ABSTRACT
Multidrug efflux systems are a major cause of resistance to antimicrobials in bacteria, including those pathogenic to humans, animals, and plants. These proteins are ubiquitous in these pathogens, and five families of bacterial multidrug efflux systems have been identified to date. By using transcriptomic and biochemical analyses, we recently identified the novel AceI (Acinetobacter chlorhexidine efflux) protein from Acinetobacter baumannii that conferred resistance to the biocide chlorhexidine, via an active efflux mechanism. Proteins homologous to AceI are encoded in the genomes of many other bacterial species and are particularly prominent within proteobacterial lineages. In this study, we expressed 23 homologs of AceI and examined their resistance and/or transport profiles. MIC analyses demonstrated that, like AceI, many of the homologs conferred resistance to chlorhexidine. Many of the AceI homologs conferred resistance to additional biocides, including benzalkonium, dequalinium, proflavine, and acriflavine. We conducted fluorimetric transport assays using the AceI homolog from Vibrio parahaemolyticus and confirmed that resistance to both proflavine and acriflavine was mediated by an active efflux mechanism. These results show that this group of AceI homologs represent a new family of bacterial multidrug efflux pumps, which we have designated the proteobacterial antimicrobial compound efflux (PACE) family of transport proteins.
IMPORTANCE
Bacterial multidrug efflux pumps are an important class of resistance determinants that can be found in every bacterial genome sequenced to date. These transport proteins have important protective functions for the bacterial cell but are a significant problem in the clinical setting, since a single efflux system can mediate resistance to many structurally and mechanistically diverse antibiotics and biocides. In this study, we demonstrate that proteins related to the Acinetobacter baumannii AceI transporter are a new class of multidrug efflux systems which are very common in Proteobacteria: the proteobacterial antimicrobial compound efflux (PACE) family. This is the first new family of multidrug efflux pumps to be described in 15 years.
Observation
Multidrug efflux is a ubiquitous mechanism of drug resistance in bacterial pathogens that is mediated by integral membrane transport proteins. These proteins are typically very promiscuous, recognizing a range of antimicrobial substrates that differ in both structure and valency. To date, five distinct families of transport proteins have been shown to include multidrug efflux systems: the major facilitator superfamily, the resistance/nodulation/division superfamily, the ATP-binding cassette superfamily, the multidrug and toxic compound extrusion family, and the small multidrug resistance family.
Recently, we identified the aceI (Acinetobacter chlorhexidine efflux) gene in Acinetobacter baumannii, which is involved in adaptive resistance to the widely used biocide chlorhexidine (1). This gene was overexpressed more than 10-fold in response to a subinhibitory shock of chlorhexidine in A. baumannii ATCC 17978. The aceI gene encodes a membrane protein that is approximately 150 amino acid residues in length and contains two tandem bacterial transmembrane pair (BTP; Pfam accession number PF05232) domains (2). Heterologous expression of aceI increased Escherichia coli resistance to chlorhexidine (1) and, conversely, deletion of aceI from the A. baumannii genome increased its susceptibility to chlorhexidine (3). The AceI protein was shown to interact directly with chlorhexidine and to mediate its efflux via an energy-dependent mechanism (1). However, resistance to other antimicrobial compounds was not observed (1).
Genes that encode BTP domain proteins homologous to aceI are carried by diverse bacterial species but are particularly common among proteobacterial lineages. Similar to A. baumannii, genes encoding BTP domain proteins were upregulated in the human pathogens Pseudomonas aeruginosa PAO1 and Burkholderia cenocepacia J2315 in response to chlorhexidine and were able to mediate resistance to this biocide (1). Furthermore, related BTP domain protein genes from the soil bacterium Acinetobacter baylyi ADP1 and the plant commensal bacterium Pseudomonas protegens Pf-5 were also shown to mediate resistance to chlorhexidine when expressed in E. coli (1). Deletion of this gene from A. baylyi ADP1 increased its susceptibility to chlorhexidine (1).
