Abstract
An increasing number of genetic variants have been implicated in autism spectrum disorders (ASD), and the functional study of such variants will be critical for the elucidation of autism pathophysiology. Here, we report a de novo balanced translocation disruption of TRPC6, a cation channel, in a non-syndromic autistic individual. Using multiple models, such as dental pulp cells, iPSC-derived neuronal cells and mouse models, we demonstrate that TRPC6 reduction or haploinsufficiency leads to altered neuronal development, morphology, and function. The observed neuronal phenotypes could then be rescued by TRPC6 complementation and by treatment with IGF1 or hyperforin, a TRPC6-specific agonist, suggesting that ASD individuals with alterations in this pathway might benefit from these drugs. We also demonstrate that MeCP2 levels affect TRPC6 expression. Mutations in MeCP2 cause Rett syndrome, revealing common pathways among ASDs. Genetic sequencing of TRPC6 in 1041 ASD individuals and 2872 controls revealed significantly more nonsynonymous mutations in the ASD population, and identified loss-of-function mutations with incomplete penetrance in two patients. Taken together, these findings suggest that TRPC6 is a novel predisposing gene for ASD that may act in a multiple-hit model. This is the first study to use iPSC-derived human neurons to model non-syndromic ASD and illustrate the potential of modeling genetically complex sporadic diseases using such cells.
Keywords: autism, induced pluripotent stem cells, disease modeling
Introduction
Autism spectrum disorders (ASDs) are complex neurodevelopmental disorders that are characterized by deficits in reciprocal social interaction and communication as well as the presence of repetitive behaviors and highly restricted interests. While the allelic ASD architecture remains unclarified, there is definitive evidence of a high degree of locus heterogeneity and a contribution from rare and de novo variants 1. However, determining a contributing role from low-frequency variants is challenging, particularly for variants that are transmitted in a non-Mendelian fashion, carry intermediate risks, and are present in conjunction with a tremendous amount of apparently neutral rare variations in the human genome 2–4.
Reprogramming somatic cells to a pluripotent state by transient over-expression of specific factors enables the development of neuronal models of genomes that are pre-disposed to human diseases 5. We recently demonstrated the utility of induced pluripotent stem cells (iPSCs) for investigating the functional consequences of mutations in the gene encoding the methyl CpG binding protein-2 (MeCP2) in neurons from patients with Rett syndrome (RTT), a syndromic form of ASD 6, 7. Neurons derived from RTT-iPSCs display several alterations compared with controls, such as increased frequency of de novo L1 retrotransposition, decreased soma size, altered dendritic spine density, and reduced excitatory synapses. Therefore, functional studies using neuronal cultures derived from iPSCs from ASD individuals are an important tool to explore the contribution of rare variants to ASD etiology. Furthermore, by capturing the genetic heterogeneity of ASDs, the iPSC model might clarify whether ASD individuals carrying distinct mutations in disparate genes share common cellular and molecular neuronal phenotypes.
Here, we characterize the breakpoints of a de novo balanced translocation t(3;11)(p21;q22) in an ASD individual that disrupts the TRPC6 gene. TRPC6, a gene not previously implicated in ASD, encodes for the canonical transient receptor potential 6 channel, a voltage-independent, Ca2+-permeable cation channel involved in dendritic spine and excitatory synapse formation 8, 9. The biological impact of the genetic alteration in the index case and its functional relationship to ASD etiology was evaluated through several analyses using the affected individual’s dental pulp cells (DPCs), mouse models, and neural cells derived from iPSCs. To test the hypothesis that different ASD-related variants can produce similar biological effects, we compared the neuronal phenotypes of iPSC-derived neurons from the TRPC6-mutant (TRPC6-mut) individual with those of patients with RTT syndrome. Finally, we conducted a large-scale case-control sequence analysis of TRPC6, which revealed a significant association of mutations in this gene with ASD.
Materials/Subjects and Methods
For More detailed information, please refer to Supplementary Methods.
Patient ascertainment
ASD individual F2749-1 (TRPC6-mutant)
The 8-year-old proband is the only child of non-consanguineous healthy parents. He was born at term after an uncomplicated pregnancy with no malformations recognized at birth. He was noted to have delayed motor skills development and poor social responsiveness and was brought for medical examination at 2 years of age. His hearing was tested and found to be normal. He did not suffer from any other chronic medical conditions, and there was no history of head trauma or seizure. On examination, the individual met the DSM-IV criteria for autistic disorder, and the diagnosis was supported by the administration of the Childhood Autism Rating Scale (CARS). The electroencephalogram and magnetic resonance imaging were normal. The individual did not have dysmorphic features, except for synophrys, which is also present in other members of the father’s family. A molecular test for Fragile-X Syndrome was normal. Karyotype analysis revealed a balanced translocation (46, XY, t[3;11] [p21;q22]) in the proband that was not found in the parents. Parenthood was confirmed through genotyping of microsatellite markers. Controls: As controls, we used six non-affected individuals that are non-related to the individual. Cells from two control individuals (USC1 and P603) were selected for reprogramming follow up studies. This project was approved by the Ethics Committees of the institutes at which the study was conducted. After a complete description of the study, written informed consent was provided by the parents.
Analysis of genomic copy number variations
Genomic DNA was hybridized to the HumanHap300 Genotyping BeadChip from Illumina (La Jolla, CA, USA) according to manufacturer’s protocol to detect possible CNVs in the ASD individual. The data were analyzed using PennCNV 10 and QuantiSNP 11 software, and the results were compared to the database of genomic variants (http://projects.tcag.ca/variation/) to classify the identified CNVs as rare or common variants.
Fluorescent in situ hybridization
Chromosomes for fluorescent in situ hybridization (FISH) analysis were prepared from colchicine-treated lymphocytes of the proband. Bacterial artificial chromosomes (BACs) encompassing the genomic regions of interest were selected from the RPCI-11 library (Roswell Park Cancer Institute) using the UCSC genome browser (http://genome.ucsc.edu/, assembly Mar. 2006, NCBI36/hg18). The BACs were fluorescently labeled by nick translation and hybridized to the metaphase spreads using standard protocols 12.
Exome sequencing
Exome sequencing and analysis were performed by BGI Tech (Shenzhen, China). Briefly, genomic DNA samples were randomly fragmented into segments with a base-pair peak of 150 to 200 bp, and library enrichment for exonic sequences was performed using Agilent SureSelect Human All Exon 51M (for individual and mother) or Agilent SureSelect Human All Exon 71M (for the father). The captured libraries were loaded on Hiseq2000, and the sequences of each individual were generated as 90-bp paired-end reads. The coverage for the three individuals was 80-fold. Burrows-Wheeler Aligner (BWA) was used for the alignment. Single nucleotide polymorphisms (SNPs) were identified by SOAPsnp, small insertion/deletion (InDels) were detected by Samtools/GATK, and single nucleotide variants (SNVs) were detected by 1/35 Varscan.
Isolation and culture of human DPCs
DPC lineages were obtained as described elsewhere 13. Briefly, dental pulp tissues were digested in a solution of 0.25% trypsin for 30 minutes at 37°C. The cells were cultivated in DMEM/F12 media (Gibco) supplemented with 15% fetal bovine serum (Hyclone, TX), 1% penicillin/streptomycin, and 1% non-essential amino acids and maintained under standard conditions (37°C, 5% CO2). The DPC control lineages used for the whole-genome expression analysis were donated by Dr. Daniela Franco Bueno and Gerson Shigueru Kobayashi of the University of São Paulo. One of the DPC control lineages used for iPSC generation was a kind gift from Dr. Songtao Shi (University of Southern California).
RNA extraction
RNA samples were extracted from lymphocytes, DPCs, and iPSCs using Trizol reagent (Invitrogen, CA) and treated with Turbo DNA-free (Ambion). Sample concentrations and quality were evaluated using a Nanodrop 1000 and gel electrophoresis.
Microarray studies
For microarray experiments, 100 ng of RNA was converted to cDNA, amplified, labeled, and hybridized to the Human Gene 1.0ST chip from Affymetrix following the manufacturer’s protocol. The chips were scanned using the GeneChip® Scanner 3000 7G System, and a quality control was processed using Affymetrix® Expression Console™ Software. The data were normalized using the robust multi-array average (RMA) method 14, and the differentially expressed genes were selected with the significance analysis of microarrays method (SAM) 15 and RankProd 16. To select DEGs, we used a p-value < 0.05 adjusted for the false discovery rate (FDR) and 3,000 permutations. Functional annotation, canonical pathways, and networks analyses were performed using Ingenuity Pathways (http://www.ingenuity.com/). The CREB target genes database (http://natural.salk.edu/CREB/search.htm 17) was used to determine whether the DEGs found are regulated by the transcription factor CREB.
Gene expression analyses by qPCR
RNA samples were reversed transcribed into cDNA using the Super Script III First Strand Synthesis System (Invitrogen, CA) according to the manufacturer’s instructions. The reactions were run on an Applied Biosystem 7500 sequence detection system using SYBR Green master mix (Applied Biosystems, CA). The primers were designed using PrimerExpress v. 2.0 software (Applied Biosystems, CA), and specificity was verified by melting curve analysis using 7500 System SDS v. 1.2 Software (Applied Biosystems, CA). Quantitative analysis was performed using the comparative threshold cycle method 18. GeNorm (www.medgen.ugent.be/genorm/) was used to determine the stability of the reference genes GAPDH, HPRT1, SDHA, and HMBS and to generate a normalization factor for the expression values of the target genes. The principles of analysis of geNorm have been described 19. Microarray validation was performed using the one-tailed unpaired t-test with Welch’s correction to compare the qPCR expression values obtained for the ASD individual and controls. A concentration of 10 µM hyperforin was used to treat the DPCs of a control sample for 15 and 30 minutes and 1, 3, 6, 24, and 48 hours. The samples were prepared in triplicate, and the results were normalized by the values obtained for an untreated sample. Primers used on this work are described in Table S1.
Western blotting
Rabbit anti-TRPC6 (ProScience, 1:250; Sigma, 1:1000); mouse anti-TRPC6 (Abcam, 1:1000); rabbit anti-CREB (Cell Signaling, 1:500); rabbit anti-P-CREB (Cell Signaling, 1:500); and mouse anti-β-actin (Ambion, 1:5000) antibodies were used as primary antibodies. Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse (Promega, 1:2000) antibodies were used as secondary antibodies. ECL Plus (Amersham) was used for signal detection. For Semiquantitative analysis of p-CREB signal, intensity was corrected with respect to CREB/β-actin relative quantification. A paired t-test analysis with a p-value < 0.05 was used to compare the control and ASD individual p-CREB signal intensity normalized data.
Cellular reprogramming
The iPSCs were obtained from the DPCs of the ASD individual and a control. Briefly, DPCs were transduced with retroviruses containing OCT4, SOX2, KFL4, and MYC to induce overexpression of these genes 5. Two days after transduction, the cells were transferred to a co-culture system with murine embryonic fibroblasts (mEFs) maintained with DMEM/F12 (Invitrogen, CA), 20% Knockout Serum Replacement (Invitrogen, CA), 1% non-essential amino acids, and 100 µM beta-mercaptoethanol and treated with 1 mM valproic acid (Sigma) for 5 days. The iPSC colonies were identified after approximately 2 weeks in this culture system, transferred to Matrigel (BD Biosciences)-coated plates, and maintained in mTeSR media (Stem Cell Technologies).
Immunocytochemistry
The cells were fixed with PBS containing 4% paraformaldehyde for 10 minutes and then incubated at room temperature for 1 hour in a blocking solution containing 5% donkey serum and 0.1% Triton X-100. The primary antibodies were incubated overnight at 4°C, followed by incubation with secondary antibodies (Jackson ImmunoResearch) for 1 hour at room temperature. Images were captured with a Zeiss microscope. The primary antibodies used included the following: Tra-1-81 (1:100, Chemicon); Nanog and Lin28 (1:500, R&D Systems); Sox2 (1:250; Chemicon); human Nestin (1:100, Chemicon); Tuj1 (1:500, Covance); MAP2 (1:100; Sigma); VGLUT1 (1:200, Synaptic Systems); GABA (1:100, Sigma); Musashi (1:200, Abcam); Ctip2 (1:200, Abcam); and Tbr1 (1:200, Abcam).
