Abstract
Members of the Transient Receptor Potential-Mucolipin (TRPML) constitute a family of evolutionarily conserved cation channels that function predominantly in endolysosomal vesicles. Whereas loss-of-function mutations in human TRPML1 were first identified as being causative for the lysosomal storage disease, Mucolipidosis type IV, most mammals also express two other TRPML isoforms called TRPML2 and TRPML3. All three mammalian TRPMLs as well as TRPML related genes in other species including C. elegans and Drosophila exhibit overlapping functional and biophysical properties. The functions of TRPML proteins include roles in vesicular trafficking and biogenesis, maintenance of neuronal development, function, and viability, and regulation of intracellular and organellar ionic homeostasis. Biophysically, TRPML channels are non-selective cation channels exhibiting variable permeability to a host of cations including Na+, Ca2+, Fe2+, and Zn2+, and are activated by a phosphoinositide species, PI(3,5)P2, that is mostly found in endolysosomal membranes. Here, we review the functional and biophysical properties of these enigmatic cation channels, which represent the most ancient and archetypical TRP channels.
Keywords: TRP channels, TRPML, mucolipins, mucolipidosis type IV, MLIV, ML4, lysosomal, lateendosomal, endosomal, endolysosomal, Ca2+, autophagy
Mucolipidosis Type IV: A Clinical Perspective
The major factors that drove researchers into characterizing the TRPML channels were that they belong to the TRP superfamily of channels, which have garnered a lot of interest recently [1], and that loss-of-function mutations in one of the human TRPML isoforms, TRPML1, results in an autosomal-recessive lysosomal storage disease (LSD) called Mucolipidosis type IV (MLIV). Originally described in 1974, MLIV was identified in patients exhibiting corneal opacity and intracellular accumulation of lysosomal inclusions [2–5]. The disease causing mutations likely originated in Lithuania around the 18th/19th centuries, and although it has now been described in the general population, the vast majority of MLIV cases involve Ashkenazi Jews, whose heterozygous carrier frequency for MLIV related mutations is ~ 1:100 [6–9]. Early descriptions of MLIV noted the accumulation of gangliosides and mucopolysaccharides and cytoplasmic autofluorescent puncta in the brain bearing similarities to those observed in ceroid-lipofuscinoses [5, 6, 10, 11]. These histological alterations still remain some of the most salient and defining features of MLIV and may be used for diagnosis of MLIV [12, 13]. Subsequent biochemical investigations revealed that patient cells accumulated 2–3 fold excess of GM3 and GD3 gangliosides, mucopolysaccharides, and phospholipids [14–17]. This abnormal accumulation of storage material in MLIV cells occurs due to their retention in acidic organelles, delayed metabolism, and diminished trafficking to the plasma membrane and Golgi [18–22]. Thus, the storage in MLIV is secondary to the trafficking and metabolic defects associated with those lipids. Indeed, the secondary defects in vesicular trafficking could underlie the heterogeneity of storage material in MLIV and other LSDs [23]. However, we cannot rule out whether some of the storage material also accumulates due to primary defects in the activity of lysosomal enzyme(s) due to alterations in trafficking of these enzymes to the lysosomes or alterations in the intraluminal environment of the lysosomes, which prevent the enzymes from functioning optimally.
Clinical features of MLIV
Often misdiagnosed as cerebral palsy [24], the characteristics of MLIV include profound psychomotor disability resulting in hypotonia (and sometimes spasticity), and severe unrelenting cognitive impairment, which becomes evident by ~1–2 years of age when patients fail to meet the normal developmental milestones [6, 12, 25–28]. Both gross and fine motor skills are severely compromised [26, 28]. As they get older, patients are unable to walk unassisted and are restricted to wheelchairs [26]. Although MLIV patients surviving into the 30s have been reported [27], the peak cognitive capacity of the more severely affected patients rarely progresses beyond that typical of 2–3 year old children. Most patients do not demonstrate significant expressive lingual ability [26, 27]. However, over the subsequent decades of life the course of MLIV is heterogeneous and slowly progressing with some symptoms showing negligible deterioration [26]. Thus, MLIV is a developmental disease with a slowly degenerative component. Indeed, head MRIs have indicated a characteristically thin corpus callosum [28, 29], which does not worsen with age—likely indicating developmental defects in axonal growth and/or myelination. However, postmortem analyses reveal that MLIV brains exhibit significant neuronal loss in the cortex and basal ganglia, and reactive astrocytosis in the brain stem [30]. The involvement of the basal ganglia in MLIV patients may underlie the deficits in cognition as well as motor control.
One exception to the overall slow time-course of MLIV progression is the rapidly progressive retinal degeneration. Even if some of the less severely affected patients do not display clear psychomotor disabilities, the ocular disturbances may still be present [31, 32]. Along with bilateral corneal opacity and optic atrophy, the retinal degeneration results in complete blindness by the end of the first decade of life [25–28, 33, 34]. Patients have also been reported to exhibit other ocular disturbances including pain, strabismus, bouts of excessive tearing and conjunctivitis, and occasional photophobia [26, 35]. Altered electroretinograms (ERGs) in MLIV patients indicate photoreceptor involvement [36]. Although other sensory modalities such as hearing are normal in MLIV patients [26], anecdotal evidence from parents suggests an elevated threshold for peripheral pain possibly suggesting diminished neuronal excitability. As opposed to some other LSDs like Gaucher’s disease and mucopolysaccharidoses, skeletal abnormalities are absent in MLIV [26, 37]. Other characteristic features of MLIV, which set this disease apart from other LSDs, include constitutive achlorhydria resulting in iron deficiency and markedly elevated serum gastrin levels—used as a diagnostic tool to detect MLIV [28, 37, 38]. The achlorhydria is likely a consequence of the large lysosomal inclusions in gastric parietal cells [39].
Identification of mutations in MCOLN1 as causative for MLIV
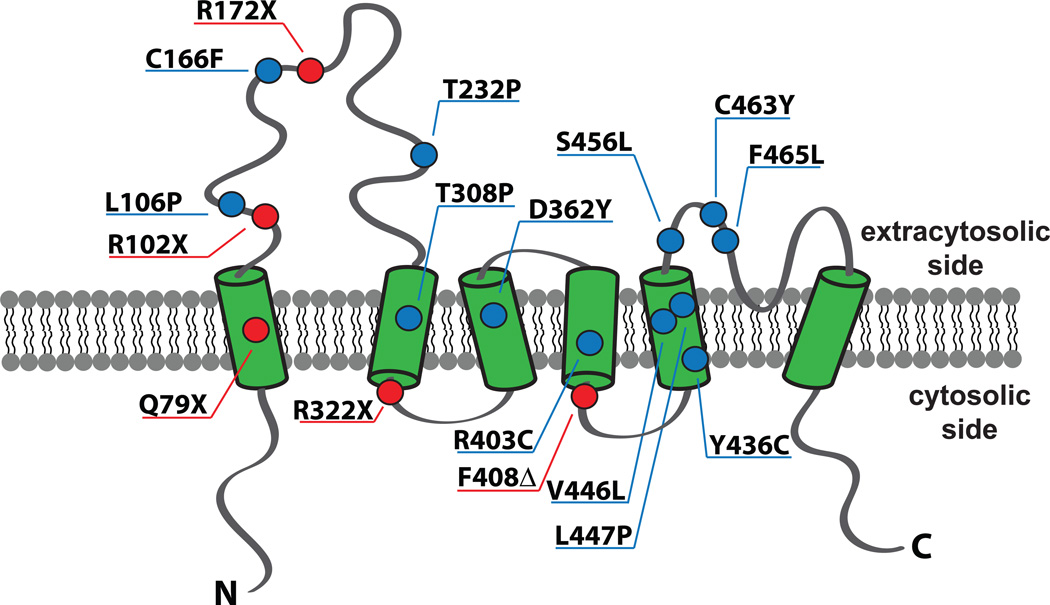
Early attempts to identify the MLIV causing mutations suggested the presence of a single mutated gene on a 1 cM region of chromosome 19p [40, 41]. Within a year, three independent studies identified mutations in MCOLN1, which encodes Mucolipin-1, as the genetic lesion underlying MLIV [8, 42, 43]. The predominant Ashkenazi Jewish haplotype (~72% of the alleles) was found to be a mutation in the splice acceptor site of the 3rd intron on MCOLN1 resulting in aberrant splicing and skipping of exons 4 and 5 [8, 42–44]. In this haplotype, a frameshift mutation results in premature termination [42]. Another Ashkenazi haplotype (albeit relatively minor, ~23% of the alleles) was identified as a ~6 kb deletion spanning the first 6–7 exons of MCOLN1 [8, 42–44]. These two mutations account for ~95% of the MLIV mutations in the MLIV population [8, 40, 42]. Other mutations included nonsense and missense mutations and an in-frame deletion in MCOLN1 (Figure 1), some of which result in relatively milder phenotypes [8, 9, 44, 45].
