Abstract
Cancer genome sequencing has revealed considerable variation in somatic mutation rates across the human genome, with mutation rates elevated in heterochromatic late replicating regions and reduced in early replicating euchromatin1-5. Multiple mechanisms have been suggested to underlie this2,6-10, but the actual cause is unknown. Here we identify variable DNA mismatch repair (MMR) as the basis of this variation. Analysing ~17 million single nucleotide variants from the genomes of 652 tumours, we show that regional autosomal mutation rates at megabase resolution are largely stable across cancer types, with differences related to changes in replication timing and gene expression. However, mutations arising after the inactivation of MMR are no longer enriched in early replicating euchromatin relative to late replicating heterochromatin. Thus, differential DNA repair and not differential mutation supply is the primary cause of the large-scale regional mutation rate variation across the human genome.
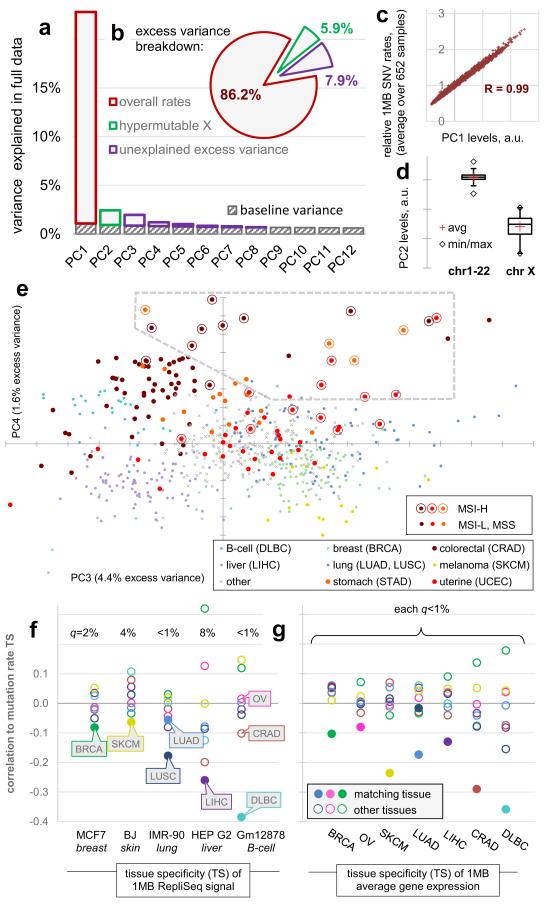
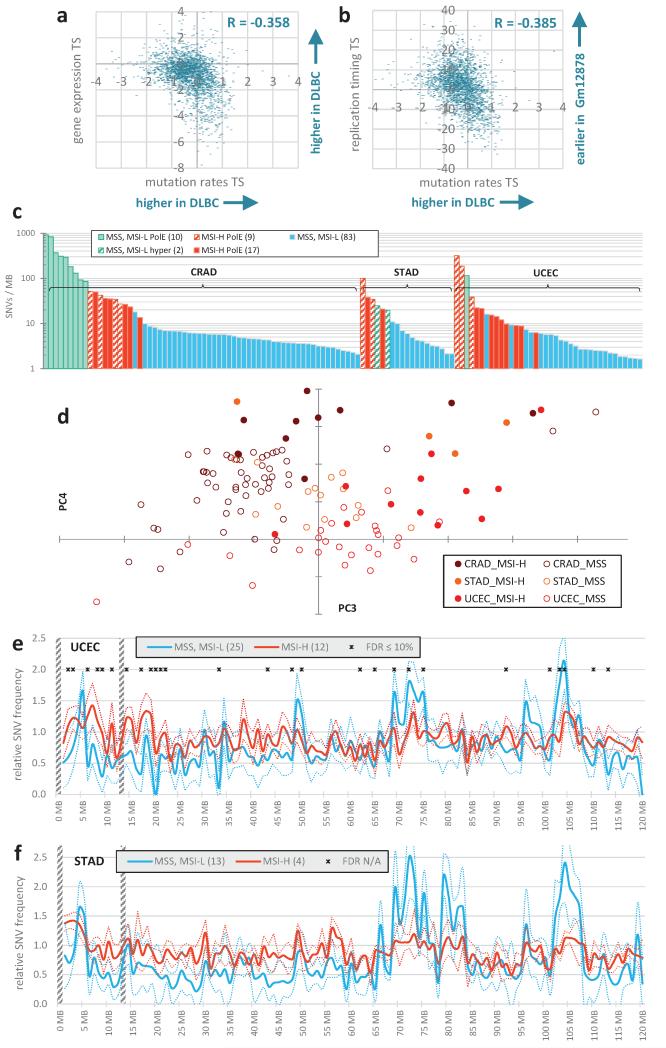
We examined 1Mb mutation densities along 652 fully sequenced human cancer genomes with >3000 SNVs (single nucleotide variants) per genome, originating from 16 tissues. This threshold enables more robust estimates of regional SNV densities in the examined samples, but it excludes cancer types with a very low mutation burden (Methods). Despite vastly different mutational loads between tissues of origin and between individual tumours11, the relative regional densities were, overall, consistent between samples. In a principal components (PC) analysis, the first PC corresponds closely to the average densities over all samples (R2=0.99) and captures 86.2% of the non-random variability between the 1Mb windows (Fig. 1a-c). This estimate of baseline variability per PC (Methods) encompasses the non-biological sources of randomness in the data (e.g. low mutation counts per bin in some cancer types) but it may also include genuine biological variability, if it is particular to individual tumour genomes. The second most prominent PCA trend (PC2, 5.9% variability; Fig 1a, d) precisely captures the known hypermutation of the X chromosome in a subset of tumours12. Across the 652 tumours, we estimate a further 7.9% of non-random variability exists that is not explained by the general pattern of regional rates or by the hypermutation of X (in PC3-8, with 4.4% in PC3 only; Fig. 1a, b).
Figure 1.
Changes in megabase-scale regional mutation rate variation between tumour samples. a-e, Principal components (PC) analysis of the 1Mb regional rates of 652 whole-genome sequences. a, Amount of variance conveyed by the prominent PCs. Baseline estimated by ‘broken stick’ method (Methods). b, Same, expressed as % above-baseline (putatively non-noise) variance. c, First PC reflects average rates. d, Second PC captures the variability in chromosome X mutation rates. e, Tumour sample loadings on PCs 3/4, highlighting cancer types significantly shifted by PC3 (Mann-Whitney test, FDR<1%), as well as STAD and UCEC. Dashed box denotes outlying samples. f, Pearson correlations of the tissue specificities (TS; Methods) of RepliSeq signal in cell lines to TS of 1Mb mutation rates in cancer types with significant PC3 shifts. q is significance of the difference of the matching vs. non-matching cancer type (Z-test, FDR corrected). g, Same, for TS of gene expression in tumour samples.