Teratoma formation
iPSC colonies from five semi-confluent 100 mm dishes (1–3 ×106 cells) were harvested after treatment with 0.5 ng/mL dispase, pelleted, and suspended in 300 µL Matrigel. The cells were injected subcutaneously into nude mice; 5 to 6 weeks after injection, teratomas were dissected, fixed overnight in 10% buffered formalin phosphate, and embedded in paraffin. The sections were stained with hematoxylin and eosin for further analysis. The protocols were approved by the University of California San Diego Institutional Animal Care and Use Committee.
Fingerprinting and karyotype
Standard G-banding karyotype and DNA fingerprinting analysis were performed by Cell Line Genetics (Madison, WI).
Neuronal differentiation
The iPSC colonies were plated on Matrigel (BD Biosciences)-coated plates and maintained for 5 days in mTSeR media (Stem Cell Technologies). On the 5th day, the media was changed to N2 media [DMEM/F12 media supplemented with 1X N2 supplement (Invitrogen) and 1 µM dorsomorphin (Tocris). After 2 days, the colonies were removed from the plate and cultured in suspension as embryoid bodies (EBs) for 2–3 weeks using N2 media with dorsomorphin during the entire procedure. The EBs were then gently dissociated with accutase (Gibco), plated on Matrigel-coated dishes, and maintained in NBF media (DMEM/F12 media supplemented with 0.5X N2, 0.5X B7 supplements, 20 ng/mL FGF and 1% penicillin/streptomycin). The rosettes that emerged after 3 or 4 days were manually selected, gently dissociated with accutase, and plated in dishes coated with 10 µg/mL poly-ornithine and 5 µg/mL laminin. This NPC population was expanded using NBF media. To differentiate the NPCs into neurons, the cells were re-plated with 10 µM ROCK inhibitor (Y-27632, Calbiochem) in the absence of FGF, with regular media changes every 3 or 4 days.
Ca2+ influx studies
Intracellular Ca2+ levels were monitored using Fluo-4 AM. The cells were incubated for 45 minutes at 37°C with 2.5 µM Fluo-4 AM and superfused for 5 minutes with HBSS buffer before the beginning of the recording. A concentration of 10 µM hyperforin (a kind gift from Dr. Willmar Schwabe GmbH & Co, Karlsruhe, Germany) was used in combination with 100 µM FFA (Sigma-Aldrich) for TRPC6 activation. Images were captured at 6-second intervals for 30 minutes using a Biorad MRC 1024 confocal system attached to an Olympus BX70 microscope. The drugs were applied at the 3rd minute using a perfusion system. A triplicate of each individual was analyzed. The average fluorescence of the individual cells was quantified and normalized to the resting fluorescence level for each cell. The plugins MultiMeasure and MeasureStacks from ImageJ software were used to measure fluorescence intensity. The analyses were performed blinded to avoid bias.
Cell cycle analysis
A total of 1×106 NPCs were harvested from a single-cell suspension with PBS washing buffer (PBS and 1% serum) and fixed in 75% EtOH for at least 2 hours at 4°C. After washing twice with washing buffer, the cells were stained with 200 µL propidium iodine (PI) solution (20 µg/mL propidium iodide, 200 µg/mL RNase A, and 0.1% Triton X-100). Multiple NPC samples from the TRPC6-mutant individual and controls were analyzed by fluorescence-activated cell sorting (FACS) on a Becton Dickinson LSRI, and cell cycle gating was examined using FLOWJO-Flow Cytometry Analysis Software.
Quantification of neuronal morphology and synaptic puncta
Neuronal tracing was performed on neurons for which the shortest dendrite was at least three times longer than the cell soma diameter using a semi-automatic ImageJ plug-in (NeuroJ). Spines and VGLUT1 puncta were quantified after three-dimensional reconstruction of z-stack confocal images. The same density of neurons was plated in each condition. Final cell density was confirmed by DAPI and Synapsin-EGFP-positive cells. Only Synapsin-EGFP positive neurons with spines were scored. Images were taken randomly for each individual and from two different experiments, using at least two different clones. Quantification was performed blind to the cell genotype. The total dendritic length includes the summed length of all dendrites per neuron and dendritic segment count represents the total number of dendritic segments per neuron. No distinction was made between different types of spines due to the unviability of this assessment using the presented method. All experiments were performed with independent clones and different controls. All analyses were performed blinded to avoid bias. For the rescue experiments, 10 ng/mL IGF1 (Peprotech) or 0.5µM hyperforin was added to neuronal cultures for 2 weeks.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed following the manufacturer’s protocol using a ChIP assay kit (Active Motif). The antibodies used were anti-MeCP2 and IgG (both from Upstate). We validated our antibody conditions for the ChIP assay with a previously characterized MeCP2 target, brain-derived neurotrophic factor (Bdnf) promoter in exon IV, and a negative region in another region of the promoter region as previously described 20, 21. The input was 5% for all samples. All ChIP assays were controlled by performing parallel experiments with either no antibody or with anti-IgG pull downs. After IP, the recovered chromatin fragments were subjected to qPCR using primers for the human TRPC6 promoter region. The primers used for human TRPC6 promoter ChIP were as follows: forward primer 1, 5’AACAGCTTGGAAACGTGGGA3’; reverse primer 1, 5’AAAGAGGCCAACAACCTGCT3’; forward primer 2, 5’TCGCAGTGACGGAAGGAAAA3’; and reverse primer 2, 5’AAACGCCAGATGTTCCCAGT3’. The qPCR values were normalized to the IgG precipitation and are shown as fold enrichment. All experiments were performed in triplicate.
Construction and characterization of retroviruses
Self-inactivating murine oncoretroviruses were engineered to express short-hairpin RNAs (shRNAs) under the control of the U6 promoter and green fluorescent protein (GFP) or the Discosoma sp. red fluorescent protein DsRed under the control of the Ef1 alpha promoter. shRNAs against TRPC6 and a non-silencing scrambled control shRNA were cloned into retroviral vectors as previously described 22. The following shRNA sequences were selected and cloned into retroviral vectors: shRNA-control, 5’-TTCTCCGAACGTGTCACGT-3’; shRNA-TRPC6-1, 5’-TCGAGGACCAGCATACATG-3’; and shRNA-TRPC6-3, 5’-CTCAGAAGATTATCATTTA-3’.
For rescue experiments, a resistant form of murine TRPC6 (TRPC6-WTR) was engineered to harbor six silent mutations in the region targeted by shRNA-TRPC6-1. The TRPC6 targeting sequence was mutated from AAT CGA GGA CCA GCA TAC ATG to AAC CGC GGC CCT GCT TAT ATG by site-directed mutagenesis. The resistant form of TRPC6 was cloned into a retroviral vector driven by the ubiquitin promoter followed by a bicistronic expression of GFP and a WPRE stabilization sequence. The specificity and efficiency of shRNA-control, shRNA-TRPC6-1, shRNA-TRPC6-3, and the TrpC6-WT constructs were verified by co-transfection into HEK-293 cells. Cell lysates were collected and analyzed by western blot analysis with anti-TRPC6 antibodies (Sigma).
Primary hippocampal cultures
Hippocampal neuronal cultures were prepared from C57BL/6 E18 embryonic mice. Briefly, hippocampi were dissected, dissociated with trypsin, and plated at a density of 300 cells/mm2 on glass coverslips coated with poly-L-lysine and laminin. The hippocampal neurons were maintained in Neurobasal medium (Gibco) supplemented with B27 (Invitrogen). Neurons were treated with either shRNA scramble control or shRNA targeting TRPC6 at DIV12-14 and were fixed for further analysis at DIV21.
In vivo stereotaxic injection of engineered retroviruses into the dentate gyrus of adult mouse hippocampus
High titers of engineered retroviruses were produced by cotransfection of retroviral vectors and vesicular stomatitis viral envelope into the 293 GP cell line as described previously 23. Supernatants were collected 24 hours post transfection, filtered through 45-µm filters, and ultracentrifuged. The viral pellet was dissolved in 14 µL of PBS and stereotaxically injected into the hilus of anesthetized mice at four sites (0.5 µL per site at 0.25 µL/minute). The following coordinates were used: posterior = 2 mm from the bregma, lateral = ±1.6 mm, ventral = ±2.5 mm; posterior = 3 mm from the bregma, lateral = ±2.6 mm, ventral = ±3.2 mm. Adult C57BL/6 mice (6–8 weeks old, female) were used for the study. All procedures followed institutional guidelines.
Immunostaining and confocal analysis
Coronal brain sections (40 µm thick) were prepared from retrovirus-injected mice. Images of GFP+ cells were acquired on a META multiphoton confocal system. Neuronal positioning was analyzed by acquiring a single-section confocal image of a GFP+ cell body stained with DAPI and assigning it to one of the four domains as illustrated. A minimum of 10 GFP+ cells were randomly chosen from the each animal, and at least three animals were used for each experimental condition, as previously described 24. Statistical significance was determined by ANOVA. Dendritic development was analyzed by via a three-dimensional reconstruction of the entire dendritic tree from Z-series stacks of confocal images. The images were converted to two-dimensional projections for analysis of dendritic length and branch number using NIH ImageJ software and the NeuronJ plugin, as described previously 24. As a measure of arborization, Sholl analysis was performed by counting the number of dendritic crossings at a series of concentric circles at 10-µm intervals from the cell body using the Sholl analysis plugin.
Slice electrophysiology
Mice housed under standard conditions were anesthetized at 3 weeks post-retroviral injection, and acute coronal slices were prepared as previously described 25. The brains were removed and placed in an ice-cold cutting solution containing the following: 110 mM choline chloride; 2.5 mM KCl; 1.3 mM KH2PO4; 25 mM NaHCO3; 0.5 mM CaCl2; 7 mM MgCl2; 10 mM dextrose; 1.3 mM sodium ascorbate; 0.6 mM sodium pyruvate; and 5 mM kynurenic acid. Slices were cut into 300-µm-thick sections with a vibratome (Leica VT1000S) and transferred to a chamber containing ACSF: 125 mM NaCl; 2.5 mM KCl; 1.3 mM KH2PO4; 25 mM NaHCO3; 2 mM CaCl2; 1.3 mM MgCl2; 1.3 mM sodium ascorbate; 0.6 mM sodium pyruvate; and 10 mM dextrose (pH 7.4, 320 mOsm), saturated with 95% O2, 5% CO2 at 35°C for 20 minutes. The slices were then maintained at room temperature for at least 45 minutes prior to placement in the recording chamber. The slices were maintained at room temperature and used for the following 4 hours. Electrophysiological recordings were performed at 34°C. Microelectrodes (4–6 MΩ) were filled with a solution containing the following: 120 mM potassium gluconate; 15 mM KCl; 4 mM MgCl2; 0.1 mM EGTA; 10.0 mM HEPES; 4 mM MgATP; 0.3 mM Na3GTP; and 7 mM phosphocreatine (pH 7.4, 300 mOsm). The whole-cell patch-clamp configuration was used in the current-clamp mode. Approximately 10–20 giga-ohm seals were obtained with borosilicate glass microelectrodes. The electrophysiological recordings were obtained at 32–34°C. Neurons and dendrites were visualized through differential interference contrast microscopy. The data were collected using an Axon Instruments 200B amplifier and acquired via a Digidata 1322A at 10 kHz.
Electrophysiology recordings using cultured human iPSC-derived neurons
Whole-cell patch clamp recordings were performed using cells cultured in the absence of astrocytes after approximately 6 weeks of differentiation. Before the recordings, the growth media were removed and replaced with a bath solution comprising the following: 130 mM NaCl; 3 mM KCl; 1 mM CaCl2; 1 mM MgCl2; 10 mM HEPES; and 10 mM glucose (pH 7.4) at room temperature (22–24°C). The electrodes for whole-cell recordings were pulled on a Flaming/Brown micropipette puller (Model P-87, Sutter Instrument, Novato, CA) from filamented borosilicate capillary glass (1.2 mm OD, 0.69 mm ID, World Precision Instruments, Sarasota, FL). The electrodes were fire polished, and the resistance values were typically 2–5 MΩ for the voltage-clamp experiments and 7–9 MΩ for the current-clamp experiments. The pipette solution contained the following: 138 mM KCl; 0.2 mM CaCl2; 1 mM MgCl2; 10 mM HEPES (Na+ salt); and 10 mM EGTA, (pH 7.4). The osmolarity of all solutions was adjusted to 290 mOsM. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), with the exception of MgCl2 (J.T. Baker, Phillipsburg, NJ). Current traces in voltage clamp were leak-subtracted. Liquid junction potentials were nulled for each individual cell with the Axopatch 1C amplifier (Molecular Devises, Sunnyvale, CA). The analyses were performed in a double-blinded manner to avoid bias.
