR, our results further show that only the sequence spanning positions 2044–2212 (F2 fragment) within the Myc 3′-UTR is functional, because both the repression of Myc by CELF1 overexpression and the stimulation of Myc by CELF1 silencing were almost completely prevented when this sequence was deleted. These findings are consistent with earlier reports that CELF1 associated with the 3′-UTRs of mRNAs encoding tumor necrosis factor (TNF), TNF-receptor 1B, c-Jun, JunB, and occludin (Vlasova et al., 2008; Zhang et al., 2008; Yu et al., 2013). In some instances, CELF1 was also shown to bind to the 5′-UTR or CR of target mRNAs (Rattenbacher et al., 2010). In this regard, we have recently reported that CELF1 associates with both the CR and 3′-UTR of the Cdk4 mRNA and represses CDK4 translation (Xiao et al., 2011).
Our results in this report also indicate that the competitive interaction of CELF1 and HuR with Myc mRNA resulted in antagonistic effects on MYC translation: HuR silencing increased CELF1 association with Myc mRNA and lowered MYC translation, while HuR overexpression lowered CELF1-Myc 3′-UTR complexes and increased MYC translation. This opposite effect of HuR and CELF1 on MYC expression was not surprising, as several studies have showed that HuR interacts functionally with other RBPs and miRNAs to remodel RNP complexes and influence the posttranscriptional fate of mRNAs. For example, HuR interacts with AUF1 to regulate the stability of mRNAs encoding JunD (Zou et al., 2010), p16INK4 (Chang et al., 2010), and phosphoenolpyruvate carboxykinase (Gummadi et al., 2012), and competes with CELF1 to modulate translation of occludin (Yu et al., 2013). Although the exact mechanism by which HuR competes with CELF1 for association with Myc mRNA is unclear, HuR and CELF1 had affinity for the same Myc 3′-UTR element (F2 fragment with both AU-rich elements [AREs] and GREs). However, we do not know at present whether CELF1 and HuR interact with the Myc 3′-UTR through a distinct nonoverlapping binding site or whether there are common sites for both CELF1 and HuR, because in some instances CELF1 also can associate with AREs (Tsuda et al., 2009; Rattenbacher et al., 2010). In the current study, we did not further characterize the specific Myc 3′-UTR nucleotides with which CELF1 and/or HuR interact, since those experiments would require more specialized biochemical, crystallographic, and molecular methods than those performed here.
The data obtained in the present study further demonstrate that CELF1 association with Myc mRNA is also tightly regulated by cellular polyamines in IECs. Polyamines have been recognized for many years as key molecules that control multiple signaling pathways (Casero and Marton, 2007; Pegg and Casero, 2011), and they are biological regulators of normal gut mucosal growth (Liu et al., 2005; Wang, 2007; Zou et al., 2010). The levels of cellular polyamines are highly regulated and depend on the dynamic balance among polyamine biosynthesis, degradation, and transport. As shown, decreasing cellular polyamines with DFMO increased CELF1 protein and induced the [CELF1-Myc mRNA] complex, which was associated with a significant decrease in MYC expression. Because both increased association of CELF1 with Myc mRNA and inactivation of MYC expression in DFMO-treated cells were completely prevented by the addition of exogenous putrescine, these observed changes in levels of [CELF1-Myc mRNA] complex and MYC expression are more likely related to polyamine depletion than to the nonspecific effect of DFMO. In polyamine-deficient cells, CELF1 silencing decreased the levels of [CELF1-Myc mRNA] complexes and enhanced MYC translation. Although the full mechanisms by which polyamines regulate CELF1 expression are not yet understood, our previous studies show that polyamine depletion decreased the levels of miR-503 that represses CELF1 translation (Cui et al., 2012). In addition, polyamine depletion also increased the cytoplasmic abundance of HuR that also directly interacts with the Celf1 3′-UTR and induces CELF1 translation (Zou et al., 2006; Xiao et al., 2011).