Figure 1. Topology of TRPML1 and MLIV-causing mutations that lead to codon change.
TRPML1 is predicted to consist of six transmembrane domains (green barrels). Reported MLIV-causing mutations cause changes to the amino acid residues located in both loops and transmembrane regions. Missense mutations causing amino acid substitution are indicated in blue circles. Mutations causing premature stop codon (X) or in-frame deletion (Δ) are indicated in red circles. N and C denote amino-terminus and carboxyl terminus respectively.
TRPMLs: Lysosomal Cation Channels
Analysis of the protein encoded by the MCOLN1 gene immediately revealed significant homology to the TRP superfamily of cation channels [8, 42, 43, 46]. Owing to the homology to TRP channels, mucolipin-1 was also named TRPML1, and was classified into the TRP superfamily of cation channels [1, 47]. Most mammals express two additional TRPML isoforms called TRPML2 and TRPML3 [47]. In contrast, zebrafish express 5 trpml related genes [48]. A single trpml gene is encoded in each of the genomes of yeast (called yvc1/trpy) [1, 49, 50], Dictostelium [51], C. elegans (called cup-5) [52], and Drosophila [53]. A detailed description of the trpml genes in these model organisms is given in a subsequent section of this review.
Subcellular localization of mammalian TRPMLs
When overexpressed in cultured cells, TRPML1 is largely a late-endosomal/lysosomal (LE/lys) transmembrane protein [21, 54–56]. One of the MLIV related variants (TRPML1T232P) is retained in the ER [54] indicating that diminished localization of TRPML1 to the lysosome can result in milder MLIV-like phenotypes. TRPML1 has two di-leucine motifs (on the N- and C-terminal cytoplasmic domains) that regulate the trafficking of TRPML1 to LE/lys [21, 55, 56]. Deletion of these di-leucine motifs causes the accumulation of TRPML1 in the plasma membrane [21, 55, 56]. Conversely, abrogation of clathrin-mediated endocytosis by the expression of a dominant-negative variant of dynamin also results in the accumulation of TRPML1 in the plasma membrane [56]. Although these findings strongly suggest that TRPML1 is trafficked to the lysosome via the cell surface, TRPML1 can also be delivered to the lysosome directly from the trans-Golgi without intermediate delivery to the plasma membrane [55, 57].
TRPMLs can also be trafficked from endolysosomes to the plasma membrane in a regulated and activity dependent manner. Constitutively active TRPML1 mutants, which mimicked the varitint-waddler (Va) mutation of mouse TRPML3 [58–61], exhibit pronounced plasma membrane localization, which is likely a consequence of lysosomal exocytosis owing to the elevated Ca2+ efflux via TRPML1 [62]. Increasing the activity of the mechanistic target of rapamycin complex-1 (MTORC1) activity in Drosophila cells also leads to elevated plasma membrane localization of fly TRPML [63]. This observation could have multiple explanations including MTORC1 decreasing TRPML endocytosis from the plasma membrane or driving TRPML exocytosis potentially by elevating TRPML channel activity.
TRPML2 and TRPML3 localize to intracellular vesicles and long-tubular structures [56, 64, 65]. Similar to TRPML1, TRPML2 has been observed in the LE/lys compartment [56, 64], and traffics via the Arf6-associated pathway and colocalizes with the MHC class I and GPI-anchored proteins [65]. In contrast, overexpressed TRPML3 localizes to multiple subcellular compartments including the early and late-endosomes [66, 67], endoplasmic reticulum [56], plasma membrane, and autophagosomes [67]. These findings indicate that TRPML3 is very dynamic in the cell, and regulates multiple steps in the endolysosomal pathway and autophagy. Moreover, the three mammalian TRPMLs are capable of forming heteromultimers [56, 68–70]. Once coupled into heteromultimers with either TRPML1 or TRPML2, the localization of TRPML3 is largely determined by either TRPML1 or TRPML2 indicating hierarchical control of TRPML subcellular distributions [56]. Moreover, the biophysical properties and regulation of the heteromultimers may be somewhat distinct from the homomultimers, which could contribute to the functional diversity of these proteins [69]. Nevertheless, despite the potential for the mammalian TRPMLs to physically interact, these interactions may have limited overall biological significance because whereas TRPML1 is ubiquitously expressed, at least TRPML2 is expressed predominantly in the lymphoid organs and kidneys [71]. Interestingly, the hierarchical control of TRPML2 by TRPML1 in these organs may also extend to transcriptional regulation because TRPML2 transcription correlates with the presence and/or activity of TRPML1 [71].
Biophysical Properties of TRPML Channels
Early studies relying on the constitutive activity of a small proportion of the overexpressed TRPML1 present on plasma membrane revealed non-selective cation conductance with permeability to Ca2+ [72–75]. The observed currents were outwardly rectifying and inhibited by low pH on the extracellular side. These features were also observed for TRPML1 reconstituted in planar lipid bilayers [76]. The outwardly rectifying TRPML1 current was inhibited by activation of protein kinase A and phosphorylation of two C-terminal serine residues S557 and S559 was shown to be critical for this regulation [77]. However, the biophysical properties described in the early studies are inconsistent with those reported in later ones, which were carried out employing rather different methods, including: (1) the use of mutant constructs that mimicked the varitint-waddler (Va) mutation of mouse TRPML3 and expressed on plasma membrane with high constitutive activities [58–61]; (2) induction of wild type TRPML2 and TRPML3 activity on plasma membrane by removal of extracellular Na+ [58, 78–80]; and (3) direct whole-lysosome recording of TRPML currents from enlarged LE/lys [50, 61]. Under these conditions, all TRPML channels show inward rectification and the current mediated by either TRPML1 or TRPML2 is enhanced by the low pH that mimics the luminal environment of lysosomes [59, 61]. On the other hand, the activity of TRPML3 is inhibited by low pH with a sigmoidal current-pH relationship and IC50 of ~ pH 6.4 for current mediated by either Na+ or Ca2+ [59, 78]. The proton-mediated inhibition appears to involve three histidines (H252, H273, and H283) in the first extracytosolic loop of TRPML3 [78]. However, for the TRPML3Va mutant (A419P), low extracytosolic pH was shown to either have no effect [78] or be inhibitory [59]. The reason for the discrepancies between the earlier and more recent studies remains obscure. Although variations in experimental conditions could account for some of the observed differences, only inward, but not outward, rectification is reported in all recent studies from different groups (see [72, 74, 81, 82]). Therefore, the nature of the outwardly rectifying currents in earlier studies remains mysterious.
Interestingly, the extracytosolic pH dependence of TRPML1Va (V432P) was found to be biphasic, showing potentiation between pH 7.5 and 4.5 and then inhibition at lower pH [81]. The optimal pH for maximal TRPML1 activity was pH 4.5, which is close to the mean pH value of the lysosomal lumen of mammalian cells. Also interesting is that Drosophila TRPML has a similar biphasic dependence on extracytosolic pH, but the optimal pH for maximal activation was shifted to pH 5.2 [81], consistent with the mean luminal pH of insect lysosomes [83].
The luminal side of the lysosomes contains mostly Na+, with relatively low amounts of Ca2+ and K+ ions [61]. In addition, the Ca2+ selectivity of TRPML3 is ~350 times that of K+ [78]. As non-selective cation channels, TRPML channels are equally permeable to Na+ and K+ [81]. The inward rectification property and the relatively positive potential in the luminal side of the lysosome [84] suggest that TRPML channels mainly mediate Na+ efflux from these organelles.
TRPML1 and TRPML2, as well as the fly TRPML, conduct Fe2+ and other divalent cations [61, 81]. However, TRPML3 appears to be relatively Fe2+-impermeable [61]. Although the divalent cation permeability of TRPML1 has been shown to be important for cellular Fe2+ and Zn2+ metabolism [61, 85], it is not known whether TRPML1 is directly permeable to Zn2+ and TRPML1 may be affecting Zn2+ metabolism indirectly without actually being permeable to the cation.
All the TRPML channels tested so far are activated by phosphatidylinositol 3, 5-bisphosphate (PI(3,5)P2)—a phosphoinositide enriched in endolysosomes [61, 81]. The findings that TRPMLs are activated by PI(3,5)P2 are consistent with the notion that TRPMLs mainly function in endolysosomes. Further supporting this idea is that TRPML1 and Drosophila TRPML are inhibited by phosphatidylinositol 4, 5-bisphosphate (PI(4,5)P2) [81, 86], the main PIP2 species in plasma membrane. Thus, although TRPML channels are trafficked to plasma membrane, they may not be functional there because of the high PI(4,5)P2 levels.