Behavioral tests in mice
The three-chamber test was used to evaluate the social behavior of TRPC6 wild type (WT), heterozygous (HET) and knockout (KO) mice. To evaluate repetitive behavior, the mice were initially observed for 10 minutes in the dark, and the time spent in grooming and freezing behavior was measured. After 5 minutes of habituation under a light condition, a small cage with a never-met animal was introduced to one side of the box, and an empty cage was introduced to the other side. The time spent in each chamber and the time spent during nose-to-nose interaction between the animals was measured. Adult mice (6–8 weeks old, male) with a C57BL/6 background were used for the study. At least 12 animals per group were utilized as biological replicates. The experimenter was blind to the genotypes. The data were analyzed using the non-parametric Kruskal-Wallis ANOVA. The analyses were performed in a double-blinded manner. All procedures followed institutional guidelines.
Mutation screening of TRPC6
Cohorts
The clinical characteristics of the Simons Simplex Collection (SSC) have previously been described in detail 26. The following exclusion criteria were used to filter the cases: 1) ineligible/ancillary status as per SSC Family Distribution List v13; 2) missing genotyping data; 3) genotyping call rate < 95%; 4) discrepancy of genotyping data with recorded gender; 5) Mendelian inconsistencies or cryptic relatedness (up to and including second-degree relatives); and 6) non-European ancestry. A total of 1041 of 1195 cases were included in the final case cohort. The National Institute of Neurological Disorders and Stroke (NINDS) Neurologically Normal Caucasian Control Panel of unrelated adult controls do not have a personal or family history (first-degree relative) of neuropsychiatric illness (http://ccr.coriell.org/Sections/Collections/NINDS/DNAPanels.aspx?Pgld=195&coll=ND). Of the 953 samples from the DNA panels NDPT020, 079, 082, 084, 090, 093, 094, 095, 096, 098, and 099, 942 passed the quality control checks described above. Additional sequence data for TRPC6 were derived from unrelated northern European (NE) adults present in an exome-sequencing database in our laboratory. Genotyping and whole-exome data were obtained for 2076 individuals, of which 1930 passed the quality control checks described above.
Mutation screening
For 1031 SSC cases and all 942 NINDS controls, amplification of the coding exons and splice sites was performed using lymphoblastoid cell line-derived genomic DNA via multiplex PCR using RainDance technology (Table S2; Lexington, MA, USA). The resulting PCR products were subjected to high-throughput sequencing on a Genome Analyzer IIx (Illumina, San Diego, CA, USA) at the Yale Center for Genomic Analysis. An in-house script was used to generate a list of variants (see Supplementary Materials for more details). Whole-exome data for 10 additional SSC cases were available and filtered for nonsynonymous singleton variants with a SAMtools SNP quality score > 50. Variant confirmation was performed on blood-derived genomic DNA for the cases because it was available and on lymphoblastoid cell line-derived genomic DNA for NINDS controls using conventional PCR and Sanger sequencing. Segregation analysis was performed on blood-derived genomic DNA for cases for which family members were available. Chromatograms were aligned and analyzed for variants using the Sequencher v4.9 program (Gene Codes, Ann Arbor, MI, USA). For the NE controls, whole-exome sequencing data were filtered by the same parameters used for the 10 SSC cases: nonsynonymous singleton variants with a SAM tools SNP quality score > 50. No read threshold was used to maximize sensitivity over specificity. These variants were not confirmed by Sanger sequencing, but the filtering parameters typically lead to a 70% confirmation rate in our experience. Therefore, we have included the maximum possible number of variants from the NE control cohort. To obtain the exome data, genomic DNA from both the 10 SSC probands and 1930 NE controls was enriched for exonic sequences using NimbleGen capture and sequenced by the Illumina Genome Analyzer IIX or HiSeq2000. The novelty and singleton status of all variants were determined by comparing all three cohorts and screening dbSNP137 and Exome Variant Server v.0.0.15 (NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, URL: http://evs.gs.washington.edu/EVS/), accessed 11/01/2012. All p values for mutation burden are two-tailed and calculated from Fisher’s exact test.
Results
Characterization of the t(3;11)(p21;q22) translocation breakpoint and exome sequencing
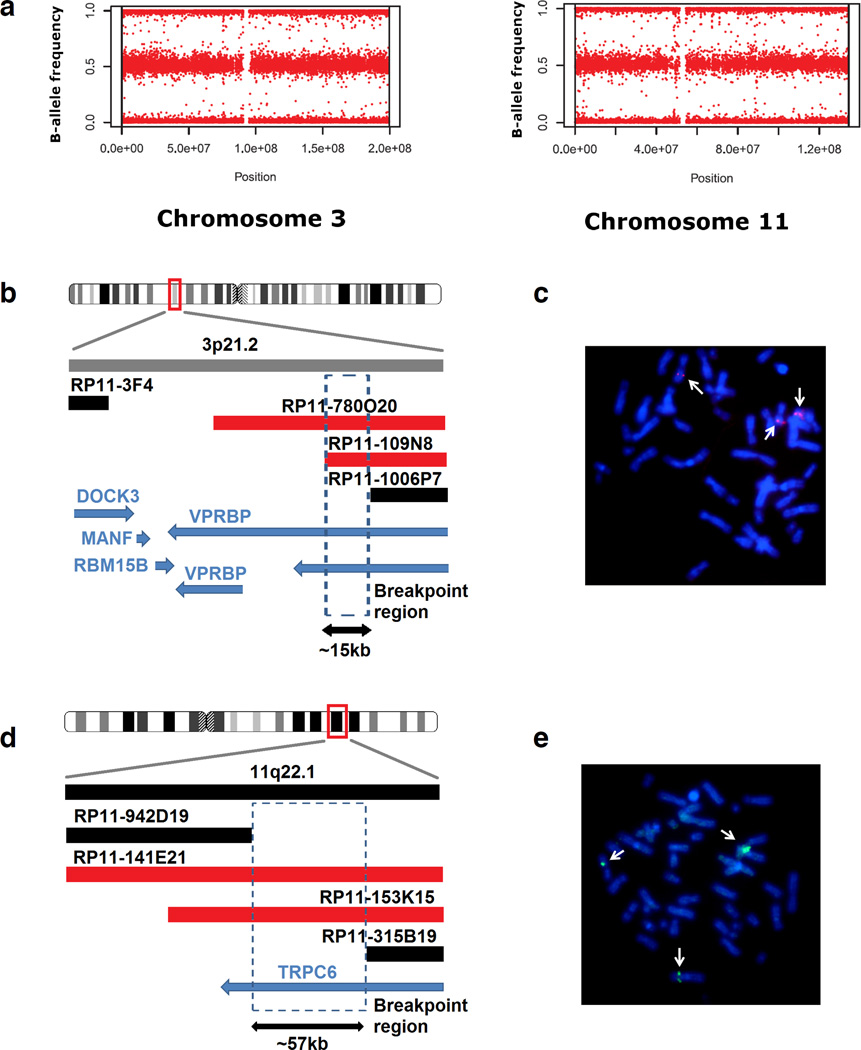
We identified an 8-year-old male autistic individual carrying a de novo 46, XY, t(3;11)(p21;q22) translocation by G-banding karyotyping of lymphoblastoid cells. No gain or loss of genetic material was observed near the breakpoint areas via a genome-wide array analysis (Figure 1a). Only a duplication (104.225.150 bp - 104.339.273 bp) on chromosome 14 was identified, which was previously shown to be a common copy number variant (CNV; http://projects.tcag.ca/variation/). Fluorescent in situ hybridization (FISH) analysis revealed that BAC probes RP11-780O20 and RP11-109N8 span the breakpoint on chromosome 3p21, while probes RP11-3F4 and RP11-1006P7 map distal and proximal to the breakpoint, respectively (Figure 1b, c). This narrowed the breakpoint to an interval of approximately 15 kb spanning the gene encoding the Vpr-binding protein (VPRBP), indicating that this gene was disrupted. Similarly, the breakpoint on chromosome 11q22 was mapped to a region spanned by probes RP11-141E21 and RP11-153K15, distal to RP11-315B9 and proximal to RP11-942D19 (Figure 1d, e), suggesting disruption of the TRPC6 gene, which was confirmed by the use of additional strategies.
Figure 1. Mapping the breakpoints in the ASD individual with the 46, XY, t(3;11)(p21;q22) karyotype.
(a) The allele frequency distribution plot for chromosomes 3 and 11 generated by SNP array genotyping showed no gain or loss of genetic material on these chromosomes. (b) The schematic view of the BAC probes used and the surrounding breakpoint area on chromosome 3. RP11 probes marked in red span the breakpoint, while the black ones do not. The shared region between probes RP11-780O20 and RP11-109N8 narrows the breakpoint area to a region inside the VPRBP gene. The blue arrows indicate open reading frames. (c) FISH imaging showing that RP11-780O20 probe (red signal) binds to normal and derivative chromosome 3 and to derivative chromosome 11, indicating that the probe spans the breakpoint (arrows). (d) A schematic view of the BAC probes used and the surrounding areas on chromosome 11. A shared region between probes RP11-153K15 and RP11-141E21 places the breakpoint in TRPC6. (e) FISH image showing the BAC probe RP11-153K15 (green signal) bound to normal chromosome 11 and both derivatives chromosomes 3 and 11 (arrows).
We first measured the expression levels of TRPC6 exons 4, 6, 12, and 13 in the lymphocytes of the ASD individual, his parents, and six non-affected control individuals by quantitative real-time PCR (qPCR) (Figure S1a). In the ASD individual’s parents and in six other individual controls, exons 6, 12, and 13 had similar expression levels as exon 4. In the ASD individual, however, the expression levels of exons 12 and 13 were reduced by 60% compared to exon 4. After sequencing all TRPC6 exons, we found that the individual was heterozygous for two common polymorphisms: one mapping to exon 6 (rs12366144) and the other to exon 13 (rs12805398). However, sequencing of cDNA from the individual’s lymphocytes revealed heterozygosity only for the polymorphism in exon 6 (Figure S1b). Parentage was confirmed through genotyping of microsatellite markers (Figure S1c). These results demonstrate that TRPC6 has biallelic expression and that the heterozygosity loss in exon 13 in the individual’s cDNA can be explained by TRPC6 disruption. Accordingly, TRPC6 is transcribed up to the breakpoint, which is located between exons 6 and 12. We did not identify any pathogenic change in TRPC6 exons upon sequencing the individual’s DNA (data not shown). We also did not identify any extra band in the protein extracts from individual’s cells using a N-terminal antibody, indicating that a truncated TRPC6 form is unlikely to be a byproduct of the translocation (Figure S1d).
Disruption of TRPC6, VPRBP, and several other unknown genes might contribute to the ASD phenotype. To identify other genetic alterations in this ASD individual, we performed exome sequencing on the individual and compared the result to those for his parents. Exome sequencing analysis revealed 50 de novo, rare, nonsynonymous variants and three frameshift insertions/deletions in the individual. Consultation with AutismKB 27 indicated that none of the other genes harboring genetic variants are associated with ASD, with the exception of the cyclic adenosine monophosphate-specific (PDE4A) gene, for which lower levels of expression have been observed in the brains of autistic individuals 28. We also observed an alteration in the ATXN3 gene, linked to the spinocerebellar ataxia-3 disease in humans. All genetic variants are presented in Table S3.