This signal can, in part, be ascribed to tissue-specific mutation rates: cancer samples from 7/16 tissues were significantly shifted in the distribution of their PC3 loadings (Mann-Whitney test, FDR<1%). Liver, colorectal and B-lymphocyte tumours were differentiated by lower PC3 loading scores, whereas melanoma, breast, ovarian and lung tumours had higher PC3 loadings (tissues highlighted in Fig. 1e). To examine if these significant regional rate changes are associated to DNA replication timing changes, we used RepliSeq data from ENCODE cell lines as a proxy for the changes occurring in the cancers of the corresponding tissues. In all cases, the correlation of the cell line-specific RepliSeq signal to the cancer type-specific 1Mb mutation rates was most prominent in the cancer type matching the cell line, with a significant difference from the non-matching cancer types (FDR<10%, Fig. 1f). Similarly, the changes in average gene expression levels in 1Mb windows in tumour samples paralleled the changes in mutation rates in the same samples with significantly stronger correlations for the matching cancer type (FDR<10%, Fig 1g; example in Extended Data Fig. 1a, b).
Visualizing the cancer samples in a PC plot revealed a group of outliers in a low-density area of the plot with extreme PC3 and PC4 loadings (box in Fig. 1e). These samples derive almost exclusively from colorectal, stomach or uterine cancers even though most samples from these three cancer types clustered elsewhere on the PC plot. One feature particular to tumours from these three tissues is that they frequently display inactivation of the mismatch repair (MMR) pathway through mutation of MMR genes or hypermethylation of the MLH1 gene promoter13-15. Inactivation of MMR results in a high incidence of small insertions and deletions (indels) at simple sequence repeats - referred to as microsatellite instability (MSI) and used to phenotypically identify MMR deficiency - but also an increased SNV load (Extended Data Fig. 1c).
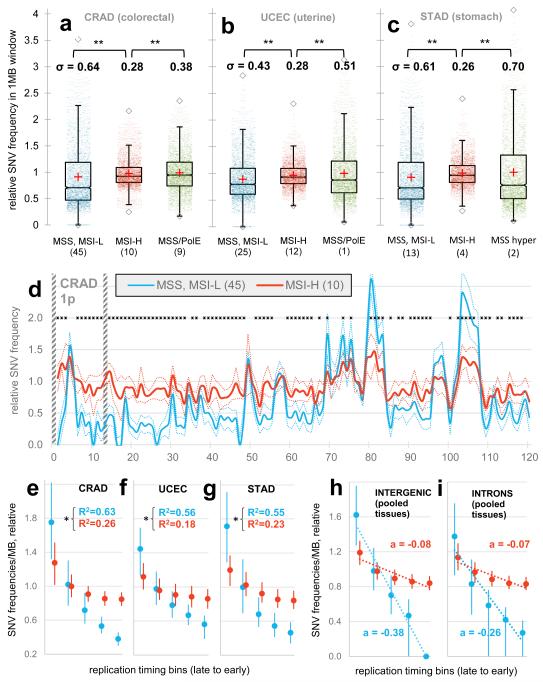
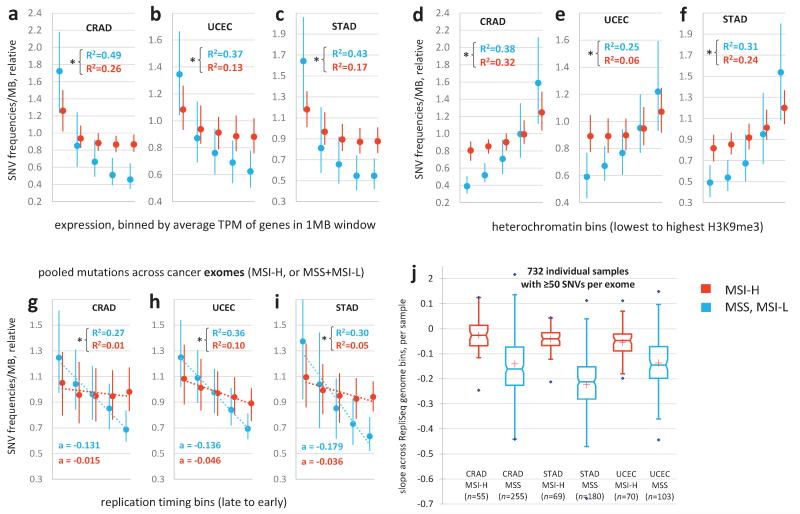
The outlier colon, gastric and uterine cancer samples on the PC plot (Fig. 1e, Extended Data Fig. 1d) were almost always MSI samples (phenotypically scored as MSI-high, MSI-H), suggesting MMR deficiency as the cause of their unusual regional mutation rate variation. To understand how the regional mutation rate differs in these samples, we plotted their mutation densities along the genome. The regional rate variation was substantially decreased in these tumours compared to in other microsatellite-stable (MSS) tumours from the same tissues (Fig. 2a-c). The change towards more homogeneous mutation rates in MSI cancers was significant across many chromosomal regions, with ~72% of the genome being affected (1977/2748 1Mb windows, Mann-Whitney test on pooled tissues, 10% FDR; examples in Fig. 2d and Extended Data Fig. 1e, f). Consistently, the regional rates in MSI cancers from all three tissues correlate poorly to replication timing (Fig. 2e-g), gene expression levels and heterochromatin (Extended Data Fig. 2a-f). Moreover, the slopes of the regression lines between binned replication timing and SNV rates revealed consistent changes in MSI cancer samples when calculated separately for intergenic and for genic (intronic) DNA in the whole genome sequences (Fig. 2h, i). Very similar trends were observed in a broader set of 950 exome sequences of colorectal, uterine and gastric cancers (Extended Data Fig. 2g-j).
Figure 2.
Reduced regional mutation rate variability in genomes of MSI cancer samples. a, b, c, Decreased variance between mutation rates of 1Mb windows in MSI samples, when compared to MSS samples (incl. MSI-L) or to ultramutated PolE/MSS samples. MSI/PolE samples are in the MSI group. In the case of STAD, comparison to PolE w.t. hypermutators. Data points in distributions are medians of relative mutation frequencies of each 1Mb window across all cancer samples in group. ** P≤0.01 by F-test for decrease in variance. d, Relative SNV frequencies across 1Mb windows of chromosome 1p in CRAD. Full/dotted lines are the median across tumour samples and its 95% C.I. For each tumour sample, relative mutation frequencies are obtained by dividing by the mean of all 1Mb windows. * FDR≤10% for rates significantly closer to unity in MSI-H samples (Mann-Whitney test). Striped bars are low alignability regions (Methods). e, f, g, Reduced correlation of regional mutation rates to replication timing in MSI cancer samples. Genomic 1Mb windows were pooled into five equal-frequency bins by the median RepliSeq signal over 11 cell lines. For each bin, median and interquartile range of relative mutation rates across 1Mb windows is shown. R2 on original (not binned) data. * P<0.01 for difference of R, after Fisher Z-transform. Prior to binning, cancer samples in a group were combined by taking the median of the relative mutation frequencies in each 1Mb window (as shown in d). h, i, Same, examined separately for genic (intronic) and intergenic regions. a is the slope of the regression line fit to binned data.