A number of small-molecule TRPML3 agonists have been identified by high throughput screening [87]. Many of them also act at TRPML2 [80], but only a few, for instance SF-22 and SF-51, also stimulate TRPML1 [50, 82, 88]. Both compounds have now been optimized and renamed MK6-83 (for SF-22) and ML-SA1 (for SF-51). These compounds have allowed detection of endogenous TRPML-like currents in fibroblasts and macrophages [50, 88, 89]. Recently, it was shown that ML-SA1 is not an agonist for Drosophila TRPML, but rather an allosteric activator that acts specifically on open TRPML channels [90]. In contrast, SF-22 has no effect on the fly channel [90].
The commonly used cation channel and/or Ca2+ channel blockers, 2APB (200 µM), SKF 96365 (50 µM), amiloride (100 µM), ruthenium red (10 µM), or nifedipine (100 µM) did not inhibit TRPML1 or TRPML3, while verapamil blocked TRPML3 only at 1 mM [59]. Recently, novel TRPML1 inhibitors, ML-SI1-3, have been described and shown to inhibit particle uptake and lysosomal exocytosis in bone marrow-derived macrophages [91], demonstrating their utility in assessing the functions of native TRPML1 channels.
TRPMLs in Animal Models
Invertebrate TRPMLs
The only TRP channel gene in the budding yeast, S. cerevisiae, is Yvc1 (also referred to as TRPY [1])—a channel that mediates release of Ca2+ from the vacuole when yeast cells are exposed to a hyperosmotic shock [49]. Multiple features of yeast Yvc1/TRPY indicate functional homology with TRPMLs. First, Yvc1/TRPY shows significant homology at a sequence level to other invertebrate and vertebrate TRPML channels (identity/similarity: with C. elegans CUP-5 = 16%/40%; with Drosophila TRPML = 15%/42%; with human TRPML1 = 16%/40%; with human TRPML2 = 16%/41%; with human TRPML3 = 16%/38%). Second, similar to vertebrate TRPML1 and Drosophila TRPML, Yvc1/TRPY is a PI(3,5)P2 activated Ca2+ release channel expressed in the membrane of the vacuole [50, 81]. The activation of TRPML1 by PI(3,5)P2 has been shown to depend on a cluster of basic amino acid residues on the N-terminus prior to the 1st transmembrane domain of the protein (amino acids residues 55–62: KFRAKGRK) [50]. A similar polybasic domain is found in the cytosolic N-terminus of Yvc1/TRPY (amino acid residues 84–91: KDKANKRK). It would be interesting to determine whether this domain mediates the activation of Yvc1/TRPY by PI(3,5)P2. Third, expression of TRPML1 in yeast cells lacking Yvc1/TRPY partially restored the hyperosmotic shock dependent vacuolar Ca2+ release [50]. Owing to these similarities of yeast Yvc1/TRPY to TRPMLs, it is likely TRPMLs represent the most ancient and archetypical TRP channels. Dictostelium TRPML was reported to be required for lysosomal Ca2+ uptake, and TRPML-deficient Dictostelium cells were characterized by reduced lysosomal Ca2+ levels, and enhanced lysosomal exocytosis [51], which is in sharp contrast to several observations made with mammalian and other invertebrate TRPMLs.
C. elegans express a single TRPML homolog called Coelomocyte Uptake Defective-5 (CUP-5) [52]. Coelomocytes are scavenger cells that line the worm body cavity and are responsible for uptake of the fluid from the body cavity and lysosomal degradation of the proteins dissolved therein. The uptake and degradation of the body cavity fluids and dissolved solutes is defective in all the CUP mutants [52]. As is the case for mammalian TRPMLs, CUP-5 was found to be a LE/lys protein and loss of the protein led to the accumulation of large lysosome-like organelles [52, 92]. In addition, the cup-5 null worms are characterized by embryonic lethality and elevated accumulation of apoptotic cells [92, 93]. Interestingly, these phenotypes are rescued by the exogenous expression of human TRPML1 or TRPML3 indicating functional redundancy between these isoforms [92, 93].
Drosophila also express a single TRPML isoform, which is a LE/lys transmembrane protein [53, 63]. Whereas the neuronal phenotypes associated with the cup-5 hypomorphs have yet to be described, trpml deficient Drosophila are characterized by several neuronal phenotypes, many of which bear remarkable congruence with the established neurological features of MLIV. The plethora of phenotypes observed following loss of Drosophila trpml include elevated accumulation of acidic organelles and autophagic vacuoles in neurons and other tissues, severely compromised autophagic flux leading to age-dependent accumulation of ubiquitinated intermediates, diminished glutamatergic synaptic transmission, elevated lethality during the pupal period, severe locomotor defects during larval and adult phases, and progressive neurodegeneration in the CNS and retina [53, 63]. The cell death in the Drosophila nervous system is incredibly precipitous, and is caused by two simultaneously occurring defects. First, defective autophagy, mitochondrial dysfunction, and elevated levels of reactive oxygen species result in cell autonomous neuronal apoptosis [53]. Second, a concomitant defect in the phagocytic clearance of these cell corpses results in a secondary “neuroinflammation”, which results in non-cell autonomous decline in neuronal viability and ultimately underlies the rapid onset of the phenotypes [53]. Thus, the expression of wild-type TRPML in the trpml mutant phagocytic cells or elevating the phagocytic clearance of the apoptotic cells significantly delays the onset of the phenotypes despite the continued cell autonomous demise of neurons [53].
Interestingly, the pupal lethality associated with the loss of Drosophila trpml was only partially suppressed by the expression of human TRPML1, which shares 38% amino acid sequence identity with Drosophila TRPML (~50% suppression with human TRPML1 compared to 100% rescue following expression of Drosophila TRPML) [53, 63, 81]. We anticipate that there could be multiple explanations for this observation. First, the optimal pH of Drosophila TRPML is ~0.6 pH units higher than that for human TRPML1 [81], which is consistent with the slightly higher pH of endolysosomes in Drosophila compared to mammals (~5.1 in Drosophila lysosomes vs. ~4.5 in mammalian lysosomes) [83, 94]. Therefore, Drosophila lysosomal pH might be optimized for Drosophila TRPML but not for human TRPML1 preventing full activation of the human channel in Drosophila cells. Second, Drosophila TRPML also shares features of mammalian TRPML2 and TRPML3 [81] indicating that expression of these other isoforms or coexpression of the three isoforms in Drosophila cell might be necessary for the complete rescue of the Drosophila trpml mutant phenotypes. Third, human TRPML1 might have a necessary subunit that is missing in Drosophila cells. For instance, human two-pore channels, which are lysosomal channels that share functional similarities with TRPML1 [89, 95–97] and may be necessary for the optimal functioning of TRPML1, are not expressed in Drosophila [98].
Vertebrate TRPMLs
The zebrafish genome encodes five TRPML genes (two TRPML1 orthologs, one TRPML2 ortholog, and two TRPML3 orthologs) [48]. Similar to mammalian TRPML1, the two TRPML1 orthologs in zebrafish are localized to LE/lys [48]. However, mutational analyses of these genes have not yet been described. Two independently generated TRPML1 knockouts in mice have been described [99, 100]. One of the lines faithfully recapitulates many of the MLIV phenotypes including accumulation of lysosome-like inclusions in the nervous system and elsewhere, progressive neurological deficits, and retinal degeneration [99]. Further evaluation of the neuropathological deficits of the MLIV mice revealed elevated ganglioside accumulation in the CNS, presence of p62 puncta characteristic of diminished autophagic flux, reduced myelination, and the presence of activated glial cells in the nervous system indicative of neuroinflammation [101]. These findings indicate that both the mouse and Drosophila MLIV models exhibit elevated neuroinflammation indicating that this may be an evolutionarily conserved outcome of loss of TRPML activity in the nervous system. Both mouse models of MLIV also exhibit achlorhydria and elevated serum gastrin levels [99, 100].