TRPC6 disruption leads to transcriptional alterations and dysregulation of CREB phosphorylation
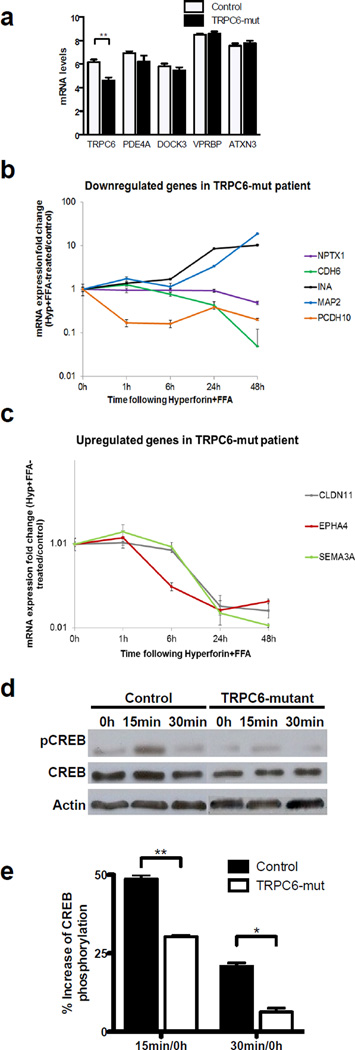
To determine gene transcription due to genetic perturbations in the ASD individual carrying the novel chromosomal translocation, we conducted a global expression analysis comparing the individual’s dental pulp cells (DPCs) to six control samples. DPCs can be easily isolated from the deciduous teeth of ASD individuals via a non-invasive procedure 29. DPCs have an ectodermic neural crest origin, express several neuronal markers, and have proven to be a useful model to study ASD 30–32. We identified 67 differentially expressed genes (DEGs) between the ASD individual and non-affected controls (P<0.05; Table S4). Functional annotation analysis revealed that 16 (24%) of these genes have a role in nervous system development and function (Table 1). We confirmed the reduction of TRPC6 expression (P<0.01) but not VPRBP (Figure 2a). The reduced level of TRPC6 expression is likely due to nonsense-mediated decay or rearrangement of regulatory elements caused by the translocation. Moreover, PDE4A, ATXN3 and DOCK3 (another neuronal gene 33 present near the break point on chromosome 3, Figure 1b) were also not differentially regulated in the individual’s DPCs (Figure 2a). TRPC6 is a Ca2+-permeable, nonselective, cation channel involved in neuronal survival, growth cone guidance, and spine and synapse formation, biological processes that have previously been implicated in ASD etiology 8, 9, 34–36. The function of VPRBP (Vpr-binding protein) is less clear and may include DNA replication, S-phase progression, and cellular proliferation 37. Given the time-consuming nature of additional functional analyses, we elected to focus on additional genetic and functional studies of TRPC6, which has not been previously associated with ASDs.
Table 1.
Selected functionally relevant genes differentially expressed between the TRPC6-mutant individual and controls.
| Gene | Fold change* | Gene Ontology | Regulation by CREB** |
qPCR validation (p value) |
|---|---|---|---|---|
| INA | −2.639988194 | Nervous system development; neurofilament cytoskeleton organization |
ChIP-on-chip | 0.0198 |
| NPTX1 | −2.855578291 | Growth of neurites; synaptic transmission; central nervous system development |
In silico | 0.0885 |
| MAP2 | −2.789671289 | Growth of neurites; development and elongation of neurites; patterning of cerebral cortex; polarization of hippocampal neurons |
ChIP-on-chip | 0.0363 |
| EPHA4 | 2.362428255 | Guidance of axons; formation of the pyramidal tract; axon guidance |
ChIP-on-chip; In silico |
0.4305 |
| CLDN11 | 4.066602785 | Axon ensheathment; calcium-independent cell-cell adhesion; migration of neuroglia |
In silico Lui et al., 2007 |
0.0005 |
| PCDH10 | −4.318180517 | Cell adhesion; establishment and function of specific cell-cell connections in the brain |
ChIP-on-chip; In silico |
0.3331 |
| CLDN1 | 4.171417178 | Calcium-independent cell-cell adhesion; myelination of cells |
In silico | |
| PTGS2 | −3.49316255 | Activation of astrocytes; activation of neuroglia; memory; positive regulation of synaptic plasticity; negative regulation of synaptic transmission, dopaminergic; positive regulation of synaptic transmission, glutamatergic |
ChIP-on-chip Gosh et al., 2007 |
|
| CDH6 | −2.675463010 | Cell-adhesion; establishment and function of specific cell-cell connections in the brain |
No evidence | 0.0418 |
| SEMA3A | 2.314408538 | Nervous system development; axonal fasciculation; regulation of axon extension involved in axon guidance; distribution of neurons; migration of neuroglia; growth of neurites; chemorepulsion of sympathetic neurons |
No evidence | 0.1828 |
| CASP1 | 2.545250054 | Activation of astrocytes; activation of neuroglia | No evidence | |
| VCAM1 | 4.546975557 | Growth of neurites; distribution of neurons; cell adhesion; guidance of axons |
No evidence | |
| ACAN | −4.199956627 | Growth of neurites; cell adhesion | No evidence | |
| CCL2 | 2.41655874 | Cell adhesion; astrocyte cell migration | No evidence | |
| HGF | 4.252390982 | Growth of neurites; complexity of dendritic trees | No evidence | |
| PCDH18 | 2.508559732 | Cell adhesion; brain development | No data available |
Logarithmic gene expression difference between ASD individual and controls
Evidence of gene transcription regulation by the transcription factor CREB according to the database http://natural.salk.edu/CREB/search.htm. Zhang and colleagues (2005) used three different strategies to identify the genes regulated by CREB: in silico analysis, chromatin co-immunoprecipitation followed by microarray analysis (ChIP-on-ChIP) and expression analysis of genes induced by forskolin (array). The genes for which no evidence of CREB regulation was found are annotated as "no evidence"; those for which no information is available in the analyzed database are annotated as "no data available".
Figure 2. TRPC6 channels regulate the expression of neuronal development genes.
(a) Differential gene expression in the controls and ASD individual cells of candidate genes located in the translocation region or detected by exome sequencing. Only TRPC6 displayed a significant reduction in mRNA levels (P<0.01). (b) Decreasing expression of candidate genes upon TRPC6 stimulation with hyperforin/FFA. (c) Genes upregulated in the TRPC6-mut genetic background after hyperforin/FFA treatment. (d) Representative western blot showing increased CREB phosphorylation after 15 and 30 minutes of hyperforin stimulation normalized to non-stimulated cells. (e) The level of CREB phosphorylation in DPCs from the TRPC6-mut individual after TRPC6 activation with hyperforin is significantly lower compared with the control sample (n = 3, P< 0.05; t-test). The error bars in all panels show the s.e.m. *P<0.05; **P<0.01; ***P<0.001.
Using the CREB-target genes database (http://natural.salk.edu/CREB/), we determined that 8 of the 16 functionally relevant DEGs are regulated by CREB, a transcription factor that is activated upon Ca2+ influx through TRPC6 8. Of the functionally relevant DEGs, we evaluated 6 CREB-target genes (INA, MAP2, NPTX1, CLDN11, PCDH10, and EPHA4) and two other genes (SEMA3A and CDH6) by quantitative PCR (qPCR) to validate the microarray experiments. We measured dysregulated expression of CDH6 (-2.68-fold, P<0.05), INA (−2.64-fold, P<0.05), MAP2 (−2.79-fold, P<0.05), and CLDN11 (4.07-fold, P<0.001) in the individual compared with controls in the same direction observed in the microarray analysis (Table 1). To validate that TRPC6 haploinsufficiency is leading to transcriptional dysregulation of these genes, we treated a control DPC culture with hyperforin plus flufenamic acid (FFA) and measured the expression levels of the same candidate genes over 48 hours (Figure 2b, c and Figure S1e). Hyperforin specifically activates TRPC6 and FFA increases the amplitude of the currents through this channel 38–40. If the candidate genes are regulated through TRPC6 signaling, we expect a change in their expression levels opposite to the observed change in the TRPC6-mut individual. After a 48-hour treatment, we observed the expected correlation for five of the eight genes. While the expression levels of SEMA3A, EPHA4, and CLDN11 were significantly reduced (−28-fold, −3.2-fold and −4.76-fold, respectively), MAP2 and INA displayed 20- and 10-fold increases in expression, respectively. These results validate the microarray data and support the hypothesis that the selected genes are regulated by the TRPC6 pathway.
We measured CREB phosphorylation in the DPCs of the individual and a control to assess the functional effect of TRPC6 disruption. Stimulation of DPCs with hyperforin plus FFA induced a significantly reduced level of increased phosphorylated CREB (p-CREB) in the individual’s DPCs (30.3±0.7%) compared to control (48.6±2.3%; P<0.005) after 15 minutes. After 30 minutes, p-CREB levels in the individual’s DPCs (6.3±2.1%) were also significantly lower compared to the control (20.9±2.1%; P<0.05) (Figure 2d and e). Taken together, these results demonstrate that several of the functionally relevant DEGs identified in the microarray studies are controlled via TRPC6 signaling, likely through CREB phosphorylation, suggesting that TRPC6 disruption influences neuronal cell function.
Generation of neural cells from ASD individuals
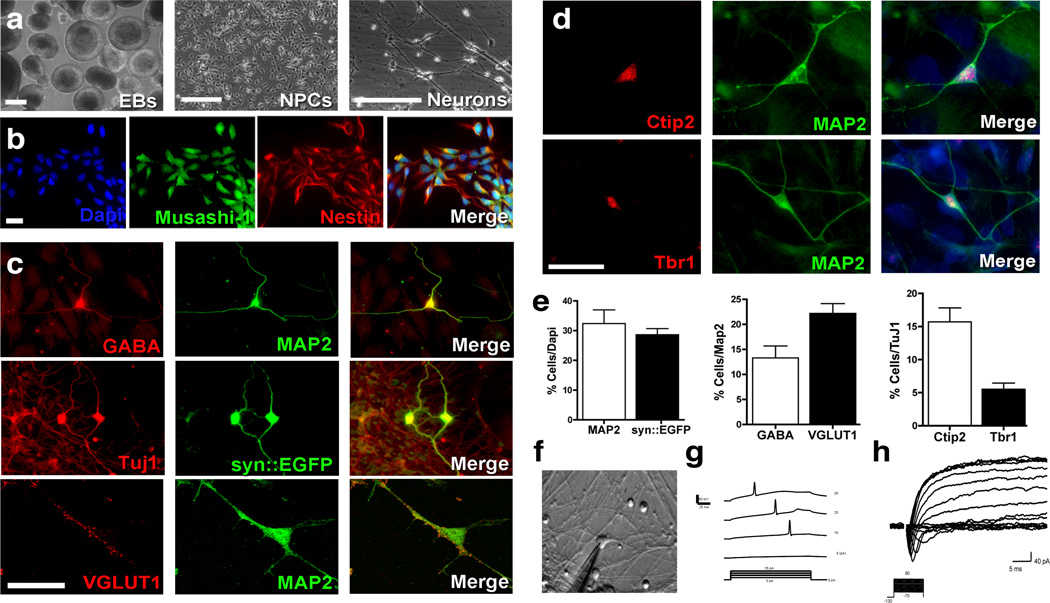
To further evaluate the effect of TRPC6 haploinsufficiency on neural cell function, we generated iPSCs from DPCs from the ASD individual and two control individuals (Figure S2 and Table S5 and S6). We chose to reprogram DPCs because these cells develop from the same set of early progenitors that generate neurons. Furthermore, the neurons derived from iPSCs generated from DPCs express higher levels of forebrain genes, many of which are implicated in ASD 41. We fully characterized three clones from each individual and used at least two different clones for follow up experiments. A summary of the clones used for each experiment can be found in Table S6. Neural progenitor cells (NPCs) and cortical neurons from iPSCs were obtained using a modified protocol from our previous publication 6. Briefly, iPSC colonies on Matrigel were treated with dorsomorphin under FGF-free conditions until confluence. Pieces of iPSC colonies were grown in suspension for 2–3 weeks as embryoid bodies (EBs) in the presence of dorsomorphin (Figure 3a). The EBs were then dissociated and plated to form rosettes. The rosettes were manually selected and expanded as NPCs (Figure 3a). These NPCs were negative for the pluripotent marker OCT4 and positive for early neural-specific markers such as Musashi-1 and Nestin (Figure 3b and Figure S3a). To obtain mature neurons, NPCs were plated with ROCK inhibitor and maintained for 3–4 additional weeks under differentiation conditions. At this stage, the cells were positive for the pan neuronal marker Tuj1 (β-III-Tubulin) and expressed the more mature neuronal markers synapsin I (SYN1) and microtubule-associated protein 2 (MAP2; Figure 3c). These cells expressed genes typically found in the cortex, including CTIP2, important for the differentiation of subcortical projecting neurons; TBR1, critical for cortical development and ABAT, a marker for GABAergic neurons, encoding for the 4-aminobutyrate aminotransferase protein and responsible for the catabolism of GABA neurotransmitter. (Figure S3a). Expression of NESTIN indicates the presence of NPCs and the expression of S100B and GFAP are indicative of glia cells, suggesting a mixed cell population at this stage (Figure S3a). In our cultures, the presynaptic SYN1 puncta were frequently adjacent to the postsynaptic marker HOMER1, suggesting the presence of developed synapses (Figure S3b). Using immunostainning, we also detected expression of the inhibitory neurotransmitter γ-aminobutyric acid (GABA) in 13% of the neurons, and 22% were positive for vesicular glutamate transporter-1 (VGLUT1), a marker for excitatory neurons, in both controls and ASD subjects (Figure 3c–e). Our protocol generated a consistent population of forebrain neurons, confirmed by the co-localization of pan-neuronal and subtype-specific cortical markers, such as 16% of Ctip2 (Layers V and VI) and 6% of Tbr1 (Layers I and VI; Figure 3d–e). Expression of peripherin and En1, markers for peripheral and midbrain neurons, respectively, was not detected. We did not observe a significant variability in these subtypes of neurons between the control and ASD backgrounds (Figure S3a). Next, we determined the functional maturation of the iPSC-derived neurons using electrophysiological methods. Whole-cell recordings were performed using cells that had differentiated for at least 6 weeks in culture. Both controls and ASD- neurons showed action potentials evoked by somatic current injections (Figure 3f–h and Figure S3c and d). Therefore, our data indicate that somatic cell reprogramming did not affect the ability of iPSC-derived neurons to mature and become electrophysiologically active.