High mutation rates in uterine and colorectal cancers can also be caused by inactivation of the proofreading domain of DNA polymerase epsilon13,14 (PolE). Proofreading is a result of a 3′-5′ exonuclease activity that enhances the accuracy of PolE by excising incorrectly placed nucleotides during synthesis. MSS PolE tumours exhibited a significantly larger spread of the regional SNV density distribution than MSI tumours (Fig. 2a, b), even though their mutational load is typically higher (Extended Data Fig. 1c). Similar conclusions are reached with stomach cancer15 hypermutators of unknown aetiology (Fig. 2c). Thus, increased mutation supply does not explain the loss of regional mutation rate variability in MSI cancers.
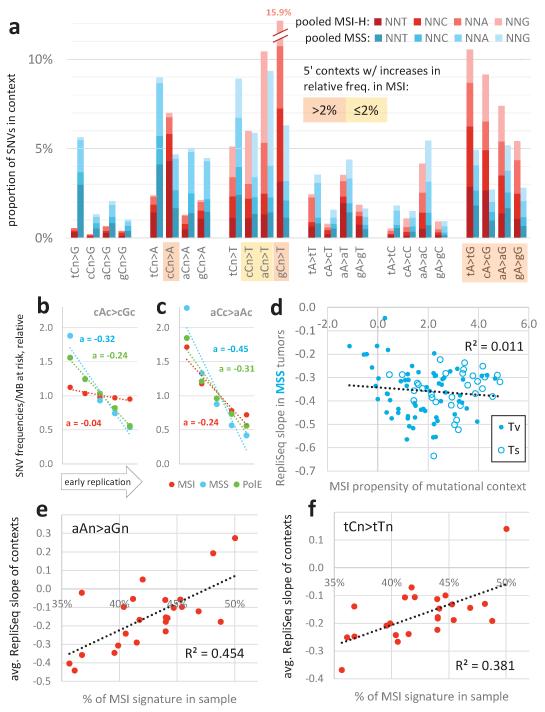
The relative frequencies of 5′ and 3′ contexts of different SNVs - the mutation spectrum - are informative of the mutational processes operative in a particular cancer type16. We observed the previously-reported17 signatures of MMR-deficiency in MSI cancers: C>T transitions in a NpCpG sequence context and C>A transversions at CpCpC (all mutations considered strand-symmetrically). In addition, we report a general increase in the relative frequency of transitions in MSI genomes, wherein A>G increases preferentially when preceded or followed by a C, and C>T clearly increases most in the GpCpN context (Extended Data Fig. 3a).
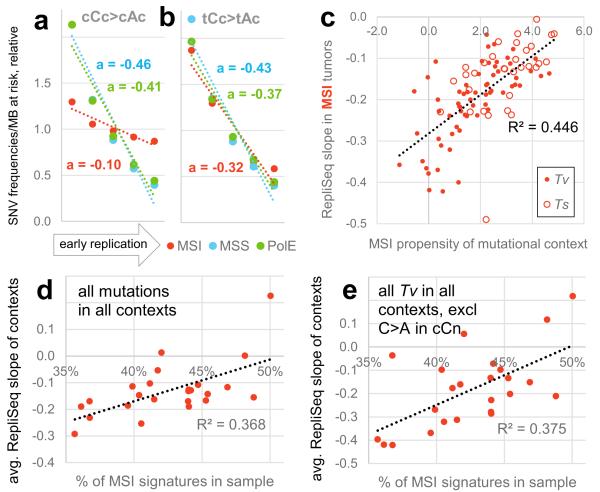
We examined how the different mutation spectra are distributed across the genome in MSI samples. The signatures most characteristic of MMR-deficiency had a considerably flatter distribution in MSI tumours than in MSS or PolE-mutated tumours whereas this was less the case for signatures not associated with MMR-deficiency (Fig. 3a, b; Extended Data Figure 3b, c). Indeed, the more abundant a mutational context becomes specifically in MSI tumours, the more uniformly it is distributed with respect to replication timing in MSI samples (Fig. 3c, R2=0.45, P<10−6) but not in MSS samples (Extended Data Fig. 3d, R2=0.01).
Figure 3.
Association of mutational signatures to MSI and to replication timing. a, b, Relative mutation rates of example MSI-associated (a) or non-MSI-associated (b) contexts across genomic replication timing bins. Dotted lines are linear fits to the bins with slope a, a measure of association to replication timing. c, Association between MSI propensity of a mutational context (log2 ratio of its frequency in MSI vs. MSS tumours) to its replication timing slope in MSI tumours. Tv, transversion. Ts, transition. d, e, Association of % MSI-specific signatures (cCn>A + gCn>T + [c/t]An>G) in a MSI tumour sample and the binned replication timing slopes for all contexts (d), or for various non-MSI transversions (e) in the same tumour sample. Slopes averaged over contexts displayed in d, e. In all panels, mutation rates were normalized to number of nucleotides at risk in a 1Mb window prior to determining the slopes. See also Extended Data Figure 3.
Notably, however, many signatures not associated with MMR-deficiency do flatten to some extent in MSI tumours. This suggests that the residual correlation to replication timing in MSI cancers (Fig. 2e-g) might derive purely from the mutations originating prior to MMR inactivation. To test this idea, we used the proportion of mutations in MSI-associated contexts to sort the MSI samples by the proportion of their history spent in a MMR-deficient state (Methods). The proportion of mutations in MSI-associated contexts significantly correlates to how flat the overall regional rates are with respect to replication timing in each sample (Fig. 3d, R2=0.37, P=0.0017). Importantly, this is also true for mutations in contexts that increase little or decrease in relative frequency in MSI genomes: all transversions (excluding C>A in CCN) and the transitions A>G in AAN and C>T in TCN (Fig. 3e, Extended Data Fig. 3e, f; R2=0.38-0.45, P<0.0015). Thus, the greater the proportion of its history a tumour has spent in a MMR-deficient state, the flatter its distribution of mutations across the genome and this flattening is observed for mutations in both MSI-associated and non MSI-associated contexts.