In addition to aceI, the A. baumannii genome harbors a second gene that encodes a BTP family protein, A1S_1503, that does not confer chlorhexidine resistance and whose expression is not induced by chlorhexidine. Similarly, P. protegens harbors a second gene encoding a BTP domain protein that appears to be nonfunctional with respect to chlorhexidine resistance (1), and the P. aeruginosa and B. cenocepacia genomes carry one or two BTP domain protein genes that are not induced by chlorhexidine (4, 5).
Here, we sought to identify alternative drug substrates for BTP domain proteins. We demonstrate that, in addition to chlorhexidine, many BTP domain proteins are able to mediate resistance to other biocides, as well as fluorescent dyes. The protein from Vibrio parahaemolyticus VP1155 provided particularly strong resistance to biocides and dyes and mediated rapid transport of acriflavine and proflavine. These results indicate that BTP domain proteins represent a new family of transport proteins that includes multidrug efflux systems, which we have designated the proteobacterial antimicrobial compound efflux (PACE) family.
BTP protein gene cloning and expression.
At the time of writing, the Pfam database (version 27.0) listed close to 800 proteins that contain BTP domains from more than 600 bacterial species (2). The majority (95%) of these proteins were predicted to have the same tandem BTP domain architecture as AceI and were encoded by Proteobacteria, particularly the gamma, beta, and alpha subdivisions (although this may be biased by the species for which genome sequence data are available). In addition to Proteobacteria, the genomes of several Veillonella and Micrococcus species (Firmicutes and Actinobacteria, respectively) also carried BTP domain protein genes. We have not, however, detected these genes in the genomes of archaeal or eukaryotic organisms.
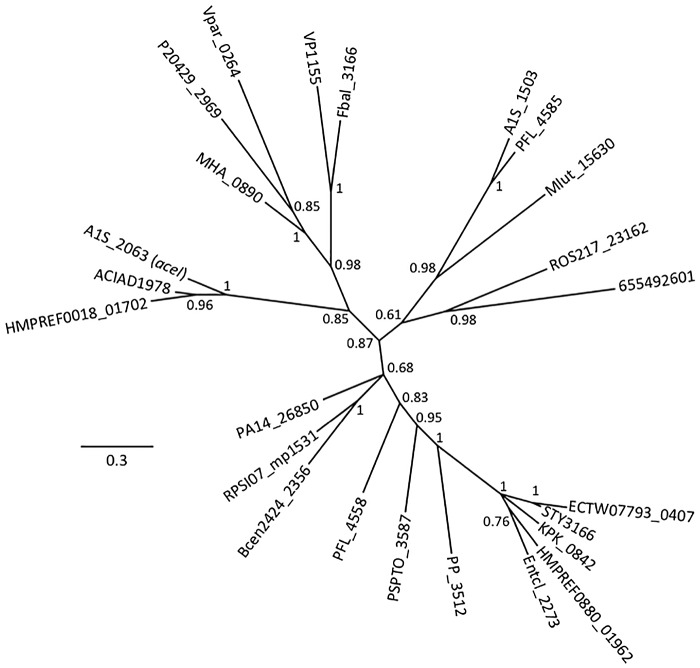
In this study, we examined the drug resistance/transport capabilities of 24 BTP domain proteins, including AceI. These proteins were selected to encompass the full spectrum of currently sampled phylogenetic diversity within this group (Fig. 1) and included 18 gammaproteobacterial proteins, 3 betaproteobacterial proteins, and 1 representative protein from each of Alphaproteobacteria, Firmicutes, and Actinobacteria (Table 1). Seven of the genes encoding these proteins were previously cloned into the E. coli pTTQ18 expression vector via conventional methods (1). The remaining 17 were synthetically designed E. coli codon-optimized sequences and were synthesized in single gBlock gene fragments (Integrated DNA Technologies) and then cloned into the pTTQ18 plasmid vector (6). With the exception of Vpar_0264, ROS217_23162 and MHA_0890, the proteins under investigation were expressed at levels detectable in Western blot assays of whole-cell lysates (see Fig. S1 in the supplemental material). This level of expression success (87.5%) is in line with our previous experiences using this expression system for the heterologous production of transport systems (7–9).
FIG 1 .
