Abstract
Release 6, the latest reference genome assembly of the fruit fly Drosophila melanogaster, was released by the Berkeley Drosophila Genome Project in 2014; it replaces their previous Release 5 genome assembly, which had been the reference genome assembly for over 7 years. With the enormous amount of information now attached to the D. melanogaster genome in public repositories and individual laboratories, the replacement of the previous assembly by the new one is a major event requiring careful migration of annotations and genome-anchored data to the new, improved assembly. In this report, we describe the attributes of the new Release 6 reference genome assembly, the migration of FlyBase genome annotations to this new assembly, how genome features on this new assembly can be viewed in FlyBase (http://flybase.org) and how users can convert coordinates for their own data to the corresponding Release 6 coordinates.
INTRODUCTION
FlyBase (http://flybase.org) is a database of Drosophila-related genetic and genomic information (1). From late-2006 until mid-2014, the reference genome assembly for the biomedical model organism, Drosophila melanogaster, has been the Berkeley Drosophila Genome Project (BDGP, http://fruitfly.org) Release 5 genome assembly (2). During this period, FlyBase has published 57 updates to the gene model annotations of this species’ genome, and has integrated many other mapped genome features, most notably those of the modENCODE project (3). In the spring of 2014, BDGP released a new assembly of the D. melanogaster genome (Release 6). This improved assembly was coordinately integrated into FlyBase and NCBI (4) and became the reference genome assembly for D. melanogaster as of the summer of 2014.
The challenge when replacing one assembly with another is the migration of anchored genomic data from the old assembly to the new one. This is especially true given that the scale of the data involved in the migration to the Release 6 genome assembly far exceeds that involved in the last such migration to the Release 5 assembly in 2006; there is an enormous amount of data in public repositories such as FlyBase, NCBI, modENCODE DCC (5), modMine (6) and the UCSC Genome Browser (7), as well as in private databases of individual laboratories that has to be updated. The new Release 6 reference genome assembly, the migration of FlyBase annotations and genome-anchored data to this new genome assembly, and a user guide on how to interrogate it and update genomic coordinates are the topics of this report.
THE RELEASE 6 REFERENCE GENOME ASSEMBLY
Description of BDGP Release 6 genome assembly
A reference genome assembly for D. melanogaster was first released in 2000 (8,9). This assembly was based on nuclear DNA sequences derived from the ‘iso-1’ reference strain: isogenic yellow (y1); cinnabar (cn1), brown (bw1), speck (sp1) (10). Since then, this iso-1-derived reference nuclear genome assembly has been revised to close gaps, improve sequence quality and add sequence from centric heterochromatin regions (2,11).
The latest BDGP Release 6 of the D. melanogaster genome assembly, still based on the iso-1 reference strain, includes a total of 143.7 Mb on 1870 scaffolds comprising 2442 contigs (Table 1). The vast majority, 137.6 Mb, of this sequence resides on seven chromosome arms: X, 2L, 2R, 3L, 3R, 4 and Y (Table 2). Additionally, there are 1862 minor scaffolds that lack precise localization. Almost half of these ‘unlocalized’ scaffolds (n = 884) have been mapped to a chromosome region by comparative genome hybridization in embryos with various chromosome deficiencies: 2CEN (centromere-proximal region of chromosome 2), 3CEN, X, Y, XY (regions mapping to both X and Y chromosomes) and rDNA (ribosomal DNA) (12). The Release 6 reference genome assembly also replaces the previous mitochondrial reference genome assembly, a composite of sequences from various D. melanogaster strains, with one derived exclusively from the iso-1 reference strain (Table 1).
Table 1. Overview of the BDGP D. melanogaster Release 6 genome assembly.