Biological Functions of TRPMLs
Role of TRPMLs in vesicular trafficking
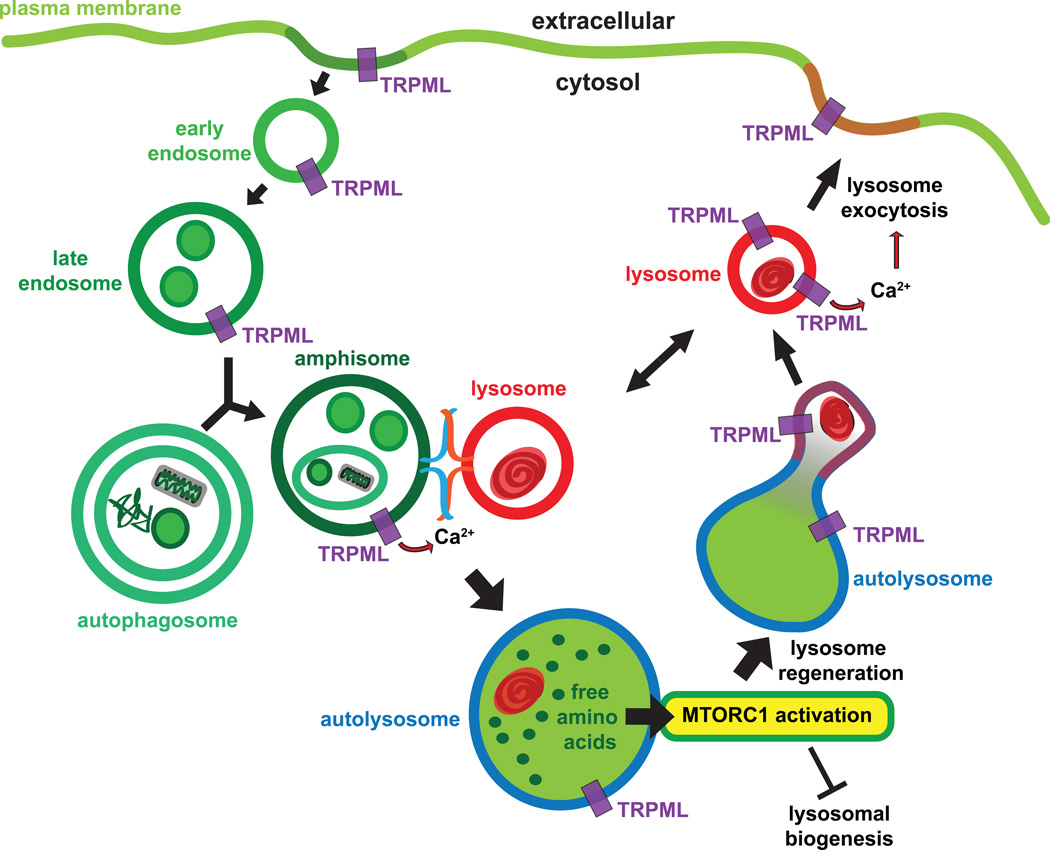
Both homotypic and heterotypic fusion of endosomal vesicles requires Ca2+ [102]. In Drosophila, the Ca2+ required for the fusion of late-endosomes and amphisomes (hybrid organelles formed by LE-autophagosome fusion) with lysosomes is provided by TRPML [63, 103] (Figure 2). These data are consistent with the findings that loss of Drosophila TRPML or mammalian TRPML1 results in the elevation of LE/lys Ca2+ levels [63, 104]. Loss of Drosophila TRPML results in the formation of “fusion-clamped” amphisomes and lysosomes, which resemble synaptic vesicles docked and primed on the presynaptic membrane, but are unable to undergo fusion resulting in diminished degradation of endocytosed and autophagic material [63, 103]. cup-5 deficient worms also exhibit diminished endolysosomal degradation of cell surface receptors [105] and loss of TRPML1 in mammalian cells results in delayed fusion of autophagic vacuoles with lysosomes [106]. Both mammalian cells and fly neurons lacking TRPMLs accumulate autophagic vacuoles, polyubiquitinated proteins, and p62—features that are characteristic of diminished autophagic flux [53, 106, 107]. These studies are consistent with early findings that the delivery of endocytosed material such as lipids is delayed to the lysosomes in MLIV cells [18–21]. However, this issue is somewhat controversial in mammalian cells because other studies have found that the delivery of endocytosed material to lysosomes is not significantly delayed or impaired following acute knockdown of TRPML1 in mammalian cells [108]. These apparently conflicting findings could be explained on the basis of alterations in kinetics as opposed to a complete block of lysosomal delivery in the TRPML1 deficient cells, whereby different experimental conditions and time-courses result in variable findings [21].
Figure 2. Biological functions of TRPMLs in different cellular compartments.
TRPML localizes to endolysosomes and facilitates Ca2+-dependent heterotypic fusion between amphisomes and lysosomes resulting in lysosomal degradation of endocytosed and autophagic material, which provides free amino acids for activation of the MTORC1. MTORC1 activation reduces lysosomal biogenesis via TFEB and promotes lysosome regeneration from autolysosomes. TRPML present on lysosome membrane is also responsible for triggering Ca2+-dependent lysosomal exocytosis, which in turn delivers TRPML to the plasma membrane.
The reformation of lysosomes from hybrid organelles formed after the fusion of late-endosomes with lysosomes is also a Ca2+-dependent process [102, 109]. Consistent with a role of TRPMLs in regulating lysosomal reformation, loss of CUP-5 results in a failure in lysosomal reformation and progressive defects in lysosomal transport in C. elegans [93, 110, 111]. However, detailed ultrastructural studies using electron microscopy, which is capable of distinguishing between lysosomes, MVBs, amphisomes, and autophagosomes, did not reveal a defect in lysosomal biogenesis/reformation in Drosophila trpml mutants [63]. Rather, the levels of electron dense lysosomes are ~10-fold elevated in the trpml deficient cells compared to wild-type controls [63]—a phenotype not consistent with diminished lysosomal reformation.
Although the biological functions of TRPML2/3 are incompletely understood, TRPML2 promotes the activation of the Arf6 pathway, and regulates the sorting of GPI-anchored proteins [65]. TRPML3, which is a non-selective cation channel that localizes to both early- and late-endosomes and is inactivated by low pH, has been shown to be involved in the regulation of endosomal Ca2+ homeostasis, luminal acidification, and fusion/trafficking of endosomal vesicles [58, 66, 67, 112, 113]. Knockdown of TRPML2 and TRPML3 also results in the accumulation of lysosome-like vesicles, indicating significant overlap in the functions of mammalian TRPMLs1-3 in regulating vesicular trafficking [68].
Similar defects in membrane and vesicular trafficking could also be occurring in other LSDs, especially since the function of TRPML proteins can be altered in these diseases. For instance, in Niemann-Pick disease cells, sphingomyelins are shown to abnormally accumulate in the lysosomes [88]. Interestingly, sphingomyelins inhibit the activity of TRPML1 leading to subsequent lysosomal storage, which can be corrected by exogenously activating TRPML1 using specific agonists [88]. These data suggest that the vesicular trafficking defects in Niemann-Pick disease and MLIV may show considerable overlap given that a decrease in TRPML1 activity underlies both diseases. It would also be useful to determine whether additional LSDs are characterized by decreased TRPML1 activity as a causal event for the vesicular storage because TRPML1 agonists could be beneficial in these diseases.
Role of TRPMLs in LE/lys and autophagosome biogenesis
The absence of TRPML genes is characterized by profound accumulation of LE/lys, as well as constitutive autophagy and elevated de novo autophagosome biogenesis [53, 63, 105–107]. Indeed, the accumulation of acidic organelles such as LE/lys is the quintessential feature of all LSDs. We recently asked: what might be the biochemical signaling pathways that underlie the accumulation of lysosomal inclusions in LSDs? Following lysosomal degradation, the amino acids derived from proteolysis are fed into anabolic pathways. Lysosomal free amino acids activate the Mechanistic Target of Rapamycin (MTOR) kinase containing complex, MTORC1 (Figure 2), which couples the levels of free amino acids to protein translation and growth [114–116]. When MTORC1 activity is elevated in the presence of abundant cellular free amino acids, the kinase complex negatively regulates both autophagosome and LE/lys biogenesis [117]. In contrast, during periods of cellular amino acid starvation, MTORC1 activity drops resulting in elevated autophagosome and LE/lys biogenesis [117]. As a consequence, increased lysosomal protein degradation ensues resulting in amino acid replenishment, reactivation of MTORC1, and cessation of autophagosome biogenesis [117, 118].