Tree showing the phylogenetic relationships of BTP family proteins included in this study. The tree was generated using MrBayes 3.2.1 (14) from a ClustalX2 alignment of protein sequences obtained from the National Center for Biotechnology Information database. Interior node values are clade credibility values (posterior probabilities) generated and assigned by MrBayes.
TABLE 1 .
Drug resistance conferred by BTP family proteinsa
| Organism | Gene or protein (locus tag) |
MIC or MIC range (µg/ml)b |
||||
|---|---|---|---|---|---|---|
| CH | DQ | BK | PF | AF | ||
| NA (negative control) | NAc (vector only) | 0.195–0.39 | 50 | 0.39–0.78 | 6.25 | 3.125 |
| Acinetobacter baumannii ATCC 17978 | A1S_2063 (aceI) | 1.56 | 12.5-25 | 0.39–0.78 | 6.25 | 3.125 |
| Acinetobacter radioresistens SH164 | HMPREF0018_01702 | 0.78 | 25 | 0.39–0.78 | 6.25 | 3.125 |
| Acinetobacter baylyi ADP1 | ACIAD1978 | 0.78 | 50 | 0.78 | 6.25 | 3.125 |
| Mannheimia haemolytica PHL213 | MHA_0890 | 0.39 | 50 | 0.78 | 6.25 | 3.125 |
| Pseudoalteromonas sp. BSi20429 | P20429_2969 | 0.195 | 50 | 0.78 | 6.25 | 6.25 |
| Veillonella parvula DSM 2008 | Vpar_0264 | 0.195 | 50 | 0.39–0.78 | 6.25 | 3.125 |
| Vibrio parahaemolyticus RIMD 2210633 | VP1155 | 1.56 | 50 | 3.125 | 25 | 12.5 |
| Ferrimonas balearica DSM 9799 | Fbal_3166 | 0.78–1.56 | 50 | 0.78 | 3.125 | 6.25 |
| Micrococcus luteus NCTC 2665 | Mlut_15630 | 0.195 | 50 | 0.39–0.78 | 6.25 | 3.125 |
| Acinetobacter baumannii ATCC 17978 | A1S_1503 | 0.195 | 25 | 0.78 | 6.25 | 6.25 |
| Pseudomonas protegens Pf-5 | PFL_4585 | 0.195 | 100 | 1.56 | 3.125–6.25 | 3.125 |
| Tepidiphilus margaritifer DSM 15129 | 655492601d | 0.195 | 50–100 | 0.78–1.56 | 1.56–3.125 | 3.125 |
| Roseovarius sp. 217 | ROS217_23162e | 0.195 | 50 | 0.78 | 6.25 | 3.125 |
| Pseudomonas aeruginosa PA14 | PA14_26850 | 0.78 | 100 | 1.56 | 6.25 | 3.125 |
| Ralstonia solanacearum PSI07 | RPS107_mp1531 | 0.195–0.39 | 50–100 | 1.56–3.125 | 3.125 | 6.25 |
| Burkholderia cenocepacia HI2424 | Bcen2424_2356 | 1.56 | 25 | 1.56–3.125 | 12.5–25 | 6.25 |
| Pseudomonas protegens Pf-5 | PFL_4558 | 1.56 | 12.5-25 | 0.39–0.78 | 3.125–6.25 | 3.125 |
| Pseudomonas syringae pv. Tomato strain DC3000 | PSPTO_3587 | 0.78 | 50 | 0.78 | 12.5–25 | 6.25 |
| Pseudomonas putida KT2440 | PP_3512 | 0.78 | 50 | 0.78–1.56 | 6.25 | 3.125 |
| Enterobacter cloacae SCF1 | Entcl_2273 | 0.39 | 25 | 1.56 | 6.25 | 3.125 |
| Yokenella regensburgei ATCC 43003 | HMPREF0880_01962 | 0.78 | 50 | 0.78–1.56 | 3.125–6.25 | 3.125 |
| Klebsiella pneumoniae 342 | KPK_0842 | 0.78 | 25 | 0.78–1.56 | 12.5 | 3.125 |
| Salmonella enterica subsp. enterica serovar Typhi strain ct18 | STY3166 | 0.195 | 25–50 | 0.39 | 6.25 | 3.125 |
| Escherichia coli TW07793 | ECTW07793_0407 | 0.39–0.78 | 50 | 0.78 | 6.25–12.5 | 6.25 |
None of the cloned BTP family genes conferred reproducible resistance to tetracycline, chloramphenicol, tetraphenylphosphonium, ethidium, Hoechst 33342, pyronin Y, acridine yellow, or 4′,6-diamidino-2-phenylindole.