Figure 3. Derivation of NPCs and neurons from iPSCs.
(a) Representative images depicting morphological changes during neuronal differentiation from control and TRPC6-mutant iPSCs. Bar = 100 µm. (b) NPCs are positive for the neural precursor markers Musashi-1 and Nestin. Bar = 50 µm. (c) Representative images of cells after neuronal differentiation. iPSC-derived neurons express neuronal markers such as GABA, MAP2, and synapsin I. (d) Examples of distinct cortical neuronal subtypes present differentiating cultures after 3 weeks. Bar = 30 µm. (e) We obtained 30% neurons in our cultures with this protocol, as measured by MAP2 staining and infection with the syn::EGFP lentiviral vector. Most MAP2-positive cells expressed VGLUT1, in contrast with 12% of neurons expressing GABA. Ctip2-positive neurons were more abundant (16%), whereas Tbr1-positive neurons were present in a small percentage in the population (6%) at the end of the differentiation protocol. (f) Morphology of neurons patched for electrophysiological recording. (g) Representative recordings of evoked action potentials in iPSC-derived neurons in response to current steps under current patch clamps. (h) Representative Na+ and K+ currents in iPSC-derived neurons. The error bars in all panels show the s.e.m.
TRPC6 disruption does not affect NPC proliferation
TRPC1, another member of the transient receptor potential channel family, is involved in NPC proliferation mediated by FGF 42. Therefore, we investigated whether reduction of TRPC6 expression levels affects the cell cycle profile. No difference was observed when comparing the percentage of cells in G1 (56.2±5.0% and 47.8±10.5%, P>0.2), S (30.6±3.0% and 36.0±6.4%, P>0.2), and G2/M (10.1±1.4% and 14.3±4.8%; P>0.2) phases between control and TRPC6-mut iPSC-derived NPCs, indicating that TRPC6 likely does not play a role in NPC proliferation, in contrast to TRPC1 (Figure S4a).
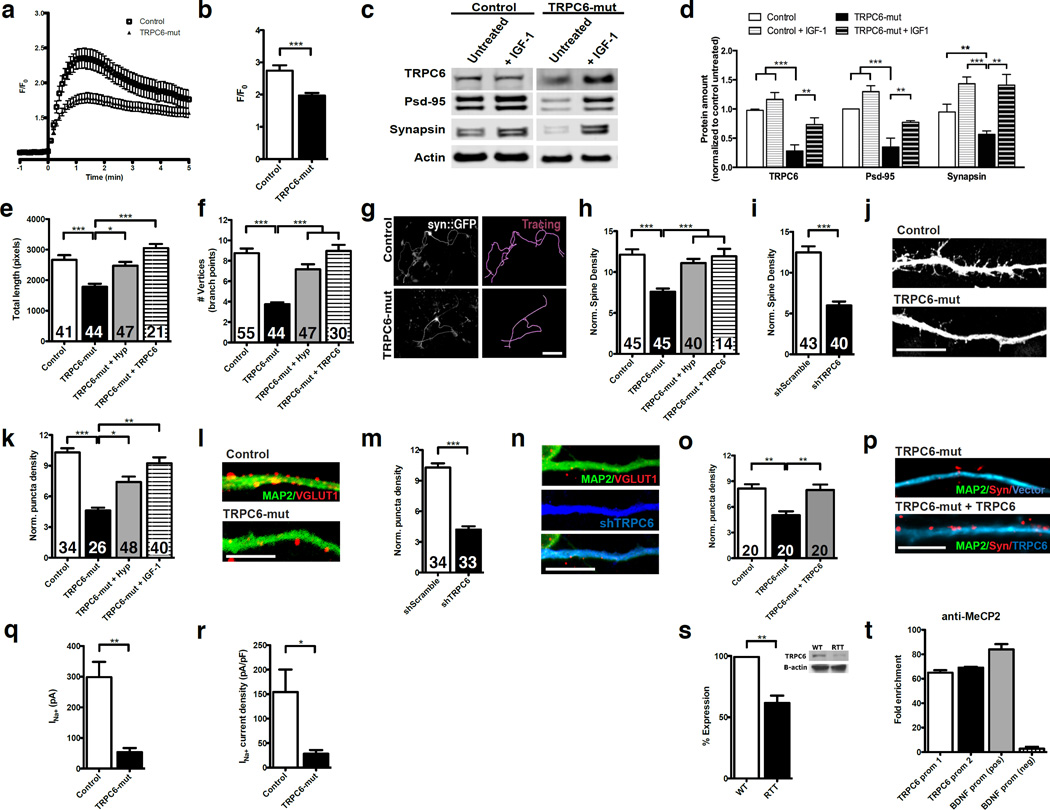
Ca2+ influx is reduced in TRPC6-mutant NPCs
The role of TRPC6 in dendritic spine formation depends on a pathway that involves Ca2+ influx through the channel 8. To test if changes in intracellular Ca2+ levels might be altered in TRPC6-mut neural cells upon TRPC6 activation, we stimulated iPSC-derived NPCs from the TRPC6-mut individual and a control with hyperforin plus FFA. This combination of drugs induced transient and repetitive increases in intracellular Ca2+ concentrations in both TRPC6-mut- and control-derived NPCs. The TRPC6 activation-induced Ca2+ oscillation peak was significantly higher in control NPCs compared with TRPC6-mut NPCs (Figure 4a). The average amplitude of the Ca2+ increase over baseline in the 100 cells analyzed was reduced by 30% in the TRPC6-mut NPCs (1.9±0.08-fold) compared with the control sample (2.7±0.2-fold; P<0.001) when stimulated with hyperforin and FFA (Figure 4b).
Figure 4. Alterations in neural cells derived from the TRPC6-mutant individual.
(a) Ca2+ influx dynamics through TRPC6 channels activated by hyperforin plus FFA were reduced in the TRPC6-mut cells. Oscillations generated by hyperforin and FFA treatment were normalized to the fluorescence of the resting level (F0), synchronized, and averaged. (b) The average peak of Ca2+ influx in the 100 cells analyzed was reduced by 30% in the TRPC6-mut NPCs compared with the control sample when the cells were stimulated with hyperforin and FFA (n=3; P<0.001; ANOVA). (c) Representative western blot of neurons derived from a clone of a control and a TRPC6-mutant iPSC line treated or not treated with IGF-1. (d) TRPC6-mutant neurons displayed low levels of TRPC6 and synaptic proteins Psd95, and synapsin I. IGF-1 treatment significantly increased the protein levels of TRPC6, Psd-95, and synapsin I in TRPC6-mutant neurons (n=3, P<0.01; t-test). (e) Bar graphs showing that the total length (microns) and (f) number of vertices (neuronal branch points) of TRPC6-mutant neurons is reduced compared with controls. Treatment with hyperforin or restoring TRPC6 expression levels rescued these defects (P<0.01; ANOVA). (g) Representative images of TRPC6-mutant and control neurons before and after neuronal tracing. The neuronal morphology was visualized using the syn::EGFP lentiviral vector. Bar = 50 µm. (h) Bar graphs showing that the spine density in TRPC6-mutant neurons was reduced compared with the controls and could be rescued after hyperforin treatment or restoring TRPC6 expression levels (P<0.01; ANOVA). (i) A specific shRNA against TRPC6 (shTRPC6) was used to confirm that the phenotype was caused by loss of TRPC6 function (P<0.01; ANOVA). (j) Representative images of neuronal spines in control and TRPC6-mutant neurons. (k) The bar graphs show that the number of glutamate vesicles in TRPC6-mutant neurons was significantly reduced compared to controls. IGF-1 and hyperforin treatment for 2 weeks increased the number of VGLUT1 puncta in TRPC6-mutant neurons (P<0.01; ANOVA). (l) Representative images of neurons stained for VGLUT1 and MAP2. (m) Control neurons expressing an shRNA against TRPC6 (shTRPC6) exhibited reduced numbers of VGLUT1 puncta compared with the neurons expressing a scrambled shRNA (shScramble). (n) Representative image of a control neuron expressing an shRNA against TRPC6. Bar = 5 µm. (o) The bar graphs show the number of synaptic puncta, as measured by synapsin I staining. Synaptic puncta counts in TRPC6-mutant neurons were reduced compared to controls. TRPC6-cDNA treatment of TRPC6-mut neurons was sufficient to increase synapses to control levels (P<0.01; n=20; ANOVA). (p) Representative image of TRPC6-mut neurons with empty vector and with vector expressing wild-type TRPC6 stained for MAP2 and synapsin I. Bar = 5µm. (q) The whole-cell Na+ current of TRPC6-mutant neurons was significantly less than that of the control (P<0.01; ANOVA). (r) The Na+ current density of TRPC6-mutant neurons was also significantly less than that of the control (P<0.01; ANOVA). (s) TRPC6 protein levels were reduced in neurons derived from an RTT iPSC clone expressing a non-functional version of MeCP2 compared with an isogenic control expressing the functional MECP2 gene. (t) Recruitment of MeCP2 on the TRPC6 promoter region by ChIP. Extracts of formaldehyde-fixed neurons were precipitated with a MeCP2 antibody and analyzed by quantitative PCR using two distinct primers for the TRPC6 promoter. The data show enrichment over the IgG control precipitation. The primers for the BDNF promoter were used as controls. The numbers of neurons analyzed (n) are shown within the bars in graphs. The error bars in all panels show the s.e.m. For the iPSC clones used in each experiment, refer to Table S4. *P<0.05; **P<0.01; ***P<0.001.
TRPC6 signaling regulates gene expression in neuronal cells
To validate our DPC findings, we examined the expression of some neuronal genes in NPCs in response to TRPC6 activation (Figure S4b). After a 48h hyperforin treatment, SEMA3A expression was reduced (0.6±0.05-fold, P<0.05), whereas INA and MAP2 again showed increased expression (2.6±0.09-fold and 1.8±0.1-fold; P<0.001). These results parallel our DPC expression analysis and support the hypothesis that TRPC6 signaling is important for the regulation of genes involved in neuronal function.