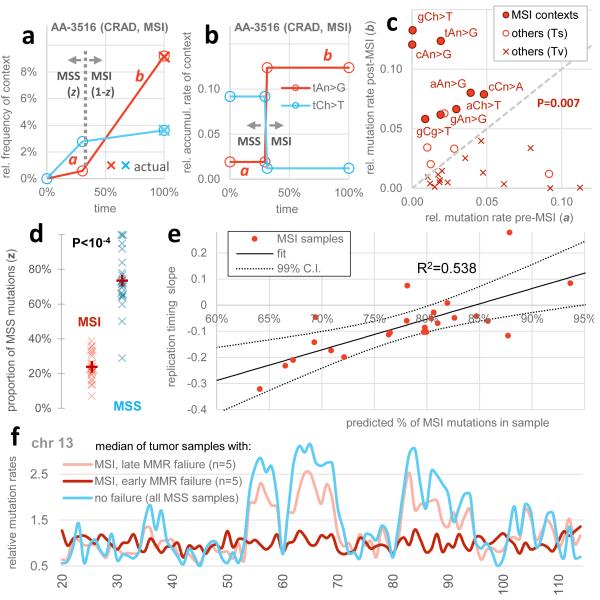
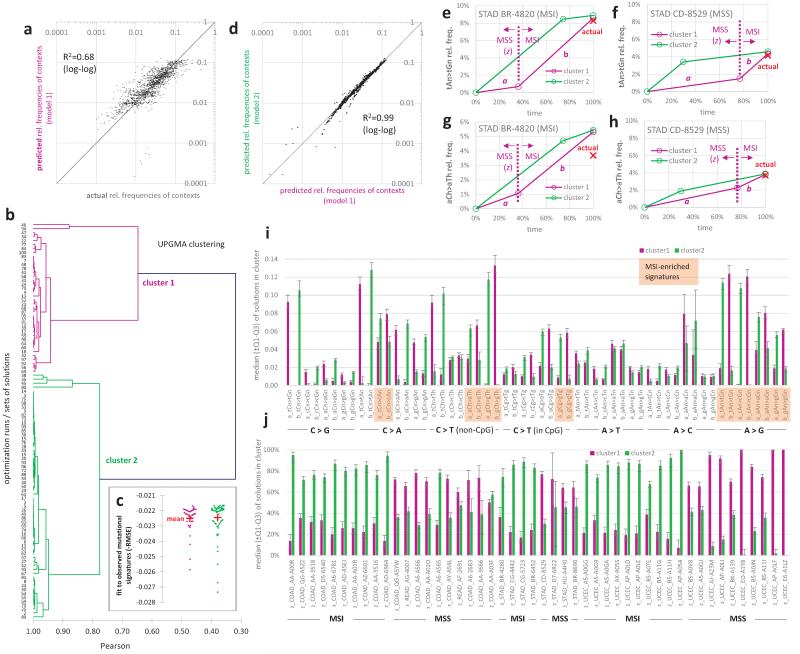
To more precisely quantify the contribution of MMR to the observed regional mutation variation across the genome, we next estimated the time spent in the MMR-proficient and MMR-deficient states in each individual tumour. We used a simple model that assumes that genome-wide mutation rates for each mutational context are equal across samples but allowing a distinct rate for each context in the MMR-proficient and MMR-deficient states (schematic in Fig. 4a, b). We employed a genetic algorithm-based optimization that minimizes the difference to observed mutational signatures (Methods). After fitting a global set of rate parameters, this simple model captures 68% of the variance in differential mutational context usage across the MSI and a set of MSS cancer samples from all three MSI-prone cancer types (Extended Data Fig. 4). Importantly, the de-convolution recapitulated the known consequences of MMR deficiency on the mutational spectrum and the MSI status of each sample (Fig. 4c, d).
Figure 4.
Inferring the time of MMR failure by a deconvolution of the mutational signatures. a, b, Examples illustrating the parameters estimated: relative mutation rates in the MSS (a) or MSI states (b), different for mutational contexts but constant across samples, and the relative time spent in MSS (z) or MSI (1-z) which can vary across samples. c, The estimated proportion of MSI mutations for the MSI vs. a set of MSS samples. P value by Mann-Whitney test, two-tailed. d, Estimated rates for mutational contexts. Significance given for increase of b over a in the eight MSI contexts (Wilcoxon test). Contexts with different 3′ flanking nucleotides were pooled, except the C>T in nCg contexts; other C>T changes are labelled nCh. Ts, transitions. Tv, transversions. e, Estimated # mutations arriving post-MMR failure correlates to the loss of variability in regional mutation rates (slope of the relative rates across replication timing bins, see Fig. 2 and 3) across the MSI samples. The 99% C.I. of the fitted line crosses zero at <100% mutations post-MSI, indicating (P<0.01) that the mutation rate landscape in MMR-deficient cells does not show replication timing-associated regional variability. f, Relative mutation rates of chromosome 13 for the median of five samples with largest % of post-MMR failure mutations (rightmost in e) vs. five MSI samples with least % post-MMR failure mutations (leftmost in e). Both groups consist of 2 CRAD, 2 UCEC and 1 STAD.
The model shows that the earlier MMR fails in the history of a tumour, the flatter its regional rate landscape is with respect to replication timing (Fig. 4e, R2=0.54, P<10−4; examples in Fig. 4f). Indeed the inferred time of MMR failure predicts the loss of regional rate variability better than a simple proportion of mutations in MSI-prone contexts (R2=0.37, in Fig. 3d). In addition, the 99% confidence interval of a linear fit to the points in Fig. 4e crosses the horizontal zero line - corresponding to a fully flat regional landscape - prior to the point where all mutations are predicted to have arrived in the MMR-deficient state. This indicates that the mutations that arose after the inactivation of MMR in these tumours are not distributed with the characteristic regional variation across the genome. In the absence of MMR, mutation rates are not reduced in early replicating euchromatic regions compared to in late replicating heterochromatin.
In summary, through an analysis of human tumours we have shown that MMR is more effective in euchromatic early-replicating regions of the human genome and that this suppresses the accumulation of mutations in these regions. MMR is known to be coupled to DNA replication, with elevated repair efficiency during S-phase18,19. Differences in DNA accessibility to the repair machinery20-22, the coupling of this machinery to the replication fork, or the time available for repair might contribute to the increased efficiency of repair in early replicating euchromatin. Across cell types, most active genes performing essential functions are euchromatic and replicated early23-25. It is thus sensible to envisage that enhanced MMR in euchromatin is a beneficial trait and one that has been selected for during evolution.