We recently found that diminished delivery of endocytosed and autophagic material to lysosomes in Drosophila cells lacking TRPML results in lower MTORC1 activity [63, 103]. Therefore, lysosomal dysfunction results in lower free amino acid levels and subthreshold MTORC1 activation. Interestingly, restoration of MTORC1 activity completely suppressed the accumulation of LE/lys in the TRPML deficient Drosophila cells [63]. These data indicate that despite the continued absence of TRPML, the accumulation of acidic vacuoles is reversed simply by the reactivation of MTORC1 [63, 103]. Although the role of MTORC1 downstream of CUP-5 has not been examined in worms, starvation due to inefficient lysosomal degradation of endocytosed embryonic yolk protein (degradation of yolk protein provides amino acids and lipids required for survival of embryos) and a ~10-fold decrease in ATP levels has been proposed to partially underlie the lethality observed in cup-5 mutants and restoration of ATP levels partially suppresses the mutant phenotypes [105]. Because elevation of cellular ATP levels should also result in increased MTORC1 activity due to suppression of AMPK activity—a negative regulator of MTORC1 activity [119], the pronounced biogenesis of autophagosomes and LE/lys and other consequence of amino acid and ATP starvation in the absence of TRPMLs is likely a consequence of diminished signaling through the MTORC1 pathway.
Role of TRPMLs in lysosomal exocytosis and vesicular delivery to the plasma membrane
Early studies of the properties of TRPML1 found that it drives the exocytosis of lysosomes [72, 73, 120]. Indeed, TRPML1 variants carrying mutations that elevate the channel activity induce constitutive lysosomal exocytosis leading to pronounced plasma membrane localization of the channels [62]. TRPML1 has also been shown to promote the formation of tubular extensions from the cell surface that are a consequence of LE/lys exocytosis via a process requiring the serine-lipase domain present in an extracytosolic domain [121]. In mouse macrophages, TRPML1 is required for the exit of endocytosed macromolecules from the lysosomes and the delivery of major histocompatibility complex II to the plasma membrane [22]. Moreover, overexpression of transcription factor EB (TFEB), a master regulator of endolysosomal biogenesis that promotes TRPML1 transcription [122], promotes lysosomal exocytosis by upregulating the expression of TRPML1 [123, 124]—yet again confirming the critical function of TRPML1 in endolysosomal exocytosis. Recently, TRPML1 was shown to be required for focal exocytosis of LE/lys to the site of phagosome formation at the plasma membrane and thereby plays a significant role in phagocytosis [91]. Therefore, loss or inhibition of TRPML1 results in a decrease in the phagocytic uptake of senescent and apoptotic cell corpses [91]. A defect in the phagocytic uptake of apoptotic neurons was also found in Drosophila lacking trpml and this defect underlies the precipitous onset of neurodegeneration in the Drosophila MLIV model [53].
Role of TRPMLs in mitochondrial function and cell viability
The concept of a “lysosome-mitochondrial” axis indicates close functional crosstalk between these two organelles [125, 126]. Indeed, unbiased protein interaction studies have found that TRPML1 physically interacts with several mitochondrial proteins [127]. Therefore, lysosomal dysfunction in cells lacking TRPML1 lead to concomitant aberrations in mitochondrial function, morphology, Ca2+ buffering capacity, and caspase-mediated cell death [128]. Knockdown of endogenous TRPML2 and TRPML3 also result in mitochondrial abnormalities [68]. Drosophila adults lacking TRPML show pronounced mitochondrial dysfunction, accumulation of mitochondria with dissipated electrochemical membrane potentials, diminished autophagic-clearance of damaged mitochondria, and increased reactive-oxygen species [53]. Together, these cytological insults result in neuronal cell loss in the brains of Drosophila adults lacking TRPML [53]. Recently, loss of TRPML1 in mammalian cells was also shown to result in loss of mitochondrial membrane potential and the buildup of reactive oxygen species [129].
Whereas the loss of TRPMLs results in mitochondrial dysfunction and eventual cell death, excessive activity of TRPMLs also results in cell death, presumably by inducing Ca2+ excitotoxicity. The varitint-waddler mice (bearing the Va mutation (A419P) in TRPML3) are characterized by deafness, vestibular alterations, and diminished pigmentation [130]. Several groups found that the Va mutation in murine TRPML3 leads to a gain of function phenotype whereby excessive Ca2+ flux through TRPML3 results in Ca2+ cytotoxicity, cell death, and auditory hair cell dysfunction [58–60, 78, 112, 131]. Similarly, introduction of the Va mutation in TRPML2 also results in elevated plasma membrane localization, Ca2+ overload, and subsequent demise of the cells [79]. Introduction of the Va mutation in conserved residues of TRPML1 or Drosophila TRPML also results in a constitutively-active channel [61, 62, 81]. These studies demonstrate that TRPML channel activity in cells needs to be tightly regulated and uncontrolled cation flux through these channels induces cellular toxicity.
Cell death in the absence of TRPMLs can also take place via mitochondria-independent mechanisms. For instance, acute knockdown of TRPML1 also results in the loss of lysosomal integrity and the release of the lysosomal protease cathepsin B into the cytosol [132]. The cytoplasmic cathepsin B contributes to the apoptosis observed in the absence of TRPML1 [132]. In this regard, decreasing the levels of LE/lys by activating MTORC1, which should also decrease the levels of cathepsin B in cell, might prove beneficial in decreasing the toxicity associated with the loss of TRPML1.
Role of TRPMLs in the regulation of lysosomal Fe2+ and Zn2+
Lysosomes and mitochondria serve as the major cellular reserve of Fe2+ and Zn2+—cations whose homeostatic regulation by TRPMLs is critical for the survival of cells [104]. Interestingly, TRPML1 serves as an endolysosomal Fe2+ channel and loss of TRPML1 results in Fe2+ accumulation in these acidic organelles [61]. The MLIV-associated point mutations affect Fe2+ flux via TRPML1 to varying extent, and this variability correlates with disease severity associated with those mutations [61]. Drosophila TRPML is also capable of fluxing Fe2+ (but not Fe3+) [81] indicating evolutionary conservation of Fe2+ fluxing capacity of TRPML channels. Loss of the TRPML1 also results in Zn2+ accumulation in acidic organelles [133]. Although the permeability of TRPML1 to Zn2+ has not been directly measured, these data might indicate a role for TRPML1 in regulating Zn2+ efflux from LE/lys. Besides mediating Zn2+ efflux directly, TRPML1 could be affecting lysosomal Zn2+ levels by mediating the exocytosis of Zn2+ laden lysosomes, trafficking of Zn2+ transporters between various subcellular locations, or the levels of Zn2+ binding proteins.
Central role for TRPMLs in LSDs
LSDs such as Niemann-Pick are also characterized by significantly reduced activity of TRPML1 [88]. In addition, overexpression of TFEB decreases the storage of acidic vacuoles in LSDs by activating TRPML1-dependent lysosomal exocytosis [122–124, 134]. Therefore, TRPML channels may have roles in multiple LSDs and activating these channels could be beneficial in a wide array of storage diseases.
A controversial role of TRPMLs in the regulation of endolysosomal pH
The impact of TRPMLs on endosomal pH has been a matter of debate and scrutiny. Early studies reported that lysosomal pH is elevated by one pH unit in MLIV cells [135]. However, subsequent studies suggested that TRPML1 might be a lysosomal H+ channel responsible for the release of H+ from the lysosomal lumen, thus a lower luminal pH in the absence of TRPML1 [74, 75]. Lower lysosomal pH following the loss of TRPML channel activity has also been reported by other studies [53, 108]. Moreover, slightly elevating the lysosomal pH using Nigericin or Chloroquine has been shown to be beneficial in MLIV cells [75]. However, another study found that Nigericin/Chloroquine treatment led to very limited attenuation of the lysosomal storage observed in MLIV cells [136]. In addition, other studies have reported that TRPML1 is not permeable to H+ [59, 61]. In summary, the exact effect of the loss of TRPML on H+ flux and endolysosomal pH, if any, is still incompletely understood.
Physical and Genetic Interactors of TRPMLs
Physical interactors of TRPMLs
A large-scale screen identified several candidate proteins that interact with TRPML1 [127]. Briefly, some of the interesting interactors of TRPML1 in this screen involved the cytoskeleton modifying small G-proteins such as Rac2 and Cdc42 (indicating a potential role of TRPML1 in cytoskeleton regulation), lysosomal enzymes such as Cathepsin B, members of the solute carrier family of transporters, the ER Ca2+ release channel IP3-receptor as well as the Ca2+ transporting ATPase (SERCA2), markers of the ER-Golgi intermediate compartments (ERGIC) and Yif1, which regulate ER to Golgi trafficking of proteins (these interactions may mediate the exit of TRPML1 and associated proteins from the ER), and peroxisome-associated Pex16 [127]. Interestingly, both approaches identified several mitochondrial proteins as TRPML1-interactors that among others include NADH dehydrogenase, ATP synthase subunits, and subunits of the mitochondrial voltage-dependent ion channel (VDAC, also known as Porin) [127].