TRPC6 disruption alters the neuronal phenotype
To determine if TRPC6 disruption influences spine formation and synaptogenesis, we investigated neurons derived from TRPC6-mut and control iPSCs. To avoid variability from reprogramming, all experiments were performed with different iPSC clones and independent experiments. All biological replicates and iPSC clones used in each experiment are summarized in Table S6. The neurons derived from this ASD individual exhibited a 60% reduction (P<0.01) in TRPC6 protein levels as measured by western blot (Figure 4c and d). We first examined neuron morphology by infecting cells with a previously described lentiviral vector containing the EGFP sequence under the control of the synapsin gene promoter (syn::EGFP) 6. By measuring the size of neurites and their ramifications, we verified that the TRPC6-mutant neurons are shorter in total length (1782±101.2 and 2666±153.7 pixels; P<0.001) and less arborized (3.7±0.2 and 8.7±0.5 vertices; P<0.001) than the controls (Figure 4e–g). Moreover, the density of dendritic spines in TRPC6-mutant neurons was reduced (7.4±0.5 spines per 20µm of dendrite length) compared with control neurons (12.9±0.8 spines; P<0.001) derived from several individuals (Figure 4h–j, Figure S3e). TRPC6 expression was previously shown to regulate spine density 8. Thus, to confirm that the alterations observed in this ASD individual were caused by TRPC6 haploinsufficiency, we downregulated TRPC6 expression in control neurons using a specific, pre-validated shRNA in a lentiviral vector. Neurons derived from control iPSCs expressing shTRPC6 exhibited a significant reduction in spine density (6.0±0.5 spines) compared with control neurons expressing a scrambled shRNA (12.5±0.7 spines; P<0.0001; Figure 4j). Even further, restoring TRPC6 expression in the TRPC6-mut neurons using a lentiviral vector expressing wild-type TRPC6 (Figure S4c–d) rescued these morphological alterations, increasing total neuronal length (3051±133.4 pixels; P<0.001), arborization (8.9±0.6 vertices; P<0.001), and dendritic spine density (11.9±0.9 spines; P<0.001) to control levels (Figure 4e, f, and h). Interestingly, the specific activation of the wild type TRPC6 in mutant neurons by hyperforin was also sufficient to rescue these morphological phenotypes in our culture conditions (Figure 4e, f and h).
TRPC6 is mainly expressed in glutamatergic synapses and its loss interferes with synapsin I cluster density in pre-synaptic sites of hippocampal neurons, suggesting that this gene has an important role in the regulation of excitatory synapse strength 9. Quantifying VGLUT1 puncta in MAP2-labeled neurons confirmed that the TRPC6-mutant neurons had a significantly lower density of VGLUT1 puncta (4.6±0.3 puncta per 20µm of dendrite length) compared with independent clones isolated from several independent controls (10.3±0.4 puncta; P<0.001) (Figure 4k–l, Figure S3g). To determine if TRPC6 haploinsufficiency contributed to the lower density of VGLUT1 puncta, we treated TRPC6-mut neurons with hyperforin to specifically stimulate TRPC6. After 2 weeks of treatment, the neurons exhibited a significant increase in the number of VGLUT1 puncta compared with untreated cells (7.4±0.5 puncta; P<0.05) (Figure 4k). Control neurons expressing shTRPC6 also exhibited a lower density of VGLUT1 puncta, indicating that loss of TRPC6 function affects the formation of glutamatergic synapses (P<0.01) (Figure 4m–n). In addition, overexpression of TRPC6 in the TRPC6-mut neurons was able to rescue synapse numbers (8.0±0.6 puncta per 20µm of dendrite length; P<0.001) to control levels, as measured by synapsin I puncta (Figure 4o–p). Finally, electrophysiological recordings revealed that the Na+ currents of TRPC6-mutant neurons (28.38±7.5 pA) were impaired compared to controls (154.4±45.9 pA; P<0.0001) (Figure 4q–r, Figure S3e).
TRPC6 and MeCP2 share a similar molecular pathway
Certain neuronal phenotypes (reduction of spine density and glutamatergic synapses) associated with TRPC6 function loss are similar to those previously described for loss of MeCP2 function in human neurons 6. MeCP2 genetic alterations have been recognized in several non-syndromic ASD individuals 43–50, and reduced MeCP2 expression in brains of autistic individuals has been reported 51, 52. In addition, two independent studies have reported that MeCP2 regulates TRPC6 expression in the mouse brain, likely through an indirect mechanism 53, 54. Thus, we investigated whether MeCP2 acts upstream of TRPC6 in human neurons. We used two iPSC clones from a female RTT patient carrying the T158M MeCP2 mutation, which results in persistent X chromosome inactivation 6. Each clone expresses a different MeCP2 allele, a wild type or mutant version of the MeCP2 gene. We then differentiated both clones into neurons and evaluated TRPC6 protein expression levels. The TRPC6 expression level was reduced by 40% in the clone carrying the non-functional version of MeCP2 compared to the wild type control clone (61.67±6.0% and 99.3±1.2%; P<0.01), indicating that MeCP2 levels affect TRPC6 expression in human neurons (Figure 4s). This observation supports MeCP2 acting upstream of TRPC6 in the same molecular pathway to affect neuronal morphology and synapse formation. We next investigated whether MeCP2 could occupy regions of the human TRPC6 promoter. Chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) revealed high levels of MeCP2 in association with the TRPC6 promoter region in human neurons, suggesting a potential mechanism of transcriptional regulation (Figure 4t, Figure S4f).
Our data suggest that the molecular pathway involving MeCP2 and TRPC6 is a rate-limiting factor in regulating glutamatergic synapse number in human neurons. Administration of insulin-like growth factor-1 (IGF-1) promotes the reversal of RTT-like symptoms in a mouse model 55 and of molecular alterations in RTT human neurons 6, and is currently in clinical trials for RTT patients 56. To investigate whether the potential convergence of molecular mechanisms underlying RTT and non-syndromic autism suggests shared therapeutic benefits, we treated TRPC6-mutant neurons with full-length IGF-1 (10 ηg/mL). Interestingly, we observed a significant increase in TRPC6 protein levels after treatment. Moreover, Psd-95 and synapsin I protein levels were also upregulated by IGF-1 (P<0.01; Figure 4c and d). IGF-1 treatment also rescued the glutamatergic synapse number in TRPC6-mutant neurons as measured by VGLUT1 puncta (9.2±0.6 puncta per 20µm of dendrite length; P<0.01), suggesting that the drug treatment could correct this neuronal phenotype (Figure 4k).
TRPC6 downregulation compromises neuronal development in vivo
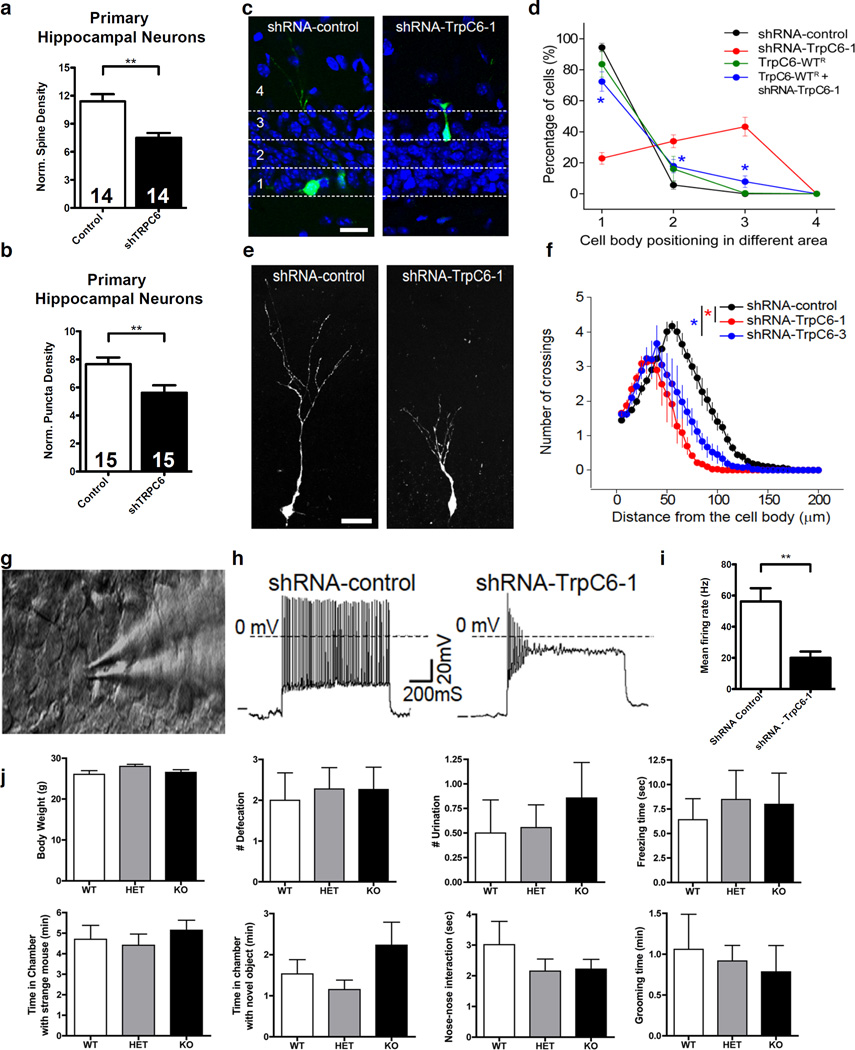
In vitro experiments in rodent primary neurons have shown that Trpc6 levels affect spine density and excitatory synapses 9, 57. To corroborate our findings from human derived neurons, we looked to examine the effect of Trpc6 loss in a rodent model. We validated two shRNAs (#1 and #3) against mouse Trpc6 by western blot analysis and used both for further experiments (Figure S4g–h). Using this shRNA targeting Trpc6, we transduced mouse primary hippocampal neurons. The neurons expressing shRNA targeting Trpc6 demonstrated reduced spine density (7.5±0.5 and 11.4±0.8 spines per 20µm of dendrite length; P<0.001) and fewer synapses (5.6±0.5 and 7.7±0.5 puncta per 20µm of dendrite length; P<0.01) versus neurons transduced with a shRNA scramble control (Figure 5a and b). Thus, as described above, we determined that TRPC6 downregulation causes similar neuronal alterations in human and rodent neurons. We next looked to validate the cell autonomous effect of TRPC6 loss of function in vivo by taking advantage of adult neurogenesis in the hippocampus 58. Using retroviruses to target newborn neurons, we delivered the shRNAs against mouse Trpc6. Trpc6 downregulation led to migration defects and reduced neuronal dendritic arborization (Figure 5c–f). Moreover, whole-cell patch clamping to record action potentials revealed a significant reduction in the firing rate of neurons expressing shRNAs against Trpc6 compared with controls (Figure 5g–i). To demonstrate the contribution of Trpc6 to these phenotypes in vivo, we rescued the migration defects by co-transfecting the shRNA with an expression construct for an shRNA-resistant form of TRPC6-WT (TRPC6-WTR; Figure 5d, Figure S4h).
Figure 5. TRPC6 regulates the neural development of adult-born neurons in the dentate gyrus of the hippocampus.