The values given are from at least two independent biological replicates, and those indicating reproducible increases in resistance are shown in boldface. Abbreviations: CH, chlorhexidine; DQ, dequalinium; BK, benzalkonium; PF, proflavine; AF, acriflavine.
NA, not applicable.
The GenBank protein ID is given for the Tepidiphilus margaritifer DSM 15129 protein.
The cloned Roseovarius sp. 217 gene ROS217_23162 contains a single base change that results in a serine-to-phenylalanine mutation at position 139 (C-terminal tail).
Chlorhexidine resistance mediated by AceI homologs.
Previously, we demonstrated that BTP domain proteins homologous to the A. baumannii AceI protein from A. baylyi ADP1 (ACIAD1978), P. aeruginosa PA14 (PA14_26850), P. protegens Pf-5 (PFL_4558), and B. cenocepacia HI2424 (Bcen2424_2356) were able to confer resistance to chlorhexidine, whereas the phylogenetically distinct (Fig. 1) proteins from A. baumannii (A1S_1503) and P. protegens (PFL_4585) did not confer resistance (1). Chlorhexidine MIC analyses were conducted as previously described (8) to gauge the level of resistance provided by the 17 newly cloned BTP domain protein homologs. These assays were conducted in medium containing 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce basal levels of expression. Among the 24 cloned genes, we saw reproducible increases in chlorhexidine resistance from 12 genes (Table 1). This result confirmed that chlorhexidine is a common substrate of this group of transporters.
BTP domain proteins are multidrug efflux systems.
To explore the possibility that BTP domain proteins represent a new family of multidrug efflux transporters, we tested their capacities to confer resistance to a range of additional antimicrobial compounds in MIC analyses. These compounds included: the biocides dequalinium, tetraphenylphosphonium, and benzalkonium, the antibiotics tetracycline and chloramphenicol, and a number of fluorescent antimicrobial dyes that are common substrates of multidrug efflux systems, such as proflavine, acriflavine, ethidium, Hoechst 33342, pyronin Y, acridine yellow, and 4′,6-diamidino-2-phenylindole (Table 1).
Resistance to a variety of antimicrobials was common, particularly among the Pseudomonas and betaproteobacterial genes (Table 1). Six BTP domain protein genes conferred reproducible resistance to the biocide benzalkonium. Additionally, two Pseudomonas proteins, PA14_26850 and PFL_4585, mediated resistance to dequalinium. Notably, PFL_4585 is a paralog of the chlorhexidine resistance protein PFL_4558, demonstrating that both BTP domain proteins encoded by P. protegens Pf-5 mediate resistance to selected biocides. Nine of the BTP domain protein genes also conferred resistance to one or both of the DNA-intercalating antimicrobial dyes acriflavine and proflavine (Table 1). Only 6 genes out of the 24 examined in this study did not provide resistance to any of the antimicrobials tested. Of these, three were not expressed at levels detectable by Western blotting (see Fig. S1 in the supplemental material). The V. parahaemolyticus gene VP1155 provided the highest and most consistent increases in resistance to the greatest number of compounds, with at least 4-fold increases in resistance to chlorhexidine, benzalkonium, proflavine, and acriflavine. To determine whether resistance mediated by these proteins was dependent on a TolC-like outer membrane, we examined resistance mediated by AceI and VP1155 in a TolC-inactivated background (10). Both of these proteins mediated resistance in this mutant strain.