Another screen found that TRPML1 physically associates with members of the lysosome-associated protein transmembrane (LAPTM) family of lysosomal transporters [137]. Knockdown of LAPTMs in mammalian cells led to the accumulation of lysosomal inclusions morphologically similar to those observed in MLIV cells [137]. These data strongly implicate the loss of interactions between TRPML1 and LAPTMs in the pathophysiology of MLIV. TRPML1 also interacts with the EF-hand containing protein, ALG-2, in a Ca2+ dependent manner and this interaction impacts endolysosomal vesicle trafficking [138].
Genetic interactors of TRPMLs
In C. elegans, loss of CUP-5 was found to be associated with elevated accumulation of a lysosomal ABC transporter, MRP-4, which is responsible for the transport of lipids into the lysosomal lumen [139]. Loss of function MRP-4 mutations suppressed the lysosomal degradation defects and the embryonic lethality observed in the CUP-5 deficient animals suggesting that the lysosomal accumulation of MRP-4 and the lipophilic molecules transported by the protein contribute to the toxicity associated with the loss of cup-5 [139]. In Drosophila, the toxicity associated with the loss of trpml is suppressed by the expression of molecular chaperones such as heat shock proteins and is enhanced by the expression of polyglutamine-stretch containing proteins [53]. These findings indicate that the toxicity associated with the loss of trpml might be related to the accumulation of macromolecular aggregates and potentially and unfolded protein response. In humans, gene expression profiles revealed that transcription of several hundred genes, many of which are involved in lysosomal function and hydrolytic activity, endolysosomal trafficking and vesicle fusion, solute transport, organellar biogenesis, and lipid metabolism, are altered in MLIV fibroblasts compared to controls [140].
Concluding Remarks
Over the last decade and a half, several in-roads have been made into understanding the biological functions and biophysical properties of TRPMLs in mammalian cells and animal models. We anticipate that in the future a host of biological functions of the TRPML channels unrelated to MLIV will also be described. However, we still face several challenges. For instance, identifying the precise nature of the neuronal dysfunction in vertebrate models of MLIV and human patients is essential before we come up with viable concepts for therapy. Another interesting question is whether lipids such as PI(3,5)P2 that activate TRPMLs are dynamically regulated in response to physiological stimuli that drive endolysosomal Ca2+ release. Also, whether or not manipulation of TRPML function can ameliorate other LSDs remains to be categorically established. Nevertheless, the future likely holds exciting new findings, which will undoubtedly drive our understanding of these unique channels and lysosomal physiology to new heights.
Acknowledgements
The work in the K.V. and M.X.Z. labs are supported by the NIH research grants R01NS081301 and R01GM092759 respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman ER, et al. Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J Pediatr. 1974;84(4):519–526. doi: 10.1016/s0022-3476(74)80671-2. [DOI] [PubMed] [Google Scholar]
- 3.Livni N, Legum C. Ultrastructure of cultured fibroblasts in mucolipidosis type IV. Exp Cell Biol. 1976;44(1):1–11. doi: 10.1159/000162848. [DOI] [PubMed] [Google Scholar]
- 4.Merin S, et al. The cornea in mucolipidosis IV. J Pediatr Ophthalmol. 1976;13(5):289–295. [PubMed] [Google Scholar]
- 5.Tellez-Nagel I, et al. Mucolipidosis IV. Clinical, ultrastructural, histochemical, and chemical studies of a case, including a brain biopsy. Arch Neurol. 1976;33(12):828–835. doi: 10.1001/archneur.1976.00500120032005. [DOI] [PubMed] [Google Scholar]
- 6.Livni N, Merin S. Mucolipidosis IV: ultrastructural diagnosis of a recently defined genetic disorder. Arch Pathol Lab Med. 1978;102(11):600–604. [PubMed] [Google Scholar]
- 7.Raas-Rothschild A, et al. Mucolipidosis type IV: the origin of the disease in the Ashkenazi Jewish population. Eur J Hum Genet. 1999;7(4):496–498. doi: 10.1038/sj.ejhg.5200277. [DOI] [PubMed] [Google Scholar]
- 8.Bargal R, et al. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 2000;26(1):118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 9.Bargal R, et al. Mucolipidosis type IV: novel MCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum Mutat. 2001;17(5):397–402. doi: 10.1002/humu.1115. [DOI] [PubMed] [Google Scholar]
- 10.Bach G, et al. Mucopolysaccharide accumulation in cultured skin fibroblasts derived from patients with mucolipidosis IV. Am J Hum Genet. 1977;29(6):610–618. [PMC free article] [PubMed] [Google Scholar]
- 11.Goldin E, et al. Cultured skin fibroblasts derived from patients with mucolipidosis 4 are auto-fluorescent. Pediatr. Res. 1995;37(6):687–692. doi: 10.1203/00006450-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Goebel HH, Kohlschutter A, Lenard HG. Morphologic and chemical biopsy findings in mucolipidosis IV. Clin Neuropathol. 1982;1(2):73–82. [PubMed] [Google Scholar]
- 13.Kohn G, et al. Mucolipidosis IV: prenatal diagnosis by electron microscopy. Prenat Diagn. 1982;2(4):301–307. doi: 10.1002/pd.1970020410. [DOI] [PubMed] [Google Scholar]
- 14.Bach G, Zeigler M, Kohn G. Biochemical investigations of cultured amniotic fluid cells in mucolipidosis type IV. Clin Chim Acta. 1980;106(2):121–128. doi: 10.1016/0009-8981(80)90164-3. [DOI] [PubMed] [Google Scholar]
- 15.Crandall BF, et al. Review article: mucolipidosis IV. Am J Med Genet. 1982;12(3):301–308. doi: 10.1002/ajmg.1320120308. [DOI] [PubMed] [Google Scholar]
- 16.Bargal R, Bach G. Phospholipids accumulation in mucolipidosis IV cultured fibroblasts. J Inherit Metab Dis. 1988;11(2):144–150. doi: 10.1007/BF01799863. [DOI] [PubMed] [Google Scholar]
- 17.Bargal R, Bach G. Phosphatidylcholine storage in mucolipidosis IV. Clin Chim Acta. 1989;181(2):167–174. doi: 10.1016/0009-8981(89)90184-8. [DOI] [PubMed] [Google Scholar]
- 18.Bargal R, Bach G. Mucolipidosis type IV: abnormal transport of lipids to lysosomes. J Inherit Metab Dis. 1997;20(5):625–632. doi: 10.1023/a:1005362123443. [DOI] [PubMed] [Google Scholar]
- 19.Jansen SM, et al. Delayed lysosomal metabolism of lipids in mucolipidosis type IV fibroblasts after LDL-receptor-mediated endocytosis. J Inherit Metab Dis. 2001;24(5):577–586. doi: 10.1023/a:1012467827719. [DOI] [PubMed] [Google Scholar]
- 20.Chen CS, Bach G, Pagano RE. Abnormal transport along the lysosomal pathway in mucolipidosis, type IV disease. Proc. Natl. Acad. Sci. USA. 1998;95(11):6373–6378. doi: 10.1073/pnas.95.11.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pryor PR, et al. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 2006;7(10):1388–1398. doi: 10.1111/j.1600-0854.2006.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson EG, et al. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 2007;8:54. doi: 10.1186/1471-2121-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samie MA, Xu H. Lysosomal exocytosis and lipid storage disorders. J Lipid Res. 2014;55(6):995–1009. doi: 10.1194/jlr.R046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakabayashi K, et al. Mucolipidosis type IV: an update. Mol Genet Metab. 2011;104(3):206–213. doi: 10.1016/j.ymgme.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zlotogora J, et al. A muscle disorder as presenting symptom in a child with mucolipidosis IV. Neuropediatrics. 1983;14(2):104–105. doi: 10.1055/s-2008-1059563. [DOI] [PubMed] [Google Scholar]
- 26.Amir N, Zlotogora J, Bach G. Mucolipidosis type IV: clinical spectrum and natural history. Pediatrics. 1987;79(6):953–959. [PubMed] [Google Scholar]
- 27.Chitayat D, et al. Mucolipidosis type IV: clinical manifestations and natural history. Am J Med Genet. 1991;41(3):313–318. doi: 10.1002/ajmg.1320410310. [DOI] [PubMed] [Google Scholar]
- 28.Altarescu G, et al. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59(3):306–313. doi: 10.1212/wnl.59.3.306. [DOI] [PubMed] [Google Scholar]
- 29.Frei KP, et al. Mucolipidosis type IV: characteristic MRI findings. Neurology. 1998;51(2):565–569. doi: 10.1212/wnl.51.2.565. [DOI] [PubMed] [Google Scholar]
- 30.Folkerth RD, et al. Mucolipidosis IV: morphology and histochemistry of an autopsy case. J Neuropathol Exp Neurol. 1995;54(2):154–164. [PubMed] [Google Scholar]
- 31.Casteels I, et al. Mucolipidosis type IV. Presentation of a mild variant. Ophthalmic Paediatr Genet. 1992;13(4):205–210. doi: 10.3109/13816819209105168. [DOI] [PubMed] [Google Scholar]
- 32.Reis S, et al. Mucolipidosis type IV: a mild form with late onset. Am J Med Genet. 1993;47(3):392–394. doi: 10.1002/ajmg.1320470319. [DOI] [PubMed] [Google Scholar]
- 33.Abraham FA, et al. Retinal function in mucolipidosis IV. Ophthalmologica. 1985;191(4):210–214. doi: 10.1159/000309589. [DOI] [PubMed] [Google Scholar]
- 34.Riedel KG, et al. Ocular abnormalities in mucolipidosis IV. Am J Ophthalmol. 1985;99(2):125–136. doi: 10.1016/0002-9394(85)90220-x. [DOI] [PubMed] [Google Scholar]