(a) Mouse primary hippocampal neurons revealed reduced spine density in neurons treated with shRNA targeting TRPC6 compared to neurons treated with shRNA scramble control (P<0.01; n=14; t-test). (b) Mouse primary hippocampal neurons demonstrated reduced synaptic puncta numbers in neurons treated with shRNA targeting TRPC6 compared to neurons treated with shRNA scramble control (P<0.01; n=15; t-test). Synaptic puncta were labeled using synapsin I antibodies and counted along MAP2+ neuronal dendrites. (c) Representative confocal images of neurons expressing shRNA-control and shRNA-TRPC6-1 at 28 dpi (days post retroviral injection). Green, GFP; blue, DAPI. Bar = 50 µm. Also shown are the divided areas of the dentate gyrus: 1, inner granule cell layer; 2, middle granule cell layer; 3, upper granule cell layer; and 4, molecular layer. (d) Summary of cell body localization of GFP+ newborn neurons under different experimental conditions at 28 dpi. The cell migration phenotype was rescued by expression of TRPC6-WTR at 14 dpi. Retroviruses co-expressing GFP and TrpC6-WTR were co-injected with retroviruses co-expressing dsRed and shRNA-TRPC6-1 into the adult mouse dentate gyri. The cell body localization of the GFP+, DsRed+, and GFP+DsRed+ neurons are quantified. The values represent the mean ± s.e.m. (n = 3; P< 0.01; ANOVA). (e) A 3-dimensional confocal reconstruction of dendritic trees of GFP+ dentate granule cells expressing shRNA-control or shRNA-TRPC6-1 at 14 dpi. Scale bar, 20 µm. (f) Sholl analysis of the dendritic complexity of GFP+ neurons at 14 dpi. Number of crossings refers to the number of dendrites intersecting concentric circles spaced 10µm apart starting from the cell body. The error bars in all panels show the s.e.m. (n = 3; P<0.05; ANOVA). (g) A sample DIC image of a newborn neuron patched in whole-cell configuration in an acute slice of the hippocampus. (h) The firing rate of repetitive action potentials of GFP+ neurons under current clamp in response to 1-s current injection steps at 21 dpi. Shown on the left is a sample trace of a GFP+ neuron expressing shRNA-control; a GFP+ neuron expressing shRNA-TRPC6#-1 is shown on the right. (i) Summary of the mean firing rate of newborn neurons. The values represent the mean ± s.d. (n = 3; P<0.01; ANOVA). A minimum of 10 GFP+ cells was randomly picked from each animal, and at least three animals (n) under each experimental condition were used. (j) Behavioral analysis of Trpc6 KO and HET mice. The mean body weight and defecation and urination episodes during the test revealed no physiological differences between the wild type (WT), heterozygote (HET), and knockout (KO) Trpc6 animals. Evaluation of time spent in freezing behavior and in grooming behavior revealed no significant differences between the groups. Social interaction was assessed by evaluating the time spent with a novel object or in nose-to-nose contact with a strange animal. Adult mice (6–8 weeks old, male) in a C57BL/6 background were used for the study. At least 12 animals per group were utilized in biological replicates. The experimenter was blind to the genotypes. The data were analyzed using non-parametric Kruskal-Wallis ANOVA. The error bars in all panels show the s.e.m. All procedures followed institutional guidelines. *P<0.05; **P<0.01; ***P<0.001.
TRPC6 knockout (KO) mice 59 display reduced exploratory activity in a square open field and elevated star maze compared with control siblings 60. Limited environmental exploration is commonly associated with ASD individuals 61. Thus, we decided to investigate whether the TRPC6 KO mouse displays other ASD-like behaviors. We assessed the social interaction and repetitive behaviors of these animals, but observed no significant differences between wild-type controls (WT) and heterozygotes (HET) or WT and KO mice (Figure 5j). Together, these data demonstrate loss or reduction of TRPC6 in a rodent model induces neuronal abnormalities paralleling our findings in the TRPC6-mut iPSC-derived neurons, such as reduced neuronal arborization, spine density and synapse numbers.
Mutation screening of TRPC6
Based on the initial observation of TRPC6 disruption by a chromosomal breakpoint, we established a narrow hypothesis focusing on TRPC6 to conduct a single gene case/control association study. We screened targeted high-throughput sequencing data from all coding exons and splice sites of TRPC6 in 1041 ASD cases from the Simons Simplex Collection (SSC) 26 and 942 ancestrally matched controls from the NINDS Neurologically Normal Caucasian Control Panel (http://ccr.coriell.org/Sections/Collections/NINDS/). A summary of the quality control metrics of the high-throughput sequencing is presented in Table S7. We focused on novel splice sites, missense, and nonsense mutations that were observed only once across all of our cohorts and not present in the dbSNP137 and 6503 exomes available from the Exome Variant Server (EVS, v.0.0.15). We reasoned that these variants were most likely to be deleterious and subject to purifying selection. Moreover, the study of variants observed only once, in combination with case-control matching for ancestry, represents a more rigorous approach to protecting against population stratification 62. Table S8 lists all such variants in TRPC6. We observed significantly more novel nonsynonymous singleton mutations in cases compared with controls (10/1041 cases versus 1/942 controls; p = 0.013, OR = 9.127, 95% CI = 1.211-191.027, Fisher exact test, two-tailed). To confirm the low mutation rate observed in this control sample, we examined the whole exome-sequencing data from an in-house database and identified an additional 1930 northern European (NE) controls who clustered tightly with the HapMap CEU cohort. We evaluated the coding exons and splice sites of TRPC6 and, to maximize sensitivity, did not set a minimum read threshold to identify all novel nonsynonymous singleton variants, which are listed in Table S5. An omnibus analysis revealed an even more significant over-representation of such variants in cases (10/1041 cases versus 4/2872 controls; p = 0.001, OR = 6.954, 95% CI = 2.008-26.321, Fisher exact test, two-tailed). Because our results indicate that TRPC6 disruption leads to haploinsufficiency of the corresponding protein, two of the case variants are particularly noteworthy: M1K, which disrupts the start codon; and Q3X, which is a very early premature stop codon. Unfortunately, live cells from these individuals were not available for follow-up functional studies. No TRPC6 mutations affecting the start codon or nonsense mutations were identified in a total of 7445 controls: 942 NINDS neurologically normal Europeans and 6503 exomes from the EVS (4300 European-American, 2203 African-American). Segregation analysis of the case variants revealed that each was inherited from an apparently unaffected parent, suggesting that these variants are incompletely penetrant, as has been previously observed for a wide range of ASD mutations such as Shank3 63 and CNTNAP2 64. Thus, although these genetic variations cannot be considered as causal mutations, they might represent risk factors for ASD. No TRPC6 CNVs have been described in ASD (http://projects.tcag.ca/autism_500k).
Discussion
A rapidly increasing number of ASD risk regions are being identified, and there is now considerable effort focused on moving from gene discovery to understanding the biological influences of these various mutations 2–4, 65–67. The development of relevant human-derived cellular models to study ASDs represents a complementary strategy to link genetic alterations to molecular mechanisms and complex behavioral and cognitive phenotypes 68. Here, we identified the disruption of the TRPC6 gene by a balanced de novo translocation in a non-syndromic ASD individual. TRPC6 is involved in the regulation of axonal guidance, dendritic spine growth, and excitatory synapse formation 8, 9, 35, processes that have been consistently implicated in ASD etiology 69–72. To explore if TRPC6 disruption could result in such neuronal alterations, we made use of several different cellular models.
Global transcriptional studies of DPCs derived from the ASD individual and expression analysis upon activation of TRPC6 in DPCs and NPCs suggested that TRPC6 signaling regulates the transcription of genes involved in neuronal adhesion, neurite growth, and axonal guidance. The abnormal dysregulation found in the ASD individual might be triggered, at least for some genes, by reduced levels of phosphorylated CREB, a transcription factor activated by TRPC6 signaling 8. CREB controls a complex regulatory network involved in memory formation, neuronal development, and plasticity in the mammalian brain, processes that are compromised in ASD 73–75.
Reprogramming the DPCs from the ASD individual to a pluripotent state allowed us to explore the functional consequences of TRPC6 disruption in human neuronal cells. Ca2+ influx was aberrant in NPCs derived from the ASD individual, suggesting that Ca2+ signaling-dependent mechanisms were compromised in these cells. Ca2+ signaling pathways have previously been implicated in ASD etiology; mutations in different voltage-gated Ca+2 channels and Ca+2-regulated signaling molecules have been identified in ASD individuals 76–79. This result, combined with the measured protein levels, reveals that disruption of TRPC6 leads to a functionally relevant haploinsufficiency, making the existence of a novel disease-relevant protein resulting from a TRPC6 and VPRBP combination unlikely.
In human neurons, TRPC6 haploinsufficiency causes other functional and morphological alterations that reflect defects in axonal and dendritic growth, such as shortening of neurites, a decrease in arborization, and a reduction in dendritic spine density. Alterations in these phenotypes were already been described for post-mortem or iPSC-derived ASD neurons 80, 81. Analysis of neurons derived from the ASD individual’s iPSCs also revealed a reduction of VGLUT1 puncta density, in agreement with previous work demonstrating that TRPC6 expression levels can modulate glutamatergic synapse formation in rat neurons 9. Alterations in glutamatergic neurotransmission have been identified in individuals with syndromic forms of ASD: dysregulation of the metabotropic glutamate receptor 1/5 (mGluR1/5) pathway has been well documented in Fragile-X syndrome, and neurons derived from RTT patient iPSCs also present a reduction in the number of VGLUT1 puncta 6, 82, 83. In addition, a reduction in glutamatergic transmission was observed in Shank3 heterozygous mice, an ASD mouse model 84. Finally, Na+ currents were also decreased in ASD individual’s neurons. This result is in agreement with previous findings that demonstrate that TRPC6 channels participate in Na+ cell entry 38. Decreased Na+ current densities have previously been reported in other ASD models 85.
Due to the high degree of locus heterogeneity, it is challenging to identify additional individuals carrying similar rare variants in the ASD population. Therefore, we used complementary functional assays such as loss-of-function experiments and mouse models to validate the observation that reduction in TRPC6 expression levels leads to abnormal neuronal phenotypes and is important for neuronal homeostasis. Moreover, we have demonstrated that several of the phenotypic alterations seen in the TRPC6-mut neurons could be rescued by both using hyperforin, which activates the channel, and expressing wild-type TRPC6. These TRPC6 loss-of-function and complementation assays underscore its importance for neuronal homeostasis. Based on the results obtained from our different cellular models, this is likely due to TRPC6 influence on Ca2+-signaling dependent mechanisms and neuronal transcriptional regulation. The common altered neuronal phenotypes shared by the TRPC6-mutant individual and RTT patients support the idea that ASD caused by different genetic mechanisms affect common pathways. Indeed, our data suggest that MeCP2 may act upstream of TRPC6, regulating its expression. Previous mouse studies 53, 54 suggested similar findings but failed to show a direct link between MeCP2 and the TRPC6 promoter through ChIP assays, likely due to the poor conservation between the promoter regions in these two species. Additional studies using large samples of idiopathic ASD individuals will help address this hypothesis.
Our findings also provide insights supporting the testing of novel drugs in ASD such as hyperforin, a drug that specifically activates TRPC6 38, 86, or IGF-1, which might increase not only TRPC6 protein levels but also other synaptic components. Therefore, individuals with alterations in this pathway might benefit from these drugs. These defects could also be rescued by activating the AKT/mTOR pathway using IGF-1. The TRPC6 KO mice exhibit reduced exploratory interest, a typical ASD-like behavior, but no impaired social interaction or repetitive movements. The lack of some ASD-like behaviors in mouse models is common and can be attributed to the inherent differences between human and mouse genetic backgrounds and neural circuits 87–90. Alternatively, other genetic alterations may be required to develop the full autistic phenotype in this mouse model. Accumulating evidence favors the multiple-hit model in a significant proportion of ASD individuals as well as in the case of the ASD individual described here 64, 91–94. In fact, while our functional data demonstrate that TRPC6 has a crucial role in synaptogenesis and is involved in pathways previously associated with ASD, our mutation screening data suggest rare TRPC6 variants may have a more moderate contribution to the disease. Our sequencing findings revealed TRPC6 loss-of-function mutations in two ASD families with incomplete penetrance of the phenotype, supporting the multi-hit hypothesis for ASD. Indeed, the individual studied here also presents other rare genetic variants, such as in the ASD associated gene PDE4A or even VPRBP, that might contribute to his phenotype. However, this does not diminish the impact of TRPC6 to the phenotype, as our experiments using hyperforin or TRPC6 complementation rescued the observed cellular alterations. This suggests that while other genetic alterations present in the individual might augment the observed phenotypes, TRPC6 disruption is the predominant contributor to the abnormal neuronal function in this ASD individual.
Thus, our results suggest TRPC6 as a novel predisposing gene for ASD that likely acts in combination with other genetic variants to contribute to autistic phenotypes. Our work demonstrates that individual-specific iPSC-derived neurons can be used to correlate novel variants in ASD individuals to the etiology of these highly complex disorders.