To explore further the substrate recognition profile of the VP1155 protein, we conducted Biolog OmniLog Phenotype microarray (PM) experiments (11). The resistance levels of E. coli BL21 carrying pTTQ18-VP1155 were compared to those of E. coli BL21 carrying pTTQ18 for 240 different antimicrobials in the PM11-20 plate series, as previously described (1, 11). These tests confirmed that VP1155 provides resistance to the substrates identified by conventional MIC assays: chlorhexidine, proflavine, and acriflavine (see Fig. S2 in the supplemental material; benzalkonium is not included in the PM11-20 panel of compounds). Furthermore, the Biolog PM tests indicated that VP1155 also provides resistance to 9-aminoacridine, domiphen bromide, 3,5-diamino-1,2,4-triazole (guanazole), and plumbagin (see Fig. S2).
Fluorimetric transport assays demonstrate efflux mediated by VP1155.
The observations of resistance to the DNA-intercalating antimicrobial dyes proflavine and acriflavine from several of the BTP domain protein genes presented the opportunity to assay directly the efflux of these substrates in real-time fluorimetric transport assays. We applied these assays to cells expressing VP1155, exploiting the capacity of this protein to mediate resistance to both dyes. The assays were conducted in the E. coli triple deletion mutant strain BW25113 (ΔacrB ΔemrE ΔmdfA::kan), which is defective in the three major E. coli multidrug efflux system genes (12) and provides a sensitive background for these assays. Cells carrying the pTTQ18-VP1155 expression plasmid were assayed both pre- and postinduction of VP1155 expression by using 0.2 mM IPTG (see Fig. S3 in the supplemental material). Cells carrying “empty” pTTQ18 treated with IPTG were also included as a negative control. The transport assays were conducted essentially as described previously (13), except that cells were grown in glycerol-supplemented medium and reenergized by using glycerol to initiate transport from substrate-loaded cells.
The fluorescence intensity of both proflavine and acriflavine is lower when intercalated into DNA, so efflux from the cell was characterized by an increase in fluorescence over time. In our transport experiments we observed a rapid increase in fluorescence in cells that expressed the VP1155 protein but not in control cells lacking this protein (Fig. 2; see also Fig. S3 in the supplemental material). These results provide additional evidence that efflux is the mechanism of resistance operating in this group of proteins.
FIG 2 .
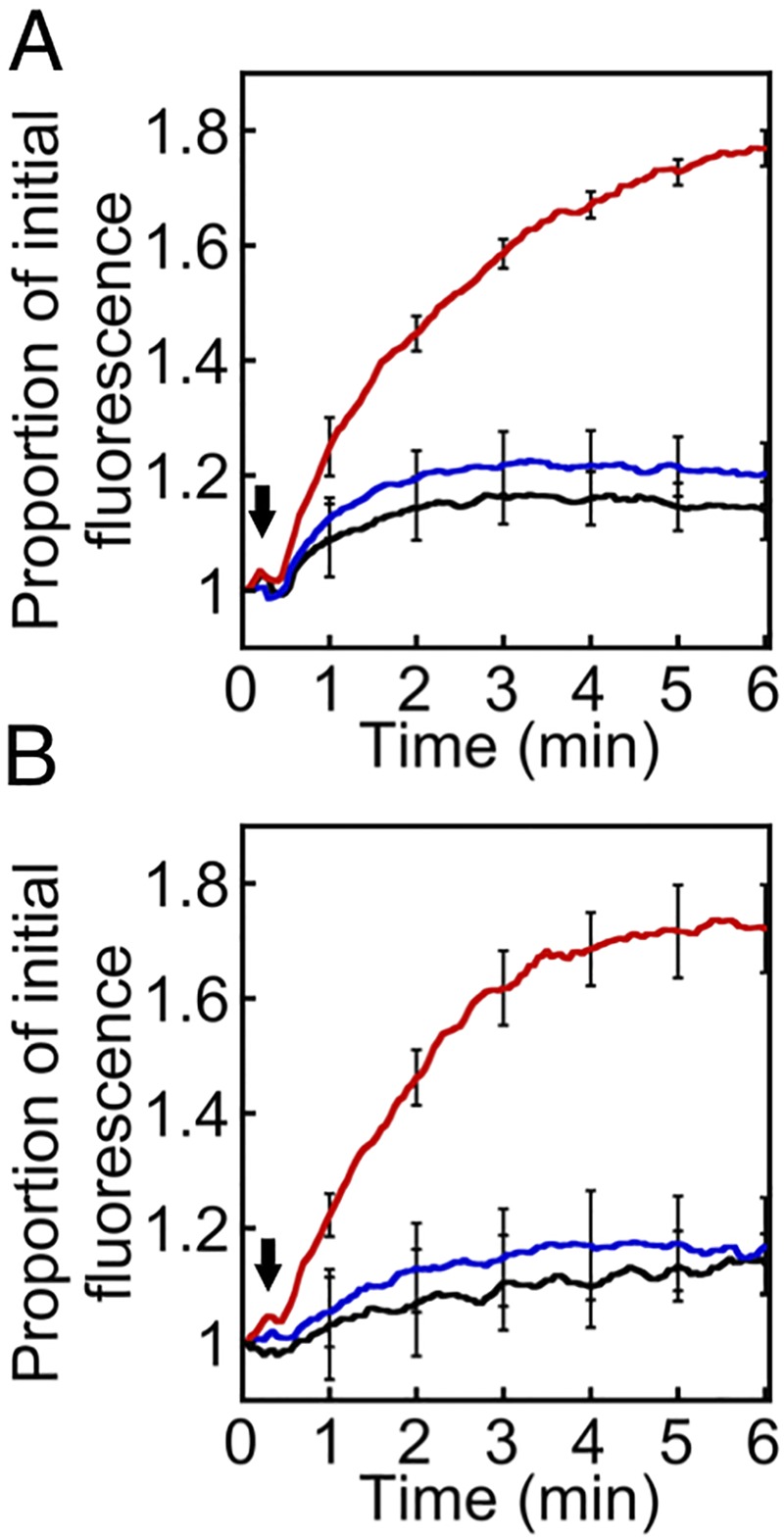