| Current release | Dmel_Release_6 |
| Data provider | BDGP |
| Collaborators | DHGP, BCM-HGSC, Celera Genomics |
| Sequenced strain | iso-1 |
| Date released | 21-JUL-2014 (FlyBase, Dmel annotation version R6.01) |
| 25-JUL-2014 (GenBank, RefSeq) | |
| NCBI accessions | Release 6 plus ISO1 MT |
| Assembly: GCA_000001215.4 | |
| RefSeq: GCF_000001215.4 | |
| BioProject: PRJNA13812 | |
| Assembly statistics | • Total sequence length = 143 726 002 bp. |
| • Total gap length = 1 152 978 bp. | |
| • Total number of scaffolds = 1870. | |
| • Seven chromosome arms (plus mitochondrial genomea): X, 2L, 2R, 3L, 3R, 4 and Y. | |
| • The vast majority of sequence, 137.6 Mb, resides on the seven chromosome arms. | |
| • 1862 ‘unlocalized’ minor scaffolds, of which 884 have been mapped cytologically or genetically to a chromosome region: X, 2CEN, 3CEN, Y, XY and rRNA. | |
| Major changes relative to Release 5 | • Release 6 is 4.2 Mb larger. |
| • Total gap length decreased by 1.5 Mb. | |
| • The majority of new sequence added to the chromosome arm scaffolds is in the heterochromatic regions, 10.0 Mb of which derives from the BDGP Release 5 scaffolds XHet, 2LHet, 2RHet, 3LHet, 3RHet and U. | |
| • The chromosome Y scaffold is vastly improved and 10 times larger at 3.1 Mb. | |
| • Most remaining gaps are in the heterochromatic regions of the assembly. | |
| • 1862 minor scaffolds replace Release 5 concatenated pseudoscaffolds (e.g. U). | |
| • 48 minor scaffolds have been modified and improved from Release 5; their names indicate their mapping (2Cen_mapped_Scaffold_10_D1684). The remaining 1814 ‘unmodified’ minor scaffolds have numeric identifiers like 2110000… | |
| • All fragmented gene annotations from Release 5 have been resolved, largely as a result of improvements to the Y and 3R scaffolds. |
aThe reference genome assembly update in Dmel R6.01 (FB2014_04) was for the nuclear genome only, maintaining the old mitochondrial genome assembly, a composite of sequences from various D. melanogaster strains (GenBank U37541.1, RefSeq NC_001709.1). With FlyBase update FB2015_01, the mitochondrial reference genome assembly was also updated, replacing the previous assembly with one derived exclusively from the iso-1 reference strain (GenBank KJ947872.2; RefSeq NC_024511.2).
Table 2. Detailed information on the BDGP D. melanogaster Release 6 genome assembly.
| Scaffold | Length (bp) | Sized gaps | Total gap size (bp) | Unsized gaps | Accessions (GenBank, RefSeq) | Comments |
|---|---|---|---|---|---|---|
| Chromosome arm scaffolds | ||||||
| X | 23 542 271 | 4 | 65 520 | 6 | AE014298.5 | • Net gain of 0.9 Mb compared to R5 X plus R5 XHet: most at scaffold end. |
| NC_004354.4 | • Central 15.4 Mb unchanged: | |||||
| R5:X:4,684,794..20,073,489 maps to R6:X:4,790,761..20,179,456. | ||||||
| • About 104 kb of new sequence added at scaffold start (14 kb from R5 U). | ||||||
| • 1 Mb of new sequence added near scaffold end, including 209 kb and 204 kb from R5 scaffolds U and XHet, respectively. | ||||||
| 2L | 23 513 712 | 0 | 0 | 2 | AE014134.6 | • Net gain of 133 kb compared to R5 2L plus R5 2LHet: all at scaffold end. |
| NT_033779.5 | • Initial 21.5 Mb unchanged: | |||||
| R5:2L:1..21,485,538 maps to R6:2L:1..21,485,538. | ||||||
| • New sequence at the end includes 293 kb and 40 kb from R5 scaffolds 2LHet and U, respectively. | ||||||
| 2R | 25 286 936 | 1 | 6600 | 7 | AE013599.5 | • Net gain of 0.9 Mb compared to R5 2R plus R5 2RHet: most at scaffold start. |
| NT_033778.4 | • Central 16.7 Mb unchanged: | |||||
| R5:2R:3,036..16,668,212 maps to R6:2R:4,115,531..20,780,707. | ||||||
| • New sequence at the start includes 2.3 Mb and 987 kb from R5 scaffolds 2RHet and U, respectively. | ||||||