Finally, our results strongly suggest that the CELF1-mediated repression of MYC translation is biologically significant and plays an important role in the regulation of the renewal of the intestinal epithelium. In biological conditions, undifferentiated IECs continuously replicate in the proliferating zone within the crypts and differentiate as they migrate up the luminal surface of the colon and the villous tips in the small intestine (Sato and Clevers, 2013; Xiao and Wang, 2014). Maintenance of the intestinal epithelial integrity depends on a dynamic balance between IEC proliferation, growth arrest, and apoptosis. The epithelium of the human small intestine experiences ∼1011 mitoses per day, and this rapid cell proliferation is tightly regulated by numerous extracellular and intracellular factors (Wang, 2007; Gunther et al., 2013). Fasting increased mucosal CELF1 abundance in the small intestine, which was associated with a significant inhibition of MYC expression and mucosal growth. In cultured IECs, ectopic overexpression of CELF1 resulted in elevation in the G1 compartment, whereas CELF1 silencing promoted cell proliferation and increased the S/G2 population. Because the basal levels of mucosal CELF1 are relatively high in the small intestine and its expression is tightly regulated by many biological modulators such as cellular polyamines, CELF1 may function as a repressor of normal intestinal mucosal growth, whereas aberrantly elevated CELF1 contributes to the pathogenesis of mucosal atrophy under certain disease conditions.
MATERIALS AND METHODS
Animal studies
C57BL/6J mice were obtained from the Jackson Laboratory and housed and handled in a specific pathogen-free animal facility at the Baltimore VA Medical Center. All experiments were performed according to animal experimental ethics committee guidelines approved by the Institutional Animal Care and Use Committee of University of Maryland School of Medicine and Baltimore VA hospital. Animals were deprived of food but allowed free access to tap water for 24 or 48 h in the fasting model. A 4-cm intestinal segment taken 0.5 cm distal to the ligament of Trietz was removed, and the mucosa was scraped from the underlying muscle with a glass microscope slide as described previously (Liu et al., 2014) and used for various measurements of the levels of mRNA and protein expression and CELF1 association with given mRNAs.
Chemicals and cell culture
Tissue culture medium and dialyzed fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA), and biochemicals were from Sigma-Aldrich (St. Louis, MO). The antibodies recognizing CELF1, HuR, Myc, GAPDH, and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Biosciences. The secondary antibody conjugated to horseradish peroxidase was purchased from Sigma-Aldrich. The IEC-6 cell line (derived from normal rat intestinal crypt cells) was purchased from the American Type Culture Collection (ATCC) at passage 13 and was maintained in T-150 flasks in DMEM supplemented with 5% heat-inactivated FBS. Passages 15–20 were used in experiments, and there were no significant changes of biological function and characterization of IEC-6 cells at passages 15–20 (Li et al., 2001; Zhang et al., 2004; Wang et al., 2010). Caco-2 cells (a human colon carcinoma cell line) were also purchased from ATCC and cultured similarly to the IEC-6 cells.
Plasmid construction
CELF1 expression vector was purchased from Origene (Rockville, MD), and the HuR expression vector was described previously (Xiao et al., 2011). The chimeric firefly luciferase reporter construct containing the Myc 3′-UTR was generated as described previously (Liu et al., 2009). The full-length Myc 5′-UTR, CR, 3′-UTR, and different GRE fragments from the Myc 3′-UTR were amplified and subcloned into the pGL3-Luc plasmid (Promega) to generate the chimeric pGL3-Luc-Myc-5′-UTR, Myc-CR, or Myc 3′-UTR reporter constructs. The sequence and orientation of the fragment in the luciferase reporter were confirmed by DNA sequencing and enzyme digestion. Transient transfections were performed as recommended by the manufacturer (Invitrogen). The luciferase reporter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega, to monitor transfection efficiencies as described previously (Ouyang et al., 2015). Forty-eight hours after transfection, luciferase activity was measured using the Dual Luciferease Assay System following the manufacturer's instructions. For measurement of translational changes, the ratio of firefly luciferase to Renilla luciferase was further normalized to the levels of Firefly and Renilla mRNAs in every experiment.
RNA interference
CELF1 or HuR was silenced by transfection with a specific siRNA as described previously (Zou et al., 2010; Xiao et al., 2011). The siRNAs specifically targeting mRNAs encoding CELF1 (siCUGBP1) or HuR (siHuR) and control-siRNA (C-siRNA) were purchased from Santa Cruz. For each 60-mm cell culture dish, 15 μl of the 20 μM stock duplex siCELF1, siHuR, or C-siRNA was used. Forty-eight hours after transfection using Lipofectamine, cells were harvested for analysis.
RT-qPCR analysis
Total RNA was isolated from cells after different treatments by using the RNeasy mini kit (Qiagen, Valencia, CA) and used in reverse transcription and PCR amplifications as described previously (Guo et al., 2002). The levels of β-actin PCR product were assessed to monitor the even RNA input in RT-qPCR samples. RT-qPCR was performed using 7500-Fast Real-Time PCR Systems (Applied Biosystems, Foster City, CA) with specific primers (Rn01328012 for CELF1; Rn01519412 for Myc), probes, and software (Applied Biosystems) by following the manufacturer's instructions.