- 35.Newman NJ, et al. Corneal surface irregularities and episodic pain in a patient with mucolipidosis IV. Arch Ophthalmol. 1990;108(2):251–254. doi: 10.1001/archopht.1990.01070040103041. [DOI] [PubMed] [Google Scholar]
- 36.Pradhan SM, et al. Electronegative electroretinogram in mucolipidosis IV. Arch Ophthalmol. 2002;120(1):45–50. doi: 10.1001/archopht.120.1.45. [DOI] [PubMed] [Google Scholar]
- 37.Geer JS, et al. Mucolipidosis type IV: a subtle pediatric neurodegenerative disorder. Pediatr Neurol. 2010;42(3):223–226. doi: 10.1016/j.pediatrneurol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffmann R, et al. Constitutive achlorhydria in mucolipidosis type IV. Proc Natl Acad Sci U S A. 1998;95(3):1207–1212. doi: 10.1073/pnas.95.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lubensky IA, et al. Lysosomal inclusions in gastric parietal cells in mucolipidosis type IV: a novel cause of achlorhydria and hypergastrinemia. Am J Surg Pathol. 1999;23(12):1527–1531. doi: 10.1097/00000478-199912000-00010. [DOI] [PubMed] [Google Scholar]
- 40.Slaugenhaupt SA, et al. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am J Hum Genet. 1999;65(3):773–778. doi: 10.1086/302549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldin E, et al. Mucolipidosis IV consists of one complementation group. Proc Natl Acad Sci U S A. 1999;96(15):8562–8566. doi: 10.1073/pnas.96.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassi MT, et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 2000;67(5):1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000;9(17):2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 44.Bach G. Mucolipidosis type IV. Mol. Genet. Metab. 2001;73(3):197–203. doi: 10.1006/mgme.2001.3195. [DOI] [PubMed] [Google Scholar]
- 45.Acierno JS, Jr., et al. A physical and transcript map of the MCOLN1 gene region on human chromosome 19p13.3-p13.2. Genomics. 2001;73(2):203–210. doi: 10.1006/geno.2001.6526. [DOI] [PubMed] [Google Scholar]
- 46.Slaugenhaupt SA. The molecular basis of mucolipidosis type IV. Curr. Mol. Med. 2002;2(5):445–450. doi: 10.2174/1566524023362276. [DOI] [PubMed] [Google Scholar]
- 47.Montell C. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci STKE. 2001;2001(90) doi: 10.1126/stke.2001.90.re1. p. re1. [DOI] [PubMed] [Google Scholar]
- 48.Benini A, et al. Characterization and expression analysis of mcoln1.1 and mcoln1.2, the putative zebrafish co-orthologs of the gene responsible for human mucolipidosis type IV. Int J Dev Biol. 2013;57(1):85–93. doi: 10.1387/ijdb.120033gb. [DOI] [PubMed] [Google Scholar]
- 49.Denis V, Cyert MS. Internal Ca(2+) release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J Cell Biol. 2002;156(1):29–34. doi: 10.1083/jcb.200111004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong XP, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lima WC, et al. Mucolipin controls lysosome exocytosis in Dictyostelium. J Cell Sci. 2012;125(Pt 9):2315–2322. doi: 10.1242/jcs.100362. [DOI] [PubMed] [Google Scholar]
- 52.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001;28(1):64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 53.Venkatachalam K, et al. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135(5):838–851. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manzoni M, et al. Overexpression of wild-type and mutant mucolipin proteins in mammalian cells: effects on the late endocytic compartment organization. FEBS Lett. 2004;567(2–3):219–224. doi: 10.1016/j.febslet.2004.04.080. [DOI] [PubMed] [Google Scholar]
- 55.Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7(3):337–353. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkatachalam K, Hofmann T, Montell C. Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J. Biol. Chem. 2006;281(25):17517–17527. doi: 10.1074/jbc.M600807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miedel MT, et al. Posttranslational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J. Biol. Chem. 2006;281(18):12751–12759. doi: 10.1074/jbc.M511104200. [DOI] [PubMed] [Google Scholar]
- 58.Kim HJ, et al. Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J Biol Chem. 2007;282(50):36138–36142. doi: 10.1074/jbc.C700190200. [DOI] [PubMed] [Google Scholar]
- 59.Xu H, et al. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc Natl Acad Sci U S A. 2007;104(46):18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grimm C, et al. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 2007;104(49):19583–19588. doi: 10.1073/pnas.0709846104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong XP, et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455(7215):992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong XP, et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J Biol Chem. 2009;284(46):32040–32052. doi: 10.1074/jbc.M109.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong CO, et al. Drosophila TRPML is required for TORC1 activation. Curr Biol. 2012;22(17):1616–1621. doi: 10.1016/j.cub.2012.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song Y, et al. TRPML cation channels regulate the specialized lysosomal compartment of vertebrate B-lymphocytes. Eur. J. Cell Biol. 2006;85(12):1253–1264. doi: 10.1016/j.ejcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Karacsonyi C, Miguel AS, Puertollano R. Mucolipin-2 localizes to the Arf6-associated pathway and regulates recycling of GPI-APs. Traffic. 2007;8(10):1404–1414. doi: 10.1111/j.1600-0854.2007.00619.x. [DOI] [PubMed] [Google Scholar]
- 66.Martina JA, Lelouvier B, Puertollano R. The calcium channel mucolipin-3 is a novel regulator of trafficking along the endosomal pathway. Traffic. 2009;10(8):1143–1156. doi: 10.1111/j.1600-0854.2009.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HJ, et al. The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10(8):1157–1167. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeevi DA, et al. A potentially dynamic lysosomal role for the endogenous TRPML proteins. J Pathol. 2009;219(2):153–162. doi: 10.1002/path.2587. [DOI] [PubMed] [Google Scholar]
- 69.Curcio-Morelli C, et al. Functional multimerization of mucolipin channel proteins. J Cell Physiol. 2010;222(2):328–335. doi: 10.1002/jcp.21956. [DOI] [PubMed] [Google Scholar]
- 70.Zeevi DA, et al. Heteromultimeric TRPML channel assemblies play a crucial role in the regulation of cell viability models and starvation-induced autophagy. J Cell Sci. 2010;123(Pt 18):3112–3124. doi: 10.1242/jcs.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samie MA, et al. The tissue-specific expression of TRPML2 (MCOLN-2) gene is influenced by the presence of TRPML1. Pflugers Arch. 2009;459(1):79–91. doi: 10.1007/s00424-009-0716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.LaPlante JM, et al. Functional links between mucolipin-1 and Ca2+-dependent membrane trafficking in mucolipidosis IV. Biochem. Biophys. Res. Commun. 2004;322(4):1384–1391. doi: 10.1016/j.bbrc.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 73.LaPlante JM, et al. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 2002;532(1–2):183–187. doi: 10.1016/s0014-5793(02)03670-0. [DOI] [PubMed] [Google Scholar]
- 74.Kiselyov K, et al. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280(52):43218–43223. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- 75.Soyombo AA, et al. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 2006;281(11):7294–7301. doi: 10.1074/jbc.M508211200. [DOI] [PubMed] [Google Scholar]
- 76.Raychowdhury MK, et al. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum. Mol. Genet. 2004;13(6):617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 77.Vergarajauregui S, et al. Mucolipin 1 channel activity is regulated by protein kinase A-mediated phosphorylation. Biochem J. 2008;410(2):417–425. doi: 10.1042/BJ20070713. [DOI] [PubMed] [Google Scholar]
- 78.Kim HJ, et al. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 2008;27(8):1197–1205. doi: 10.1038/emboj.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lev S, et al. Constitutive activity of the human TRPML2 channel induces cell degeneration. J Biol Chem. 2010;285(4):2771–2782. doi: 10.1074/jbc.M109.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grimm C, et al. Constitutive activity of TRPML2 and TRPML3 channels versus activation by low extracellular sodium and small molecules. J Biol Chem. 2012;287(27):22701–22708. doi: 10.1074/jbc.M112.368876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng X, et al. Drosophila TRPML forms PI(3,5)P2-activated cation channels in both endolysosomes and plasma membrane. J Biol Chem. 2014;289(7):4262–4272. doi: 10.1074/jbc.M113.506501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen CC, et al. A small molecule restores function to TRPML1 mutant isoforms responsible for mucolipidosis type IV. Nat Commun. 2014;5:4681. doi: 10.1038/ncomms5681. [DOI] [PubMed] [Google Scholar]
- 83.Swetha MG, et al. Lysosomal membrane protein composition, acidic pH and sterol content are regulated via a light-dependent pathway in metazoan cells. Traffic. 2011;12(8):1037–1055. doi: 10.1111/j.1600-0854.2011.01214.x. [DOI] [PubMed] [Google Scholar]