| 3L | 28 110 227 | 4 | 117 660 | 5 | AE014296.5 | • Net gain of 1.0 Mb compared to R5 3L plus R5 3L Het: all at scaffold end. |
| NT_037436.4 | • Initial 24.5 Mb unchanged, except an unsized gap is now sized at 7kb: | |||||
| R5:3L:1..24,523,740 maps to R6:3L:1..24,530,640. | ||||||
| • New sequence at the end includes 2.3 Mb, 328 kb and 150 kb from R5 scaffolds 3LHet, 2RHet and U, respectively. | ||||||
| 3R | 32 079 331 | 9 | 22 772 | 18 | AE014297.3 | • Net gain of 1.7 Mb compared to R5 3R plus R5 3RHet: all at scaffold start. |
| NT_033777.3 | • Last 27.9 Mb of unchanged: | |||||
| R5:3R:1..27,905,053 maps to R6:3R:4,174,278..32,079,331. | ||||||
| • New sequence at the end includes 2.2 Mb and 1.0 Mb from R5 scaffolds 3RHet and U, respectively. | ||||||
| 4 | 1 348 131 | 1 | 17 000 | 0 | AE014135.4 | • Net loss of 3.7 kb compared to R5 scaffold 4. |
| NC_004353.4 | • Replacement of R5 scaffold start: | |||||
| 24.1 kb of sequence removed (some moved to R6 3R, X and Y) and replaced with 3.4 kb from the R5 scaffold U. | ||||||
| • Change in start of R6 scaffold 4 completes the JYalpha gene annotation. | ||||||
| • The remaining sequence is unchanged, but the unsized gap in R5 is now sized at 17kb. | ||||||
| Y | 3 667 352 | 61 | 242 633 | 150 | CP007106.1 | • Net gain of 3.3 Mb compared to R5 YHet: over a 10-fold increase. |
| NC_024512.1 | • 232.3 kb carried over from R5 scaffold YHet. | |||||
| • New sequence includes 702.5 kb and 84.2 kb from R5 scaffolds U and 3LHet, respectively. | ||||||
| Mitochondrial scaffold | ||||||
| M | 19 524 | 0 | 0 | 0 | KJ947872.2 | • Derived from iso-1 reference strain. |
| NC_024511.2 | ||||||
| Minor unordered scaffold groups (number of scaffolds per group) | ||||||
| Comments on scaffold group | ||||||
| X (446) | 1 005 345 | 26 | 72 915 | 0 | • Chromosome X mapped. | |
| 2CEN (28) | 222 873 | 20 | 60 073 | 1 | • Chromosome 2 centromere-proximal region. | |
| 3CEN (144) | 729 966 | 26 | 41 429 | 10 | • Chromosome 3 centromere-proximal region. | |
| Y (199) | 860 223 | 39 | 63 081 | 24 | • Chromosome Y mapped. | |
| XY (66) | 209 541 | 4 | 806 | 1 | • Scaffolds map to both X and Y chromosomes. | |
| rDNA (1) | 76 973 | 2 | 16 500 | 0 | • Ribosomal DNA (RefSeq, NW_007931121.1). | |
| unmapped (978) | 3 053 597 | 139 | 402 989 | 11 | • Unmapped. Now represented by separate scaffolds instead of a concatenated pseudoscaffold. | |
| • Net loss of 7.0 Mb, compared to R5 U, due to movement of sequences to chromosome arms or to mapped minor scaffold groups. | ||||||
Major improvements in the Release 6 genome assembly compared to Release 5
Release 6 has a number of important changes from Release 5 (Tables 1 and 2). Release 6 is 4.2 Mb larger, even as the total assembly gap length has been reduced to 1.2 Mb, a decrease of 1.5 Mb. The main areas of improvement are to the centric heterochromatin regions of the major chromosome arm scaffolds (X, 2L, 2R, 3L, 3R, 4), which have incorporated over 10 Mb of sequence from minor Release 5 scaffolds (XHet, 2LHet, 2RHet, 3LHet, 3RHet and U). The chromosome Y scaffold has been improved dramatically, increasing over 10-fold in size to 3.1 Mb. Almost all remaining gaps in the chromosome arm scaffolds are in the heterochromatic regions. Small, unmapped scaffolds are now represented individually, instead of being concatenated into pseudoscaffolds (e.g. Release 5 pseudoscaffolds U, 3LHet).
Access to the Release 6 and Release 5 genome assemblies
The BDGP Release 5 and Release 6 genome assemblies have been deposited at NCBI. Full reports are available at NCBI (4), which provides global statistics and quality metrics for each assembly, as well as a ‘full sequence report’ and links to GenBank (13) and RefSeq (14) accessions for individual scaffolds (Table 3).