Methods
TCGA genomes, calling somatic mutations
We downloaded aligned short reads (to hg19/GRCh37) for the available whole-genome sequences of tumours (n=630) and the matched normal tissue from the TCGA repository at CGHub. We then called somatic single-nucleotide variants (SNVs) in each tumour-normal pair using Illumina’s Strelka 1.0.6 workflow26. Strelka is a highly accurate caller, with a low false positive rate of SNVs at the default settings27,28. We further increased the stringency of Strelka’s post-call filtering to prevent spurious mutation calls. By default Strelka requires that the overall confidence score (QSS_NT) is ≥15, that the fraction of filtered basecalls at the site (BCNoise) is <40%, and also that the fraction of reads crossing site with spanning deletions (SpanDel) is <75%. Here, we allow very few filtered or gapped reads at the site: BCNoise and SpanDel must both be <3% for the tumour sample and <10% for the normal tissue. Exceptionally, for extremely high confidence calls (QSS_NT≥45), BCNoise for the tumour sample may be <6%. For TCGA leukaemia (LAML), we downloaded the called somatic SNVs from the corresponding publication29 (n=50 samples).
Other whole genome sequences
We downloaded the previously called somatic SNVs for whole genome sequences (n=507) from the online supplementary material of Alexandrov et al.16; these samples did not overlap the TCGA dataset. Next, we downloaded the somatic SNVs from the whole genome sequences in the ICGC v15.1 database, in case the same genome sets were not already available in Alexandrov et al. or TCGA; this encompasses the ICGC projects RECA-EU, MALY-DE and EOPC-DE (n=150 genomes). Finally, we removed all samples of B-cell lymphoma from the Alexandrov et al. set due to a suspected partial overlap with a broader set in ICGC MALY-DE.
Filtering and dividing the genome into 1Mb windows
To rule out errors due to misalignment of short reads, we masked out all regions in the genome defined in the “CRG Alignability 36” track30, requiring a 36-mer to be unique in the genome even after allowing for two differing nucleotides. Next, we masked out the regions in the UCSC Browser blacklists (Duke and DAC), chosen for often exhibiting anomalous signal in next-generation sequencing experiments (http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=wgEncodeMapability). Finally, we discarded the exons (plus flanking 2nt) of the UCSC gene set to avoid the signal stemming from selection on gene coding regions. Then we divided the genome into non-overlapping 1Mb windows and discarded those with <250 kb of DNA passing the above filters; we also exclude the remaining windows on chromosome Y (n=7) as RepliSeq data was not available for them (see below). This yielded 2748 1Mb windows with an average of 664 kb highly alignable, non-blacklisted, non-exonic DNA per window. When determining SNV densities, the SNV counts in each window were divided by its effective length (after masking) to obtain frequencies per MB. The choice of 1Mb resolution is a trade-off between inclusion of low mutation burden tumour samples/types (better with coarser resolution) and the level of detail in describing the regional variability (better with finer resolution). Furthermore, replication timing is known to be organized in megabase-scale domains23,25.
Conditions for inclusion in the final genome set
We merged the three sets of genomes (TCGA, Alexandrov et al. and ICGC) and discarded the genomes that had less than 3000 SNVs in the alignable regions of the genome in order to obtain more reliable regional mutation density estimates. This filter will completely remove the cancer types with very low SNV loads, such as TCGA thyroid, prostate, leukaemia, or kidney chromophobe. We also removed four cancer types represented by only a single genome with ≥3000 SNVs (LGG in TCGA, ALL and medulloblastoma in Alexandrov et al., and EOPC-DE in ICGC). Three TCGA uterine cancer UCEC genomes (AP-A0L0, B5-A1MY, EY-A1GS) exhibited a high number of C>A changes at low clonal frequencies, consistent with known artefacts arising due to 8-oxoG in oxidized DNA samples31 and were thus excluded, arriving to a final set of 652 genomes and 16,962,503 SNVs in the alignable, non-blacklisted, non-exonic regions of said genomes (data sources listed in Supplementary Information Table 1).
Microsatellite instability status of samples
The MSI-H, MSI-L or MSS status were taken from the clinical data files on the TCGA FTP site repository, named nationwidechildrens.org_clinical_patient_xxxx.txt or nationwidechildrens.org_auxiliary_xxxx.txt, where xxxx is one of coad, read, stad, or ucec, corresponding to colon, rectal, stomach or uterine cancer. The methods used for PCR phenotyping are described in the corresponding TCGA publications13-15. Given previous reports stating that MSI-L samples are much more similar to MSS than to MSI-H in clinical and genomic features32,33, we grouped MSI-L with MSS in all analyses.
For 10 (of 64) colorectal cancers with the whole-genome sequence available, the MSI annotations were not supplied in the TCGA and we thus inferred them from them from the overall load of somatic SNVs and introns called by Strelka (see above). In particular, AD-A5EJ and QG-A5Z2 were putatively labelled as MSI tumours (both having >15k small indels and >40k SNVs) and QG-A5YW, QG-A5YX, A6-A56B, QG-A5YV, A6-A566, QG-A5Z1, A6-A567 and A6-A565 were putative MSS tumours (all with <1.5k small indels and <15k SNVs). In case of uterine and stomach cancers, all samples with whole genome sequences were assigned a MSI/MSS label by the TCGA.
MSI and MSS polymerase ε (PolE) mutants
The PolE status of samples was inferred by requiring a nonsynonymous somatic mutation called in the PolE gene. Consistent with previous reports32, the MSI tumours with a PolE mutation have an overall mutational load similar to MSI PolE w.t. samples, unlike the PolE mutated MSS samples which are ultramutated13,14. We thus grouped the MSI PolE mutants with MSI PolE w.t. samples, except in the case of 2 (out of 3) MSI PolE mutants in uterine cancer (UCEC: AX-A0J1 and AP-A051). These samples had a much higher mutational load than other MSI UCEC samples (leftmost UCEC columns in Extended Data Fig. 1c). Moreover, they also had a mutational signature not consistent with other MSI - in particular, the relative abundance of A>G transitions was low in these samples (11% and 13%), much less than in 10 other UCEC MSI (avg. 34%, range: 23-60%). We thus omitted these two MSI+PolE UCEC samples from all analyses that involved examining mutational signatures (corresponding to Fig. 3 and 4, and Extended Data Fig. 3 and 4), thus forming the set of 24 MSI samples used therein, of which 10 CRAD, 10 UCEC and 4 STAD.
Cancer exome sequences
The called somatic mutations from exome sequencing (MAF files) was downloaded from the TCGA for the COAD/READ (=CRAD), UCEC and STAD cancer types in October 2014. For each tumour sample (TCGA patient), we selected the newest available MAF that had mutation data for that sample, and we did not load further mutation data for that sample from other MAFs (files listed in Supplementary Information Table 1). Next, we assigned MSI-H, MSI-L or MSS status of the exomes from the same data sources used for the whole genomes (see above) and discarded samples where the MSI status was not known. This left 950 samples, of which 195 MSI-H, and the rest MSS or MSI-L (pooled together). PolE status was not inferred for exomes. The genomic mask for the exome analysis was constructed differently than for whole genomes: we similarly used the “CRG Alignability 36” filter and the two UCSC blacklists. However, we excluded all DNA except the protein coding exons (but not exons of commonly mutated cancer genes, which were also excluded). Finally, we retained only the 1Mb windows with at least 5 kb of alignable, non-backlisted exonic DNA. This reduced the initial set of 2748 windows (for the whole genome analysis) to a 1709 windows for the exome analysis. Density of mutations was expressed per Mb of available DNA in each window, and again normalised by dividing by the average of all windows in a sample. For the analysis where we considered each exome separately (Extended Data Fig. 2j), we limited the analysis to exomes with ≥50 SNVs in the selected genomic windows.