Western blot analysis
Whole-cell lysates were prepared using 2% SDS, sonicated, and centrifuged (12,000 rpm) at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS–PAGE (7.5% acrylamide). After proteins were transferred onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing Myc, CELF1, or HuR; following incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Analysis of newly translated protein
New synthesis of MYC protein was measured by l-[35S]methionine and l-[35S]cysteine incorporation assays as described previously (Liu et al., 2009). Cells were incubated with 1 mCi (1 Ci = 37 GBq) of l-[35S]methionine and l-[35S]cysteine per 60-mm plate for 20 min, whereupon cells were lysed using radio-IP assay buffer. IPs were carried out for 1 h at 4°C by using either a polyclonal antibody recognizing MYC or IgG1 (BD Biosciences PharMingen, San Diego, CA). After extensive washes in TNN buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, and 0.5% NP-40), the immunoprecipitated material was resolved by 10% SDS–PAGE, transferred onto polyvinylidene difluoride filters, and visualized with a PhosphorImager (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Polysome analysis was performed as described (Chen et al., 2008). Briefly, cells at ∼70% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide, lifted by scraping in 1 ml of polysome extraction buffer, and lysed on ice for 10 min. Nuclei were pelleted, and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm using a UV-6 detector (ISCO, Louisville, KY). After the RNA in each fraction was extracted, the levels of each individual mRNA were quantified by RT-qPCR in each of the fractions.
Biotin pull-down assays
For synthesis of RNA transcripts, cDNA from IEC-6 cells was used as a template for PCR amplification of the CR and 3′-UTR of Myc mRNA. The 5′ primers contained the T7 RNA polymerase promoter sequence (T7): 5′-CCAAGCTTCTAATACGACTC-ACTATAGGGAGA-3′. For preparation of the Myc 5′-UTR template (spanning positions 55–536), oligonucleotides 5′-GCCTCCTGCCTCCAAAAG-3′ and GCTTCAAATAACGCGAGGAG-3′ were used. For preparation of the Myc CR template (spanning positions 537–1898), oligonucleotides (T7)5′-TCTGCGACGAGGAAGAGAAT-3′ and 5′-TGCTCATCTGCTTGAACGGA-3′ were used. For preparation of the Myc 3′-UTR template (spanning positions 1899–2355), oligonucleotides (T7)5′-ACTTACTGAGGAAACGGCGA-3′ and 5′-TAAGAGAAGGCTCAATTATATTT-3′ were used. For preparation of the Myc 3′-UTR fragment-1 (F1) template (spanning positions 1899–2071), oligonucleotides (T7)5′-TGCATAAACTGACCGGAAGTGAGGA-3′ and 5′- AGTTCTTTTATGCCTTAACTTTGAGGCA-3′ were used. For preparation of the Myc 3′-UTR fragment-2 (F2) template (spanning positions 2044–2212), oligonucleotides (T7)5′-TGCCTCAAAGTTAAGGCATAAAAGAACT-3′ and 5′-ATCTTGTATAACTGTTATAA-ACGTTTTATTAAAGT-3′ were used. For preparation of the Myc 3′-UTR fragment-3 (F3) template (spanning positions 2205–2355), oligonucleotides (T7)5′-TACAAGATTTTAAGACATGTATG-ATAAACCATAA-3′ and 5′-TAAGAGTTGGCTCAATTATATTTTTTCCA-3′ were used. PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 polymerase in the presence of biotin-cytidine 5′-triphosphate as previously described (Liu et al., 2009). For biotin pull-down assays, biotinylated transcripts (6 μg) were incubated with 120 μg cytoplasmic lysate for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by Western blotting.
RNP IP assays
For assessment of the association of endogenous CELF1 or HuR with endogenous Myc mRNA, IP of RNP complexes was performed as previously described (Zou et al., 2006; Yu et al., 2013). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody against CELF1 or HuR, or IgG1 (negative control). RNA in the IP materials was used in RT followed by RT-PCR and RT-qPCR analyses to detect the presence of Myc and Gapdh mRNAs.
Assay for ODC enzyme activity and polyamine analysis
ODC activity was determined by a radiometric technique in which the amount of 14CO2 liberated from L-[1-14C]ornithine was estimated. Sample collection and the assay procedure were carried out as described in our previous publications (Wang and Johnson, 1994). Enzymatic activity was expressed as picomoles of CO2 per milligram of protein per hour. The cellular polyamine content was analyzed by high-performance liquid chromatography (HPLC) analysis as previously described (Liu et al., 2006). Briefly, after 0.5 M perchloric acid was added, the cells were frozen at −80°C until ready for extraction, dansylation, and HPLC analysis. The standard curve encompassed 0.31–10 μM. Values that fell > 25% below the curve were considered undetectable. The results are expressed as nanomoles of polyamines per milligram of protein.