Table 3. NCBI accessions for various BDGP D. melanogaster genome assembly releases.
| BDGP release | NCBI accessions (assembly) (RefSeq) | NCBI release date | 1st release at FlyBase | Retired at FlyBase | Comments |
|---|---|---|---|---|---|
| 5 | GCA_000001215.2, | 2007/10/22 | Dmel R5.1 | Dmel R5.57 | |
| GCF_000001215.2 | (FB2006_01) | (FB2014_03) | |||
| 6 plus MT | GCA_000001215.3, GCF_000001215.3 | 2014/07/25 | Dmel R6.01 (FB2014_04) | Dmel R6.03 (FB2014_06) | • Only the nuclear genome assembly was updated. |
| 6 plus ISO1 MT | GCA_000001215.4, GCF_000001215.4 | 2014/08/01 | Dmel R6.04 (FB2015_01) | •The mitochondrial genome assembly was updated. |
Beginning with FlyBase update FB2014_04 (released July 25, 2014), all FlyBase data are now reported with respect to the Release 6 genome assembly. However, Release 5 versions of FlyBase, including the last FlyBase update before Release 6 (FB2014_03, R5.57), will remain accessible from the ‘Archives’ menu in the blue navigation bar at the top of most FlyBase pages (‘Other Archives’ sub-menu, http://flybase.org/static_pages/downloads/archivedata3.html). Thus, through these Archives, all Release 5 sequence locations described in gene and genomic feature reports will remain available, as will tools such as BLAST (15) and GBrowse (16) using Release 5. All versions of FlyBase on the Release 5 genome assembly are also available for download on the FlyBase FTP site, available from the ‘Files’ menu of the FlyBase navigation bar (ftp://ftp.flybase.net/releases/).
DATA MIGRATION TO THE RELEASE 6 REFERENCE GENOME ASSEMBLY
Feature migration
FlyBase represents as many features located on the genome as possible. Primary among these are the gene model annotations, represented by localized exon features that are joined into one or more models representing transcript isoforms. In addition to the gene models, FlyBase contains numerous other types of localized features including ones that are part of the sequenced genome (e.g. protein-binding sites, enhancers), reagents and variations reported in the literature (e.g. aberrations, mutant alleles, mobile element insertion sites) and discretely aligned features (cDNAs, protein similarities, gene model predictions, RNA-Seq splice junctions). The replacement of the Release 5 genome assembly with Release 6 required migration of millions of discrete features and over a hundred RNA-Seq datasets from the old to the new assembly.
Assembly-to-assembly alignments between Release 5 and Release 6 were generated by NCBI (4), using previously described methods (http://www.ncbi.nlm.nih.gov/genome/tools/remap/docs/alignments), and provided to FlyBase. Using the information provided in these alignments, a map was generated that identified coordinate spans in Release 5 that correspond to equivalent unchanged regions in the new Release 6 assembly. Because the Release 6 assembly contains some regions that are inverted relative to their orientation in Release 5, and other regions that have moved to different scaffolds, the mapping file and necessary coordinate transformations needed to take these types of changes into account. The mapping file underlies the coordinates converter tool (see below) and is available at FlyBase (http://flybase.org/reports/FBrf0225389.html). This map was used to perform the coordinate conversions necessary to update the location of the features from their old Release 5 coordinates and localize them to the new scaffolds of the Release 6 assembly.
Realignment of high-throughput datasets
For many types of high-throughput datasets, direct mapping of reads to the new genome assembly is preferable to the migration process described above, since it allows alignment to newly sequenced regions. However, direct mapping is not always available from the original data producers. Therefore, our policies are as follows.
When available, we will replace any FlyBase-migrated dataset with a new analysis directly mapped to the Release 6 genome assembly.
For a limited time, we will accept genome datasets mapped to the Release 5 assembly and migrate them to the Release 6 assembly for presentation in FlyBase.
With respect to aligned nucleotide and protein database records presented in FlyBase as evidence, GenBank has provided new Release 6 alignments of cDNA, expressed sequence tag (EST) and proteins using a combination of BLAST (15), ProSplign and Splign (17). GenBank is also producing a Release 6 gene prediction set using their eukaryotic genome annotation pipeline (14).