Acriflavine and proflavine efflux mediated by VP1155. E. coli BW25113 ΔacrB ΔemrE ΔmdfA::kan cells carrying pTTQ18 only (black line) or pTTQ18-VP1155 (blue [noninduced] and red [induced]) were grown in Luria-Bertani medium containing 0.5% glycerol to an optical density at 600 nm of 0.6. Samples of pTTQ18-VP1155 cells were taken and assayed as noninduced controls (blue lines). The cultures were then grown for a further 1 h, after which 0.2 mM IPTG was added to induce expression directed by the Ptac promoter in pTTQ18. The cells were washed in assay buffer (HEPES, pH 7.0) and loaded with 20 µM acriflavine (A) or proflavine (B) in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). The loaded cells were again washed and suspended in assay buffer (37°C). Transport was initiated with the addition of 1% glycerol at the point marked with an arrow. The assays were performed in biological triplicates, and the error bars show the standard errors of the means at 1-min intervals. Expression of the RGSH6-tagged VP1155 protein in the samples was assessed by Western blotting (see Fig. S3 in the supplemental material) and was positively correlated with efflux.
Conclusions.
In this study, we examined a large panel of genes encoding proteins related to the AceI chlorhexidine efflux system for their ability to confer resistance to a set of 12 different biocides, antibiotics, and antimicrobial dyes. To facilitate this broad survey of phylogenetically diverse proteins, we adopted a synthetic cloning approach; the majority of genes were codon optimized for expression in E. coli and chemically synthesized for cloning into our expression system. Of the 24 transport proteins studied, 21 were expressed at levels detectable in Western blot assays of whole-cell lysates, and 18 conferred resistance to one or more antimicrobial compounds.
Our results demonstrate that this group of proteins is a new family of bacterial multidrug efflux systems, which we have designated the proteobacterial antimicrobial compound efflux (PACE) family. The PACE family is only the sixth family of bacterial multidrug efflux systems to have been described and the first new family for more than 15 years. Multidrug efflux systems encoded by nosocomial pathogens are particularly problematic. Antimicrobial pressures selecting for increased expression of multidrug exporters can promote resistance to not only the selecting compound but also to a swath of otherwise-effective compounds.
Using a radiolabeled substrate, we previously demonstrated that the AceI protein within the PACE family is able to mediate the active efflux of chlorhexidine. In this work, we have identified fluorescent substrates for several members of the PACE family that facilitate the development of rapid fluorimetric efflux assays. These assays will be highly valuable in future studies to define the molecular transport mechanism operating in members of this family, including the mode of energization, which is likely to involve an electrochemical gradient.