Gene expression data
The expression levels in tumours were downloaded from the TCGA RnaSeqV2 data sets34 where they are expressed as TPM (transcript-per-million35) values for each gene. For 15 TCGA cancer types that had RnaSeqV2 data available, we downloaded TPM levels for those tumour samples where we had called whole-genome somatic SNVs, in total 8-92 RnaSeqV2 samples per cancer type (average=29). The TPM levels of genes overlapping (incl. partially) each 1Mb window were averaged for a tumour sample, and we then averaged over all samples of each cancer type to get the final expression level for that 1Mb window in that cancer type; if lower than 0.01 TPM, it was adjusted to 0.01 TPM. The overall, cross-tissue 1Mb expression levels (in Extended Data Fig. 2a-c) are then the medians across 15 cancer types.
The “tissue specificity” (TS) of gene expression of 1Mb windows for a particular cancer type (in Fig. 1g) is the log2 ratio of TPM in that cancer type and the average TPM across all cancer types. In comparing the TS of gene expression to TS of mutation rates, we limited the analyses to the 2442/2748 1Mb windows that were at least somewhat expressed (>0.01 TPM) in at least one examined cancer type. Moreover, we excluded chromosome Y for consistent treatment with the replication timing TS analysis (see below). Finally, we considered only the 8 cancer types (7 tissues) with significant shifts in the PC3 of regional mutation rates.
Replication timing data
We downloaded the RepliSeq measurements36 (as wavelet-smoothed signal37) of ENCODE cell lines from the UCSC Genome Browser (also available in NCBI GEO as GSE34399). To avoid biasing the sample, we excluded multiple lymphoblastoid cell lines and retained Gm12878 as a representative. We computed the average RepliSeq signal within 1Mb genome windows of the remaining 11 cell lines, except chromosome Y which was unavailable in the original data. The resulting values ranged from 0-100, where higher values indicate earlier replication. The overall, cross-tissue replication timing signal (used for genome binning in Fig. 2, Fig. 3 and Extended Data Figs 2 and 3) is the median value across the 11 cell lines.
The “tissue specificity” (TS) of replication timing in a cell line (in Fig. 1f) is the difference between the RepliSeq signal of that cell line and the average signal across all cell lines. For the TS analysis, we considered those cell lines that (i) could be matched to a cancer type, based on their tissue of origin and that (ii) corresponded to one of the 8 cancer types found to have a significant shift in PC3 of the regional mutation rates (Fig. 1e). In particular, replication timing TS in MCF7 cells served as a proxy for BRCA (breast cancer), BJ cells for SKCM (melanoma), IMR-90 cells for LUSC/LUAD (lung cancers), HEP G2 cells for LIHC (liver cancer) and Gm12878 for DLBC (lymphoma). For consistency with the gene expression TS analyses, we considered the same set of 1Mb windows (see above) in the replication timing TS analysis.
Heterochromatin data
We downloaded RoadMap epigenomics H3K9me3 ChIP-seq signal for a diverse set of healthy tissues or cell lines from NCBI GEO, encompassing: GSM621651 (adult kidney), GSM537710 (adult liver), GSM670028 (mesenchymal stem cells), GSM772917 (CD4 naïve), GSM669939 (fetal lung), GSM450266 (H1 cell line), GSM521914 (IMR90 cell line). In addition, we include the H3K9me3 levels from Barski et al.38 that were previously shown to have a strong correlation to regional mutation rates in cancer2. We calculated the mean H3K9me3 signal in 1Mb windows for each sample and trimmed the distribution at the 99.9th percentile. Then we log-transformed the values and found the median over 8 tissues/cell lines to get the overall H3K9me3 levels used for binning (in Extended Data Fig. 2d-f).
Slopes over genomic bins
We used the overall, cross-tissue RepliSeq signal to create five equal-frequency (same number of 1Mb windows) genome bins for further analyses. Additional sets of bins were created also for the gene expression and for the heterochromatin signal (Extended Data Fig. 2a-f). The slope of a regression line fitted through the average 1Mb relative mutation rates of each bin is measure of association of mutation rates to replication timing (Figs 2-4). This measure has the desirable property of being robust to differences in the overall mutation load between smaller groups of tumour samples or individual genomes, or between mutational signatures.
For the analysis where intergenic and genic regions were examined separately in MSI and MSS cancers, we used the UCSC gene set to define these regions. Since the gene exons had already been excluded in a genome preprocessing step, the genic regions effectively consist only of introns. Only for the purposes of this intergenic vs. genic analysis, we relaxed the requirement of ≥250 kb (of total alignable DNA) per 1Mb window to ≥100 kb (either intronic or intergenic) alignable DNA per 1Mb. When calculating relative mutation rates, regardless if analyzing intergenic or intronic mutations, each sample was always normalised by dividing by the mean of the aggregate (intergenic plus intronic DNA) SNVs rates across all its windows. The five RepliSeq bins were the same as in the whole-genome analysis.
Statistical analysis – general
Principal components analysis (PCA) was performed in R 3.1.1 (R Core Team, Vienna, Austria) and in XLStat 2014.2 (Addinsoft, Paris, France) on the relative 1Mb mutation rates of each tumour sample, where samples were features (columns in data table), and 1Mb windows were examples (rows). To find the relative rates, first the SNV densities per Mb of alignable, non-exonic, non-blacklisted DNA (see above) were determined for each 1Mb window in every tumour sample. Then, these densities in each sample were normalised by dividing by the mean SNV density of all windows in that sample. Thus, the relative mutation frequencies >1 correspond to above-average SNV densities in a tumour sample, and <1 to below-average densities. The approximate 95% confidence intervals of the median (across tumour samples: Fig. 2d, Extended Data Fig. 1e, f) for the 1Mb windows were estimated using the formula ±1.58*IQR/sqrt(n_samp), as defined in the R function boxplot.stats and references therein. In the PCA, the tumour samples were features (columns) and the 1Mb windows were examples (rows); therefore, the PCs will be linear combinations of tumour samples, and the loadings of the samples on PCs 3 and 4 are shown in Fig. 1e. The expected (baseline) percent variance in each PC stemming from noise in data was estimated using the ‘broken stick’ method, found to outperform related approaches39. The cancer types were tested for shifts in PC3 loadings using a Mann-Whitney test (two-tailed) where the loadings of samples in one cancer type were contrasted to the loadings of samples in all other types.