The modENCODE Transcription Group (http://www.modencode.org/celniker) has also provided FlyBase with de novo alignments of modENCODE RNA-Seq coverage data to the Release 6 genome, including the original developmental profile (18) and the most recent profiles for tissues, treatments and cell lines (19). The new Release 6-aligned datasets will be available on FlyBase by the public update FB2014_06. In the interim, these and other RNA-Seq coverage datasets have been migrated to the Release 6 genome assembly based on the same mapping file used for feature migration (see above). RNA-Seq coverage datasets are displayed in GBrowse and are used to calculate RPKM gene expression values (20). These RPKM values are presented in the High-Throughput Expression sections of gene reports and can be queried extensively using the RNA-Seq Search tool (http://flybase.org/static_pages/rna-seq/rna-seq_search.html). A more detailed description of the FlyBase RPKM calculation is available (http://flybase.org/reports/FBrf0221009.html).
Summary of changes to gene models and other features
The large majority of data were migrated to the new assembly. Unsurprisingly, most of the features that failed to automatically migrate were originally mapped to unlocalized scaffolds (pseudoscaffold U) or to heterochromatic arms that were represented as separate entities in Release 5 (e.g. 2RHet, 3RHet).
Considering gene model annotations, there were 42 gene models that did not automatically migrate to the new assembly because of alterations to the sequence that fell within the gene span. These annotations were manually reviewed and 12 of them were re-annotated on the new assembly. The 30 remaining annotations that failed to migrate had all been located to unlocalized scaffolds in Release 5; 18 of these were ribosomal RNA gene fragments, 11 were Suppressor of Stellate (Su(Ste)) multicopy non-coding RNAs and one was a pseudogene. Additionally, there were 35 annotations that were automatically migrated, but for which the coordinate conversion to Release 6 suggested changes within the annotation. After manual review, 21 of these automatically migrated annotations were accepted without additional changes, 13 were modified to reflect a new CDS and one was deleted (a ribosomal RNA gene fragment).
The improvement of the assembly in heterochromatic regions also led to the resolution of 11 previously fragmented gene annotations. These improvements resulted from relocation of sequence from the Release 5 pseudoscaffold U and unordered ‘Het’ scaffolds (e.g. 3RHet) to the Release 6 chromosome arm scaffolds: Y (six cases), 3R (two cases), 2R, 3L and 4 (one case each). The annotation of these gene models as complete, intact genes is part of the D. melanogaster annotation version R6.02 (FB2014_05), and documented in FlyBase's Release 6 Sequence Assembly Notes, available from the ‘Documents’ menu of the FlyBase navigation bar (http://flybase.org/static_pages/docs/release_notes.html).
Other types of annotated features that did not migrate to the new assembly included 25 annotated mobile elements, seven origins of replication and 54 transcription factor binding sites. In addition, in regions with significant changes, there were aligned and analysis features like ESTs, protein similarities, RNA-Seq alignments and RNA-Seq-derived exon junctions that failed to migrate. While we migrated as many of these types of analysis features as possible, our ultimate goal is to obtain new alignments and analysis on the Release 6 assembly (see above).
Conversion tools for users
Many researchers will want to convert the Release 5-based coordinates for their own data into the corresponding Release 6 coordinates. For small sets of data, FlyBase provides the Coordinates Converter tool, which converts genomic coordinates based on a specific genome assembly release to those for a later release. For each set of coordinates converted, the tool also reports if there have been any changes to the genomic sequence in that region between the assembly releases. For example, for the input Release 5 coordinates ‘2LHet:16,945..17,066’, this tool returns the corresponding Release 6 coordinates ‘2L:23,158,471..23,158,593’, with the notes ‘includes 1 area of change; different scaffold.’ The Coordinates Converter may be accessed from the ‘Tools’ menu on the navigation bar, and offers a number of input and output options (Figure 1). A separate Release 6-to-Release 5 backwards converter is also provided (Figure 1).
Figure 1.