Notably, PACE family proteins are encoded in the core genomes of many proteobacterial species that are separated by hundreds of millions of years of evolution. Given that the substrates we have now defined for these efflux systems—chlorhexidine, dequalinium, benzalkonium, proflavine, and acriflavine—are synthetic biocides that have only been widely used within the last century, it seems unlikely that these compounds are the physiological substrates of these transporters. Nonetheless, as with other multidrug efflux systems, the substrate promiscuity of PACE efflux systems is likely to have enhanced the success of proteobacterial pathogens in clinical settings.
SUPPLEMENTAL MATERIAL
Western blots showing expression of BTP domain family proteins in E. coli BL21. Cells carrying the respective pTTQ18 expression plasmids with cloned BTP domain protein genes were grown to an optical density at 600 nm (OD600) of 0.8 in LB broth. Expression of the BTP domain proteins was induced using 0.2 mM IPTG for 2 h. Whole-cell lysates were run on a 4-to-20% SDS-PAGE gel, and the overexpressed proteins were detected with an anti-RGSH6–horseradish peroxidase conjugate (Qiagen). Download
Kinetic response curves paralleling bacterial growth for Biolog phenotype microarray antimicrobial tests in which the pTTQ18-VP1155 plasmid conferred an advantage. Curves for E. coli BL21 cells carrying pTTQ18 are shown in red, curves for BL21 cells carrying pTTQ18-VP1155 are shown in green, and regions of overlap in the response curves of these two strains are shown in yellow. Cells were grown in the presence of 0.05 mM IPTG to promote expression of the cloned gene. The curves depict the color intensity of a redox-active dye (y axis) over time (x axis; 30 h). (A) Plate PM14, well A3 (acriflavine); (B) plate PM14, well B3 (9-aminoacridine); (C) plate PM15, well D6 (domiphen bromide); (D) plate PM18, well G7 (3,5-diamino-1,2,4-triazole [guanazole]); (E) plate PM18, well H12 (plumbagin); (F) plate PM19, well C4 (chlorhexidine); (G) plate PM20, well D3 (proflavine). Download
Western blot showing expression levels of VP1155 in E. coli BW25113 ΔacrB ΔemrE ΔmdfA::kan cells carrying pTTQ18 or pTTQ18-VP1155, which were used in fluorimetric transport assays (see Fig. 2). Expression of the VP1155 protein was determined by using an anti-RGSH6–horseradish peroxidase antibody (Qiagen). Lane 1 (vector only) contained lysate from cells carrying “empty” pTTQ18 plasmid and were grown in the same way as the VP1155-induced cells. Lane 2 (VP1155 noninduced) contained lysate from cells carrying pTTQ-VP1155 that were grown to an optical density at 600 nm (OD600) of 0.6 in medium lacking IPTG. Lane 3 (VP1155) contained lysate from cells carrying pTTQ-VP1155 that were grown to an OD600 of 0.6 in Luria-Bertani medium and then supplemented with 0.2 mM IPTG for 1 h to induce expression of VP1155. Download
ACKNOWLEDGMENTS
This work was supported by a project grant from the Australian National Health and Medical Research Council (1060895) to I.T.P., K.A.H., and P.J.F.H. and the award of a Leverhulme Trust emeritus fellowship to P.J.F.H.
We thank Shimon Schuldiner from the Hebrew University of Jerusalem for the gift of E. coli strain BW25113 ΔacrB ΔemrE ΔmdfA::kan.
Footnotes
Citation Hassan KA, Liu Q, Henderson PJF, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6(1):e01982-14. doi:10.1128/mBio.01982-14.