Mutational signatures – general
As in previous work16, the mutational signatures are defined as relative frequencies of SNVs at different nucleotides in all possible 5′ and 3′ nucleotide contexts. The mutations are counted strand-symmetrically, thus six possible changes exist: C>G, C>A, C>T, A>T, A>C and A>G. These are equivalent, respectively, to: G>C, G>T, G>A, T>A, T>G and T>C. Each of the 6 changes has four possible 5′ and four 3′ neighbouring nucleotides, which amounts to 6×4×4=96 contexts. Extended Data Fig. 3a shows their relative usage, as % of SNVs observed in each context. The MSI propensities of contexts (in Fig. 3 and Extended Data Fig. 3) were defined as the log2 ratio of the absolute mutation frequency (per MB) of a context in the MSI samples to its mutation frequency in the MSS samples.
DNA word frequencies are not necessarily equally frequently occurring in genomes; for instance, the CpG dinucleotide is rare. Moreover, they may vary in frequency across the genome, as is most evident in the global G+C content variation. The outcome of analyses that compare how mutational context usage co-varies with replication timing (Figs 3 and 4, Extended Data Figs 3 and 4) may be affected by this. Therefore, we normalised the mutation frequencies in different contexts by dividing each by the number of corresponding nucleotides-at-risk in each 1MB window (only nucleotides passing the alignability mask described above). On data normalised thusly, we determined the strength of association to replication timing via the slope of the line fitted to RepliSeq bins, as described above. (Of note, as with the full set of mutations, here also all 1Mb windows values were divided by the genome average prior to binning.) In case different contexts needed to be combined, the RepliSeq slopes were determined for each context separately and then the slopes were averaged for the combined context.
Determining time of MMR failure in samples
We aimed to determine the relative time spent in the MMR-proficient and MMR-deficient states for each of the 24 MSI samples: 10 CRAD, 4 STAD and 10 UCEC (see “PolE mutants” section above) by deconvoluting their mutational signatures into MSI and MSS components. To these 24 MSI samples, we added a further 24 MSS samples, matched by cancer type, while excluding the PolE ultramutators (in CRAD/UCEC) or otherwise hypermutated (in STAD) samples.
To reduce the number of parameters that need to be estimated, we pooled the contexts with different 3′ nucleotides together (this corresponds to how bars are drawn in Extended Data Fig. 3a). An exception were the NCG>NTG changes (deamination of CpG dinucleotides) that were kept separate from other NCN>NTN changes, which are here denoted as NCH (H = A, C or T). This yields a total of 28 contexts, for each of which we estimated a pre-MMR failure relative mutation rate (a) and a post-MMR failure relative mutation rate (b). The rates were assumed to be constant across samples and in time. Each sample may, however, spend a different fraction of time in the pre-MMR-failure state (z). Thus the total number of parameters to estimate is 28 (a for contexts) + 28 (b for contexts) + 24 (z for MSI samples) + 24 (z for MSS samples) = 104.
For a set of 104 parameters, the expected relative frequency of a context ctx in a sample samp may be calculated as actx*Zsamp + bctx*(1-Zsamp). A measure of goodness-of-fit for a candidate set of parameters was the negative root-mean-square difference between these expected relative frequencies, and the observed relative frequencies of use of 28 different contexts across the 48 samples. We used a genetic algorithm-based optimization to maximize the fit to the observed mutational spectra, as implemented in the rgenoud 5.7-12 package40 in R. The parameters were at defaults, except max.generations=500, gradient.check=FALSE, wait.generations=100 and BFGS=FALSE. The starting populations of solutions were generated with rnorm(1, 0.2, 0.2) for the rates a, rnorm(1, 0.5, 0.3) for the rates b and rnorm(1, 0.5, 0.3) for the times z.
The optimization was run 100 times with different random seeds for function genoud. The algorithm always converged to one of two clusters of very similar solutions with very similar goodness-of-fit scores and that predicted nearly identical mutational spectra for the 48 tumour samples (Extended Data Fig. 4bcd). Upon examining the coefficients of the solutions and schematic diagrams of the accumulation of mutations in time (Extended Data Fig. 4e-j), it becomes evident that the two sets of solutions are equivalent, with symmetrical a and b mutation rates, and also the pre-MMR failure time (z) and the post-MMR failure time (1-z) across all samples. As the representative solution used in further analysis, we take the median a, b and z coefficients across solutions of the cluster where the MSI-specific mutational signature GCH>GTH is predicted to increase in rate after MMR failure. In no other way was the algorithm aware of which signatures are specific to MSI tumours, or which of the 48 samples were MSI and which were MSS. The algorithm could, in principle, be used to deconvolute other mutational processes which have a distinct time of (de)activation during carcinogenesis. Its practical use would, however, likely be limited to those processes which, like MSI, have a very distinct mutational signature.
Extended Data
Extended Data Figure 1.
Overall mutational burden and megabase-scale regional rate variability in tumour samples of MSI-prone cancer types. a, b, Correlations of tissue specificity (TS, see Methods) in regional mutation rates of diffuse large B-cell lymphoma (DLBC) with TS of gene expression in DLBC (a), or with TS of replication timing in the Gm12878 lymphoblastoid cell line (b). c, Overall mutational load, as SNVs per Mb of alignable genomic DNA (Methods) for MSI-H, MSS (includes MSI-L), polymerase ε (PolE) mutant tumours, or otherwise hypermutated tumour samples. d, Principal components plot with PCs 3 and 4, as in Fig. 1e, but showing only tumour samples for colorectal (CRAD), uterine (UCEC) and stomach (STAD) cancers for visual emphasis. e, f, Relative SNV frequencies across 1Mb windows of chromosome 1p in UCEC and STAD. Full/dotted lines are the median across tumour samples and its 95% C.I. For each tumour sample, relative mutation frequencies are always obtained by dividing by the mean of all 1Mb windows. MSI/PolE samples are in the MSI-H group; hyper/ultramutators are not in the MSS group. * FDR≤10% for rates significantly closer to unity in MSI-H samples (Mann-Whitney test; not applicable to STAD because of too few MSI-H samples).
Extended Data Figure 2.