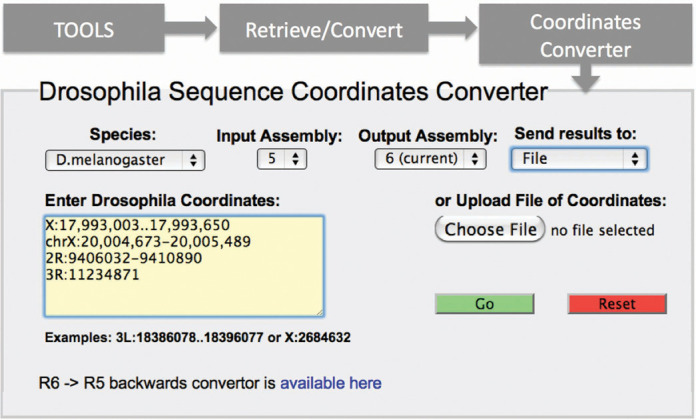
Coordinates Converter. This tool can be accessed by the ‘Tools’ menu in the blue navigation bar found at the top of FlyBase pages, under both the ‘Retrieve/Convert’ sub-menu (shown) and the ‘Genomic/Map Tools’ sub-menu. A variety of input formats are accepted, and input lists may be uploaded from a text file or entered directly into the input box; output may be to a browser view or a downloadable file (menu at upper right). By default, the input assembly is set to Release 5 and the output to Release 6, but other forward conversions can be specified. For conversion from Release 6 back to Release 5, an analogous Coordinates Converter tool is provided (link at lower left).
While the FlyBase Coordinates Converter tool can be used for lists of genomic coordinates, the NCBI Genome Remapping Service is recommended for the conversion of large datasets (www.ncbi.nlm.nih.gov/genome/tools/remap). For D. melanogaster, users of this remapping service can upload files and convert coordinates between Releases 5 and 6 (in either direction). A variety of file formats are supported for input as well as output. In addition to the main ‘Annotation Data’ file, summary and mapping reports are provided.
ACCESS TO DATA ON FLYBASE
Download of the Release 6 genome assembly sequence
Sequence data for the D. melanogaster genome assembly can be downloaded from the FlyBase FTP site, accessible from the ‘Files’ menu in the FlyBase navigation bar (ftp://ftp.flybase.net/). Various cuts of the data are provided in multiple formats, ranging from GFF3 (http://www.sequenceontology.org/gff3.shtml), FASTA (21) and GTF (http://mblab.wustl.edu/GTF2.html) files to Chado XML (22) data sliced into portions such as scaffolds, gene models, etc. These data are provided for the current genome release and all older releases. The latest, most up-to-date D. melanogaster genome can be found in the /genomes/Drosophila_melanogaster/current subdirectory; within that location, raw scaffold data are in the /dna directory (e.g. dmel-raw_scaffolds-r6.xx.tar.gz). The FlyBase FTP site requires an anonymous login in passive mode; these are the defaults for most FTP clients and browsers, but if you experience any difficulties please check your settings.
BLAST
FlyBase provides a stand-alone BLAST (15) server for 50 different arthropod genomes (http://flybase.org/blast/). With each FlyBase release, the BLAST tool is updated with the latest genome and annotation data. Thus, as of FB2014_04, Release 6 data have been underlying BLAST alignments for D. melanogaster. Archived versions of FlyBase may be used to perform BLAST alignments against Release 5 genomic sequence and gene model annotation sets. FlyBase BLAST offers standard BLAST options (such as TBLASTN) as well as advanced options where users can specify the matrix, word size, codon bias and complexity filters. When using the ‘Genome Assembly’ database option, GBrowse buttons will appear in the results next to each BLAST hit, allowing users to directly map their hits onto the D. melanogaster genome. This feature is also available for the other 11 Drosophilid genomes that are represented in FlyBase GBrowse. This allows users to see the BLAST hit within the context of the surrounding genome, which can be particularly useful when assessing whether a hit is biologically significant or whether to study a candidate gene further.
GBrowse 2
For the past decade, FlyBase has used the generic genome browser, GBrowse (16), for the display of genome annotations and genome-aligned evidence on the reference genome assembly. GBrowse allows users to navigate to a region of interest using coordinates or a landmark (e.g. a gene name or feature identifier), and from that entry point, zoom or scroll along the genome. The feature types displayed are user-selected. Genomic sequence for the region in view can be downloaded in a variety of formats. Concurrent with the introduction of Release 6, FlyBase has also upgraded to GBrowse 2 (23). GBrowse 2 retains the same functionality described above for GBrowse. However, GBrowse 2 contains some important upgrades, particularly its ability to handle much more data, which has allowed FlyBase to offer all D. melanogaster data options on a single view. GBrowse 2 also offers users easier track customization (vertical placement, open or hidden views) and limited smooth-track panning (side-to-side sliding). A new lasso feature can be used to select a region of interest, and either navigate to that region or download the associated genomic sequence (Figure 2). GBrowse 2 is accessible from the homepage and various FlyBase report pages.