Statistics
Values are means ± SEM from three to six samples. Autoradiographic results and studies of immunofluorescence staining were repeated three times. The significance of the difference between means was determined by analysis of variance (ANOVA). The level of significance was determined by using Duncan's multiple-range test (Harter, 1960).
Supplementary Material
Acknowledgments
This work was supported by Merit Review Awards (to J.-Y.W. and J.N.R.) from the U.S. Department of Veterans Affairs; grants from the National Institutes of Health (DK57819, DK61972, and DK68491 to J.-Y.W.); and funding from the National Institute on Aging–Intramural Research Program (to M.G.). J.-Y.W. is a Senior Research Career Scientist, Biomedical Laboratory Research & Development Service, U.S. Department of Veterans Affairs.
Abbreviations used:
- AREs
AU-rich elements
- CDK4
cyclin-dependent kinase 4
- CELF1
ELAV-like family member 1
- CR
coding region
- CUGBP1
CUG-binding protein 1
- DFMO
d, l-α-difluoromethylornithine
- FBS
fetal bovine serum
- FL
firefly luciferase
- GREs
GU-rich elements
- GST
glutathione S-transferase
- HPLC
high-performance liquid chromatography
- IECs
intestinal epithelial cells
- IgG1
immunoglobulin G1
- IP
immunoprecipitation
- miRNAs
microRNAs
- ODC
ornithine decarboxylase
- RBPs
RNA-binding proteins
- RL
Renilla luciferase
- RNP
ribonucleoprotein
- RT-qPCR
reverse transcription followed by real-time quantitative PCR
- siCELF1
siRNA targeting CELF1
- siHuR
siRNA targeting HuR
- siRNA
small interfering RNA
- TNF
tumor necrosis factor
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1500) on March 25, 2015.
*These authors contributed equally to this work.
†Present address: Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China.
REFERENCES
- Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Cao S, Xiao L, Ra JN, Zou T, Liu L, Zhang D, Turner DJ, Gorospe M, Wang JY. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell. 2014;25:1234–1243. doi: 10.1091/mbc.E13-09-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardani R, Bugiardini E, Renna LV, Rossi G, Colombo G, Valaperta R, Novelli G, Botta A, Meola G. Overexpression of CUGBP1 in skeletal muscle from adult classic myotonic dystrophy type 1 but not from myotonic dystrophy type 2. PLoS One. 2013;8:e83777. doi: 10.1371/journal.pone.0083777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J. 2012;446:113–123. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Claperon A, Chretien Y, Rey C, Scatton O, Soubrane O, Conti F, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- Choi WT, Folsom MR, Azim MF, Meyer C, Kowarz E, Marschalek R, Timchenko NA, Naeem RC, Lee DA. C/EBPβ suppression by interruption of CUGBP1 resulting from a complex rearrangement of MLL. Cancer Genet Cytogenet. 2007;177:108–114. doi: 10.1016/j.cancergencyto.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HK, Rao JN, Zou T, Liu L, Xiao L, Gu H, Turner DJ, Yang P, Wang JY. Jnk2 deletion disrupts intestinal mucosal homeostasis and maturation by differentially modulating RNA-binding proteins HuR and CUGBP1. Am J Physiol Cell Physiol. 2014;306:C1167–C1175. doi: 10.1152/ajpcell.00093.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummadi L, Taylor L, Curthoys NP. Concurrent binding and modifications of AUF1 and HuR mediate the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in kidney cells. Am J Physiol Renal Physiol. 2012;303:F1545–F1554. doi: 10.1152/ajprenal.00400.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C, Neumann H, Neurath MF, Becker C. Apoptosis, necrosis and necroptosis: cell death regulation in the intestinal epithelium. Gut. 2013;62:1062–1071. doi: 10.1136/gutjnl-2011-301364. [DOI] [PubMed] [Google Scholar]
- Guo X, Rao JN, Liu L, Rizvi M, Turner DJ, Wang JY. Polyamines regulate β-catenin tyrosine phosphorylation via Ca2+ during intestinal epithelial cell migration. Am J Physiol Cell Physiol. 2002;283:C722–C734. doi: 10.1152/ajpcell.00054.2002. [DOI] [PubMed] [Google Scholar]
- Harter JL. Critical values for Duncan's new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Uchida H, Yokote T, Ohtake K, Kobayashi J. Fasting-induced intestinal apoptosis is mediated by inducible nitric oxide synthase and interferon-γ in rat. Am J Physiol Gastrointest Liver Physiol. 2010;298:G916–G926. doi: 10.1152/ajpgi.00429.2009. [DOI] [PubMed] [Google Scholar]