- 84.Cang C, et al. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell. 2013;152(4):778–790. doi: 10.1016/j.cell.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kukic I, et al. Zinc-dependent lysosomal enlargement in TRPML1-deficient cells involves MTF-1 transcription factor and ZnT4 (Slc30a4) transporter. Biochem J. 2013;451(2):155–163. doi: 10.1042/BJ20121506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc Natl Acad Sci U S A. 2012;109(28):11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grimm C, et al. Small molecule activators of TRPML3. Chem Biol. 2010;17(2):135–148. doi: 10.1016/j.chembiol.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X, et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151(2):372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng X, et al. Differential mechanisms of action of the mucolipin synthetic agonist, ML-SA1, on insect TRPML and mammalian TRPML1. Cell Calcium. 2014 doi: 10.1016/j.ceca.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Samie M, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev Cell. 2013;26(5):511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hersh BM, Hartwieg E, Horvitz HR. TheCaenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. USA. 2002;99(7):4355–4360. doi: 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Treusch S, et al. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101(13):4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 95.Calcraft PJ, et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459(7246):596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pitt SJ, et al. TPC2 is a novel NAADP-sensitive Ca2+ release channel, operating as a dual sensor of luminal pH and Ca2+ J Biol Chem. 2010;285(45):35039–35046. doi: 10.1074/jbc.M110.156927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jha A, et al. Convergent regulation of the lysosomal two-pore channel-2 by Mg(2)(+), NAADP, PI(3,5)P(2) and multiple protein kinases. EMBO J. 2014;33(5):501–511. doi: 10.1002/embj.201387035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patel S, Marchant JS, Brailoiu E. Two-pore channels: Regulation by NAADP and customized roles in triggering calcium signals. Cell Calcium. 2010;47(6):480–490. doi: 10.1016/j.ceca.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Venugopal B, et al. Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. Am. J. Hum. Genet. 2007;81(5):1070–1083. doi: 10.1086/521954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chandra M, et al. A role for the Ca2+ channel TRPML1 in gastric acid secretion, based on analysis of knockout mice. Gastroenterology. 2011;140(3):857–867. doi: 10.1053/j.gastro.2010.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Micsenyi MC, et al. Neuropathology of the Mcoln1(−/−) knockout mouse model of mucolipidosis type IV. J. Neuropathol. Exp. Neurol. 2009;68(2):125–135. doi: 10.1097/NEN.0b013e3181942cf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pryor PR, et al. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149(5):1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Venkatachalam K, Wong CO, Montell C. Feast or famine: role of TRPML in preventing cellular amino acid starvation. Autophagy. 2013;9(1):98–100. doi: 10.4161/auto.22260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kiselyov K, et al. TRPML: transporters of metals in lysosomes essential for cell survival? Cell Calcium. 2011;50(3):288–294. doi: 10.1016/j.ceca.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schaheen L, Dang H, Fares H. Basis of lethality in C. elegans lacking CUP-5, the Mucolipidosis Type IV orthologue. Dev. Biol. 2006;293(2):382–391. doi: 10.1016/j.ydbio.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 106.Vergarajauregui S, et al. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet. 2008;17(17):2723–2737. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Curcio-Morelli C, et al. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol Dis. 2010;40(2):370–377. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miedel MT, et al. Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 2008;205(6):1477–1490. doi: 10.1084/jem.20072194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luzio JP, Bright NA, Pryor. PR. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem Soc Trans. 2007;35(Pt 5):1088–1091. doi: 10.1042/BST0351088. [DOI] [PubMed] [Google Scholar]
- 110.Piper RC, Luzio JP. CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 2004;14(9):471–473. doi: 10.1016/j.tcb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 111.Campbell EM, Fares H. Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol. 2010;11:40. doi: 10.1186/1471-2121-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagata K, et al. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci U S A. 2008;105(1):353–358. doi: 10.1073/pnas.0707963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lelouvier B, Puertollano R. Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J Biol Chem. 2011;286(11):9826–9832. doi: 10.1074/jbc.M110.169185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18(9):524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laplante JM, et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 2006 doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 121.LaPlante JM, et al. The cation channel mucolipin-1 is a bifunctional protein that facilitates membrane remodeling via its serine lipase domain. Exp Cell Res. 2011;317(6):691–705. doi: 10.1016/j.yexcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 123.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Medina DL, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21(3):421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem. Biol. Interact. 2006;163(1–2):29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 126.Terman A, Gustafsson B, Brunk UT. Mitochondrial damage and intralysosomal degradation in cellular aging. Mol. Aspects Med. 2006;27(5–6):471–482. doi: 10.1016/j.mam.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 127.Spooner E, et al. Systematic screens for proteins that interact with the mucolipidosis type IV protein TRPML1. PLoS One. 2013;8(2):e56780. doi: 10.1371/journal.pone.0056780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jennings JJ, Jr., et al. Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem. 2006;281(51):39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 129.Coblentz J, St Croix C, Kiselyov K. Loss of TRPML1 promotes production of reactive oxygen species: is oxidative damage a factor in mucolipidosis type IV? Biochem J. 2014;457(2):361–368. doi: 10.1042/BJ20130647. [DOI] [PubMed] [Google Scholar]
- 130.Di Palma F, et al. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA. 2002;99(23):14994–14999. doi: 10.1073/pnas.222425399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van Aken AF, et al. TRPML3 mutations cause impaired mechano-electrical transduction and depolarization by an inward-rectifier cation current in auditory hair cells of varitint-waddler mice. J Physiol. 2008;586(Pt 22):5403–5418. doi: 10.1113/jphysiol.2008.156992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Colletti GA, et al. Loss of lysosomal ion channel transient receptor potential channel mucolipin-1 (TRPML1) leads to cathepsin B-dependent apoptosis. J Biol Chem. 2012;287(11):8082–8091. doi: 10.1074/jbc.M111.285536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eichelsdoerfer JL, et al. Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J Biol Chem. 2010;285(45):34304–34308. doi: 10.1074/jbc.C110.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spampanato C, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 2013;5(5):691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bach G, Chen CS, Pagano. RE. Elevated lysosomal pH in Mucolipidosis type IV cells. Clin Chim Acta. 1999;280(1–2):173–179. doi: 10.1016/s0009-8981(98)00183-1. [DOI] [PubMed] [Google Scholar]
- 136.Kogot-Levin A, et al. Mucolipidosis type IV: the effect of increased lysosomal pH on the abnormal lysosomal storage. Pediatr Res. 2009;65(6):686–690. doi: 10.1203/PDR.0b013e3181a1681a. [DOI] [PubMed] [Google Scholar]
- 137.Vergarajauregui S, Martina JA, Puertollano R. LAPTMs regulate lysosomal function and interact with mucolipin 1: new clues for understanding mucolipidosis type IV. J Cell Sci. 2011;124(Pt 3):459–468. doi: 10.1242/jcs.076240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vergarajauregui S, Martina JA, Puertollano R. Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of mucolipin-1. J Biol Chem. 2009;284(52):36357–36366. doi: 10.1074/jbc.M109.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schaheen L, Patton G, Fares H. Suppression of the cup-5 mucolipidosis type IV-related lysosomal dysfunction by the inactivation of an ABC transporter in C. elegans. Development. 2006;133(19):3939–3948. doi: 10.1242/dev.02575. [DOI] [PubMed] [Google Scholar]
- 140.Bozzato A, Barlati S, Borsani G. Gene expression profiling of mucolipidosis type IV fibroblasts reveals deregulation of genes with relevant functions in lysosome physiology. Biochim Biophys Acta. 2008;1782(4):250–258. doi: 10.1016/j.bbadis.2008.01.002. [DOI] [PubMed] [Google Scholar]