Figure 2.
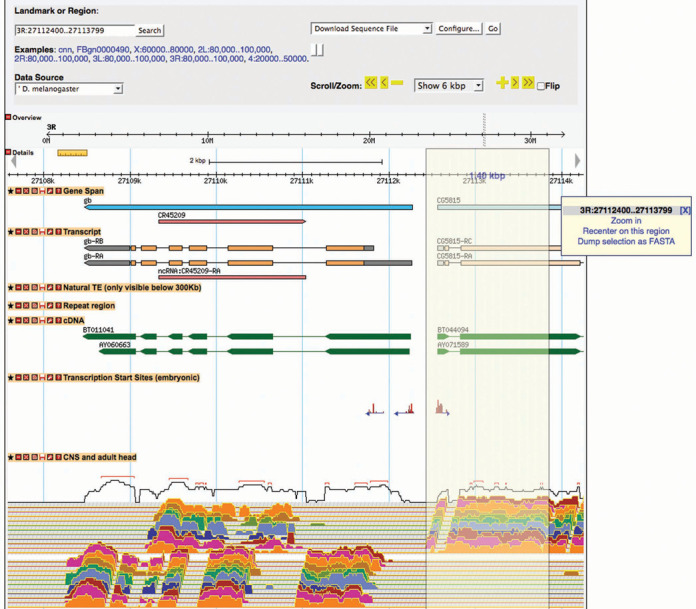
GBrowse 2. Like the original GBrowse, GBrowse 2 allows users to navigate to a region of the genome using coordinates or a landmark, and zoom or scroll along the genome to browse annotated features and aligned evidence; the data tracks shown are user-selected; genomic sequence for the region in view can be downloaded by selecting ‘Download Sequence File’ in the download option menu at the top right. However, GBrowse 2 can handle more data and has convenient new features. Tracks can be moved simply by dragging the track title bar vertically, and hidden or removed by clicking on the appropriate boxes within the track title bar. In GBrowse 2, one can also download sequence for a smaller region within view by lassoing the region (as shown for a 1.4-kb region within the 6-kb view) and selecting ‘Dump selection as FASTA’ from the resulting pop-up menu (far right).
CONCLUSIONS
Recommendations for referencing public D. melanogaster genome data
With the replacement of the reference genome assembly, as well as the frequent changes to annotation sets, it is important that researchers, during publication and when submitting to public data repositories, indicate clearly the versions of genome assembly and FlyBase annotation datasets that were employed in a particular analysis. Genome assemblies can be unambiguously identified by the corresponding NCBI accession (Table 3). FlyBase annotation sets can be unambiguously identified by the FlyBase release number or the gene model annotation set version number. For example, the September 2014 public update of FlyBase (FB2014_05) includes ‘release notes’ that specify Release 6 as the reference genome assembly, and version R6.02 as the FlyBase D. melanogaster gene model annotation set for this update. Citing either FB2014_05 or FlyBase R6.02 is sufficient to uniquely point to this public dataset. The release notes are accessible through the ‘Documents’ menu in the FlyBase navigation bar (http://flybase.org/static_pages/docs/release_notes.html). All FlyBase pages indicate the FlyBase version in the header; in GBrowse 2, the header also indicates the FlyBase annotation version in view (e.g. R6.02). Archived release notes for previous FlyBase updates (and the release dates) are available through the navigation bar's ‘Archives’ menu (‘Other Archives’ sub-menu), so that researchers can go back to identify the relevant FlyBase version from which data were obtained on a given date.
While FlyBase updates gene model annotations and other molecular features with each bimonthly public database update, some users prefer to update less frequently. For such users, FlyBase recommends using the frozen datasets that we submit annually to GenBank and RefSeq at NCBI; the current FlyBase submission to NCBI corresponds to FlyBase D. melanogaster annotation set version R6.01.