Reduced correlation of regional mutation rates to gene expression, heterochromatin and replication timing in genomes and exomes of MSI tumours. a, b, c, The 1Mb windows in the genome were pooled into five equal-frequency bins by the average gene expression levels (log2 TPM) in each window. The median and interquartile range of relative mutation rates across 1Mb windows is shown for each bin. R2 always determined on original (not binned) data. * P<0.01 for difference of R after Fisher Z-transform. Gene expression levels are medians over RnaSeq TPM across 15 cancer types. Relative SNV frequencies of each tumour sample were obtained by normalizing by the average SNV density of all genomic 1 Mb windows of that sample. Prior to binning the windows, cancer samples in a group were combined by taking the median of the relative mutation frequencies in for each 1Mb window, as illustrated for CRAD in Fig. 2d. PolE/MSI samples are in the MSI group; ultramutators are not in the MSS group. MSI-L samples are pooled with MSS. d, e, f, Same, but for five heterochromatin bins (median H3K9me3 signal over 8 tissues and cell lines). g, h, i, Regional mutation rates in exome sequences of a broader set of 195 MSI-H tumour samples. The 1709 genomic 1 Mb windows with at least 5 kb alignable protein-coding DNA each were grouped into five equal-frequency bins by the median RepliSeq signal over 11 cell lines (Methods). Mutations were pooled across all samples in one cancer type with a known MSI-H or MSS status (Methods). a is the slope of the regression line fit to binned data. j, Slopes a determined for individual cancer exomes with a sufficient number of mutations (≥50 SNVs). Number of samples n shown below each group. For all cancer types, MSI-H samples have significantly less negative slopes than MSS (P<0.01, Mann-Whitney test, one tailed). MSI-H also includes the MSI-H/PolE mutant samples, and MSS includes the MSI-L samples. In the exome analyses, ultramutators were not considered separately.
Extended Data Figure 3.
Association of mutational signatures to microsatellite instability and to replication timing. Related to Figure 3. a, Relative frequencies of the 96 mutation contexts (strand-symmetric) in MSI versus MSS cancers; the MSS group includes MSI-L samples but not MSS/PolE ultramutators. Mutations were pooled across samples of MSI-prone tissues (CRAD, UCEC and STAD). b, c, Similar to Fig. 3a and 3b, showing two additional examples of mutational contexts with different MSI propensities and their relative mutation rates across across five genomic replication timing bins. d, Lack of correlation between the MSI propensity of a mutational context to its replication timing slope in MSS tumours samples (compare to Fig. 3c, which shows slopes in MSI samples). Tv, transversion. Ts, transition. e, f, Association of % MSI-specific signatures (cCn>A + gCn>T + [c/t]An>G) across cancer samples and the binned replication timing slopes for two non-MSI Ts signatures in same samples. Slopes averaged over contexts displayed in each plot. In all panels except a, mutation rates were normalized to number of nucleotides-at-risk in a 1Mb window prior to determining the replication timing slopes.
Extended Data Figure 4.
The deconvolution of MSI mutational spectra robustly converges onto two equivalent solutions. Related to Figure 4. a, Agreement of the observed relative frequencies of mutational contexts in each tumour sample with the predictions of model 1 (having median a, b and z coefficients across all solutions in cluster 1). b, Sets of best-fit solutions determined in a hundred optimization runs initialized with different starting conditions. The solutions cluster into two homogeneous clusters (Pearson R>0.9 between >90% of the solutions within a cluster, in UPGMA clustering). c, d, Solutions within both clusters have similar fit to observed data (c) and make extremely similar predictions for mutation spectra in tumour samples (d). e-h, Example mutation accumulation diagrams for two mutation contexts typical of MSI tumours, shown for a MSI tumour (e,g) and for a MSS tumour (f,h). i, j, Values of the parameters in two solution clusters, with medians and interquartile ranges (shown as whiskers). Each solution encompasses 104 parameters: relative mutation rates a and b for each of 28 mutational contexts (i), and the relative pre-MMR failure time z for each tumour sample of the 24 MSI and 24 MSS samples (j).
Supplementary Material
Acknowledgements
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (BFU2011-26206 and ‘Centro de Excelencia Severo Ochoa 2013-2017’ SEV-2012-0208), an ERC Consolidator Grant IR-DC (616434), AGAUR, the EMBO Young Investigator Program, the EMBL-CRG Systems Biology Program, FP7 project 4DCellFate (277899), FP7 project MAESTRA (ICT-2013-612944) and by Marie Curie Actions.
Footnotes
The authors declare no competing financial interests.
References
- 1.Hodgkinson A, Chen Y, Eyre-Walker A. The large-scale distribution of somatic mutations in cancer genomes. Hum. Mutat. 2012 doi: 10.1002/humu.21616. doi:10.1002/humu.21616. [DOI] [PubMed] [Google Scholar]
- 2.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 3.Woo YH, Li W-H. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat. Commun. 2012;3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 4.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat. Commun. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc. Natl. Acad. Sci. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu TC. A possible function of constitutive heterochromatin: the bodyguard hypothesis. Genetics. 1975;79(Suppl):137–150. [PubMed] [Google Scholar]
- 9.Sima J, Gilbert DM. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr. Opin. Genet. Dev. 2014;25:93–100. doi: 10.1016/j.gde.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-L, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jäger N, et al. Hypermutation of the Inactive X Chromosome Is a Frequent Event in Cancer. Cell. 2013;155:567–581. doi: 10.1016/j.cell.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TCGA Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.TCGA Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.TCGA Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013 doi: 10.1038/nature12477. doi:10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch Repair, But Not Heteroduplex Rejection, Is Temporally Coupled to DNA Replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelbrock MA, Kaliyaperumal S, Williams KJ. DNA mismatch repair efficiency and fidelity are elevated during DNA synthesis in human cells. Mutat. Res. 2009;662:59–66. doi: 10.1016/j.mrfmmm.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amouroux R, Campalans A, Epe B, Radicella JP. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri S, Wyrick JJ, Smerdon MJ. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:1690–1700. doi: 10.1093/nar/gkp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiratani I, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubelsky Y, et al. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24:1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiratani I, et al. Global Reorganization of Replication Domains During Embryonic Stem Cell Differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods’ references
- 26.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 27.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts ND, et al. A comparative analysis of algorithms for somatic SNV detection in cancer. Bioinformatics. 2013;29:2223–2230. doi: 10.1093/bioinformatics/btt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.TCGA Research Network Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derrien T, et al. Fast Computation and Applications of Genome Mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67–e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T-M, Laird PW, Park PJ. The Landscape of Microsatellite Instability in Colorectal and Endometrial Cancer Genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlik TM, Raut CP, Rodriguez-Bigas MA. Colorectal Carcinogenesis: MSI-H Versus MSI-L. Dis. Markers. 2004;20:199–206. doi: 10.1155/2004/368680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 36.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917–927. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barski A, et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Jackson DA. Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology. 1993;74:2204. [Google Scholar]
- 40.Mebane WR, Sekhon JS. Genetic Optimization Using Derivatives: The rgenoud package for R. J. Stat. Softw. In. 2010:473–487. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.