Supplementary Material
Acknowledgments
The work was supported by grants from the California Institute for Regenerative Medicine (CIRM) TR2-01814, the National Institutes of Health through the NIH Director's New Innovator Award Program (1-DP2-OD006495-01), A NARSAD Independent Investigator Grant to A.R.M.; a NIH predoctoral training grant T32 GM008666 to A.A.; Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and by NIH (NS047344) and SAFRI to H.S.; NIH (NS048271, HD069184), NARSAD, and MSCRF to G.L; NIH (K08MH087639) to A.R.G; and NIH (RC2MH089956) to MWS. We would like to thank the ASD individuals and their families, Daniela Franco Bueno and Gerson Shigeru Kobayashi for the DPC control samples, Kristin Rose for technical support, and Dr. Willmar Schwabe, GmbH & Co, Karlsruhe, Germany, for providing hyperforin.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14(12):1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai Y, Feng S, Ge R, Du W, Zhang X, He Z, et al. TRPC6 channels promote dendritic growth via the CaMKIV-CREB pathway. J Cell Sci. 2008;121(Pt 14):2301–2307. doi: 10.1242/jcs.026906. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Du W, Zhou K, Tai Y, Yao H, Jia Y, et al. Critical role of TRPC6 channels in the formation of excitatory synapses. Nat Neurosci. 2008;11(7):741–743. doi: 10.1038/nn.2127. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35(6):2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, et al. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 13.Beltrao-Braga PI, Pignatari GC, Maiorka PC, Oliveira NA, Lizier NF, Wenceslau CV, et al. Feeder-free derivation of induced pluripotent stem cells from human immature dental pulp stem cells. Cell Transplant. 2011 doi: 10.3727/096368911X566235. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 21.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 22.Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63(6):761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130(6):1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72(4):559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439(7076):589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68(2):192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2012;40(Database issue):D1016–D1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun NN, Reutiman TJ, Lee S, Folsom TD, Fatemi SH. Expression of phosphodiesterase 4 is altered in the brains of subjects with autism. Neuroreport. 2007;18(17):1841–1844. doi: 10.1097/WNR.0b013e3282f16dca. [DOI] [PubMed] [Google Scholar]
- 29.Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, et al. Stem cell properties of human dental pulp stem cells. J Dent Res. 2002;81(8):531–535. doi: 10.1177/154405910208100806. [DOI] [PubMed] [Google Scholar]
- 30.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A. 2003;100(10):5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.d'Aquino R, De Rosa A, Laino G, Caruso F, Guida L, Rullo R, et al. Human dental pulp stem cells: from biology to clinical applications. J Exp Zool B Mol Dev Evol. 2009;312B(5):408–415. doi: 10.1002/jez.b.21263. [DOI] [PubMed] [Google Scholar]
- 32.Griesi-Oliveira K, Sunaga DY, Alvizi L, Vadasz E, Passos-Bueno MR. Stem cells as a good tool to investigate dysregulated biological systems in autism spectrum disorders. Autism Res. 2013;6(5):354–361. doi: 10.1002/aur.1296. [DOI] [PubMed] [Google Scholar]
- 33.Kashiwa A, Yoshida H, Lee S, Paladino T, Liu Y, Chen Q, et al. Isolation and characterization of novel presenilin binding protein. J Neurochem. 2000;75(1):109–116. doi: 10.1046/j.1471-4159.2000.0750109.x. [DOI] [PubMed] [Google Scholar]
- 34.Tai Y, Feng S, Du W, Wang Y. Functional roles of TRPC channels in the developing brain. Pflugers Arch. 2009;458(2):283–289. doi: 10.1007/s00424-008-0618-y. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Jia YC, Cui K, Li N, Zheng ZY, Wang YZ, et al. Essential role of TRPC channels in the guidance of nerve growth cones by brain-derived neurotrophic factor. Nature. 2005;434(7035):894–898. doi: 10.1038/nature03477. [DOI] [PubMed] [Google Scholar]
- 36.Jia Y, Zhou J, Tai Y, Wang Y. TRPC channels promote cerebellar granule neuron survival. Nat Neurosci. 2007;10(5):559–567. doi: 10.1038/nn1870. [DOI] [PubMed] [Google Scholar]
- 37.McCall CM, Miliani de Marval PL, Chastain PD, 2nd, Jackson SC, He YJ, Kotake Y, et al. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol Cell Biol. 2008;28(18):5621–5633. doi: 10.1128/MCB.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leuner K, Kazanski V, Muller M, Essin K, Henke B, Gollasch M, et al. Hyperforin--a key constituent of St. John's wort specifically activates TRPC6 channels. Faseb J. 2007;21(14):4101–4111. doi: 10.1096/fj.07-8110com. [DOI] [PubMed] [Google Scholar]
- 39.Muller M, Essin K, Hill K, Beschmann H, Rubant S, Schempp CM, et al. Specific TRPC6 channel activation, a novel approach to stimulate keratinocyte differentiation. J Biol Chem. 2008;283(49):33942–33954. doi: 10.1074/jbc.M801844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leuner K, Heiser JH, Derksen S, Mladenov MI, Fehske CJ, Schubert R, et al. Simple 2,4-diacylphloroglucinols as classic transient receptor potential-6 activators--identification of a novel pharmacophore. Mol Pharmacol. 2010;77(3):368–377. doi: 10.1124/mol.109.057513. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Lin M, Foxe JJ, Pedrosa E, Hrabovsky A, Carroll R, et al. Transcriptome comparison of human neurons generated using induced pluripotent stem cells derived from dental pulp and skin fibroblasts. PLoS One. 2013;8(10):e75682. doi: 10.1371/journal.pone.0075682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiorio Pla A, Maric D, Brazer SC, Giacobini P, Liu X, Chang YH, et al. Canonical transient receptor potential 1 plays a role in basic fibroblast growth factor (bFGF)/FGF receptor-1-induced Ca2+ entry and embryonic rat neural stem cell proliferation. J Neurosci. 2005;25(10):2687–2701. doi: 10.1523/JNEUROSCI.0951-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cukier HN, Lee JM, Ma D, Young JI, Mayo V, Butler BL, et al. The Expanding Role of MBD Genes in Autism: Identification of a MECP2 Duplication and Novel Alterations in MBD5, MBD6, and SETDB1. Autism Res. 2012 doi: 10.1002/aur.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cukier HN, Rabionet R, Konidari I, Rayner-Evans MY, Baltos ML, Wright HH, et al. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11(3):291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuwano Y, Kamio Y, Kawai T, Katsuura S, Inada N, Takaki A, et al. Autism-associated gene expression in peripheral leucocytes commonly observed between subjects with autism and healthy women having autistic children. PLoS One. 2011;6(9):e24723. doi: 10.1371/journal.pone.0024723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campos M, Jr, Pestana CP, dos Santos AV, Ponchel F, Churchman S, Abdalla-Carvalho CB, et al. A MECP2 missense mutation within the MBD domain in a Brazilian male with autistic disorder. Brain Dev. 2011;33(10):807–809. doi: 10.1016/j.braindev.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Piton A, Gauthier J, Hamdan FF, Lafreniere RG, Yang Y, Henrion E, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16(8):867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dotti MT, Orrico A, De Stefano N, Battisti C, Sicurelli F, Severi S, et al. A Rett syndrome MECP2 mutation that causes mental retardation in men. Neurology. 2002;58(2):226–230. doi: 10.1212/wnl.58.2.226. [DOI] [PubMed] [Google Scholar]
- 49.Lam CW, Yeung WL, Ko CH, Poon PM, Tong SF, Chan KY, et al. Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet. 2000;37(12):E41. doi: 10.1136/jmg.37.12.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28(3):205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 51.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1(4):e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samaco RC, Nagarajan RP, Braunschweig D, LaSalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet. 2004;13(6):629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- 53.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, Calfa G, Larimore J, Pozzo-Miller L. Activity-dependent BDNF release and TRPC signaling is impaired in hippocampal neurons of Mecp2 mutant mice. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1205271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106(6):2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khwaja OS, Ho E, Barnes KV, O'Leary HM, Pereira LM, Finkelstein Y, et al. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc Natl Acad Sci U S A. 2014;111(12):4596–4601. doi: 10.1073/pnas.1311141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leuner K, Li W, Amaral MD, Rudolph S, Calfa G, Schuwald AM, et al. Hyperforin modulates dendritic spine morphology in hippocampal pyramidal neurons by activating Ca(2+) -permeable TRPC6 channels. Hippocampus. 2012 doi: 10.1002/hipo.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70(4):687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dietrich A, Mederos YSM, Gollasch M, Gross V, Storch U, Dubrovska G, et al. Increased vascular smooth muscle contractility in TRPC6−/− mice. Mol Cell Biol. 2005;25(16):6980–6989. doi: 10.1128/MCB.25.16.6980-6989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beis D, Schwarting RK, Dietrich A. Evidence for a supportive role of classical transient receptor potential 6 (TRPC6) in the exploration behavior of mice. Physiol Behav. 102(2):245–250. doi: 10.1016/j.physbeh.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Pierce K, Courchesne E. Evidence for a cerebellar role in reduced exploration and stereotyped behavior in autism. Biol Psychiatry. 2001;49(8):655–664. doi: 10.1016/s0006-3223(00)01008-8. [DOI] [PubMed] [Google Scholar]
- 62.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44(3):243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chailangkarn T, Acab A, Muotri AR. Modeling neurodevelopmental disorders using human neurons. Curr Opin Neurobiol. 2012 doi: 10.1016/j.conb.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sbacchi S, Acquadro F, Calo I, Cali F, Romano V. Functional annotation of genes overlapping copy number variants in autistic patients: focus on axon pathfinding. Curr Genomics. 2010;11(2):136–145. doi: 10.2174/138920210790886880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cruz-Martin A, Crespo M, Portera-Cailliau C. Delayed stabilization of dendritic spines in fragile X mice. J Neurosci. 2010;30(23):7793–7803. doi: 10.1523/JNEUROSCI.0577-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chapleau CA, Larimore JL, Theibert A, Pozzo-Miller L. Modulation of dendritic spine development and plasticity by BDNF and vesicular trafficking: fundamental roles in neurodevelopmental disorders associated with mental retardation and autism. J Neurodev Disord. 2009;1(3):185–196. doi: 10.1007/s11689-009-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34(3):371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 74.Balschun D, Wolfer DP, Gass P, Mantamadiotis T, Welzl H, Schutz G, et al. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23(15):6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dworkin S, Mantamadiotis T. Targeting CREB signalling in neurogenesis. Expert Opin Ther Targets. 2010;14(8):869–879. doi: 10.1517/14728222.2010.501332. [DOI] [PubMed] [Google Scholar]
- 76.Hemara-Wahanui A, Berjukow S, Hope CI, Dearden PK, Wu SB, Wilson-Wheeler J, et al. A CACNA1F mutation identified in an X-linked retinal disorder shifts the voltage dependence of Cav1.4 channel activation. Proc Natl Acad Sci U S A. 2005;102(21):7553–7558. doi: 10.1073/pnas.0501907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krey JF, Dolmetsch RE. Molecular mechanisms of autism: a possible role for Ca2+ signaling. Curr Opin Neurobiol. 2007;17(1):112–119. doi: 10.1016/j.conb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 78.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 79.Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281(31):22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- 80.Belichenko PV, Oldfors A, Hagberg B, Dahlstrom A. Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5(12):1509–1513. [PubMed] [Google Scholar]
- 81.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 2011;20(R2):R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hagerman R, Hoem G, Hagerman P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1(1):12. doi: 10.1186/2040-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27(7):370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 84.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1(1):15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han S, Tai C, Westenbroek RE, Yu FH, Cheah CS, Potter GB, et al. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489(7416):385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muller WE. Current St John's wort research from mode of action to clinical efficacy. Pharmacol Res. 2003;47(2):101–109. doi: 10.1016/s1043-6618(02)00266-9. [DOI] [PubMed] [Google Scholar]
- 87.Wohr M, Silverman JL, Scattoni ML, Turner SM, Harris MJ, Saxena R, et al. Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res. 2012 doi: 10.1016/j.bbr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu JY, Xia QQ, Xia J. A review on the current neuroligin mouse models. Sheng Li Xue Bao. 2012;64(5):550–562. [PubMed] [Google Scholar]
- 90.Oddi D, Crusio WE, D'Amato FR, Pietropaolo S. Monogenic mouse models of social dysfunction: Implications for autism. Behav Brain Res. 2013 doi: 10.1016/j.bbr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 91.Poot M, Beyer V, Schwaab I, Damatova N, Van't Slot R, Prothero J, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010;11(1):81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 92.Poot M, van der Smagt JJ, Brilstra EH, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011;135(3–4):228–240. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]
- 93.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS genetics. 2012;8(2):e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.