Adoption of the Release 6 reference genome assembly by the Drosophila research community
Uniform adoption of the new Release 6 reference genome assembly by researchers and the resources they use will be a gradual process. It is our understanding that the Release 6 assembly will have replaced Release 5 at the UCSC genome browser (7) and Ensembl (24) by January 2015 or earlier. Time lines for similar updates at other public D. melanogaster resources are not known to us. Additionally, many ongoing research projects are surely based on the Release 5 reference genome assembly. We advise researchers to be mindful of this issue when using a publicly available dataset or resource, and to identify the relevant reference genome assembly and FlyBase gene model annotation set version used for a particular analysis.
Acknowledgments
We would like to thank the BDGP for advance access to the Release 6 genome assembly and associated metadata. We would like to thank the PIs, curators and developers of FlyBase for their comments on the manuscript, and Susan St. Pierre for help in preparing the figures. The current members of the FlyBase Consortium are: William Gelbart, Nicholas H. Brown, Thomas Kaufman, Maggie Werner-Washburne, Richard Cripps, Kris Broll, Madeline Crosby, Gilberto dos Santos, David Emmert, L. Sian Gramates, Kathleen Falls, Beverley B. Matthews, Susan Russo, Andrew Schroeder, Pinglei Zhou, Mark Zytkovicz, Boris Adryan, Helen Attrill, Marta Costa, Steven Marygold, Peter McQuilton, Gillian Millburn, Laura Ponting, Raymund Stefancsik, Susan Tweedie, Josh Goodman, Gary Grumbling, Victor Strelets and Jim Thurmond.
Footnotes
The members of the FlyBase Consortium are listed in the Acknowledgements.
ACCESSION NUMBERS
GCA_000001215.4, GCF_000001215.4, PRJNA13812, U37541.1, NC_001709.1, KJ947872.2, NC_024511.2, AE014298.5, NC_004354.4, AE014134.6, NT_033779.5, AE013599.5, NT_033778.4, AE014296.5, NT_037436.4, AE014297.3, NT_033777.3, AE014135.4, NC_004353.4, CP007106.1, NC_024512.1, GCA_000001215.2, GCF_000001215.2, GCA_000001215.3, GCF_000001215.3.
FUNDING
National Human Genome Research Institute at the National Institutes of Health [U41 HG00739 to W.G., PI]; Medical Research Council (UK) [G1000968 to N.B., PI]. Funding for open access charge: National Human Genome Research Institute at the National Institutes of Health [U41 HG00739 to W.G., PI].
Conflict of interest statement. None declared.
REFERENCES
- 1.St Pierre S.E., Ponting L., Stefancsik R., McQuilton P., the FlyBase Consortium FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoskins R.A., Carlson J.W., Kennedy C., Acevedo D., Evans-Holm M., Frise E., Wan K.H., Park S., Mendez-Lago M., Rossi F., et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celniker S.E., Dillon L.A., Gerstein M.B., Gunsalus K.C., Henikoff S., Karpen G.H., Kellis M., Lai E.C., Lieb J.D., MacAlpine D.M., et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washington N.L., Stinson E.O., Perry M.D., Ruzanov P., Contrino S., Smith R., Zha Z., Lyne R., Carr A., Lloyd P., et al. The modENCODE Data Coordination Center: lessons in harvesting comprehensive experimental details. Database. 2011;2011:bar023. doi: 10.1093/database/bar023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contrino S., Smith R.N., Butano D., Carr A., Hu F., Lyne R., Rutherford K., Kalderimis A., Sullivan J., Carbon S., et al. modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012;40:D1082–D1088. doi: 10.1093/nar/gkr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The Genome Sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 9.Myers E.W., Sutton G.G., Delcher A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2203. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 10.Brizuela B.J., Elfring L., Ballard J., Tamkun J.W., Kennison J.A. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celniker S.E., Wheeler D.A., Kronmiller B., Carlson J.W., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0079. RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B., Caudy A., Parsons L., Rosebrock A., Pane A., Raj S., Wieschaus E. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genome Res. 2012;22:2507–2519. doi: 10.1101/gr.137406.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2014;42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapustin Y., Souvorov A., Tatusova T., Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol. Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L., Artieri C.G., van Baren M.J., Boley N., Booth B.W., et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M., et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. U.S.A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mungall C.J., Emmert D.B. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 23.Stein L.D. Using GBrowse 2 to visualize and share next-generation sequence data. Brief. Bioinform. 2013;14:162–171. doi: 10.1093/bib/bbt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]