- Jones K, Timchenko L, Timchenko NA. The role of CUGBP1 in age-dependent changes of liver functions. Ageing Res Rev. 2012;11:442–449. doi: 10.1016/j.arr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lalles JP, David JC. Fasting and refeeding modulate the expression of stress proteins along the gastrointestinal tract of weaned pigs. J Anim Physiol Anim Nutr (Berl) 2011;95:478–488. doi: 10.1111/j.1439-0396.2010.01075.x. [DOI] [PubMed] [Google Scholar]
- Li L, Rao JN, Bass BL, Wang JY. NF-κB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G992–G1004. doi: 10.1152/ajpgi.2001.280.5.G992. [DOI] [PubMed] [Google Scholar]
- Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Kyoung Chung H, Yang H, Gorospe M, Kontoyiannis D, Wang JY. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25:3308–3318. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Guo X, Rao JN, Zou T, Marasa BS, Chen J, Greenspon J, Casero RA, Jr, Wang JY. Polyamine-modulated Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem J. 2006;398:257–267. doi: 10.1042/BJ20060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li L, Rao JN, Zou T, Zhang HM, Boneva D, Bernard MS, Wang JY. Polyamine-modulated expression of c-Myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am J Physiol Cell Physiol. 2005;288:C89–C99. doi: 10.1152/ajpcell.00326.2004. [DOI] [PubMed] [Google Scholar]
- Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Santora R, Rao JN, Guo X, Zou T, Zhang HM, Turner DJ, Wang JY. Activation of TGF-β-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1056–G1067. doi: 10.1152/ajpgi.00151.2003. [DOI] [PubMed] [Google Scholar]
- Marcu KB, Bossone SA, Patel AJ. Myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Ouyang M, Su W, Xiao L, Rao JN, Jiang L, Li Y, Turner DJ, Gorospe M, Wang JY. Modulation by miR-29b of intestinal epithelium homeostasis through the repression of menin translation. Biochem J. 2015;465:315–323. doi: 10.1042/BJ20141028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE, Casero RA., Jr Current status of the polyamine research field. Methods Mol Biol. 2011;720:3–35. doi: 10.1007/978-1-61779-034-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleo F, Arvanitakis M, Van Gossum A, Preiser JC. Gut failure in the ICU. Semin Respir Crit Care Med. 2011;32:626–638. doi: 10.1055/s-0031-1287871. [DOI] [PubMed] [Google Scholar]
- Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG pre-mutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwar S, Balasubramanian S, Sundaramurthy S, House R, Wilusz CJ, Kuppuswamy D, D'Silva N, Gillespie MB, Hill EG, Palanisamy V. Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells. RNA Biol. 2013;10:277–286. doi: 10.4161/rna.23315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA. Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood. 2008;112:2305–2317. doi: 10.1182/blood-2007-09-111856. [DOI] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids. 2007;33:241–252. doi: 10.1007/s00726-007-0518-z. [DOI] [PubMed] [Google Scholar]
- Wang JY, Johnson LR. Expression of protooncogenes c-fos and c-myc in healing of gastric mucosal stress ulcers. Am J Physiol Gastrointest Liver Physiol. 1994;266:G878–G886. doi: 10.1152/ajpgi.1994.266.5.G878. [DOI] [PubMed] [Google Scholar]
- Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J. 2010;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildhaber BE, Yang H, Teitelbaum DH. Total parenteral nutrition-induced apoptosis in mouse intestinal epithelium: modulation by keratinocyte growth factor. J Surg Res. 2003;112:144–151. doi: 10.1016/s0022-4804(03)00160-4. [DOI] [PubMed] [Google Scholar]
- Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24:3038–3046. doi: 10.1091/mbc.E13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Wang JY. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol. 2014;19:46–53. doi: 10.1016/j.coph.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers. 2014;2:e28320. doi: 10.4161/tisb.28320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Rao JN, Guo X, Liu L, Zou T, Turner DJ, Wang JY. Akt kinase activation blocks apoptosis in intestinal epithelial cells by inhibiting caspase-3 after polyamine depletion. J Biol Chem. 2004;279:22539–22547. doi: 10.1074/jbc.M314337200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–7919. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.