REFERENCES
- 1.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJ, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye T, Van Acker H, Peeters E, Sass A, Buroni S, Riccardi G, Mahenthiralingam E. 2011. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother 55:1912–1919. doi: 10.1128/AAC.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nde CW, Jang HJ, Toghrol F, Bentley WE. 2009. Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ Sci Technol 43:8406–8415. doi: 10.1021/es9015475. [DOI] [PubMed] [Google Scholar]
- 6.Stark MJ. 1987. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 7.Saidijam M, Bettaney KE, Szakonyi G, Psakis G, Shibayama K, Suzuki S, Clough JL, Blessie V, Abu-Bakr A, Baumberg S, Meuller J, Hoyle CK, Palmer SL, Butaye P, Walravens K, Patching SG, O’Reilly J, Rutherford NG, Bill RM, Roper DI, Phillips-Jones MK, Henderson PJ. 2005. Active membrane transport and receptor proteins from bacteria. Biochem Soc Trans 33:867–872. doi: 10.1042/BST0330867. [DOI] [PubMed] [Google Scholar]
- 8.Hassan KA, Brzoska AJ, Wilson NL, Eijkelkamp BA, Brown MH, Paulsen IT. 2011. Roles of dha2 family transporters in drug resistance and iron homeostasis in Acinetobacter spp. J Mol Microbiol Biotechnol 20:116–124. doi: 10.1159/000325367. [DOI] [PubMed] [Google Scholar]
- 9.Hassan KA, Xu Z, Watkins RE, Brennan RG, Skurray RA, Brown MH. 2009. Optimized production and analysis of the staphylococcal multidrug efflux protein QacA. Protein Expr Purif 64:118–124. doi: 10.1016/j.pep.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackie AM, Hassan KA, Paulsen IT, Tetu SG. 2014. Biolog phenotype microarrays for phenotypic characterization of microbial cells. Methods Mol Biol 1096:123–130. doi: 10.1007/978-1-62703-712-9_10. [DOI] [PubMed] [Google Scholar]
- 12.Tal N, Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A 106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner RJ, Taylor DE, Weiner JH. 1997. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother 41:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. BioInformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blots showing expression of BTP domain family proteins in E. coli BL21. Cells carrying the respective pTTQ18 expression plasmids with cloned BTP domain protein genes were grown to an optical density at 600 nm (OD600) of 0.8 in LB broth. Expression of the BTP domain proteins was induced using 0.2 mM IPTG for 2 h. Whole-cell lysates were run on a 4-to-20% SDS-PAGE gel, and the overexpressed proteins were detected with an anti-RGSH6–horseradish peroxidase conjugate (Qiagen). Download
Kinetic response curves paralleling bacterial growth for Biolog phenotype microarray antimicrobial tests in which the pTTQ18-VP1155 plasmid conferred an advantage. Curves for E. coli BL21 cells carrying pTTQ18 are shown in red, curves for BL21 cells carrying pTTQ18-VP1155 are shown in green, and regions of overlap in the response curves of these two strains are shown in yellow. Cells were grown in the presence of 0.05 mM IPTG to promote expression of the cloned gene. The curves depict the color intensity of a redox-active dye (y axis) over time (x axis; 30 h). (A) Plate PM14, well A3 (acriflavine); (B) plate PM14, well B3 (9-aminoacridine); (C) plate PM15, well D6 (domiphen bromide); (D) plate PM18, well G7 (3,5-diamino-1,2,4-triazole [guanazole]); (E) plate PM18, well H12 (plumbagin); (F) plate PM19, well C4 (chlorhexidine); (G) plate PM20, well D3 (proflavine). Download
Western blot showing expression levels of VP1155 in E. coli BW25113 ΔacrB ΔemrE ΔmdfA::kan cells carrying pTTQ18 or pTTQ18-VP1155, which were used in fluorimetric transport assays (see Fig. 2). Expression of the VP1155 protein was determined by using an anti-RGSH6–horseradish peroxidase antibody (Qiagen). Lane 1 (vector only) contained lysate from cells carrying “empty” pTTQ18 plasmid and were grown in the same way as the VP1155-induced cells. Lane 2 (VP1155 noninduced) contained lysate from cells carrying pTTQ-VP1155 that were grown to an optical density at 600 nm (OD600) of 0.6 in medium lacking IPTG. Lane 3 (VP1155) contained lysate from cells carrying pTTQ-VP1155 that were grown to an OD600 of 0.6 in Luria-Bertani medium and then supplemented with 0.2 mM IPTG for 1 h to induce expression of VP1155. Download